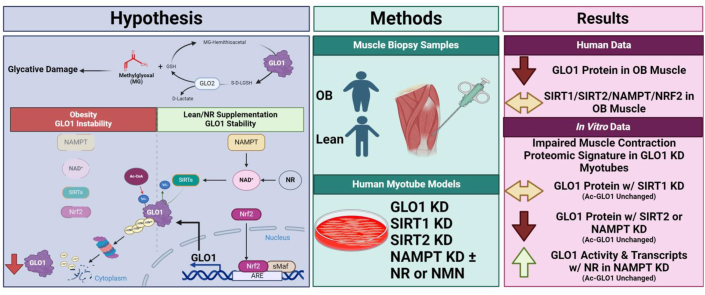
Abstract
Glyoxalase I (GLO1) is the primary enzyme for detoxification of the reactive dicarbonyl methylglyoxal (MG). Loss of GLO1 promotes accumulation of MG resulting in a recapitulation of diabetic phenotypes. We previously demonstrated attenuated GLO1 protein in skeletal muscle from individuals with type 2 diabetes (T2D). However, whether GLO1 attenuation occurs prior to T2D and the mechanisms regulating GLO1 abundance in skeletal muscle are unknown. GLO1 expression and activity were determined in skeletal muscle tissue biopsies from 15 lean healthy individuals (LH, BMI: 22.4 ± 0.7) and 5 individuals with obesity (OB, BMI: 32.4 ± 1.3). GLO1 protein was attenuated by 26 ± 0.3 % in OB compared to LH skeletal muscle (p = 0.019). Similar reductions for GLO1 activity were observed (p = 0.102). NRF2 and Keap1 expression were equivocal between groups despite a 2-fold elevation in GLO1 transcripts in OB skeletal muscle (p = 0.008). GLO1 knock-down (KD) in human immortalized myotubes promoted downregulation of muscle contraction and organization proteins indicating the importance of GLO1 expression for skeletal muscle function. SIRT1 KD had no effect on GLO1 protein or activity whereas, SIRT2 KD attenuated GLO1 protein by 28 ± 0.29 % (p < 0.0001) and GLO1 activity by 42 ± 0.12 % (p = 0.0150). KD of NAMPT also resulted in attenuation of GLO1 protein (28 ± 0.069 %, p = 0.003), activity (67 ± 0.09 %, p = 0.011) and transcripts (50 ± 0.13 %, p = 0.049). Neither the provision of the NAD+ precursors NR nor NMN were able to prevent this attenuation in GLO1 protein. However, NR did augment GLO1 specific activity (p = 0.022 vs NAMPT KD). These perturbations did not alter GLO1 acetylation status. SIRT1, SIRT2 and NAMPT protein levels were all equivocal in skeletal muscle tissue biopsies from individuals with obesity and lean individuals. These data implicate NAD+-dependent regulation of GLO1 in skeletal muscle independent of altered GLO1 acetylation and provide rationale for exploring NR supplementation to rescue attenuated GLO1 abundance and activity in conditions such as obesity.
Keywords: Dicarbonyl stress, Nicotinamide riboside, NAD+, Methylglyoxal
Graphical abstract
1. Introduction
As of 2016, 13 % of the world's adults are classified as obese (body mass index [BMI] ≥ 30 kg/m2), nearly triple the prevalence of 1975 [1]. This proportion is likely diminished by the inclusion of impoverished and malnourished countries. In well developed countries the rate is significantly higher such as in the United States, where one third of the adult population is classified as having obesity. BMI of this magnitude increases the risk of type 2 diabetes (T2D) 10–23-fold depending on the severity of obesity. The development of skeletal muscle insulin resistance is a primary driver of T2D development in the context of obesity as this tissue is responsible for >85 % of insulin stimulated glucose disposal [2].
A myriad of mechanisms have been attributed to the development of skeletal muscle insulin resistance in the context of obesity. There is mounting evidence that the accumulation of dicarbonyl stress may also be contributing to this process and should be investigated as a potential driver of insulin resistance in skeletal muscle [3]. Methylglyoxal (MG) is the most abundant dicarbonyl in vivo which is formed in proportion to glycolytic flux primarily through triosephosphate degradation. Accumulation of MG with obesity and diabetes has been previously reported in animals [4,5] and humans [[6], [7], [8], [9]]. Aberrant MG accumulation leads to direct protein damage by modifying lysine and arginine residues to form irreversible adducts. In mouse models of MG accumulation, MG impairs insulins sensitivity by directly modify insulin signaling proteins such as IRS-1 in skeletal muscle [[10], [11], [12]]. The glyoxalase enzymatic system consisting of glyoxalase I (GLO1) and GLO2 is a highly efficient detoxification mechanism to prevent the accumulation of MG.
Deletion of GLO1 results in the development of diabetes and insulin resistance in drosophila [13], zebrafish [14] and mice [15]. In addition, our lab has previously demonstrated that protein of GLO1 is attenuated in skeletal muscle from individuals with obesity and T2D and positively correlates with insulin sensitivity [9]. However, it is still unclear if attenuation of skeletal muscle GLO1 expression occurs with obesity prior to diabetes development. In addition, the mechanisms by which GLO1 protein expression is regulated in skeletal muscle are not known.
Interestingly, APOE null mice fed a high fat diet develop obesity, metabolic-associated fatty liver disease (MAFLD), and have attenuated liver GLO1 protein expression [16]. Treating hepatocytes with palmitate to model the metabolic milieu of MAFLD leads to hyperacetylation of GLO1, leading to its ubiquitination and degradation thereby lowering GLO1 protein abundance in these cells. In addition, more recent screens have also suggested that GLO1 acetylation may regulate its activity and protein abundance [17,18]. In an acetylome peptide screen GLO1 was deacetylated by the NAD+-dependent deacetylase sirtuin 2 (SIRT2) which like GLO1 primarily resides in the cytosol [17]. SIRT1 also has been implicated in regulation of GLO1 in an endothelial cell model of glycemic stress [19]. SIRT1 and SIRT2 have been reported to attenuate the development of insulin resistance in the context of obesity [[20], [21], [22], [23], [24], [25]]. Further, obesity has also been reported to down regulate SIRT1 and SIRT2 abundance and activity potentially via the loss of NAD+, which has been linked to impairments in the NAD+ scavenging enzyme nicotinamide phosphoribosyl transferase (NAMPT) [[26], [27], [28]]. These observations are further supported by studies in which provision of NAD+ precursors attenuated the development of insulin resistance in these contexts [[29], [30], [31]].
Therefore, the goals of this study were twofold: 1) Determine if GLO1 protein expression is altered in skeletal muscle from individuals with obesity compared to age-matched individuals without obesity. 2) Determine the effects of SIRT1, SIRT2, NAMPT, and the NAD precursors nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN) on GLO1 protein abundance, acetylation, activity, and gene expression in human immortalized myotubes. We hypothesized that skeletal muscle GLO1 protein abundance and activity would be lower in individuals with obesity compared to age-matched individuals without obesity. We also hypothesized knockdown (KD) of SIRT1, SIRT2 or NAMPT in human immortalized myotubes would demonstrate hyperacetylated GLO1 along with lower GLO1 protein abundance and activity which in the case of NAMPT KD could be prevented by NR co-treatment.
2. Methods
2.1. Participants
The current investigation is a retrospective analysis of a larger study that consisted of four sequential visits as previously described [32,33]. All experimental protocols were approved by the Institutional Review Board at the University of Illinois of Chicago (IRB Approval #: 2015-0127), and all subjects gave consent for biological specimens to be kept by the investigator (JMH) for use in future research. Prior to enrollment, all participants provided verbal and written informed consent. Baseline measures including height, weight, body fat percentage via dual x-ray absorptiometry (DXA), and maximal aerobic capacity (VO2Max) were collected on the first visit of the study. These baseline physiologic data are represented in Table 1 and include n = 18 LH and n = 6 OB individuals. These numbers may differ from the data represented in certain figures due to limitations in biospecimen availability. For example, 2 of the LH participants and 1 participant in the OB group did not have sufficient protein sample to perform Western blotting for GLO1 but did have sufficient cDNA for ddPCR. Apart from these demographic data, all data in this current reporting were derived from the baseline skeletal muscle biopsy [32]. Three days before the visit participants completed diet and physical activity logs, which they were asked to replicate in the days leading up to their subsequent visits. Participants were also instructed to abstain from vigorous exercise, and alcohol consumption 48 h prior to each visit, and caffeine consumption 24 h prior to each visit. Participants were asked to arrive at each visit by sedentary means (i.e. car, public transportation etc.) and having fasted for at least 12 h. Testing of all participants was done in the early morning (0700–0900 h) to account for any diurnal variations in outcome measures and to minimize the participants’ burden of having to be fasted for the study.
Table 1.
Participant characteristics.
| Variable, Units | LH, n = 18 | OB, n = 6 | p |
|---|---|---|---|
| Age, years | 25 ± 0.9 | 28 ± 1.8 | 0.202 |
| Sex, %F | 44 % | 40 % | – |
| Weight, kg | 68.2 ± 2.3 | 94.3 ± 5.9 | <0.0001 |
| BMI, kg/m2 | 22.4 ± 0.5 | 32.6 ± 1.1 | <0.0001 |
| Body Fat, % | 23.8 ± 1.4 | 38.3 ± 2.8 | <0.0001 |
| VO2Max, ml/kg/min | 47.3 ± 1.8 | 37.8 ± 3.0 | 0.012 |
| Glucose, mg/dL | 91.5 ± 2.1 | 89.1 ± 4.2 | 0.585 |
| Insulin, mU/L | 4.9 ± 0.5 | 11.6 ± 3.4 | 0.005 |
| HOMA-IR | 1.1 ± 0.1 | 2.5 ± 0.7 | 0.006 |
Baseline anthropometric and metabolic characteristics of lean healthy (LH) individuals and individuals with obesity (OB) are presented. Data are presented as mean ± SEM and were compared via unpaired students T-Test. Significance set at p < 0.05). Sample sizes for all outcomes except glucose, insulin, and HOMA-IR are n = 18 for LH and n = 6 for OB. Sample sizes for glucose, insulin, and HOMA-IR are n = 15 for LH and n = 5 for OB.
2.2. Skeletal muscle biopsy and muscle tissue homogenization
Skeletal muscle biopsies were taken from the m. vastus lateralis under local anesthetic (Lidocaine, 2 % without epinephrine) as previously described [[32], [33], [34], [35]]. The extracted skeletal muscle tissue was quickly cleaned of all visible connective tissue, fat, and blood. Portions of the samples designated for protein extraction for Western blotting experiments were then flash frozen in liquid nitrogen. Portions of the samples designated for RNA isolation were placed in RNALater (Invitrogen) prior to freezing in liquid nitrogen. Samples were then stored at −80 °C until analysis. Approximately 10 mg of skeletal muscle tissue was weighed in a custom-built freezer and homogenized with ceramic beads (Lysing Matrix D; FastPrep®-24; MP Bio, Santa Ana, CA) in 20 vol of ice cold 1X Cell Lysis Buffer (#9803, Cell Signaling Technology, Danvers, MA) supplemented with 1X Protease/Phosphatase Inhibitor Cocktail (#5872, Cell Signaling Technology, Danvers, MA). Protein concentration for each sample homogenate was determined by a commercially available bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL).
2.3. Human immortalized myotube model
These cells were developed at the University of Texas Southwestern in the lab of Dr. Woodring Wright as previously described [36]. To develop this immortal cell line biopsy explants from a deltoid biopsy were cultured to isolate myoblasts. After expansion, myoblasts were then infected with pBabe vectors containing cyclin-dependent kinase 4 (CDK4) and human telomerase (hTERT) as well as flanking LoxP sites to allow for excision of either or both expression cassettes. These immortalized cells were chosen to avoid issues with myoblast senescence that occurs rapidly in human primary myoblasts (∼8–10 passages) while also being able to probe these mechanisms in human derived tissues to preserve translatability [37].
2.4. Cell culture conditions
In attempt to closely mimic the physiologic environment and limit the oxidative stress of canonical tissue culture conditions of growing cells under physiological hyperoxic conditions (∼20 % O2), we cultured the human immortalized myoblasts and differentiated myotubes at oxygen tensions more closely mimicking those experienced by skeletal muscle in vivo (∼5 %). To produce this physiological environment, cells were placed in low-oxygen chambers and the ambient air was displaced by purging low oxygen gas (2 % O2, 5 % CO2, N2 balance) for 4 min at ∼5 psi through re-sealable inlets consistent with previous published descriptions of this method [38]. Chambers were subsequently sealed and placed in a humidified tissue culture incubator. This process achieves a final oxygen tension of 2–5% within the incubation chamber. We pre-determined that there was no effect of performing tissue culture at the desired 2–5% O2 tension on NAD concentration, GLO1 protein or NAMPT protein (Data Not Shown).
