Summary
Spatial transcriptomics enables a single-cell resolution view of gene expression patterns in tissues, providing insight into their biological functions. However, applying this approach to the skin presents inherent challenges. Here, we present a protocol for preparing mammalian skin samples encompassing hair follicles for spatial transcriptomics. We describe steps for sample preparation, embedding, acquisition of frozen slices, RNA quality control, tissue mounting, fixation, staining, and imaging. We then detail procedures for permeabilization, reverse transcription, and cDNA collection.
For complete details on the use and execution of this protocol, please refer to Chen et al.1
Subject areas: Genetics, Genomics, Model Organisms, Molecular Biology
Graphical abstract
Highlights
-
•
Steps for processing samples and removing fat to improve mammalian skin processing
-
•
Guide for skin tissue preparation, orientation, and sectioning in spatial transcriptomics
-
•
In-depth information and critical steps on spatial transcriptomics for skin research
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Spatial transcriptomics enables a single-cell resolution view of gene expression patterns in tissues, providing insight into their biological functions. However, applying this approach to the skin presents inherent challenges. Here, we present a protocol for preparing mammalian skin samples encompassing hair follicles for spatial transcriptomics. We describe steps for sample preparation, embedding, acquisition of frozen slices, RNA quality control, tissue mounting, fixation, staining, and imaging. We then detail procedures for permeabilization, reverse transcription, and cDNA collection.
Before you begin
Institutional permissions
Human tissue samples used for scientific research must be collected from volunteers or patients only after receiving their written informed consent and obtaining official ethical approval from the relevant authorities. The human hair follicle samples used in this protocol were obtained from the Department of Plastic Surgery, at Hangzhou First People’s Hospital with informed consent and received approval from the BGI Ethics Committee, license number BGI-IRB 23117. Additionally, the rat skin sample collection was approved by the BGI Ethics Committee under the license number BGI-IRB A23034.
Main points of designing, selecting, and collecting samples
The quality of mammalian skin and hair follicle samples is the most critical step for successful spatial transcriptomics processing, including RNA integrity (RIN) and section integrity. Here are the primary factors impacting sample quality during the pre-preparation step.
-
1.
The size of skin including hair follicle samples for spatial transcriptomics should be a minimum of 2 cm × 2 cm. Some skin sections must be prepared for RIN detection and section adjustment.
-
2.
The structure of the samples must be maintained to meet the biological research objectives, ensuring that all relevant structures for analysis are present. For skin samples, this typically involves including the main layers epidermis, dermis, as well as muscles, while excluding fat to avoid interference with slicing. To preserve the hair follicle biological information, it is crucial to preserve the entire structure including the dermal papilla, bulb, bulge, arrector pili muscle and sebaceous gland.
-
3.
The RNA integrity number needs to be more than 7 for spatial transcriptomics. An increase in sample collection time will lead to a decrease in RNA integrity; therefore, the time between isolating the sample from the body and embedding must be less than 30 min to ensure the maintenance of RNA integrity.
Note: RNA integrity number and RNA quality number are both quality controls for RNA quality from different testing equipment, but the results are universal.
-
4.
Water or other liquids on the sample surface should be thoroughly dried or wiped off before the embedding step.
Equipment preparation
Timing: 10 min
-
5.
Set one thermostat to 37°C for chip drying and permeabilization.
-
6.
Set another thermostat to 42°C for reverse transcription.
-
7.
Set a third thermostat to 55°C for tissue removal and cDNA release.
-
8.
Set the water bath-slide dryer to 37°C.
-
9.
Place the STOmics commercial kits for spatial transcriptomics experiments at room temperature.
Note: Room temperature is between 23°C–27°C.
Alternatives: The STOmics commercial kits utilize the Stereo-seq technology, which is one of the methods for spatial transcriptomics. These kits can also be substituted by other alternatives for using other spatial transcriptomics techniques, such as Visium, Slide-seq and more.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Nuclease free water (NF-water) | Ambion | AM9937 |
| Anhydrous ethanol | Xilong Scientific Co., Ltd. | 72188–01 |
| AMPure XP | Agencourt | A63882 |
| HCl | Sigma-Aldrich | 2104-50ML |
| Glycerol | Solarbio | G8190 |
| 20 × SSC | Ambion | AM9770 |
| Methanol | Sigma | 34860-1L-R |
| Other | ||
| Cryotome | Dakewe | CT520 |
| Fluorescence microscope (Z-capture function) | Stereo-OR 100 | SCI-01-016 |
| Metal bath | BIOBASE | BJPX-DB2 |
| Centrifuge | Kylin-Bell | LX-200B |
| Whirlpool mixer | Kylin-Bell | QL-901 |
| T100 Thermal Cycler | Bio-Rad∗ | 1861096 |
| ProFlex 3 × 32-well PCR system | ABI∗ | 4483636 |
| Slide spinner | Labnet | C1303-T |
| NEBNext Magnetic separation rack | NEB | S1515S |
| DynaMag-2 magnet | Thermo Fisher Scientific | 12321D |
| Qubit 3.0 fluorometer | Thermo Fisher Scientific | Q33216 |
| 4200 TapeStation system | Agilent | G2991BA |
| Water bath-slide dryer | Kedee | KD-H |
| Bio-Fragment analyzers | BiOptic | Qsep100 |
| Stereo-seq Transcriptomics T Kit | STOmics/BGI | 111KT114 |
| Stereo-seq Chip T slide (1 cm × 1 cm) | STOmics/BGI | 210CT114 |
| Sakura Tissue-Tek O.C.T. compound (OCT) | Sakura | 4583 |
| RNeasy Mini Kit | QIAGEN | 74104 |
| Hematoxylin and eosin (H&E) | Solarbio | G1120 |
| VAHTS DNA Clean Beads | ∗Vazyme | N411-02 |
| Qubit ssDNA Assay Kit | Invitrogen | Q10212 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Q32854 |
| Clear adhesive film | MicroAmp | 4306311 |
| Compound microscope | Olympus Corporation | BX53 |
| Petri dish | BD | 353003 |
| Surgical sponge | Dynarex | DY3242-P |
| Coverslip | CITOGLAS | 10227105P |
| 24-well plate | Corning | 3513 |
Materials and equipment
Stereo-seq Transcriptomics T Kit
| Reagent | Final concentration | Amount |
|---|---|---|
| RI | N/A | 300 μL ×1 |
| PR Enzyme | N/A | 10 mg × 1 |
| PR Rinse Buffer | N/A | 880 μL ×1 |
| Glycerol | N/A | 50 μL ×1 |
| RT Reagent | N/A | 720 μL ×1 |
| RT Oligo | N/A | 1 OD ×1 |
| RT Additive | N/A | 44 μL ×1 |
| Reverse Enzyme | N/A | 44 μL ×1 |
| TR Buffer | N/A | 1725 μL ×1 |
| cDNA Release Enzyme | N/A | 88 μL ×1 |
| cDNA Release Buffer | N/A | 1725 μL ×1 |
| cDNA Primer | N/A | 36 μL ×1 |
