Abstract
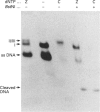
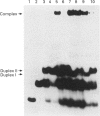
The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by the mechanism-based inhibitor 5-azacytosine has remained unclear, mainly because of the unavailability of a substrate in which the inhibitor, but not normal cytosine, is present at the target site. We synthesized an oligonucleotide duplex containing a single target site for the EcoRII methyltransferase, in which the target base is 5-azacytosine. This substrate formed a stable covalent complex with EcoRII methyltransferase in the absence and in the presence of the cofactor S-adenosylmethionine. The complex formed in the presence of the cofactor was resistant to SDS and moderate heat treatment, and a methyl group was incorporated into the complex. Enzyme titration and kinetic studies of inhibition suggest that methyl transfer to the complex occurred only during the first turnover of the reaction. These results suggest that, when the enzyme binds to 5-azacytosine in the presence of the cofactor, a methyl group is transferred to the N-5 position of the base, resulting in the inactivation of the enzyme.
Full text
PDF


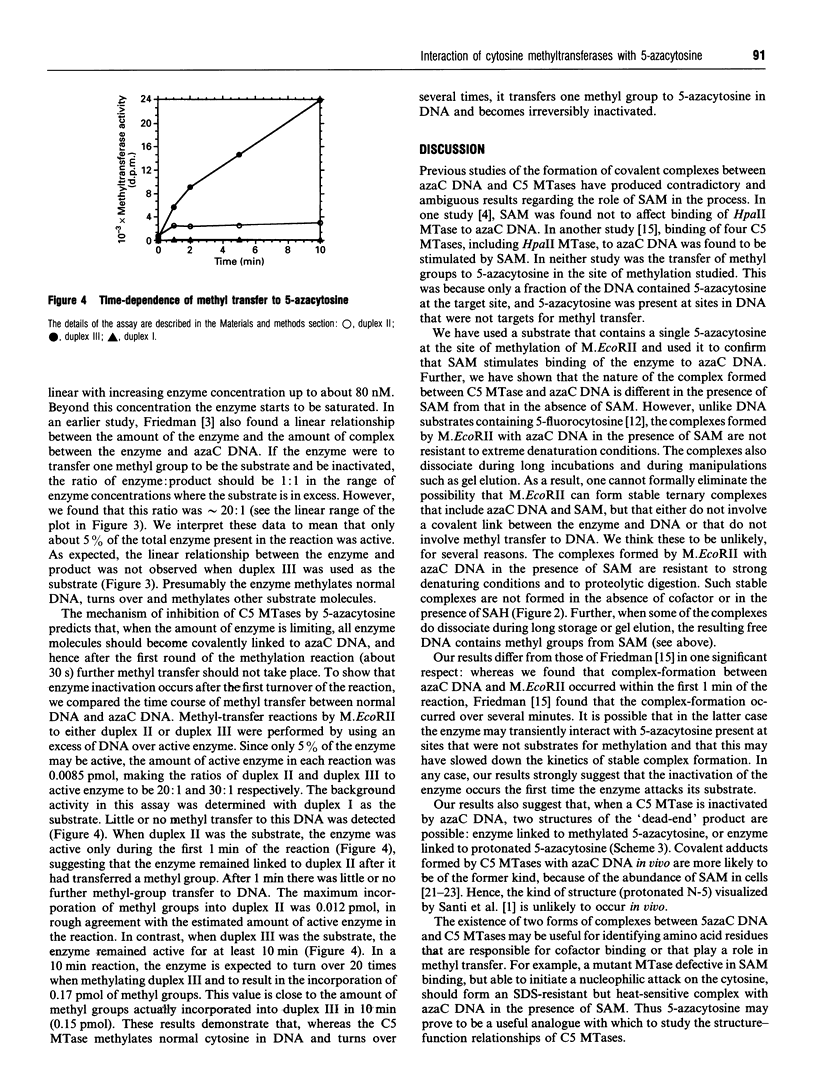

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Butkus V., Klimasauskas S., Kersulyte D., Vaitkevicius D., Lebionka A., Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985 Aug 26;13(16):5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. Binding of the EcoRII methylase to azacytosine-containing DNA. Nucleic Acids Res. 1986 Jun 11;14(11):4543–4556. doi: 10.1093/nar/14.11.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Friedman S. The irreversible binding of azacytosine-containing DNA fragments to bacterial DNA(cytosine-5)methyltransferases. J Biol Chem. 1985 May 10;260(9):5698–5705. [PubMed] [Google Scholar]
- Gabbara S., Bhagwat A. S. Interaction of EcoRII endonuclease with DNA substrates containing single recognition sites. J Biol Chem. 1992 Sep 15;267(26):18623–18630. [PubMed] [Google Scholar]
- Greene R. C., Hunter J. S., Coch E. H. Properties of metK mutants of Escherichia coli K-12. J Bacteriol. 1973 Jul;115(1):57–67. doi: 10.1128/jb.115.1.57-67.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanck T., Schmidt S., Fritz H. J. Sequence-specific and mechanism-based crosslinking of Dcm DNA cytosine-C5 methyltransferase of E. coli K-12 to synthetic oligonucleotides containing 5-fluoro-2'-deoxycytidine. Nucleic Acids Res. 1993 Jan 25;21(2):303–309. doi: 10.1093/nar/21.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C., Friedman S. Inhibition of recA-mediated strand exchange by adducts of azacytosine-containing DNA and the EcoRII methylase. J Biol Chem. 1991 Sep 15;266(26):17424–17429. [PubMed] [Google Scholar]
- Lu L. J., Randerath K. Long term instability and molecular mechanism of 5-azacytidine-induced DNA hypomethylation in normal and neoplastic tissues in vivo. Mol Pharmacol. 1984 Nov;26(3):594–603. [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh L. P., Zielinski W. S., Kalisch B. W., Pfeifer G. P., Sprinzl M., Drahovsky D., van de Sande J. H., Jovin T. M. Synthesis and characterization of poly[d(G-z5C)]. B-Z transition and inhibition of DNA methylase. Biochemistry. 1985 Aug 27;24(18):4806–4814. doi: 10.1021/bi00339a014. [DOI] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Norment A., Garrett C. E. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satishchandran C., Taylor J. C., Markham G. D. Novel Escherichia coli K-12 mutants impaired in S-adenosylmethionine synthesis. J Bacteriol. 1990 Aug;172(8):4489–4496. doi: 10.1128/jb.172.8.4489-4496.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M., Constantinides P. A., Jones P. A. 5-Azacytidine, DNA methylation, and differentiation. Curr Top Microbiol Immunol. 1984;108:115–127. doi: 10.1007/978-3-642-69370-0_8. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982 Dec 15;162(3):679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Kubareva E. A., Romanova E. A., Oretskaya T. S., Gromova E. S., Shabarova Z. A., Bhagwat A. S. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993 Jan 25;21(2):295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]