Cells were cultured in custom growth media designed to accommodate optimum, healthy growth of immortalized myoblasts. The growth media was made in batches of 500 mL and consisted of 4:1 DMEM (SH30021FS, Hyclone): M199 (SH3025301, Hyclone) (referred to as Media X or MX), 15 % fetal bovine serum (FBS) (10437028, Fisher), 50 mM HEPES buffer (15630-080, Invitrogen), 0.03 μg/mL Zing Sulfate (Zn2SO4) (S68-500, Fisher Scientific), 1.4 μg/mL Vitamin B12 (V2786, Sigma), and 55 ng/mL Dexamethasone (72092, Stem Cell Technology). Media was brought up to a final glucose concentration of 25 mM by adding d-glucose and was then sterile filtered through a 0.22 μm vacuum driven filtration system into 50 mL aliquots (Steriflips, SCGP00525, Sigma). Hepatocyte growth factor (HGF) (GF116, Chemicon International) and basic fibroblast growth factor (bFGF) (HRP-0011, BioPioneer) were added to 50 mL aliquots of sterile media at final concentrations of 2.5 ng/mL and 10 ng/mL respectively on a weekly basis to avoid the degradation of the growth factors in media.
Prior to initiating the tissue culture, tissue culture plates were coated with 0.1 % pigskin gelatin. To initiate tissue culture, cryopreserved cells were seeded on 10 cm2 plates at least 1.5 × 106 cells per plate. In anticipation of ∼70 % attrition following cryopreservation, this would result in a seeding density of ∼1.0 × 106 live cells per 100 mm plate (1 × 104 live cells per mm) which was the seeding density used when passaging cells into subsequent 100 mm plates. The day after seeding, the growth media was replaced with 10 mL of fresh, warmed growth media. Growth media was changed every 48 h from then on until cells reached at least 80 % confluence at which point they were either passaged, cryopreserved, or differentiated depending on the need.
Differentiation of myoblasts into myotubes was performed by switching the cells to MX with 2 % Horse Serum (26050088, Fisher) and 25 mM glucose when they reached at least 85 % confluency. Cells were left in differentiation media for 5 days and replaced with fresh media every 48 h during that period.
2.5. Cell experimental conditions
After differentiation transfection of siRNAs were performed in MX + 10 % FBS with 25 mM Glucose supplemented with scRNAs or siRNAs against SIRT1, SIRT2, NAMPT, or GLO1 and RNAiMax (13778150, Fisher) diluted in OptiMEM (31985062, Fisher). 0.25 mL of diluted scRNA or siRNA with RNAiMax were added dropwise onto appropriate wells containing 1.75 mL of pre-warmed MX + 10 % FBS and 25 mM Glucose. Cells were then incubated for 48 (SIRT1, SIRT2, and NAMPT siRNA) or 72 (GLO1 siRNA) hours as indicated before being harvested as described previously. For TMT proteomic analyses, siRNA and scRNA treatments (GLO1 and NAMPT) were 72 h. For NR rescue experiments in NAMPT siRNA cells, 50 mM of NR (NC1627048, Cayman Chemical) or NMN (N3501, Millipore Sigma) dissolved in water was added to cells in the last 24 h of being treated with NAMPT siRNA.
2.6. Cell lysis for protein extraction for Western blotting
Frozen cell pellets were lysed in ice-cold 1X Cell Lysis Buffer (CST), supplemented with 1X protease/phosphatase inhibitor cocktail (CST), as well as 10 mM nicotinamide (NAM, N06336-100G, Sigma Aldrich), and 1 μM Trichostatin A (TSA, T8552-1 MG, Sigma Aldrich) to inhibit sirtuin and HDAC activity respectively to preserve the acetylation state of proteins at the time of sample collection. Following resuspension of pellets in lysis buffer, samples were frozen in liquid N2 and then thawed on ice for cryo-lysis. Next, samples were sonicated on ice two times for 30 s. Lastly, samples were centrifuged at 14,000 X G for 10 min at 4o C. The supernatant was transferred to a new tube to be used for downstream Western blotting and immunoprecipitation (IP) applications. An aliquot of the supernatant was diluted and assayed for protein concentration via BCA (Pierce). This cell lysis procedure was also used for samples analyzed for GLO1 content via ELISA and GLO1 activity except for the buffer used. For these applications the GLO1 activity assay buffer provided with the purchased kit was supplemented with protease/phosphatase inhibitors, 10 mM NAM and 1 μM TSA.
2.7. Western blotting
Aliquots of human skeletal muscle homogenate or cell lysates containing 10–20 μg of total protein were diluted in equal volumes of 2x Laemmli Buffer (BioRad, 1610737, Hercules, CA) with 5 % β-mercaptoethanol (βME) (BioRad, 1610710) prior to heating at 90 °C for 10 min. Denatured samples were brought to room temperature, loaded onto 10 % pre-cast Criterion TGX gels (5671033, BioRad) and separated by SDS-PAGE at 200 V for 50 min. Separated proteins were then transferred to nitrocellulose membranes via Transblot Turbo semi-dry transfer system for 11 min (BioRad). Membranes were then blocked with Protein-Free Blocking Buffer (PFBB, 92780003, Li-Cor, Lincoln, NE) for 1 h at room temperature with gentle rocking. After blocking, membranes were cut and probed with primary antibodies against SIRT1 (1:500, #9475, Cell Signaling Technology), NAMPT (1:1000, A0256, ABclonal), and GLO1 (1:1000, SC-133144, Santa Cruz Biotechnology, Dallas, TX), NRF2 (1:500, AB-62352, ABCAM, Cambridge, UK), and KEAP1 (1:1000, A1820, ABclonal, Woburn, MA), or SIRT2 (1:1000, #12650S, Cell Signaling Technology) depending on the predicted molecular weight of the target. The next day, membranes were removed from primary antibody solutions, washed and incubated with an 800 nm (Green) fluorophore-conjugated secondary antibody (1:20,000, Li-Cor, Lincoln, NE) diluted in PFBB + 0.1 % Tween-20 for 1 h at room temperature with gentle rocking while protected from light. The secondary antibody solution was subsequently removed, and the membranes were washed before scanning on an Odyssey CLx Imaging Systemchannel(Li-Cor). After imaging was completed for the proteins of interest, membranes were incubated for 30 s in 5 mL of REVERT total protein stain (Li-Cor). Total protein images were scanned on the 680 nm (red) channel using imaging cabinet (BioRad). Fluorescent signals were both quantified on Image Studio software (V4.0.21; Li-Cor) using the Western blot quantification function. All target protein signals were made relative to total protein stain acquired from the REVERT staining.
2.8. TMT LC-MS/MS
Frozen cell pellets were lysed in ice-cold 1X RIPA buffer (CST) as described above for other cell samples. The resultant supernatants were aliquoted to be used for downstream Western blotting, and TMT-LC-MS/MS applications. An aliquot of the supernatant was diluted and protein concentration was adjusted to 2 mg/mL. Samples of 75 μg were lysed and labeled with TMT 18-plex via manufacturer's protocol (Thermo Fisher Scientific). Briefly, upon reduction (5 mM DTT, for 30 min at 45 C) and alkylation (15 mM 2-chloroacetamide, for 30 min at room temperature) of cysteines, the proteins were precipitated by adding 6 vol of ice-cold acetone followed by overnight incubation at −20 °C. Precipitates were spun down, and the pellets were allowed to air dry. The pellet was re-suspended in 0.1 M TEAB and overnight (∼16 h) digestion with trypsin/Lys-C mix (1:25 protease:protein; Promega) at 37 °C was performed with constant mixing using a thermomixer. The TMT 18-plex reagents were dissolved in 41 μl of anhydrous acetonitrile and labeling was performed by transferring the entire digest to TMT reagent vial and incubating at room temperature for 1 h Reaction was quenched by adding 8 μl of 5 % hydroxyl amine and further 15 min incubation. Labeled samples were mixed together, and dried using a vacufuge. An offline fractionation of the combined sample (∼200 μg) into 8 fractions was performed using high pH reversed-phase peptide fractionation kit according to the manufacturer's protocol (Pierce; Cat #84868). Fractions were dried and reconstituted in 9 μl of 0.1 % formic acid/2 % acetonitrile in preparation for LC-MS/MS analysis. Table 2 depicts the isobaric channel assigned to each sample. To obtain superior quantitation accuracy, we employed multinotch-MS3 (McAlister GC) which minimizes the reporter ion ratio distortion resulting from fragmentation of co-isolated peptides during MS analysis. Orbitrap Fusion (Thermo Fisher Scientific) and RSLC Ultimate 3000 nano-UPLC (Dionex) was used to acquire the data. Two μl of the sample was resolved on a PepMap RSLC C18 column (75 μm i.d. x 50 cm; Thermo Fisher Scientific) at the flow-rate of 300 nl/min using 0.1 % formic acid/acetonitrile gradient system (2–22 % acetonitrile in 150 min; 22–32 % acetonitrile in 40 min; 20 min wash at 90 % followed by 50 min re-equilibration) and directly spray onto the mass spectrometer using EasySpray source (Thermo Fisher Scientific). Mass spectrometer was set to collect one MS1 scan (Orbitrap; 120K resolution; AGC target 2 × 105; max IT 100 ms) followed by data-dependent, “Top Speed” (3 s) MS2 scans (collision induced dissociation; ion trap; NCE 35; AGC 5 × 103; max IT 100 ms). For multinotch-MS3, top 10 precursors from each MS2 were fragmented by HCD followed by Orbitrap analysis (NCE 55; 60K resolution; AGC 5 × 104; max IT 120 ms, 100–500 m/z scan range) [39].
Table 2.
Isobaric Tagging Scheme for GLO1 and NAMPT siRNA Proteomic Experiments.
| Sample ID | TMT Isobaric Tag |
|---|---|
| scRNA1 | 126 |
| GLO1 siRNA1 | 127 N |
| NAMPT siRNA1 | 127C |
| GLO1 siRNA2 | 128 N |
| NAMPT siRNA2 | 128C |
| scRNA2 | 129 N |
| NAMPT siRNA + NR1 | 129C |
| GLO1 siRNA3 | 130 N |
| NAMPT siRNA + NR2 | 130C |
| GLO1 siRNA4 | 131 N |
| NAMPT siRNA + NR3 | 131C |
| scRNA3 | 132 N |
| NAMPT siRNA + NMN1 | 132C |
| NAMPT siRNA3 | 133 N |
| NAMPT siRNA3 | 133C |
| NAMPT siRNA + NMN2 | 134 N |
| NAMPT siRNA4 | 134 N |
| NAMPT siRNA + NMN3 | 134C |
| scRNA4 | 135 N |
Isobaric tagging of human immortalized myotubes for 18-plex TMT assay used for LC-MS/MS proteomic analysis.
2.9. Acetyl-lysine immunoprecipitation
125 μg of cell lysates were incubated with unconjugated protein A agarose beads (Santa Cruz Biotechnology, Dallas, TX) for 1 h at 4 °C with rotation to preclear the lysates of any potential interfering or non-specific interacting substances from the lysates. Following incubation with unconjugated beads, samples were centrifuged at 21,000 X G for 1 min and the pre-cleared lysates were saved and the unconjugated beads were discarded. The pre-cleared lysates were then incubated with a 50 μL suspension of agarose beads conjugated with anti-Acetyl Lysine antibody (ICP03880, ImmuneChem, Burnaby, BC) overnight while rotating at 4 °C. After incubation, the samples were centrifuged at 21,000 x G for 1 min. The supernatant was then removed and saved in a new microcentrifuge tube. The beads containing the antigen-antibody complex were washed three times with ice cold 1X PBST and two times with 1X PBS before the suspension was moved to a new tube. Following the last wash, the beads were resuspended in 60 μL 1X Laemmeli Buffer (BioRad, Hercules, CA) with without βME and heated at 90 °C for 5 min to elute the proteins from the beads. Supernatant (35 μL) was combined with equal volume of 2X Laemmeli Buffer with βME and heated at 95o C for 5 min 30 μL of the eluates and supernatants were then loaded onto a 10 % polyacrylamide gel, separated by SDS-PAGE, and transferred to a nitrocellulose membrane via semi-dry transfer for 11 min with a Transblot Turbo (BioRad). Membranes were blocked in PFBB for 1 h followed by being cut and incubated with anti-GLO1 (1:1000, sc-133144, Santa Cruz Biotechnology), and anti-P53 (1:1000, sc-126, Santa Cruz Biotechnology) overnight at 4 °C. Membranes were washed with TBST and were incubated with anti-mouse secondary antibody conjugated to IRDye 800CW (green channel) (1:20,000, 926–32212, LiCor) in PFBB supplemented with 0.1 % Tween-20 for 1 h. The membranes were then prior to being scanned and quantified on the Odyssey CLx Imaging System (Li-Cor). Next, membranes were probed for acetylated proteins by incubating them with an anti-acetyl lysine antibody (1:1000) (Ab21623, Abcam) overnight at 4o C with gentle rocking. Following primary incubation, the membranes were washed as described above and incubated with an anti-rabbit secondary antibody conjugated to IRDye 680RD (red channel) (1:20,000) (926–68071, LiCor) in PFBB with 0.1 % Tween-20 for 1 h. Following this incubation, the membranes were washed, imaged, and analyzed. For quantitation, the GLO1 or p53 signal in the eluate was quantified as the acetylated versions of these proteins and was normalized to the total acetyl lysine signal in the eluate. In addition, the un-normalized data (i.e. quantitation of the acetylated target alone) is presented in Supplementary Fig. 2.