| cDNA Amplification Mix | N/A | 220 μL ×1 |
Store at 4°C for up to 1 year
5 × SSC
| Reagent | Final concentration | Amount |
|---|---|---|
| 20 × SSC | N/A | 5 mL |
| Nuclease Free Water | N/A | 15 mL |
| Total | N/A | 20 mL |
Store at 4°C for up to 1 month
1 × SSC
| Reagent | Final concentration | Amount |
|---|---|---|
| 5 × SSC | N/A | 5 mL |
| Nuclease Free Water | N/A | 20 mL |
| Total | N/A | 25 mL |
Store at 4°C for up to 1 month
Tissue ssDNA staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 5 × SSC | N/A | 94.5 μL |
| Qubit ssDNA Reagent | N/A | 0.5 μL |
| RNase inhibitor | 2 U/μL | 5 μL |
| Total | N/A | 100 μL |
Store at room temperature for up to 1 week
10 × Permeabilization Reagent Stock Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PR Enzyme powder (Stereo-seq Transcriptomics T Kit) | 10 × | 100 μL |
| HCl | 0.01 M | 900 μL |
| Total | N/A | 1 mL |
Store at −20°C for up to 3 months
1 × Permeabilization Reagent Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 10 × Permeabilization Reagent Stock Solution | 1 × | 100 μL |
| HCl | 0.01 M | 900 μL |
| Total | N/A | 1 mL |
Store at −20°C for up to 1 week
RT Mix
| Reagent | Final concentration | Amount |
|---|---|---|
| RI | N/A | 5 μL |
| RT Oligo | N/A | 5 μL |
| Reverse T Enzyme | N/A | 5 μL |
| RT Reagent | N/A | 80 μL |
| RT Additive | N/A | 5 μL |
| Total | N/A | 100 μL |
Store at −20°C for up to 1 week
cDNA Release Mix
| Reagent | Final concentration | Amount |
|---|---|---|
| cDNA Release Buffer | N/A | 380 μL |
| cDNA Release Enzyme | N/A | 20 μL |
| Total | N/A | 400 μL |
Store at −20°C for up to 1 week
Qubit dsDNA Mix
| Reagent | Final concentration | Amount |
|---|---|---|
| Invitrogen Qubit dsDNA HS Buffer | N/A | 199 μL |
| Qubit dsDNA HS Reagent 200 × | 1 × | 1 μL |
| Total | N/A | 200 μL |
Store at 4°C for up to 1 week
Step-by-step method details
Sample preparation and embedding
Timing: 1 h
Timing: no more than 30 min (for step 1)
Timing: no more than 30 min (for step 2)
Preparation of rat skin and human scalp samples with hair follicles, fat removal and embedding. The step of sample preparation is critical for spatial transcriptomics experiments, especially skin tissues. If the tissue RNA integrity is greater than 7 and the section is intact, the spatial transcriptomics experimental success rate can be very high. And how to protect RNA integrity and section status is a new challenge in skin tissue.
-
1.
Sample requirements.
Human scalp sample processing (30 min).-
a.Human scalp sample collection and processing (15 min).
-
i.Obtain ethically approved human scalp samples (with hair follicles) from the Affiliated Hangzhou First People’s Hospital.
-
ii.Place samples in a dry Petri dish and put them on ice within 5 min. Keep them on ice for no more than 10 min until the next step.
-
i.
-
b.Fat removal (15 min).The skin section embedded after fat removal appears more intact, preserving the skin structures when compared to the one containing adipose tissue (Figure 1).
-
i.Depending on the experimental requirements, use surgical scissors to cut tissues of appropriate sizes for processing.
-
ii.Transfer tissues from the Petri dish to a surgical sponge under the Compound Microscope for no more than 5 min.
-
iii.Secure the hair follicles with tweezers and gently remove the subcutaneous fat from the scalp sample with curved tissue scissors or scalpels for no more than 10 min (Figure 2).
-
iv.Immediately transfer the fat-removed scalp tissue back to ice and wait for embedding.Note: Ensuring complete immersion of the scalp tissue in OCT is crucial during the frozen embedding process. If any portion of the tissue remains exposed or the surrounding OCT layer is excessively thin, it is necessary to add more OCT before it solidifies fully. Typically, the embedding process involves adding an OCT layer 2–3 mm tick above the tissue, followed by placing the sample on a frozen platform until it is thoroughly solidified.
 CRITICAL: The angle at which the tissue is embedded on OCT determines the direction of subsequent sections. It is recommended that the direction of hair follicle growth should be parallel to the direction of the embedding cassette, and the direction should be recorded and photographed.
CRITICAL: The angle at which the tissue is embedded on OCT determines the direction of subsequent sections. It is recommended that the direction of hair follicle growth should be parallel to the direction of the embedding cassette, and the direction should be recorded and photographed.
-
i.
Rat skin sample collection and processing (30 min).-
c.Rat skin sample collection (15 min).
-
i.Euthanize the rats following the approved ethical procedures performed in the First Affiliated Hospital of Naval Medical University.
-
ii.Select the specific location on the rat skin to collect samples based on the experimental purpose.
-
iii.Put the samples in a dry Petri dish on ice and perform the next steps for no more than 15 min.
-
iv.Shave the hair from the corresponding area of the rat skin.
-
v.Use tweezers to pinch and pull the rat skin to facilitate surgical removal.
-
vi.Use surgical scissors to cut out the appropriate size of the rat skin.
-
i.
-
d.Fat removal (15 min).
-
i.Transfer the fat-removed skin tissue from the Petri dish to a surgical sponge under a microscope for no more than 5 min.
-
ii.Secure the tissue with tweezers, and gently remove the subcutaneous fat from the skin sample with curved tissue scissors or scalpels for no more than 10 min.
-
iii.Rinse the sample with the pre-prepared 1 × PBS.
-
iv.Blot excess liquids from the sample surface with dust-free paper.
-
v.Immediately transfer the fat-removed skin to ice and wait for embedding.Note: The maximum allowable time for tissue processing, from cutting to embedding, is 30 min.
-
i.
-
a.
-
2.Frozen embedding.
-
a.Preparing the embedding cassette. Pre-coat the bottom of the embedding cassette with OCT at room temperature to a thickness of approximately 2 mm, and the thickness could be estimated by the embedding cassette. Place it inside a cryostat or on a freezing stage with dry ice for cooling.Note: Depending on the research requirements and the size of the skin or scalp tissue select the size of the embedding cassette. We regularly use 17 × 17 × 5 mm embedding cassettes. Label the cassette with the sample information beforehand.
-
b.Preparing the tissue. Use dust-free paper to absorb any liquids from the tissue surface.
-
c.Positioning the tissue. Carefully lay the tissue sample on the frozen OCT layer that has solidified at the bottom of the cassette.
-
d.Adding OCT. Gently pour another layer of room-temperature OCT over the tissue. Ensure the skin or scalp tissue is completely covered with OCT.Note: Be cautious to avoid generating air bubbles while embedding the sample in OCT. If air bubbles form, use a syringe to carefully remove them from around the tissue sample.