2.10. RNA extraction and reverse transcription
RNA extraction was performed using Qiagen's RNeasy kit following the kit protocol with modifications to increase yield for fibrous tissues such as skeletal muscle. Briefly, RNAlater-fixed samples were homogenized in RLT buffer with BME via bead homogenization. Samples were treated with 10 μL Proteinase K (Qiagen), incubated at 55 °C for 10 min and then centrifuged at 10,000 X G for 3 min at room temperature. Supernatants were transferred to a sterile microfuge tube and 450 μL of ethanol was added to each sample and transferred on to the RNeasy spin columns. Ethanol extracts were collected via centrifugation at 9000 X G for 30 s at room temperature and the protocol provided by Qiagen was then followed thereafter. 2 μL of each extraction was analyzed for RNA concentration via Nanodrop (ThermoFisher). Average RNA concentration was 128 ± 9.9 ng/uL (mean ± SEM) for LH and 118 ± 17.7 ng/uL for OB samples. Reverse transcription was performed with iScript Advanced reverse transcriptase kit (BioRad) via manufacturer protocols to generate 150 ng of cDNA which was then diluted 1:4 with nuclease-free water.
2.11. Droplet digital PCR
Droplet digital PCR (BioRad) was used to quantify transcripts of GLO1, NRF2, KEAP1, SIRT1, SIRT2, and NAMPT [40]. Primers were designed for each target using Roche's universal probe library (UPL) assay design center (lifescience.roche.com). In brief, ddPCR assays were performed by combining 2 μL of cDNA (3.75 ng) with ddPCR mix for probes no dUTP (BioRad), along with appropriate probes, primers and nuclease free water yielding a 20 μL reaction. To control for a small amount of background signal in the ddPCR, a triplicate of no-template negative controls in which the cDNA was substituted for nuclease-free water was performed alongside each of the assays and subtracted from the final signal. The 20 μL reaction was then combined with 70 μL of droplet generation oil for probes (BioRad) and droplets were generated using a droplet generator (BioRad). The resultant droplet suspension (40 μL) was then carefully pipetted onto a 96 well plate which was then sealed and placed in a thermocycler where 40 cycles of PCR were performed. The droplets were then analyzed using a QX200 Droplet Digital PCR system (BioRad) by counting the droplets positive for FAM fluorescent probes. Transcript copy number was corrected for non-specific signal by subtracting the number of copies detected in the NTC samples. All transcript data are presented as copy number per ng cDNA input.
2.12. GLO1 activity
To determine GLO1 activity, 10 μL aliquots of skeletal muscle homogenate and cell lysates prepared in GLO1 activity assay buffer supplemented with 10 mM NAM and 1 μM TSA were analyzed in triplicate via a colorimetric enzymatic activity assay (K591, BioVision, Milpitas, CA).
This assay was performed according to the manufacturer protocol except for the homogenates being made in the cell lysis buffer as previously described rather than the GLO1 activity buffer provided with the kit to avoid the need for using more precious sample for a separate homogenization. This activity kit uses the same principle for determination of GLO1 activity as described by Thornalley et al. [41].
To determine GLO1 specific activity (Units/mg GLO1 protein), the activity data from the GLO1 activity assay was made relative to the amount of GLO1 in the sample. GLO1 input was determined by performing a GLO1 ELISA (MBS2021816, MyBioSource, San Diego, CA) on another aliquot of same samples analyzed for GLO1 activity. Samples were assayed according to the manufacturer protocol using aliquots of cell lysates containing 20 μg of protein.
2.13. Statistics
Figures were made using matplotlib package in Python (Jupyter Notebooks available upon reasonable request) or GraphPad Prism (Version #9.4.0). Statistical analyses were run in GraphPad Prism and (Version #9.4.0). Individual data points are provided for transparency and bars represent the mean values and error bars represent the standard error of the mean (SEM). Un-paired student's T-Tests or Mann-Whitney U-Tests were used to compare LH and OB outcomes as well as scRNA and SIRT1 and SIRT2 siRNA treated myotubes where appropriate. One-way ANOVA with post-hoc analyses utilizing Bonferroni correction were used to analyze NAMPT siRNA and NR Rescue experiments. Data were checked for normality using Shapiro-Wilk's test and appropriate parametric or non-parametric tests were used for correlative analysis depending on the normality of the outcomes assessed. The p value for statistical significance was set at p < 0.05.
Proteome Discoverer (v2.4; Thermo Fisher) was used for data analysis of TMT 18-plex experiment. MS2 spectra were searched against SwissProt human protein database (20,353 entries; download date: 6/17/2020) using the following search parameters: MS1 and MS2 tolerance were set to 10 ppm and 0.6 Da, respectively; carbamidomethylation of cysteines (57.02146 Da) and TMT labeling of lysine and N-termini of peptides (229.16293 Da) were considered static modifications; oxidation of methionine (15.9949 Da) and deamidation of asparagine and glutamine (0.98401 Da) were considered variable. Identified proteins and peptides were filtered to retain only those that passed ≤1 % FDR threshold. Quantitation was performed using high-quality MS3 spectra (Average signal-to-noise [S/N] ratio of 6 and < 50 % isolation interference).
Normalized factors were calculated using the Gygi method for TMT quantification. Briefly, S/N of all proteins in each channel were summed to quantify total protein and then divided by the channel with the largest total protein value. The unique channel normalization factor was then added to each protein identified to create normalized abundance of protein. These normalized abundances were then used to calculate Log2 fold change scores using the scRNA samples (n = 4) as the references by which GLO1 siRNA (n = 4), NAMPT siRNA (n = 4), NAMPT siRNA with NR co-treatment (n = 3), and NAMPT siRNA with NMN co-treatment (n = 3) were all compared. Z-scores for all protein abundance ratios were generated for each experiment to perform principal component analyses (PCA).
A T-Tests were used to compare protein expression differences between groups. Resultant p values were the adjusted via Benjamini-Hochberg correction method. For volcano plots, heat maps, and dot plots, adjusted p values were converted to -Log10 p. For heat maps with hierarchical clustering and gene ontology enrichment analyses, only proteins that significantly changed with GLO1 siRNA (-Log10 p < 1.3) were used for analysis. Uniprot accessions of significantly up regulated proteins and down regulated proteins were entered into DAVID independently. Proteins were searched against the human gene ontology biological process annotations. Pathway fold enrichments were generated by DAVID based on the number of highly associated proteins identified determined by Fisher's exact test.
3. Results
3.1. GLO1 protein is reduced in skeletal muscle from individuals with obesity
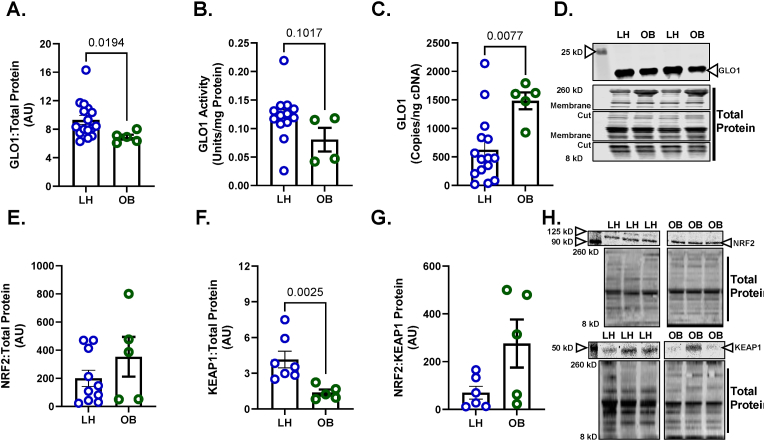
Our lab has previously demonstrated a lower GLO1 protein abundance in skeletal muscle tissue from individuals with T2D which also negatively correlated with BMI and positively correlated with whole body insulin sensitivity, determined by hyperinsulinemic-euglycemic clamp [9]. However, whether skeletal muscle GLO1 protein abundance is attenuated with obesity alone is unknown. Therefore, we measured GLO1 protein abundance in skeletal muscle biopsy samples from young LH individuals (BMI 22.4 ± 0.5), and age-matched individuals with obesity (BMI: 32.6 ± 1.1) (Table 1) and found GLO1 protein abundance was reduced by 26 ± 0.03 % (p = 0.019) in OB compared to LH skeletal muscle (Fig. 1A). We also observed a similar non-significant trend for reduced GLO1 activity (34 ± 0.17 %, p = 0.1017) in OB skeletal muscle samples (Fig. 1B). However, GLO1 transcripts were elevated in OB compared to LH skeletal muscle (Fig. 1C) despite there being no differences in one of GLO1's transcription factors NRF2 (Fig. 1E). The negative regulator of NRF2, KEAP1, was suppressed in muscle from OB individuals (p = 0.0025, Fig. 1F) although the balance of NRF2:KEAP1 protein was not different (Fig. 1G). Given the mismatch in GLO1 transcripts and protein abundance in OB skeletal muscle, we next set out to determine if post translational mechanisms could regulate GLO1 protein and activity in skeletal muscle. In light of previous work suggesting GLO1 acetylation precedes its degradation in a liver MAFLD model [16], we sought to determine the potential role of the NAD+-dependent deacetylases SIRT1 and SIRT2 on GLO1 acetylation, protein abundance and activity in skeletal muscle.
Fig. 1.
GLO1 is Attenuated in Skeletal Muscle from Individuals with Obesity
Human skeletal muscle biopsy samples (vastus lateralis) were analyzed for A) GLO1 protein abundance, B) GLO1 activity, and C) GLO1 transcripts. A representative Western blot for GLO1 and total protein used for normalizing signal quantification of target proteins are displayed in panel D. Protein level of GLO1's transcription factor NRF2 (E), its negative regulator KEAP1 (F), and their ratio (G) were measured via Western blot. A representative Western blot for NRF2, KEAP1, and total protein are displayed in panel H. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test or Mann-Whitney U test, depending on normality, with significance set to p < 0.05.
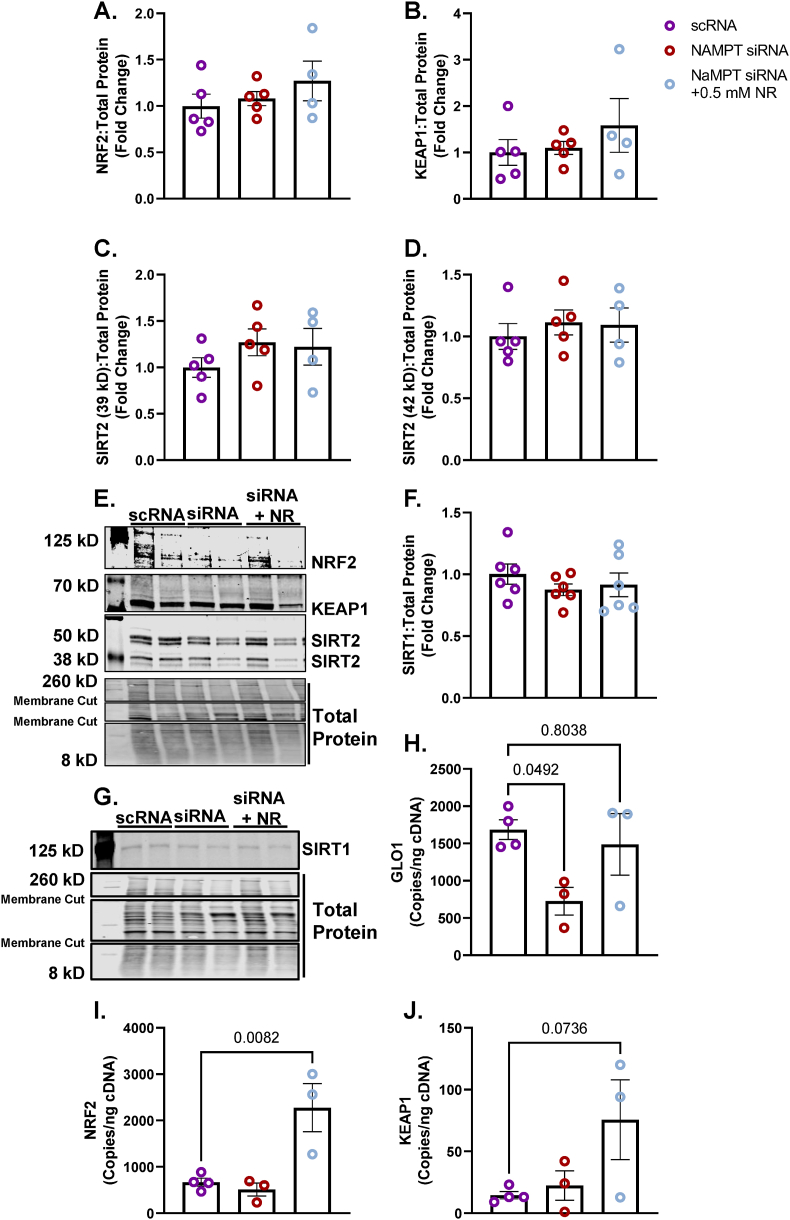
3.2. Loss of SIRT2 but not SIRT1 attenuates GLO1 protein expression in human immortalized myotubes
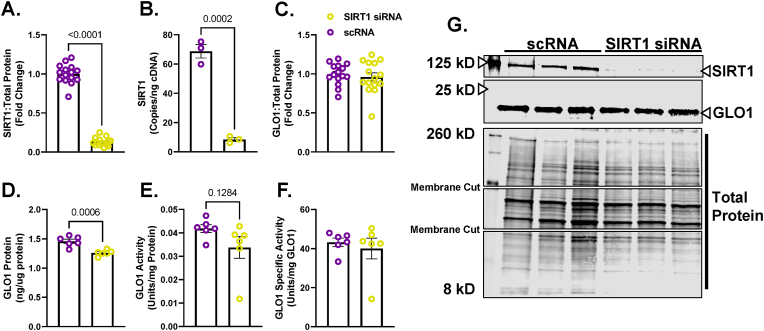
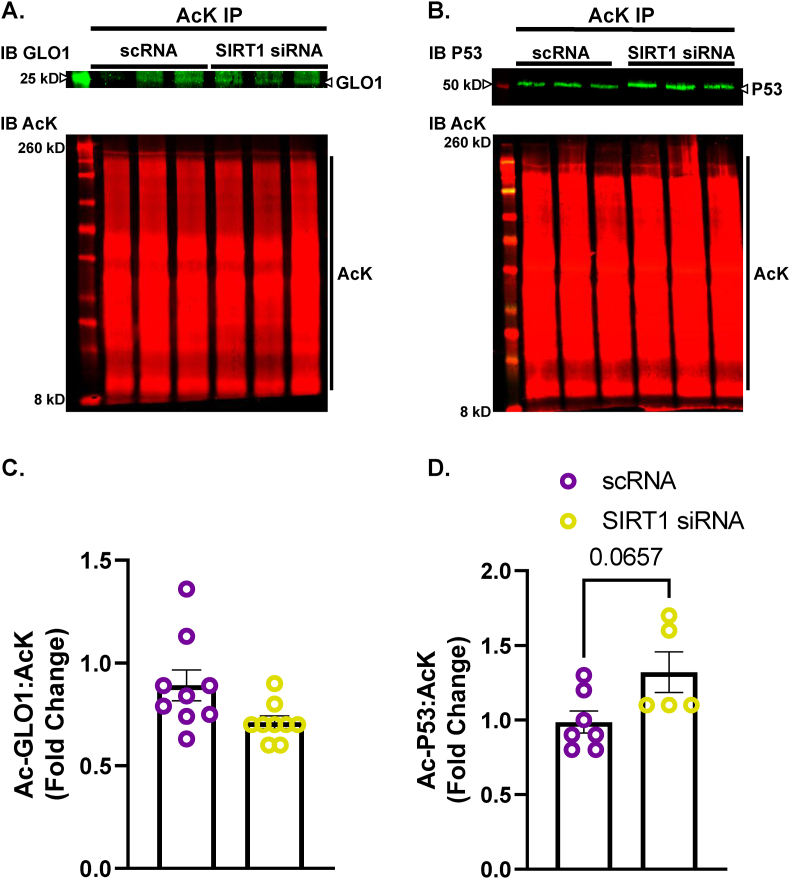
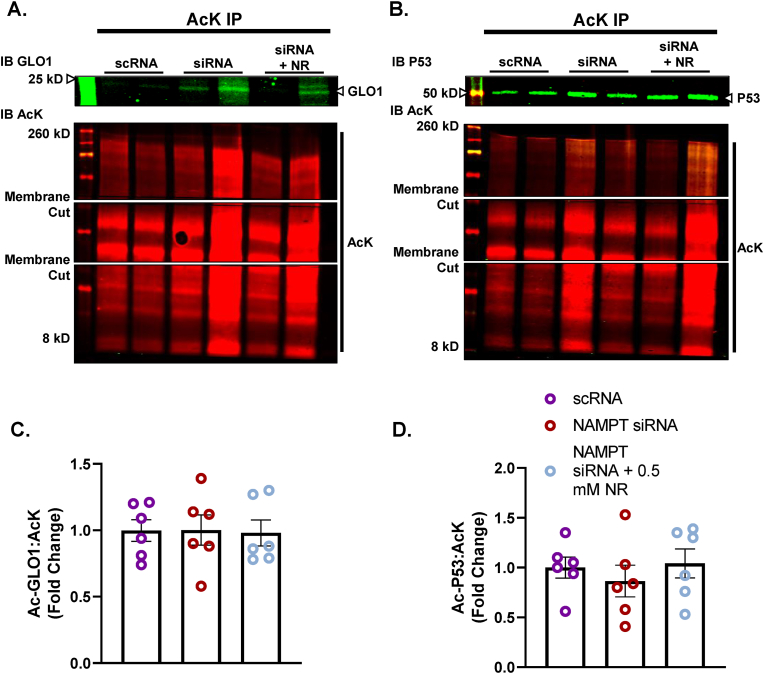
To explore the potential role of SIRT1 on regulating GLO1 protein abundance and activity, we transfected human immortalized myotubes with siRNA against SIRT1 for 48 h which resulted in an 89 ± 0.015 % reduction in SIRT1 protein (p < 0.0001, Fig. 2A), and an 88 ± 0.018 % reduction in SIRT1 transcripts (p = 0.0002, Fig. 2B). When measured via Western blot, SIRT1 knock down (KD) did not alter GLO1 protein abundance (Fig. 2C) and when measured via ELISA, GLO1 protein was modestly reduced by 17 ± 0.016 % (p = 0.0006, Fig. 2D). Knock down of SIRT1 did not attenuate GLO1 activity (U/mg protein; 20 ± 0.12, p = 0.12, Fig. 2E) or specific activity (Fig. 2F). In addition, GLO1 transcripts were not altered by SIRT1 KD (Supplemental Fig. 1A), nor was there any difference in NRF2 or KEAP1 protein (Supplemental Figs. 1B and C) or transcripts (Supplemental Figs. 1G and H). GLO1 acetylation was assessed by immunoprecipitating acetylated proteins and then immunoblotting for GLO1. SIRT1 KD did not affect GLO1 acetylation as assessed by this method (Fig. 3A–C). P53 acetylation was also assessed using this method and demonstrated a modest trend for increased ratio of acetylated P53: total protein acetylation (Fig. 3B–D; p = 0.0657). However, acetylated P53 was significantly increased in SIRT1 KD samples if left un-normalized to total acetylated proteins (p = 0.026, Supplemental Fig. 2B) which may be relevant given SIRT1 KD should affect the abundance of acetylated proteins in general. These data suggest a modest reduction in SIRT1 activity with our siRNA treatment.
Fig. 2.
Effect of SIRT1 Knock Down On GLO1 in Human Immortalized Myotubes
Human Immortalized Myotubes transfected with SIRT1 siRNA for 48 h resulting in depletion of SIRT1 protein (A), and transcripts (B). The effect of SIRT1 KD on GLO1 protein abundance was determined via Western blot (C) and ELISA (D). GLO1 specific activity (F) was determined by making GLO1 activity (E) relative to GLO1 abundance in each sample which was determined via ELISA. A representative Western blot for GLO1, SIRT1 and total protein used for normalizing signal quantification of target proteins are displayed in panel G. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test with significance set to p < 0.05.
Fig. 3.
Effect of SIRT1 Knock Down In Human Immortalized Myotubes on GLO1 Acetylation
Lysine-acetylated proteins (AcK) were immunoprecipitated from Human Immortalized Myotubes transfected with SIRT1 siRNA for 48 h and subsequently immunoblotted (IB) for GLO1 (A,C), and P53 (B,D). Signal quantification of acetylated GLO1 (C), and P53 (D) were made relative to the signal of acetylated proteins that were immunoprecipitated to account for any technical variance in the IP procedure. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test with significance set to p < 0.05.
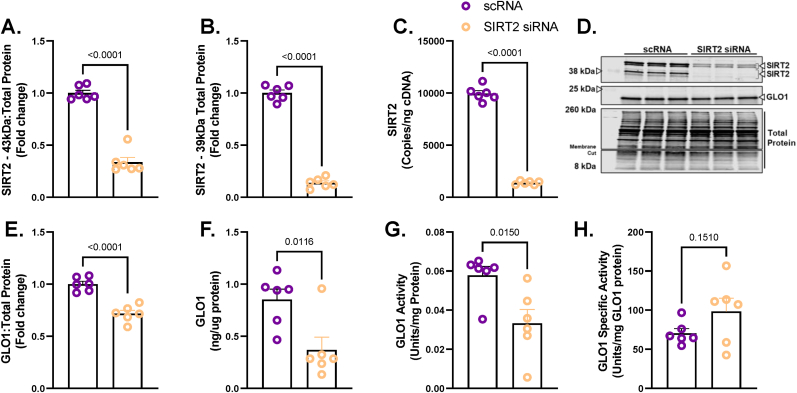
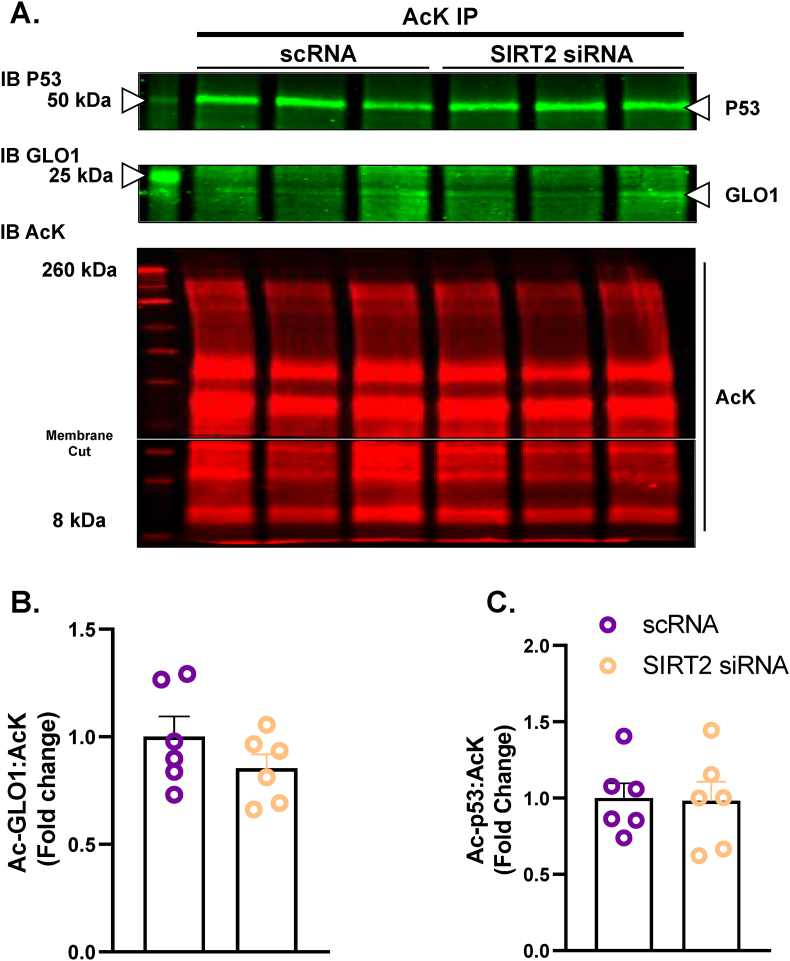
Unlike SIRT1, SIRT2 is primarily localized in the cytosol where GLO1 resides. Therefore, we explored the potential role of SIRT2 to regulate GLO1 by using the same siRNA strategy as described for SIRT1. SIRT2 KD resulted in a 66 ± 0.055 % reduction in the 43 kDa SIRT2 isoform (p < 0.0001, Fig. 4A), an 86 ± 0.017 % reduction in the 39 kDa SIRT2 isoform (p < 0.0001, Fig. 4B) and an 86 ± 0.006 % reduction in SIRT2 transcripts (p < 0.001, Fig. 4C). SIRT2 KD reduced GLO1 protein as determined by Western blot (28 ± 0.29 % reduction, p < 0.0001, Fig. 4E), and ELISA (63 ± 0.13 % reduction, p = 0.0116, Fig. 4F). GLO1 activity was also significantly reduced in SIRT2 KD cells (p = 0.015, Fig. 4G) although this effect was lost with GLO1 specific activity. Similar to SIRT1 KD, SIRT2 KD did not affect GLO1 transcripts or NRF2/KEAP1 protein or transcripts (Supplemental Fig. 3). SIRT2 KD did not affect GLO1 acetylation nor P53 acetylation (Fig. 5 and Supplemental Fig. 2) suggesting that SIRT2 deacetylase activity was not fully suppressed or P53 deacetylation was compensated for by an alternative mechanism (e.g. SIRT1).