-
e.Freezing the tissue. Place the tissues in a cryostat or ice box containing dry ice for rapid freezing (Figure 3).
-
f.Storage. Wrap the embedding cassette securely in aluminum foil. Store the OCT-embedded tissue samples in a −80°C freezer.Note: Tiny air bubbles that are closer to the tissue need to be removed, while those that are farther away are acceptable. Avoid creating large air bubbles adjacent to the tissue, as these can cause the cryosection to break.Note: The tissues may be stored at −80°C for up to 3 years.
-
a.
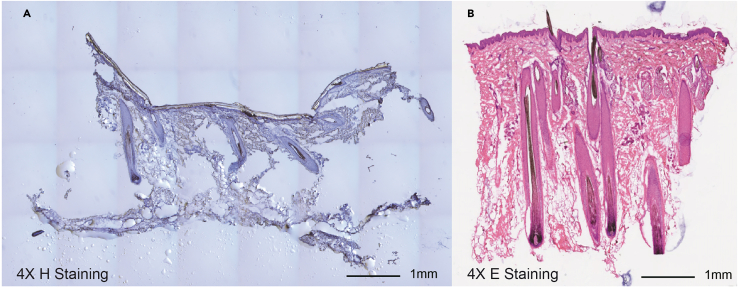
Figure 1.
Human scalp section H&E staining
H&E staining images of human scalp sections before (A) and after (B) fat removal. This comparison highlights the benefits of the fat removal preparatory step. For rapid identification of tissue integrity, simple dyes can be used to stain the structure such as (A) using Hematoxylin and (B) mainly using Eosin with a small amount of Hematoxylin.
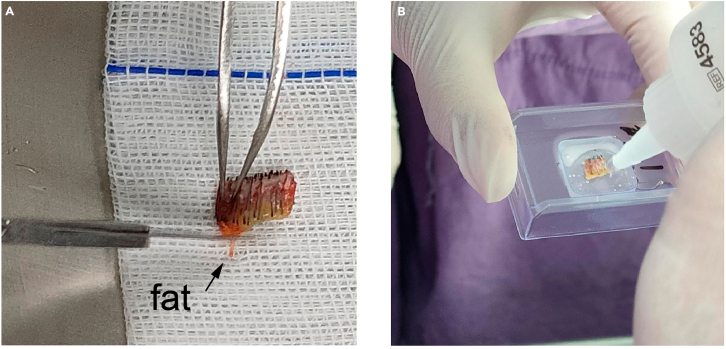
Figure 2.
Scalp tissue with removed fat and OCT embedding
(A) Remove fat from the human scalp using tweezers.
Remove most of the fat below the scalp, leaving a small 1–2 mm size section of fat to protect the structure of hair follicles.
(B) Perform OCT embedding for human scalp.
Figure 3.
The embedding stage of rat skin sample in OCT before freezing
The sample stabilizes in the middle of the OCT at an angle present by the individual, and sample information and embedding angle are marked.
Acquisition of frozen slices and RNA quality control
Timing: 2 h
Timing: 40 min to 1 h (for step 3)
Timing: 1 h–1.2 h (for step 4)
This step involves obtaining cryosections of tissues for Spatial Transcriptomics experiments and conducting RNA quality control. RNA quality control is necessary for the beginning of the batch experiment. However, if following the successful steps and producing a high RNA quality number of tissue consistently (more than 7), detect 1-2 tissues obtained from each batch, instead of detecting each sample.
-
3.Cryosection Preparation.
-
a.Setting the slide drier. Adjust and preheat the Water Bath-Slide Dryer to 37°C.Note: A Water Bath-Slide Drier can be substituted with a Metal bath.
-
b.Cryotome settings. Set the temperature of the specimen holder to −15°C and the cryotome chamber temperature to −20°C.Note: An over-cooled or overheated specimen holder may lead to tissue section cracking or severe wrinkling during sectioning. If using the cryotome without the dual temperature control (head and chamber), set the cryotome temperature to −17°C first, and adjust it in the range from −15°C to −20°C depending on the status of sections in real-time.
-
c.Preparing tools. Place and pre-cool clean forceps, brushes, and razor blades inside the cryostat chamber.
-
d.Tissue acclimatization: Transfer the OCT-embedded tissue sample from the −80°C freezer to the cryostat chamber. Allow it to equilibrate to the cryostat chamber temperature for about 30 min.
-
e.Trimming the tissue block. Remove the aluminum foil covers from the sample. Use scalpel blades to gradually trim the embedded tissue block along the perimeter of the tissue to the desired size. Such as, if the tissue size is 1 × 1 cm, trim the tissue block to 1.5 × 1.5 cm first, and then slowly trim the tissue block to 1.2 × 1.2 cm or 1.1 × 1.1 cm. The tissue surrounding OCT must be left about 1–2 mm.Note: To optimize tissue mounting on a 1 × 1 cm Stereo sequencing chip, at a later step, it is advisable to trim the sectioning area to smaller than 0.9 × 1 cm.
-
f.Mounting the tissue block. Securely affix the embedded tissue block onto the cryostat chucks with OCT.
-
a.
-
4.RNA quality control.
-
a.Cryosectioning. Open the Specimen Chuck Release Lever to allow the Cryotome Chuck to insert into the Specimen Holder, and then adjust the angle of Cryotome Chuck and close the Specimen Chuck Release Lever to fix Cryotome Chuck.
-
b.Trimming. Carefully trim the excess OCT to expose the tissue (Figure 4). Then, proceed to cut 20 consecutive sections.
-
c.Transfer all sections to a 1.5 mL centrifuge tube. These samples will be used for subsequent measurements of tissue RNA integrity (RIN).
-
d.RNA integrity (RIN) assessment: Extract tissue RNA using the RNeasy Mini Kit. Assess the quality and fragment distribution of cDNA with Qsep100 (Figure 5).
-
a.
Note: For the RNeasy Mini Kit protocol visit Qiagen’s website: https://www.qiagen.com/zh-us/products/discovery-and-translational-research/dna-rna-purification/rna-purification/total-rna/rneasy-kits.
Note: Evaluate the RNA quality and fragment distribution of cDNA using a Qsep100 Bio-Fragment Analyzer. More information can be found at the Bioptic website: (https://www.bioptic.com.cn/ProductDetail/5133859.html).
Alternatives: Other kits and analytical equipment may be used as alternatives for RNA extraction and testing.