Fig. 4.
Effect of SIRT2 Knock Down On GLO1 in Human Immortalized Myotubes
Human Immortalized Myotubes transfected with SIRT2 siRNA for 48 h resulting in depletion of both isoforms of SIRT2 protein at 43 kDa (A) and 39 kDa (B), and SIRT2 transcripts (C). The effect of SIRT2 KD on GLO1 protein abundance was determined via Western blot (D and E) and ELISA (F). GLO1 specific activity (H) was determined by making GLO1 activity (G) relative to GLO1 abundance in each sample which was determined via ELISA. A representative Western blot for GLO1, SIRT2 and total protein are displayed in panel H. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test with significance set to p < 0.05.
Fig. 5.
Effect of SIRT2 Knock Down In Human Immortalized Myotubes on GLO1 Acetylation
Lysine-acetylated proteins (AcK) were immunoprecipitated from Human Immortalized Myotubes transfected with SIRT2 siRNA for 48 h and subsequently immunoblotted (IB) for GLO1 (A,B), and P53 (A,C). Signal quantification of acetylated GLO1 (B), and P53 (C) were made relative to the signal of acetylated proteins that were immunoprecipitated to account for any technical variance in the IP procedure. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test with significance set to p < 0.05.
3.3. NAMPT KD attenuates GLO1 protein, and activity which is partially rescued by NR
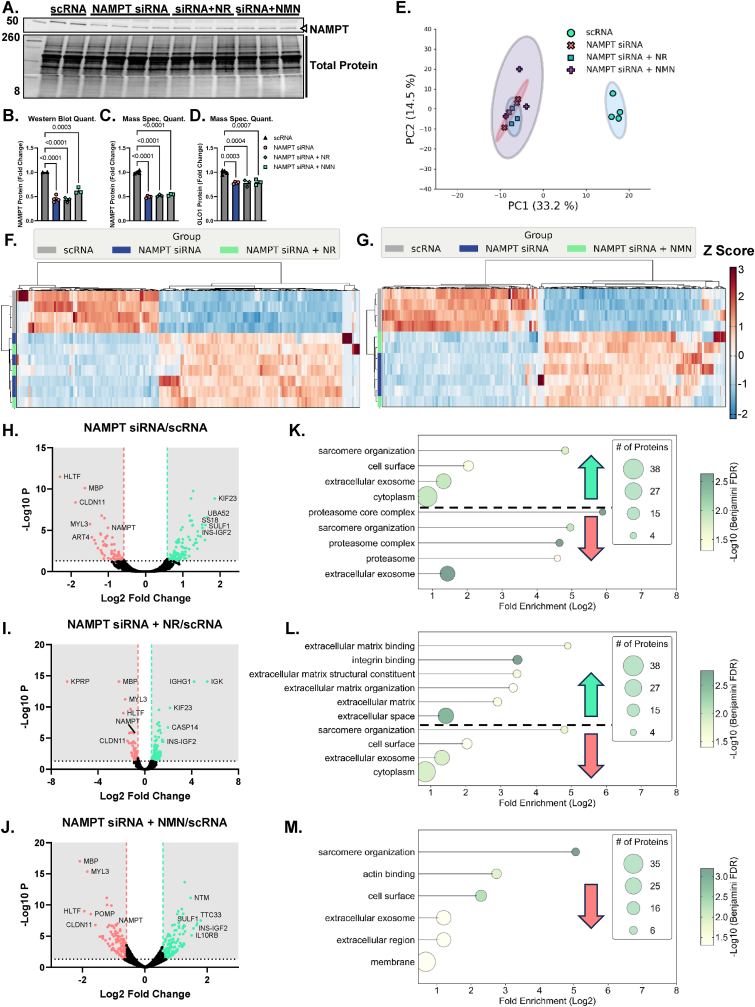
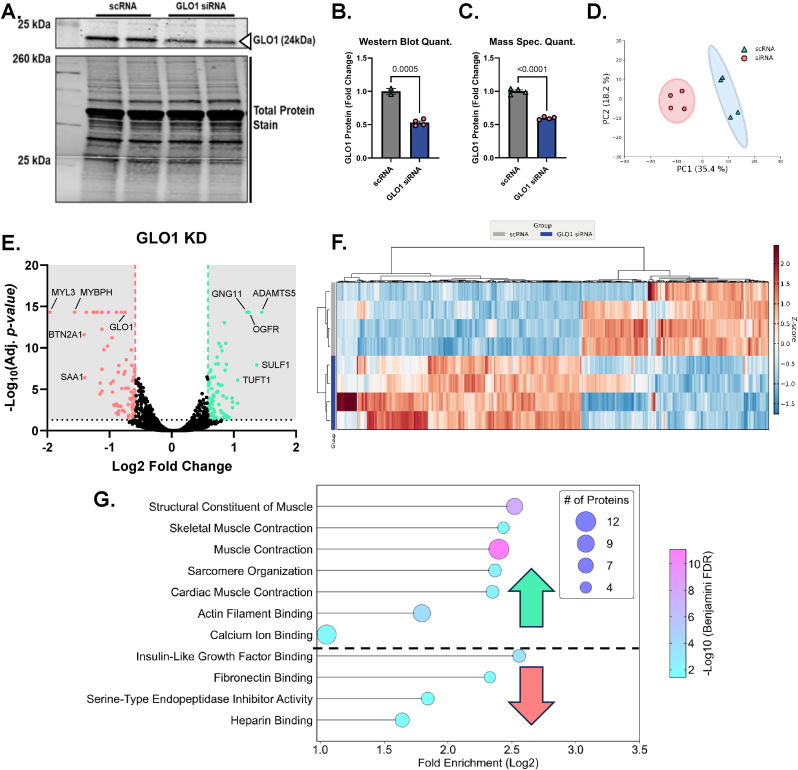
NAMPT is the rate limiting enzyme in NAD+ scavenging pathway and is therefore upstream of NAD-dependent sirtuins (Supplemental Fig. 4A). To test if loss of NAD scavenging ability and thereby loss of downstream global sirtuin activity, we treated cells with NAMPT siRNA for 48 or 72 h. We also co-treated NAMPT KD cells with the NAD+ precursor NR or NMN for the final 24 h of siRNA treatment to rescue the NAD+ depletion of the NAMPT KD. NAMPT siRNA at 72 h resulted in 55 ± 0.03 % reduction of NAMPT protein abundance (p < 0.0001, Fig. 6B) which was similar to the quantitation from our proteomics data (p < 0.0001, Fig. 6C). NAMPT KD also reduced GLO1 protein measured by proteomics (Fig. 6D). This effect was significant with a one-way ANOVA (p = 0.0003) but did not survive the FDR adjustment applied to our proteomics data set. Neither NR nor NMN co-treatment altered GLO1 protein abundance quantified in our proteomics data set (Fig. 6D). PCA analysis demonstrated good and distinct clustering of the scRNA and siRNA treated samples with no distinction in clustering when cells were co-incubated with NR or NMN (Fig. 6E). Heat map and hierarchical clustering analysis also suggest large proteomic changes with NAMPT KD but minimal effect on the proteomes of both treatment conditions (Fig. 6F and G). This inability of NR and NMN to rescue the proteomic effects of NAMPT KD are exemplified by volcano plots (Fig. 6H–J) which demonstrate several of the same proteins remain similarly changed in NAMPT KD and rescue experiments. Gene ontology and KEGG pathways (Fig. 6K–M) were also similarly enriched between all experiments whereby pathways involved in skeletal muscle contraction and organization are perturbed across all conditions although these pathways were annotated to be both enriched and depleted with NAMPT siRNA with different proteins contributing to the annotation in each case (Fig. 6K–M). In each condition, with the exception of NMN co-treatment, pathways involved in extracellular structure and function were similarly altered (Fig. 6K–M). These data suggest that while NAMPT protein is likely critical for skeletal muscle function, provision of NAD precursors is not sufficient to restore GLO1 or the proteome at large to normal levels.
Fig. 6.
Proteomic Assessment of NAMPT Knockdown and NR or NMN Co-Treatment in Human Immortalized Myotubes.
Given that NAMPT protein is upstream of both SIRT1 and SIRT2 activity by virtue of their reliance on NAD, we next knocked down NAMPT in human myotubes as confirmed by western blots and quantification by TMT mass spec. A - C). GLO1 was also found to be ∼30 % lower in myotubes treated with NAMPT siRNA which was not rescued by either NR or NMN co-treatment. Globally, PCA plots in E) and hierarchical clustering in F and G) demonstrate a largely ineffectiveness of both NR and NMN to rescue the proteomic changes of NAMPT siRNA treatment. Volcano plots comparing NAMPT siRNA (H), NAMPT siRNA + NR (I), and NAMPT siRNA + NMN (J) to scRNA treated cells also demonstrates similar protein changes in the attempted rescue experiments as the siRNA alone. This is further exemplified in dot plots (K–M) depicting enrichment for similar GO and KEGG pathway terms across all 3 experiments.
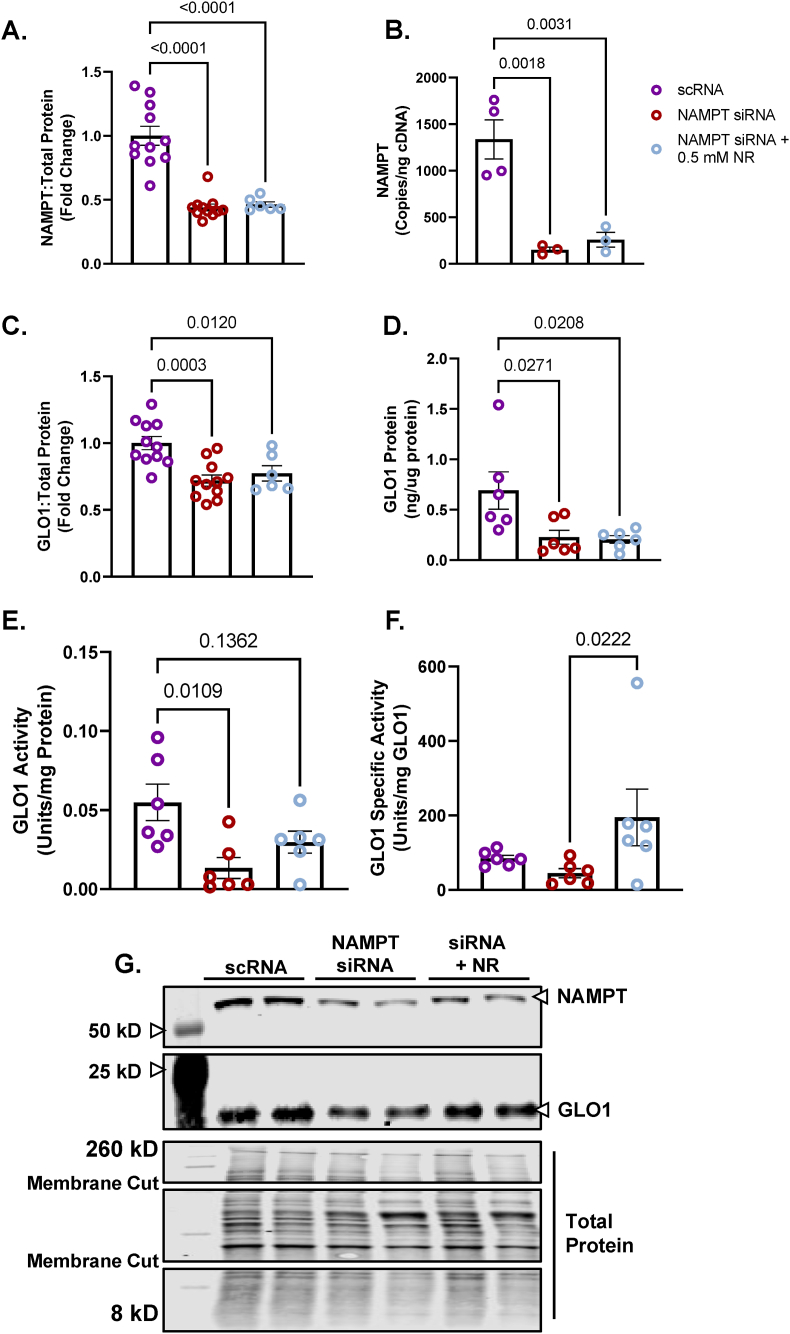
We followed up on these experiments by exploring the effect of NAMPT KD and NR on other aspects of GLO1 such as activity, transcriptional regulation and acetylation. For these experiments we treated cells with NAMPT siRNA for 48 h and NR for the last 24 h 48 h of NAMPT siRNA resulted in a similar reduction of NAMPT protein that we observed with 72h siRNA treatment (56 ± 0.02 %, p < 0.0001, Fig. 7A) and a 91 ± 0.03 % (p < 0.0001, Fig. 6B) reduction in NAMPT transcripts. Neither NAMPT protein nor transcripts were rescued by co-treatment with NR (Fig. 7A and B). Similar to our Western blots and TMT proteomic quantitation from our 72-h experiments, NAMPT KD similarly attenuated GLO1 protein measured via Western blot (28 ± 0.069 %, p = 0.0003, Fig. 7C) and ELISA (67 ± 0.092 %, p = 0.0271, Fig. 7D), neither of which were rescued by 24-h co-treatment with 0.5 mM NR. GLO1 Activity was significantly reduced by NAMPT KD (76 ± 0.07 %, p = 0.0109, Fig. 7E) which was rescued by NR co-treatment (Fig. 7E). Specific activity was not significantly attenuated by NAMPT KD but was able to be augmented by NR co-treatment (Fig. 7F).
Fig. 7.
Effect of NAMPT Knock Down and NR Rescue On GLO1 Protein Expression and Activity in Human Immortalized Myotubes
Human Immortalized Myotubes transfected with NAMPT siRNA for 48 h, or siRNA for 48 h with 0.5 mM nicotinamide riboside (NR) treatment for the final 24 h (NAMPT siRNA + 0.5 mM NR). NAMPT siRNA depleted myotubes of NAMPT protein (A), and transcripts (B) which were not rescued by NR. The effects of NAMPT KD and NR rescue on GLO1 protein abundance was determined via Western blot (C) and ELISA (D). GLO1 specific activity (F) was determined by making GLO1 activity (E) relative to GLO1 abundance in each sample which was determined via ELISA. A representative Western blot for GLO1, NAMPT and total protein used for normalizing signal quantification of target proteins are displayed in panel G. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via One-way ANOVA using Bonferroni Post Hoc analysis with significance set to p < 0.05.
Neither NAMPT KD nor NR co-treatment affected NRF2, KEAP1, SIRT1, or either of the SIRT2 isoforms (Fig. 8A–F). However, NAMPT KD did attenuate GLO1 transcripts which was rescued by NR co-treatment (Fig. 8H). In addition, while NRF2 and KEAP1 proteins were not altered by the experimental conditions, NR co-treatment augmented NRF2 transcripts (p = 0.0082) and trended towards increasing KEAP1 transcripts (p = 0.0736) (Fig. 8I and J). Lastly, GLO1 and P53 acetylation assessed by acetyl-lysine immunoprecipitation were not affected by NAMPT KD or NR co-treatment when assessing by ratio of each acetylated target to total acetyl lysine (Fig. 9). However, given that NAMPT KD may considerably affect global acetylation of proteins, we also assessed the signal of acetylated GLO1 and P53 without adjusting for total acetyl lysine (Supplemental Figs. 2E and F). This representation of the data revealed a modest (∼30 %) non-significant increase in acetylated P53 with NAMPT siRNA which was not altered by NR co-treatment.
Fig. 8.
Effect of NAMPT Knock Down and NR Rescue On GLO1 Transcripts in Human Immortalized Myotubes
Human Immortalized Myotubes transfected with NAMPT siRNA for 48 h, or siRNA for 48 h with 0.5 mM nicotinamide riboside (NR) treatment for the final 24 h (NAMPT siRNA + 0.5 mM NR). NRF2 (A), KEAP1 (B), SIRT2 (C,D), and SIRT 1 (F) proteins were unaltered by NAMPT knockdown and NR. Representative western blots for protein targets are provided in panels E and G. GLO1 transcripts were attenuated by NAMPT siRNA and rescued by NR (H). Copies of GLO1's transcription factor NRF2 and its negative regulator KEAP1 were not affected by NAMPT siRNA but NRF2 copies were enhanced by co-treatment with NR (I–J). Data are presented as mean ± SEM with individual data points superimposed. Data were compared via One-way ANOVA using Bonferroni Post Hoc analysis with significance set to p < 0.05.
Fig. 9.
Effect of NAMPT Knock Down and NR Rescue In Human Immortalized Myotubes on GLO1 Acetylation
Lysine-acetylated proteins (AcK) were immunoprecipitated from Human Immortalized Myotubes transfected with NAMPT siRNA for 48 h, or siRNA for 48 h with 0.5 mM nicotinamide riboside (NR) treatment for the final 24 h (NAMPT siRNA + 0.5 mM NR) and subsequently immunoblotted (IB) for GLO1 (A,C), and P53 (B,D). Signal quantification of acetylated GLO1 (B), and P53 (C) were made relative to the signal of acetylated proteins that were immunoprecipitated to account for any technical variance in the IP procedure. Data are presented as mean ± SEM with individual data points superimposed. Data were compared one-way ANOVA using Bonferroni Post Hoc analysis with significance set to p < 0.05.
3.4. Potential role for NAMPT, SIRT1 and SIRT2 to regulate GLO1 protein in OB skeletal muscle
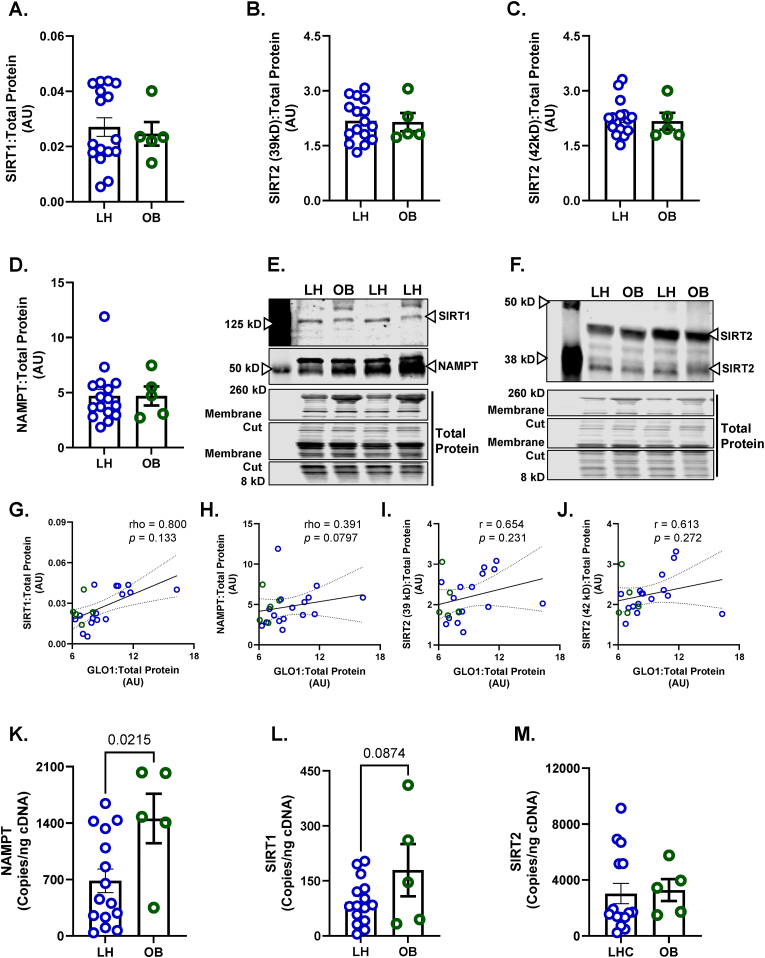
Given that we had identified SIRT2 and NAMPT, but not SIRT1, as potential mechanisms regulating GLO1 protein and activity, we revisited our human skeletal muscle biopsy samples to probe for these targets. SIRT1, SIRT2 and NAMPT protein expressions were not different in OB vs LH skeletal muscle samples (Fig. 10A–D). GLO1 protein expression was not found to be correlated with SIRT1 (rho = 0.800, p = 0.133, Fig. 10G), NAMPT (rho = 0.391, p = 0.0797, Fig. 10H) or either isoform of SIRT2 protein (r = 0.654, p = 0.231, Fig. 10I; r = 0.613, p = 0.272, Fig. 10J). Similar to the mismatch observed between GLO1 protein and transcripts, both NAMPT and SIRT1 transcripts were higher in OB compared to LH skeletal muscle whereas SIRT2 transcripts were not different between groups (Fig. 10K–M).
Fig. 10.
Role of SIRT1, SIRT2 and NAMPT on GLO1 in Skeletal Muscle from Individuals with Obesity
Human skeletal muscle biopsy (vastus lateralis) samples were analyzed for A) SIRT1, B,C) SIRT2 isoforms, and D) NAMPT protein abundance. Representative western blots for target proteins and total protein used for normalizing signal quantification of target proteins are displayed in panels E and F. GLO1 protein abundance was positively correlated with SIRT1 protein (G) and NAMPT protein (H) but not SIRT2 protein isoforms (I, J). NAMPT (K), SIRT1 (L), and SIRT2 (M) transcripts were assessed via ddPCR. Data are presented as mean ± SEM with individual data points superimposed. Data were compared via independent student's T Test with significance set to p < 0.05.
3.5. Proteomic consequence of GLO1 knock down in human immortalized myotubes
Finally, we set out to explore the proteomic consequences of human skeletal muscle cells with attenuated GLO1 protein abundance, given our observation that this is the molecular milieu of individuals with obesity. While the consequences of dicarbonyl stress as a result of cells, or animals lacking GLO1 have been described, little is known about the global proteomic consequence of GLO1 attenuation in skeletal muscle and there is no data on this topic in human skeletal muscle. To fill this gap, we knocked down GLO1 in human immortalized myotubes and performed proteomic analyses. GLO1 KD resulted in an approximately 50 % reduction in GLO1 protein as assessed by both Western blotting and TMT quantitation (Fig. 11A–C). PCA analysis of the proteome demonstrated distinct clustering of scRNA and siRNA-treated samples (Fig. 11D). This was corroborated by unsupervised hierarchical clustering of significantly altered proteins (Fig. 11F). GLO1 KD resulted in the upregulation of proteins involved in extracellular matrix remodeling and fibrosis and subsequent downregulation of proteins involved in sarcomere structure and skeletal muscle contractile function such as skeletal muscle myosin complex (Fig. 11G). These data demonstrate the importance of GLO1 for skeletal muscle structure and function.
Fig. 11.
Proteomic Consequences of GLO1 Knock Down in Human Immortalized Myotubes.