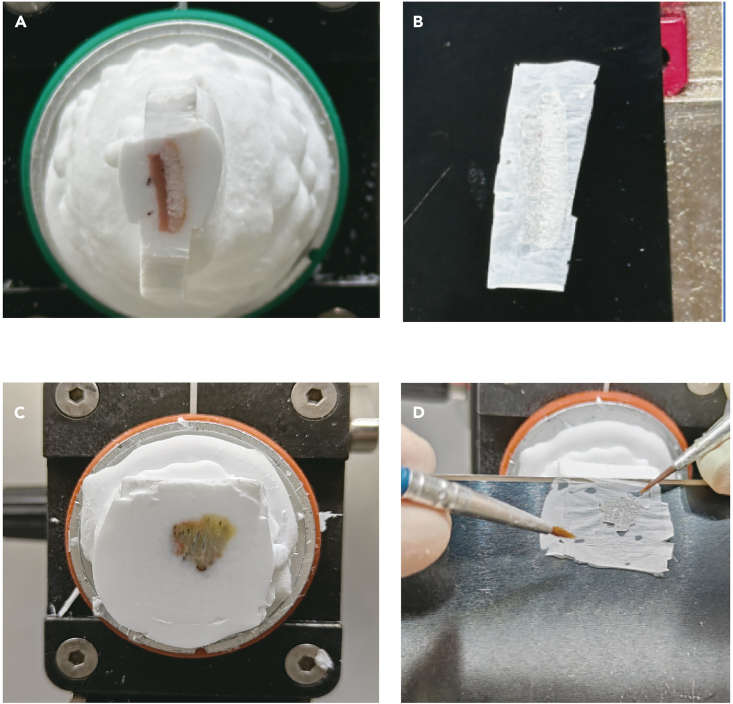
Figure 4.
Mounting the rat and human tissue on the holder and taking cryo-section
(A) and (B) are the process for the rat skin tissue, and (C) and (D) are for the human scalp tissue.
The section status is critical for spatial transcriptomics experiment which needs the unbroken section. But if the tissue sections are only used for RNA quality detection, the section status is not affected by the detection results whether broken or unbroken. However, try to trim the excess of OCT around the tissue which significantly influences the RNA extraction and detection using kits. Leave 3–5 mm OCT outside the tissue.
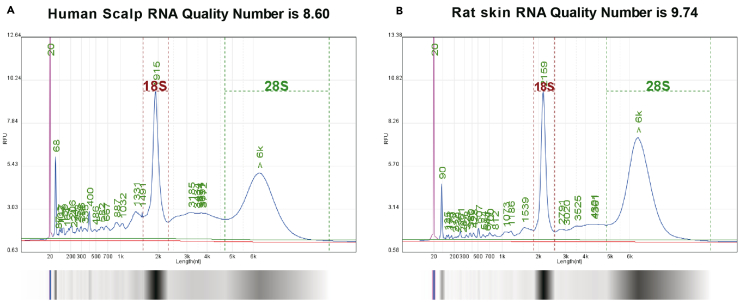
Figure 5.
NA quality number detection results
RNA quality number (RQN) of rat skin (A) and human scalp tissue samples (B). The RQN value exceeding 7 is considered satisfactory for quality.
Tissue mounting, fixation, staining, and imaging
Timing: 2–4 h
Timing: 10–20 min (for step 5)
Timing: 40–60 min (for step 6)
Timing: 20–30 min (for step 7)
Timing: 1–2 h (for step 8)
Timing: no more than 30 min (for step 9)
Conduct spatial transcriptomics experiments, which encompass tissue mounting, fixation, cell staining and imaging. In this protocol, we use the STOmics/BGI reagents and kits and change some details depending on skin tissues.
-
5.Tissue mounting.
-
a.Stereo-seq chip preparation. Allow the Stereo-seq chip, from the kit of choice, to acclimate to room temperature for 3 min.
-
b.Chip rinsing. Rinse the chip with 100 μL of nuclease-free (NF)-water twice using a pipette (Figure 6).
-
c.Absorbing excess liquid. Place dust-free paper under the chip to absorb any excess liquid blown out during the rinsing process.
-
d.Drying the chip. Position the chip on the surface of a Water Bath-Slide Drier for 3 min at 37°C to dry it.Note: The chip is ready for tissue mounting only when it is completely dry and dust-free from any wavy white residues by eye inspection.
-
e.Anhydrous Methanol preparation for fixation. Pour 3 mL of anhydrous methanol into a 24-well plate, ensuring the volume is sufficient to submerge the chip entirely.
-
f.Cover the 24-well plate with the lid and pre-cool it for at least 5 min at −20°C.
-
g.Positioning and orientation of the specimen on the cryostat. Securely affix the tissue-mounted cryostat chuck onto the cryostat specimen holder, adjusting the angle as necessary to ensure obtaining aligned structures, such as oriented hair follicles (Figure 2).
 CRITICAL: Orientation is one of the most important, hard and time-consuming steps. Usually, there are two directions including longitudinal section and horizontal section of hair follicles. Taking the longitudinal section as an example, the specimen angle in OCT is known in advance (Figure 1 Critical). Try to place the tissue-mounted cryostat chuck onto the cryostat specimen holder in the desired direction. Subsequently, perform cryosection and attach to a microscope slide, and observe and check the angle of tissue or targeted structure such as hair follicles under the microscope for the first sections until have aligned full-length sections of hair follicles. If the structure is hard to observe, stain the section with a dye such as hematoxylin. Next, adjust the angle of tissue onto the cryostat specimen holder, repeat staining and adjusting until the angle is desired.
CRITICAL: Orientation is one of the most important, hard and time-consuming steps. Usually, there are two directions including longitudinal section and horizontal section of hair follicles. Taking the longitudinal section as an example, the specimen angle in OCT is known in advance (Figure 1 Critical). Try to place the tissue-mounted cryostat chuck onto the cryostat specimen holder in the desired direction. Subsequently, perform cryosection and attach to a microscope slide, and observe and check the angle of tissue or targeted structure such as hair follicles under the microscope for the first sections until have aligned full-length sections of hair follicles. If the structure is hard to observe, stain the section with a dye such as hematoxylin. Next, adjust the angle of tissue onto the cryostat specimen holder, repeat staining and adjusting until the angle is desired. -
h.Cryosectioning. Proceed to perform cryosection and obtain tissue sections. Gently flatten the tissue sections by touching the surrounding OCT with pre-cold brushes.Note: The optimal thickness for skin cryosections is 10 μm, while hair follicle cryosections should be 12 μm due to their tendency to fracture. Additionally, these thicknesses will allow the proper hybridization of the sample RNA to the chip library.
 CRITICAL: For the Spatial transcriptomics experiment, the section should be unbroken and flat. If the tissue is curly, it can be unfolded. If it breaks, it needs to be re-sectioned. The status of the section is critical for spatial transcriptomics due to the location of cells and genes being the main detection target.
CRITICAL: For the Spatial transcriptomics experiment, the section should be unbroken and flat. If the tissue is curly, it can be unfolded. If it breaks, it needs to be re-sectioned. The status of the section is critical for spatial transcriptomics due to the location of cells and genes being the main detection target. -
i.Mounting sections onto Stereo-seq chips.
-
i.The tissue section is laid flat on the cryostat operating table (−20°C) (Figure 7 Step 1).
-
ii.Take the edge of the chip with a pair of tweezers (room temperature).
-
iii.Mount the tissue section onto the chip by gently pressing the section with the front side of the chip (Figure 7 Step 2).
-
iv.As the OCT embedding compound melts, it facilitates the attachment of the sample to the chip, without the need for an additional coating matrix such as gelatine or polysine.
-
v.Drying the mounted chip. Flip the Stereo-seq Chip and immediately dry it with the front side facing up on the Water Bath-Slide Drier at 37°C for 6 min (Figure 7 Step 3).