To explore the proteomic consequences of decreased GLO1 abundance in human immortalized myotubes, we knocked down GLO1 by treating myotubes with GLO1 siRNA which resulted in an approximately 50 % reduction in GLO1 protein as confirmed by Western blotting (B–C) and quantification via TMT mass spec (C). PCA demonstrates the unique clustering of samples in the scRNA and siRNA-treated samples and significantly altered proteins are visually demonstrated in the volcano plot in panel E. F) Hierarchical clustering of a heat map of significantly changed proteins with siRNA identified several unique families of similarly altered proteins within the siRNA-treated group. This is confirmed and visually represented in the dot plot in G) demonstrating that many pathways involving muscle contraction and organization are attenuated in human myotubes with attenuated GLO1 protein, exemplifying the importance of GLO1 in the muscle cell.
4. Discussion
To our knowledge, this is the first study demonstrating attenuated skeletal muscle GLO1 protein abundance in individuals with obesity compared to age-matched controls. GLO1 is the rate limiting enzyme for the detoxification of the reactive dicarbonyl, MG. MG and MG-derived protein adducts are increased in circulation with obesity in both the post absorptive [8] and the post prandial states [6,7]. Skeletal muscle is the predominant site for insulin-mediated glucose disposal in the post-prandial state and therefore, a loss of GLO1 may leave the skeletal muscle tissue vulnerable to increased metabolic flux and contribute to the development of skeletal muscle insulin resistance [[9], [10], [11],[42], [43], [44], [45]] [][[9], [10], [11], [42], [43], [44], [45]][]. Indeed, skeletal muscle cell culture models have demonstrated the ability of increased MG to directly impair insulin signaling [11,42,43], and preclinical models have shown that genetic deletion of GLO1 promotes insulin resistance and a T2D phenotype [11,13,14]. Herein, we demonstrated that GLO1 KD in human immortalized myotubes instigated the expression of extracellular matrix proteins and fibrosis, such as fibronectin binding. These findings are similar to a previous study in rat L6 myotubes [46] which observed an increase in collagen proteins following GLO1 KD. In addition, pathways involved in skeletal muscle contraction and sarcomere organization were downregulated with GLO1 KD. These data suggest augmenting skeletal muscle GLO1 in states of reduced protein abundance, or activity, may have broad positive effects on skeletal muscle function.
NRF2 is the putative transcription factor for GLO1 [47]. However, in skeletal muscle from individuals with obesity, the protein expressions of NRF2 and its negative regulator KEAP1 were found to be no different from that of the LH group in the current investigation. In addition, GLO1 transcripts were elevated in the OB compared to LH group, indicating the potential for post-translational regulation of GLO1. Several lines of evidence have suggested acetylation as a potential regulator of GLO1's protein expression [[16], [17], [18]] and for SIRT1 to regulate its acetylation status [12,19]. For example, hyperacetylation of GLO1 promoted its ubiquitination and degradation in liver cells treated with high concentration palmitate to model MAFLD [16]. A peptide acetylome screen also identified SIRT1 and SIRT2 to deacetylate GLO1 in vitro [17]. However, these mechanisms have never been explored in human skeletal muscle.
Interestingly, SIRT1 KD in human immortalized myotubes did not robustly or reproducibly affect GLO1 protein or activity whereas KD of SIRT2 produced a substantial and reproducible attenuation of GLO1 protein and activity, without affecting GLO1 acetylation. However, it is possible that SIRT1 and SIRT2 may compensate for one another [48] and thus both may need to be reduced in order to observe changes in GLO1 acetylation in our model. P53 acetylation is often used as a surrogate measure of sirtuin activity. Notably, we attempted to quantify both the acetyl-P53 and acetyl-lysine signals following IP to make a ratio of Ac-P53:AcK. We did this to be as rigorous as possible given that it is likely that each replicate of the IP resulted in slightly different efficiency of acetyl-lysine IP. With this approach, we observe a non-significant approximately 1.3-fold enrichment of acetylated P53 (p = 0.0657) with SIRT1 KD which is what we would expect a priori. However, it is also possible that by normalizing to total acetylated lysines we are actually confounding our data given that total acetylated proteins would also be expected to change with KD of these enzymes (SIRT1, SIRT2, NAMPT). Therefore, we also assessed the un-normalized quantitation of acetylated GLO1 and P53 (Supplemental Fig. 2) which demonstrates that while none of our perturbations alter GLO1 acetylation regardless of the quantitation, P53 is significantly enriched in SIRT1 KD and non-significantly enriched with NAMPT KD.
The nuance of SIRT1's observed role in skeletal muscle should also be taken into account. For example, data from Simon Schenk's lab where skeletal muscle specific knockout of SIRT1 did not increase the abundance of Ac-P53 compared to wild-type mice [49] suggests that in skeletal muscle, the redundancies of other sirtuins may be able to compensate for loss of SIRT1. We hypothesized that SIRT2, is the most likely to perform this function as it is more highly expressed in skeletal muscle compared to SIRT1 which is the rationale for our downstream experiments. However, similar to SIRT1 KD, KD of SIRT2 and the upstream NAMPT enzyme did not alter acetylation of GLO1 or P53 despite 60–80 % depletion of SIRT2 protein isoforms and NAMPT protein. Therefore, while we can be fairly certain that SIRT1 activity was attenuated at least modestly in our model we cannot be certain in regards to SIRT2 and NAMPT leaving the mechanism of attenuated GLO1 protein via SIRT2 and NAMPT KD open for speculation and further investigation in the future.
SIRT2 KD also did not affect NRF2 or KEAP1 protein or transcripts. NRF2 is a transcription factor for GLO1 and has previously been shown to translocate to the nucleus upon deacetylation [[50], [51], [52]]. Unfortunately, we were unable to detect acetylated NRF2 in our IP samples although if NRF2 hyperacetylation were contributing to reduced GLO1 following SIRT2 KD, one would expect a subsequent decrease in GLO1 transcripts which was not observed. Therefore, the mechanism by which SIRT2 KD attenuates GLO1 protein is left unanswered. Another question that remains is why KD of SIRT2 but not SIRT1 promoted this reduction in GLO1 protein? A peptide screen previously showed that SIRT2 was more effective at deacetylating a GLO1 peptide than SIRT1 in vitro [17]. Another factor may be the cellular location of SIRT2, like GLO1, is predominantly cytosolic whereas SIRT1 shuttles between the nucleus and the cytosol. Lastly, sirtuins have other activities other than deacetylase activities which may be at play. For example, SIRT1 and SIRT2 also appears to possess NAD+-dependent demyristoylase activity [53] and SIRT2 was recently described to possess delactoylase activity [54]. The same group also describes a mechanism whereby lactoylation of glycolysis enzymes impairs their activity [55]. Perhaps lactoylation of GLO1, by its product S-d-lactoglutathatione, similarly downregulates it's activity and protein abundance - which can be reversed by SIRT2 activity to delactoylate GLO1. Indeed, previous work demonstrated that depletion of cellular NAD+ by inhibiting NAMPT with FK866 in human and mouse myotubes resulted in increased lactate to pyruvate ratio as well as accumulation of substrates upstream of GAPDH [56]. It is therefore reasonable to hypothesize that NAD-dependent de-lactoylation of GLO1 by SIRT2 may be a mechanism to regulate GLO1 abundance and activity in parallel with glycolytic flux. However, we did not observe significant changes to the abundance of proteins in the glycolytic pathways, including GAPDH, in the proteomics data from our NAMPT KD experiments. Future work is needed to test this potential mechanism for SIRT2-mediated de-lactoylation to regulate GLO1 and its relationship to glycolytic flux.
We show here that KD of the NAD+ scavenging enzyme NAMPT also reduces GLO1 protein and activity to a similar extent as SIRT2 KD, which was rescued by provision of the NAD+ precursor NR. These data suggest a role for NAD+-mediated regulation of GLO1 protein. However, the mechanisms involved in the NAMPT KD model appear specific to this perturbation whereby the NAMPT KD reduced GLO1 transcripts which was rescued by NR. However, NAMPT KD did not affect NRF2 or KEAP1 protein despite NR-mediated increase in NRF2 transcripts. These findings implicate complex mechanistic regulation of GLO1 protein and activity via SIRT2, NAMPT, and NAD+. In context of human skeletal muscle tissue, we observed no difference in SIRT1, SIRT2, NAMPT, NRF2, or KEAP1 protein in the OB compared to LH group. We began exploring these mechanisms in skeletal muscle tissue based on our observations of reduced GLO1 protein abundance with obesity and published literature in liver cells demonstrating GLO1 acetylation leading to its degradation [16] We chose not to recapitulate the lipotoxicity model used by Spanos et al., 2018 [16] or use a cell culture model of obesity (i.e. hyperinsulinemia with palmitate) given that we have previously observed that myotubes treated with insulin increases GLO1 protein abundance (unpublished observation). Insulin likely facilitates the glyoxalase enzymatic system to help the cell compensate for increased glycolytic flux and subsequent increased MG generation. We also found that unlike liver cells, proteomics experiments conducted on myotubes treated with palmitate or other models of lipotoxicity did not result in decreased GLO1 protein abundance [57]. While the precise mechanisms of attenuated GLO1 protein abundance in vivo remain to be fully elucidated, the ability of NR to augment GLO1 activity is promising for therapeutic intervention.
Future work is needed to further explore the link between loss of skeletal muscle GLO1 and the development of insulin resistance and other skeletal muscle dysfunction (e.g. MG accumulated with age and age-related skeletal muscle dysfunction). The relative tissue contribution of MG generation with obesity is also unknown. However, Maessen et al. describe rapid and robust increases in MG and MG-H1 in the time course of oral glucose tolerance testing that parallel the hyperglycemic excursion [6]. These data point to skeletal muscle tissue as a potential candidate tissue for MG generation given that skeletal muscle accounts for greater than 80 % of insulin stimulated glucose disposal. Certainly, more work is also needed to elucidate the complex mechanisms that appear to be governing GLO1 abundance and activity in skeletal muscle. However, the findings presented herein demonstrating that NR can promote GLO1 transcripts, and activity, as well as NRF2 are perhaps the most exciting data which warrant further exploration of NR as a potential therapy for the prevention and treatment of metabolic dysfunction. The area of NAD+ precursors as potential health therapies has gained much attention recently and several clinical trials have been published with mixed results to their efficacy [31,[58], [59], [60], [61], [62], [63]]. However, this field still has a long way to go in terms of optimizing the form of the NAD+ precursor, its dose, timing and perhaps most importantly the mode delivery.
In conclusion, GLO1 is decreased in skeletal muscle tissue from individuals with obesity prior to the development of T2D. In vitro modeling of this molecular milieu via GLO1 KD in human immortalized myotubes suggests that skeletal muscle lacking GLO1 is characterized by skeletal muscle dysfunction. Mechanistic experiments in human myotubes implicate SIRT2, NAMPT, and NAD+ as potential regulators of GLO1 protein and activity and suggest that NR or other NAD precursors may be able to augment GLO1. Future work is needed to further elucidate the mechanisms described here and determine the relevance of these mechanisms and potential efficacy of NAD+-targeted interventions in vivo to attenuate dicarbonyl stress and promote insulin sensitivity.
CRediT authorship contribution statement
Edwin R. Miranda: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Pallavi Varshney: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. Corey E. Mazo: Data curation, Formal analysis, Investigation, Writing – review & editing. James Shadiow: Data curation, Formal analysis, Investigation, Writing – review & editing. Andrew T. Ludlow: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing – review & editing. Jacob M. Haus: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Venkatesha Basrur, Ph.D., and the Proteomics Resource Facility at the Department of Pathology, University of Michigan, for the experimental processing of the samples and analytic assistance. We thank Shinya Watanabe for help developing the python scripts for analysis and visualization of proteomics data. This work was supported by NIH grants R01 DK109948 and R21 AG081916.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103300.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.WHO. 2020. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight
- 2.DeFronzo R.A., Ferrannini E., Sato Y., Felig P., Wahren J. Synergistic interaction between exercise and insulin on peripheral glucose uptake. J. Clin. Invest. 1981;68:1468–1474. doi: 10.1172/JCI110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda E.R., Haus J.M. Glyoxalase I is a novel target for the prevention of metabolic derangement. Pharmacol. Therapeut. 2023;250 doi: 10.1016/j.pharmthera.2023.108524. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y., Wang P., Sang S. Dietary genistein inhibits methylglyoxal-induced advanced glycation end product formation in mice fed a high-fat diet. J. Nutr. 2019;149:776–787. doi: 10.1093/jn/nxz017. [DOI] [PubMed] [Google Scholar]
- 5.Maessen D.E., et al. Delayed intervention with pyridoxamine improves metabolic function and prevents adipose tissue inflammation and insulin resistance in high-fat diet–induced obese mice. Diabetes. 2016;65:956–966. doi: 10.2337/db15-1390. [DOI] [PubMed] [Google Scholar]
- 6.Maessen D.E., et al. Post-glucose load plasma alpha-dicarbonyl concentrations are increased in individuals with impaired glucose metabolism and type 2 diabetes: the CODAM study. Diabetes Care. 2015;38:913–920. doi: 10.2337/dc14-2605. [DOI] [PubMed] [Google Scholar]
- 7.Van Den Eynde M.D.G., et al. Diet-induced weight loss reduces postprandial dicarbonyl stress in abdominally obese men: secondary analysis of a randomized controlled trial. Clin. Nutr. 2021;40:2654–2662. doi: 10.1016/j.clnu.2021.03.042. [DOI] [PubMed] [Google Scholar]
- 8.Rabbani N., Thornalley P.J. Glyoxalase 1 modulation in obesity and diabetes. Antioxidants Redox Signal. 2019;30:354–374. doi: 10.1089/ars.2017.7424. [DOI] [PubMed] [Google Scholar]
- 9.Mey J.T., et al. Dicarbonyl stress and glyoxalase enzyme system regulation in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018;314:R181–R190. doi: 10.1152/ajpregu.00159.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remor A.P., et al. Differential effects of insulin on peripheral diabetes-related changes in mitochondrial bioenergetics: involvement of advanced glycosylated end products. Biochim. Biophys. Acta. 2011;1812:1460–1471. doi: 10.1016/j.bbadis.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Riboulet-Chavey A., et al. Methylglyoxal impairs the insulin signaling pathways independently of the formation of intracellular reactive oxygen species. Diabetes. 2006;55:1289–1299. doi: 10.2337/db05-0857. [DOI] [PubMed] [Google Scholar]
- 12.Cai W., et al. Oral advanced glycation endproducts (AGEs) promote insulin resistance and diabetes by depleting the antioxidant defenses AGE receptor-1 and sirtuin 1. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15888–15893. doi: 10.1073/pnas.1205847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodd E., et al. The combination of loss of glyoxalase1 and obesity results in hyperglycemia. JCI Insight. 2019;4 doi: 10.1172/jci.insight.126154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moraru A., et al. Elevated levels of the reactive metabolite methylglyoxal recapitulate progression of type 2 diabetes. Cell Metabol. 2018;27:926–934.e928. doi: 10.1016/j.cmet.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher D., et al. Compensatory mechanisms for methylglyoxal detoxification in experimental & clinical diabetes. Mol. Metabol. 2018;18:143–152. doi: 10.1016/j.molmet.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanos C., et al. Proteomic identification and characterization of hepatic glyoxalase 1 dysregulation in non-alcoholic fatty liver disease. Proteome Sci. 2018;16 doi: 10.1186/s12953-018-0131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauh D., et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 2013;4 doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 18.Cortizo F.G., et al. The activity of glyoxylase 1 is regulated by glucose-responsive phosphorylation on Tyr136. Mol. Metabol. 2022;55 doi: 10.1016/j.molmet.2021.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santini, et al. SIRT1-Dependent upregulation of antiglycative defense in HUVECs is essential for resveratrol protection against high glucose stress. Antioxidants. 2019;8:346. doi: 10.3390/antiox8090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feige J.N., et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metabol. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Sun C., et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metabol. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H.H., et al. SIRT1 attenuates high glucose-induced insulin resistance via reducing mitochondrial dysfunction in skeletal muscle cells. Exp. Biol. Med. (Maywood, NJ, U. S.) 2015;240:557–565. doi: 10.1177/1535370214557218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora A., Dey C.S. SIRT2 negatively regulates insulin resistance in C2C12 skeletal muscle cells. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2014;1842:1372–1378. doi: 10.1016/j.bbadis.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Lantier L., et al. SIRT2 knockout exacerbates insulin resistance in high fat-fed mice. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemos V., et al. The NAD+-dependent deacetylase SIRT2 attenuates oxidative stress and mitochondrial dysfunction and improves insulin sensitivity in hepatocytes. Hum. Mol. Genet. 2017;26:4105–4117. doi: 10.1093/hmg/ddx298. [DOI] [PubMed] [Google Scholar]
- 26.Kitada M., et al. Calorie restriction in overweight males ameliorates obesity-related metabolic alterations and cellular adaptations through anti-aging effects, possibly including AMPK and SIRT1 activation. Biochim. Biophys. Acta Gen. Subj. 2013;1830:4820–4827. doi: 10.1016/j.bbagen.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Cantó C., et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi S.-E., et al. Elevated microRNA-34a in obesity reduces NAD+levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12:1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantó C., et al. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metabol. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uddin G.M., Youngson N.A., Sinclair D.A., Morris M.J. Head to head comparison of short-term treatment with the NAD+ precursor nicotinamide mononucleotide (NMN) and 6 Weeks of exercise in obese female mice. Front. Pharmacol. 2016;7 doi: 10.3389/fphar.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshino M., et al. Nicotinamide mononucleotide increases muscle insulin sensitivity in prediabetic women. Science. 2021;372:1224–1229. doi: 10.1126/science.abe9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaves A.B., et al. Exercise reduces the protein abundance of TXNIP and its interacting partner REDD1 in skeletal muscle: potential role for a PKA-mediated mechanism. J. Appl. Physiol. 2022;132:357–366. doi: 10.1152/japplphysiol.00229.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazo C.E., Miranda E.R., Shadiow J., Vesia M., Haus J.M. High intensity acute aerobic exercise elicits alterations in circulating and skeletal muscle tissue expression of neuroprotective exerkines. Brain Plasticity Preprint. 2022:1–14. doi: 10.3233/BPL-220137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda E.R., Shahtout J.L., Funai K. Chicken or egg? Mitochondrial phospholipids and oxidative stress in disuse-induced skeletal muscle atrophy. Antioxid. Redox Signal. 2023;38(4–6):338–351. doi: 10.1089/ars.2022.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shadiow J., et al. Exercise-induced changes to the fiber type-specific redox state in human skeletal muscle are associated with aerobic capacity. J. Appl. Physiol. 2023;135:508–518. doi: 10.1152/japplphysiol.00662.2022. 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadler G., et al. Establishment of clonal myogenic cell lines from severely affected dystrophic muscles - CDK4 maintains the myogenic population. Skeletal Muscle. 2011;1:12. doi: 10.1186/2044-5040-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorley M., et al. Skeletal muscle characteristics are preserved in hTERT/cdk4 human myogenic cell lines. Skeletal Muscle. 2016;6 doi: 10.1186/s13395-016-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright W.E., Shay J.W. Inexpensive low-oxygen incubators. Nat. Protoc. 2006;1:2088–2090. doi: 10.1038/nprot.2006.374. [DOI] [PubMed] [Google Scholar]
- 39.McAlister G.C., et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayed M.E., et al. Catalysis-dependent inactivation of human telomerase and its reactivation by intracellular telomerase-activating factors (iTAFs) J. Biol. Chem. 2019;294:11579–11596. doi: 10.1074/jbc.RA118.007234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thornalley P.J. Modification of the glyoxalase system in human red blood cells by glucosein vitro. Biochem. J. 1988;254:751–755. doi: 10.1042/bj2540751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiory F., et al. Methylglyoxal impairs insulin signalling and insulin action on glucose-induced insulin secretion in the pancreatic beta cell line INS-1E. Diabetologia. 2011;54:2941–2952. doi: 10.1007/s00125-011-2280-8. [DOI] [PubMed] [Google Scholar]
- 43.Nigro C., et al. Methylglyoxal impairs endothelial insulin sensitivity both in vitro and in vivo. Diabetologia. 2014;57:1485–1494. doi: 10.1007/s00125-014-3243-7. [DOI] [PubMed] [Google Scholar]
- 44.Rodrigues T., et al. Methylglyoxal-induced glycation changes adipose tissue vascular architecture, flow and expansion, leading to insulin resistance. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-01730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rai A.K., et al. Fructose-induced AGEs-RAGE signaling in skeletal muscle contributes to impairment of glucose homeostasis. J. Nutr. Biochem. 2019;71:35–44. doi: 10.1016/j.jnutbio.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Stratmann B., Goldstein B., Thornalley P., Rabbani N., Tschoepe D. Intracellular accumulation of methylglyoxal by glyoxalase 1 knock down alters collagen homoeostasis in L6 myoblasts. Int. J. Mol. Sci. 2017;18:480. doi: 10.3390/ijms18030480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue M., et al. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem. J. 2012;443:213–222. doi: 10.1042/BJ20111648. [DOI] [PubMed] [Google Scholar]
- 48.van Leeuwen I.M., et al. Modulation of p53 C-terminal acetylation by mdm2, p14ARF, and cytoplasmic SirT2. Mol. Cancer Therapeut. 2013;12:471–480. doi: 10.1158/1535-7163.MCT-12-0904. [DOI] [PubMed] [Google Scholar]
- 49.A. Philp et al., Sirtuin 1 (SIRT1) Deacetylase Activity Is Not Required for Mitochondrial Biogenesis or Peroxisome Proliferator-Activated Receptor-Gamma Coactivator-1alpha (PGC-1alpha) Deacetylation Following Endurance Exercise. [DOI] [PMC free article] [PubMed]
- 50.Huang K., et al. Polydatin promotes Nrf2-ARE anti-oxidative pathway through activating Sirt1 to resist AGEs-induced upregulation of fibronetin and transforming growth factor-beta1 in rat glomerular messangial cells. Mol. Cell. Endocrinol. 2015;399:178–189. doi: 10.1016/j.mce.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Huang K., et al. Sirt1 resists advanced glycation end products-induced expressions of fibronectin and TGF-beta1 by activating the Nrf2/ARE pathway in glomerular mesangial cells. Free Radic. Biol. Med. 2013;65:528–540. doi: 10.1016/j.freeradbiomed.2013.07.029. [DOI] [PubMed] [Google Scholar]
- 52.Ma F., et al. P53/NRF2 mediates SIRT1's protective effect on diabetic nephropathy. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1272–1281. doi: 10.1016/j.bbamcr.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Madsen A.S., et al. Investigating the sensitivity of NAD+-dependent sirtuin deacylation activities to NADH. J. Biol. Chem. 2016;291:7128–7141. doi: 10.1074/jbc.M115.668699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.E. A.-O. Jennings et al., Sirtuin 2 Regulates Protein LactoylLys Modifications. [DOI] [PMC free article] [PubMed]
- 55.Gaffney D.O., et al. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem. Biol. 2020;27:206–213.e206. doi: 10.1016/j.chembiol.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oakey L.A., et al. Metabolic tracing reveals novel adaptations to skeletal muscle cell energy production pathways in response to NAD (+) depletion. Wellcome Open Res. 2018;3:147. doi: 10.12688/wellcomeopenres.14898.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deshmukh A.S., Cox J., Jensen L.J., Meissner F., Mann M. Secretome analysis of lipid-induced insulin resistance in skeletal muscle cells by a combined experimental and bioinformatics workflow. J. Proteome Res. 2015;14:4885–4895. doi: 10.1021/acs.jproteome.5b00720. [DOI] [PubMed] [Google Scholar]
- 58.Conze D., Brenner C., Kruger C.L. Safety and metabolism of long-term administration of NIAGEN (nicotinamide riboside chloride) in a randomized, double-blind, placebo-controlled clinical trial of healthy overweight adults. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dollerup O.L., et al. A randomized placebo-controlled clinical trial of nicotinamide riboside in obese men: safety, insulin-sensitivity, and lipid-mobilizing effects. Am. J. Clin. Nutr. 2018;108:343–353. doi: 10.1093/ajcn/nqy132. [DOI] [PubMed] [Google Scholar]
- 60.Dollerup O.L., et al. Nicotinamide riboside does not alter mitochondrial respiration, content or morphology in skeletal muscle from obese and insulin‐resistant men. J. Physiol. 2020;598:731–754. doi: 10.1113/JP278752. [DOI] [PubMed] [Google Scholar]
- 61.Remie C.M.E., et al. Nicotinamide riboside supplementation alters body composition and skeletal muscle acetylcarnitine concentrations in healthy obese humans. Am. J. Clin. Nutr. 2020;112:413–426. doi: 10.1093/ajcn/nqaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trammell S.A.J., et al. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 2016;7 doi: 10.1038/ncomms12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Membrez M., et al. Trigonelline is an NAD(+) precursor that improves muscle function during ageing and is reduced in human sarcopenia. Nat. Metab. 2024;6:433–447. doi: 10.1038/s42255-024-00997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.