-
i.
-
a.
-
6.Tissue Fixation.
-
a.Methanol fixation. Immediately immerse the tissue-mounted Stereo-seq Chip into the pre-cooled methanol and incubate for 30 min at −20°C, ensuring complete submersion of all tissue sections.
-
b.After fixation, relocate the 24-well plate containing the chips to a fume hood for safe handling of the specimens and chemicals.
-
c.Chip retrieval. Carefully remove the Stereo-seq Chip from the methanol using clean tweezers.
-
d.Residue removal. Gently wipe away excess methanol from the back of the chips and surrounding areas with dust-free paper, taking care not to touch the front surface of the chip where the tissue is mounted.
-
e.Methanol evaporation. Leave the Stereo-seq Chip in the fume hood for 4–6 min at room temperature to allow complete evaporation of the methanol.Note: Prepare Tissue ssDNA staining solution (100 μL/Chip) in advance while waiting for the methanol to evaporate.
-
f.Chip transfer. After the methanol has fully evaporated, immediately place the Stereo-seq Chip into a clean 10 cm Petri dish.
-
a.
-
7.Fluorescent Staining.
-
a.Addition of the tissue ssDNA staining solution. Add a total of 100 μL of the tissue staining solution per chip depositing one droplet at each corner of the chip. Then, add the rest of the solution to the center to merge all the droplets, ensuring the entire chip is covered with the staining solution.
-
b.Incubation. Ensure that the chip, especially the tissue section, is completely covered with the staining solution. Incubate the chip at room temperature on the bench for 5 min in a shaded area.
-
c.Washing steps. Slightly tilt the Stereo-seq Chip and carefully aspirate the staining solution from one corner using a pipette, avoiding contact with the chip. Apply 100 μL of Wash Buffer to each chip and aspirate it, as described for the staining solution.
-
d.Drying the chip. Place the chip onto dust-free paper and use a power dust remover to blow away any residual liquid, moving from one side to the other.
-
e.Slide preparation. Place 1 μL of nuclease-free (NF)-water on a glass slide. Carefully transfer the chip onto the glass slide using tweezers and adhere the back of the chip to the slide with the water droplet (Figure 8).Note: Slide preparation is only to facilitate the transfer of the chip to the microscope for observation and removal. So, this is an optional step.
-
f.Mounting the coverslip. Gently pipette 5 μL of glycerol onto the center of the tissue on the chip without introducing air bubbles. Using tweezers, place a coverslip onto the chip. Place on the edge while holding the other end, then gradually lower and cover the entire chip with the coverslip. Ensure that the chips are completely covered by glycerol and the coverslip.Note: To avoid the fluorescent signal decreasing, proceed to capture the fluorescent images as soon as possible, ideally within 1 h.
-
a.
-
8.
Fluorescence Imaging.
Fluorescent Staining is essential for delineating cell boundaries to facilitate cell segmentation (Figure 9; Figure 10).-
a.Stereo-seq chip positioning. Securely place the Stereo-seq Chip onto the imaging platform of choice.
-
b.Area Selection. Remove the light shield and select the specific chip area as the target for imaging. The imaging version is 1.1.60 in Stereo-seq OR 100 which can be replaced by a fluorescence microscope with a Z-capture function.
-
c.Image capture. Capture fluorescence images using the microscope with the following settings: FITC channel (488 nm), 10× objective lens, 200 ms exposure time, 3 W power LED lights, and a full scan of the capture area.
- d.
-
e.Chip separation. After imaging, carefully slide the chip with the coverslip using a pair of tweezers to detach the chip from the glass slide.
-
f.Coverslip removal. Hold the edge of the coverslip with a pair of tweezers and gently slide it off the Stereo-seq Chip until they are completely separated.Note: If there is a concern regarding potential damage to the tissue on the chip, incubate the chip with the coverslip in 5 × SSC at room temperature for 8 min. This allows for sufficient permeation and facilitates an easier separation of the chip from the coverslip.
-
a.
-
9.
H&E staining.
H&E staining is used for observing the organizational form and pathological features.-
a.Stain glass slides with 500 μL of hematoxylin for 2 min.
-
b.Rinse tissue slowly with tap water for 2 min.
-
c.Add 500 μL of Differentiation Solution dropwise and differentiate the stained tissue for 1 min, ensuring that the Differentiation Solution covers all areas of the tissue evenly.
-
d.Repeat step 9.2.
-
e.Apply approximately 500 μL of eosin staining solution dropwise for 1 min, ensuring that the dye evenly covers all tissue areas.
-
f.Repeat step 9.2.
-
g.Place the stained slides in a dryer for 3 min until the moisture on the surface of the slides is dried.
-
h.Add approximately 8 μL of glycerin to the glass slide, cover and seal the glass slide.
-
i.Record and name the sample number.
-
a.
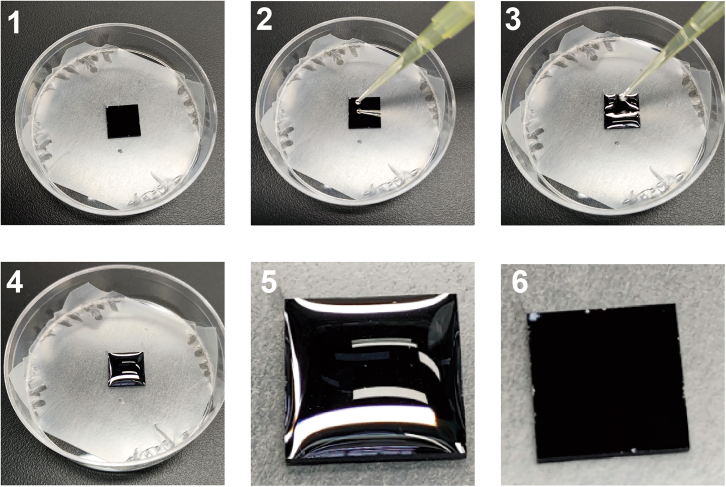
Figure 6.
Rinse the chip with NF-water using a pipette
Put the chip in a dry Petri dish (Step 1). Add 100 μL of NF-water to the edge of the chip (Step 2 and 3). Ensure that the surface of the chip is completely covered by NF-water (Step 4, Zoom-in in Step 5). Remove the liquid from the surface of the chip to complete a cleaning (Step 6).
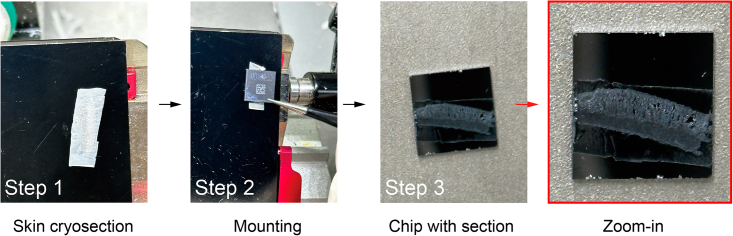
Figure 7.
Workflow of tissue mounting
For Step 1, perform cryosection with the desired angle to obtain an unbroken section, and then mount the section on the surface of the chip which needs the targeted structure is on the middle of the chip, such as in Step 3. In Step 3, provide a zoom-in image to see the tissue orientation on the chip.
Figure 8.
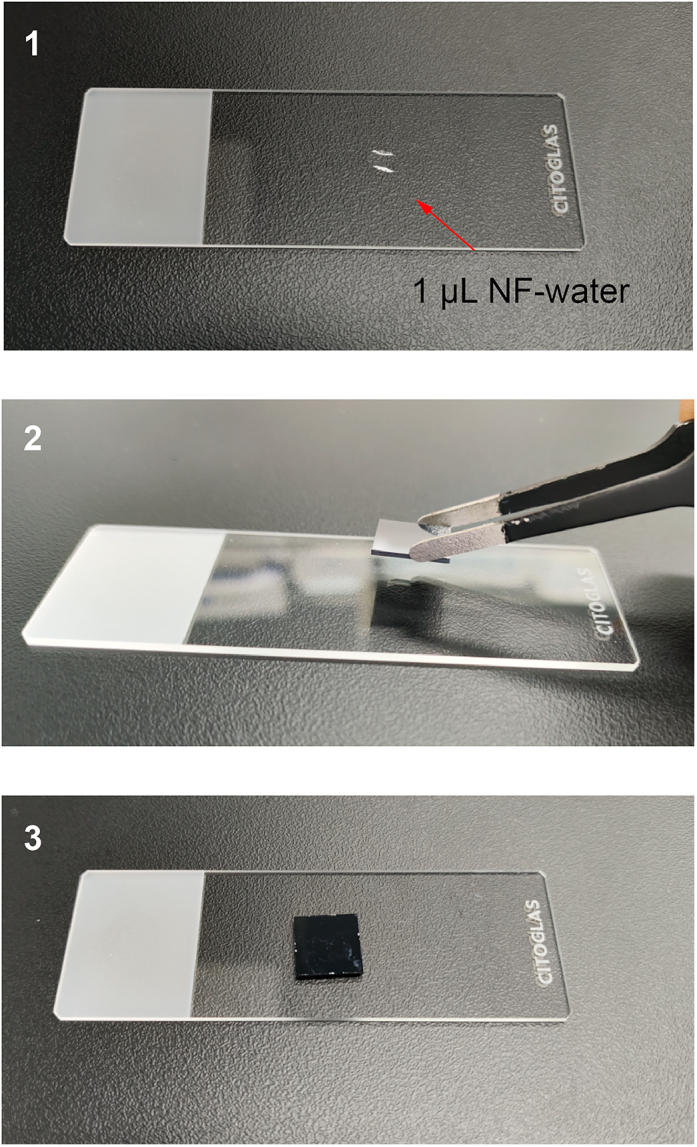
Transfer the chip on the glass slide to prepare imaging
Place 1 μL of NF-water on a glass slide (Step 1). Transfer the chip onto the glass slide using tweezers (Step 2) and adhere the back of the chip to the slide with the water droplet (Step 3).
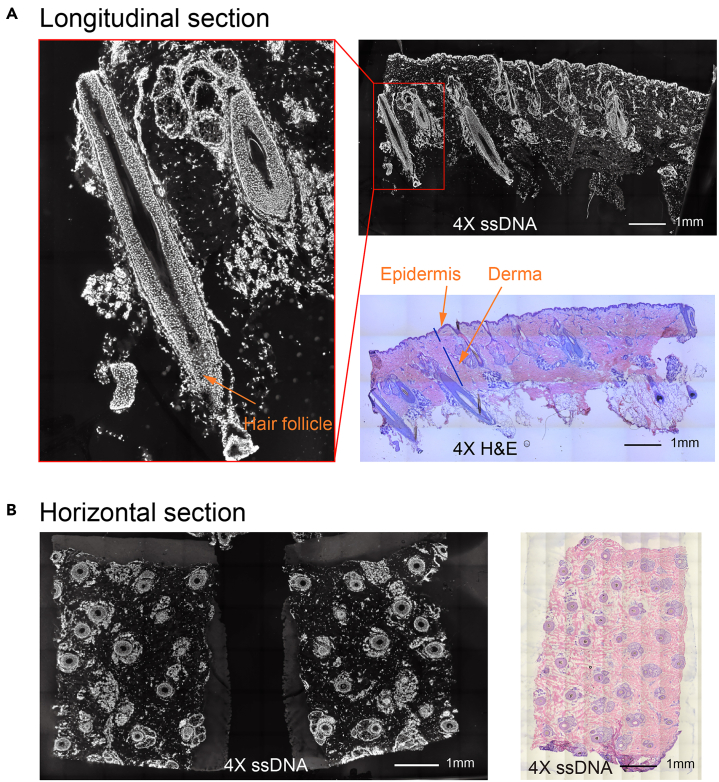
Figure 9.
ssDNA and H&E imaging of human scalp with Longitudinal section and Horizontal section
(A) Longitudinal section of the human scalp. The longitudinal section is made in the direction of hair follicle growth which allows us to observe the whole hair follicle structure such as marked in the left imaging. The skin structure mainly includes the epidermis and derma marked in the right H&E imaging except for hair follicles. The ssDNA and H&E staining are the adjacent sections, 4X magnification.
(B) Horizontal section of the human scalp. The horizontal section is made perpendicular to the direction of hair follicle growth which allows us to observe the cross-section of hair follicles. The ssDNA and H&E staining are the adjacent sections, 4X magnification.
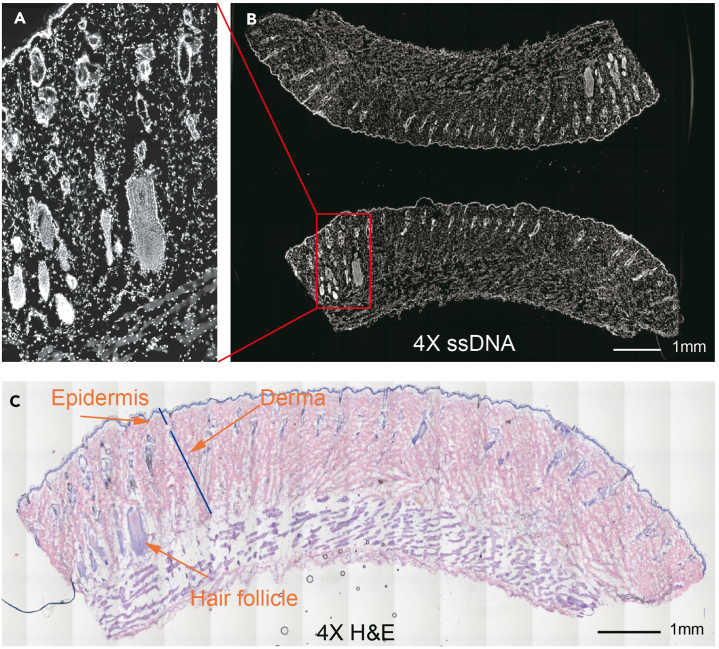
Figure 10.
ssDNA and H&E imaging of rat skin
Longitudinal section of rat skin. The ssDNA (A, B) and H&E (C) imaging are the adjacent sections, 4X magnification. The hair follicle zoomed in (A), and the epidermis and derma are marked.
Permeabilization, reverse transcription and cDNA collection
Timing: 1 day
Timing: 20–30 min (for step 10)
Timing: 3–5 h (for step 11)
Timing: no more than 30 min (for step 12)
Timing: 3–5 h (for step 13)
Timing: 6–12 h (for step 14)
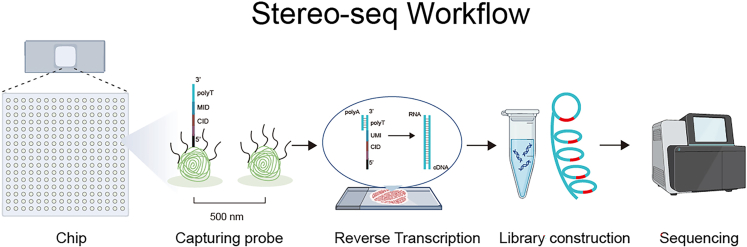
This step involves conducting the Spatial transcriptomics experiment, which includes permeabilizing the tissue section for probe hybridization with RNA using the imaging chip of Stereo-seq, reverse transcription to cDNA and collection for sequencing (Figure 11).
Note: Step Four is mainly from the Stereo-seq protocol and, it is changed the details depending on skin tissues. Stereo-seq can be replaced by other spatial transcriptomics, but the main steps are the same including Tissue Permeabilization, Reverse Transcription, Tissue Removal, cDNA release, amplification and sequencing.
-
10.Tissue Permeabilization.
-
a.Permeabilization solution. Gently apply 150 μL of 1 × Permeabilization Reagent Solution onto the chip. Start by pipetting one droplet at each corner and then add the remaining solution to the center to combine all droplets.Note: Ensure that the chip is entirely covered with the 1 × Permeabilization Reagent Solution and avoid drying the specimen during the process.
-
b.Incubation temperature and time. Place the chip in the thermostat set to 37°C and allow it to incubate for 12 min.
-
c.Solution removal. Carefully tilt the Stereo-seq chip and aspirate the 1 × Permeabilization Reagent Solution as much as possible from the corner using a pipette, avoiding contact with the chip surface.
-
d.Washing. Gently rinse the chip with 150 μL of 0.1 × SSC and then aspirate the solution in the same manner described in the previous step. Ensure the chip retains some moisture.Note: Proceed immediately with the reverse transcription step to prevent RNA degradation.
-
a.
-
11.Reverse Transcription.
-
a.RT mix application. Carefully dispense 100 μL of the pre-prepared RT Mix onto the chip in the same manner as previously described, ensuring the tissue is fully covered.
-
b.Incubation temperature and time. Place the chip with the RT Mix onto it in the thermostat set to 42°C for 3 h.
-
c.Solution removal. After incubation, remove the chip and aspirate the solution in the same manner as described before, being careful not to touch the chip surface.
-
d.Washing. Rinse once with 100 μL 0.1 × SSC gently and transfer the chip to a 24-well plate.
-
a.
-
12.Tissue Removal
-
a.Tissue release. Add 400 μL of TR Buffer to each well containing the chips transfer the 24-well plate to the thermostat set at 55°C, and incubate for 10 min.
-
b.Transfer the chip to an empty well within the same 24-well plate.
-
c.Residual tissue removal. Add 400 μL 0.1 × SSC to the new well and gently pipette up and down to help detach any remaining tissue from the chip.
-
d.Solution removal. Slightly tilt the 24-well plate and aspirate the 0.1 × SSC solution from the corner of the well, ensuring it does not contact the chip surface.
-
a.
-
13.cDNA Release.
-
a.Chip relocation. Transfer the Stereo-seq chip into a new well of the 24-well plate.
-
b.cDNA release. Add 400 μL of cDNA release reagent to the well. Seal the 24-well plate with Clear Adhesive Film to prevent evaporation.
-
c.Incubation temperature and time. Place the 24-well plate in the thermostat set to 55°C and incubate for 3 h.
-
d.cDNA collection. After completing the cDNA release step, remove the 24-well plate from the thermostat. Collect the solution from the well and transfer it to a 1.5 mL Eppendorf tube.
-
e.Residual cDNA collection. Add 350 μL of NF-Water to the well. Using a pipette, combine it with the previously collected 400 μL solution. Add NF-water to achieve a final volume of 750 μL.
-
a.
-
14.
cDNA Purification Amplification and sequencing.
Figure 11.
The Stereo-seq workflow from RNA capturing to sequencing
The capturing probe contains polyT, molecular identity (MID) and coordinates identity (CID). PolyT is used for capturing polyA sequences on the 3′ RNA, CID and MID are spatial barcodes for alignment RNA location. After tissue RNA is captured by probes, reverse transcription to extend and obtain the complementary DNA. Collect cDNA for amplification and sequencing.
The detailed methodologies are outlined in the STOmics kit protocols, which are standard for such experiments. Therefore, specific details are not provided here, and the readers are referred to the manufacturer’s instructions.
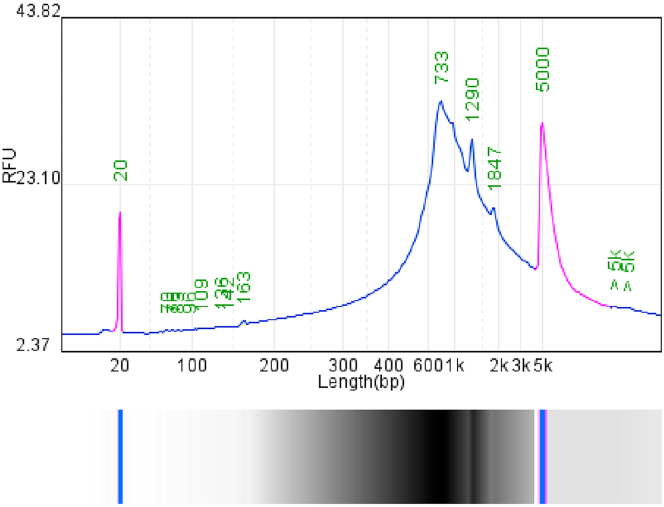
The quality control of cDNA includes the fragment distribution which is mainly from 600-1200 bp (Figure 12) and the cDNA output is greater than 20 ng using normal amplification protocol.
Figure 12.
The cDNA fragment range of skin tissue
After cDNA purification and amplification, the existing cDNA needs to be done quality control mainly detecting the main fragment and peak of cDNA. Qualified qualities include the peak being in the range of 600–1200 bp (Here is 733 bp) while the main fragment is in the range of 600-2000 bp.
Send the purified and amplified cDNA for Stereo-sequencing. In this protocol, we sent it to the China National GeneBank (CNGB) facilities.
CRITICAL: Based on the tissue coverage, it is recommended that the cDNA library of each tissue be sequenced for more than 5G total reads.
Expected outcomes
The protocol establishes successful, learnable processes of mammalian skin for spatial transcriptomics, here using Stereo-seq to perform the experiment and quality control including:
-
•
Tissue integrity. The skin tissue sections should be intact and unbroken, with the structure of the skin preserved and not deformed.
-
•
RNA quality. The RNA quality, as indicated by the RNA Quality Number (RIN), should best be higher than 7. The tissue RNA value between 5 and 7 is a risk organization, and these types of tissues also can be performed in this protocol with lower expectations on gene number.
-
•
ssDNA staining. Each cell should be distinctly visible in the ssDNA staining imaging and the quality control of ssDNA imaging will be done by a Stereo-seq automatic imaging control system (https://en.stomics.tech/).
-
•
H&E staining. Each cell and structure should be distinctly visible in the H&E-stained sections and captured images.
-
•
cDNA fragmentation and value. The cDNA fragment peak should range between 600-1200 bp and the total product yield should exceed 20 ng.
Based on test results, the success of the experiment hinges on the tissue section integrity, RNA quality, and cDNA fragmentation. Additionally, ssDNA staining can significantly impact subsequent analyses, particularly cell segmentation. It’s important to note that H&E imaging primarily serves histomorphological observations and does not interfere with spatial transcriptomics in Stereo-seq.
In practice, adhering to the protocol, particularly regarding tissue section integrity and RNA quality, has consistently led to successful Stereo-seq experiments (more than 4k genes in every 100 μm × 100 μm within tissue areas), almost 100%. Before implementing these measures, obtaining intact sections and high RNA quality posed a significant challenge, resulting in a lack of sequencing results for all experiments.
Limitations
Although this protocol details steps for enhancing the preparation of mammalian skin including hair follicles for spatial transcriptomics, the success rate is not guaranteed to be 100% due to tissue heterogeneity, especially in human skin. And the expected success rate is more than 80%. Consequently, critical stages such as tissue preparation and sectioning necessitate considerable practice by researchers. Additionally, the methods primarily focus on human and rat skin; therefore, the protocol may need further adaptation and testing for other species.
Troubleshooting
Problem 1
The hair follicles of the human scalp vary in thickness, resulting in differing hardness among adjacent hair follicles. This variability can lead to difficulties in obtaining intact sections, as thick hair may resist being cut cleanly.
Potential solution
(Step 8, Figure 9, Longitudinal section and Horizontal section).
-
•
In such cases, consider adjusting the orientation of the tissue. Generally, when the blade is parallel to the hair follicle, challenges may arise due to the impact of thicker hair follicles on the blade. By orienting the blade perpendicular to the hair follicle, you can minimize the damage and enhance the overall integrity of the tissue section.
Problem 2
The RNA quality of the skin tissue is not adequately preserved, leading to suboptimal data quality with low RIN of less than 7 and cDNA fragments of insufficient length. Contributing factors include.
-
•
Prolonged sampling and embedding time for skin and hair follicle samples, with exposure to air exceeding 3 h, can accelerate RNA degradation.
-
•
Storage temperatures of the embedded samples are above −60°C and the storage duration exceeds 3 years.
-
•
Repeated freezing and thawing of stored samples.
Potential solution
(Step 1).
-
•
Optimize the dissection time and sample processing. The Fat removal step should be completed within 1 h. With sufficient expertise, strive to finish within 30 min to ensure the freshness of the sample and prevent RNA degradation.
-
•
Optimize storage conditions. Maintain the storage temperature of the embedded samples at −80°C and do not store them for more than 3 years to preserve RNA integrity.
-
•
Sample handling. Avoid repeated freezing and thawing of stored samples to minimize RNA degradation.
Problem 3
In cases where the cDNA amplification product is below 20 ng or the fragment distribution does not concentrate within the 600–1200 bp range, experimental issues may arise, leading to an inability to sequence or yielding poor sequencing quality.
Potential solution
(Steps 1, 5, 14).
Address the issue based on the specific circumstance.
-
•
Adequate fragment size, insufficient total RNA. If the cDNA amplification product is less than 20 ng but the fragment distribution is concentrated within the 600–1200 bp range, the RNA length is normal, but the total amount is insufficient. Begin by examining the size of the tissue area attached to the 1 cm × 1 cm Stereo-seq chip. If it is too small, the total RNA quantity will naturally be low. Ensure the tissue area is greater than 6 mm × 6 mm or consider affixing multiple smaller tissues to a single chip. If the tissue size is adequate but the total RNA amount remains low, consider increasing the number of PCR cycles, generally by increments of up to 10 cycles at a time.
-
•
Inadequate fragment size and total RNA. When the cDNA amplification product is less than 20 ng and the fragment distribution is not within the 600–1200 bp range, this indicates both short RNA length and insufficient total amount. Assess the fragment distribution after several rounds of PCR cycles. If the fragments remain short and low in quantity, this stage of the process fails. Check the RIN value of the sample and review the previously mentioned steps in the experimental process for any issues.
Resource availability
Lead contact
Chuangyu Liu (liuchuanyu@genomics.cn).
Technical contact
Yujia Jiang (yujia.jiang@bio.ku.dk; jiangyujia@genomics.cn).
Materials availability
All the materials used in this protocol are commercially available. For specifics regarding Stereo-seq, visit www.stomics.tech/sap/.
Data and code availability
Detailed information on the Stereo-seq experimental protocol is available in the publication by Chen et al.1
Acknowledgments
This research was supported by the Guangdong Provincial Key Laboratory of Genome Read and Write (no. 2017B030301011) and Shenzhen Key Laboratory of Single-Cell Omics (no. ZDSYS20190902093613831).
We thank all members of the Center for Digitizing Cells from the Institute of SuperCells (BGI) for their helpful comments. Additionally, we thank the China National GeneBank for providing sequencing services for this project.
We thank the members of the Department of Burn Surgery, the First Affiliated Hospital of Naval Medical University, Shanghai, China for providing rat skin tissues, including Dayuan Xu, Jianyu Lu, and Yushu Zhu.
We thank Minjuan Wu who is from the Department of Histology and Embryology, Naval Military Medical University, Shanghai, China for providing suggestions about the structures of rat skin.
The graphical abstract was created using BioRender.
Author contributions
Y.J. and C.L. conceived the idea. Y.J., R.L., and Y.L. conducted the experiments. Y.J. wrote the manuscript with input from R.L. and Y.L. J.X. and J.F. assisted with the writing. C.L., X.W., and M.P.-M. supervised the study and provided substantial revisions to the manuscript. R.H., W.Z., S.X. and J.Z. provided rat skin tissues, and Y.X., Z.Z. and J.Z. provided human skin tissues. All authors reviewed and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xiaoyu Wei, Email: weixiaoyu@genomics.cn.
Chuanyu Liu, Email: liuchuanyu@genomics.cn.
Reference
- 1.Chen A., Liao S., Cheng M., Ma K., Wu L., Lai Y., Qiu X., Yang J., Xu J., Hao S., et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022;185:1777–1792.e21. doi: 10.1016/j.cell.2022.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Detailed information on the Stereo-seq experimental protocol is available in the publication by Chen et al.1