Abstract
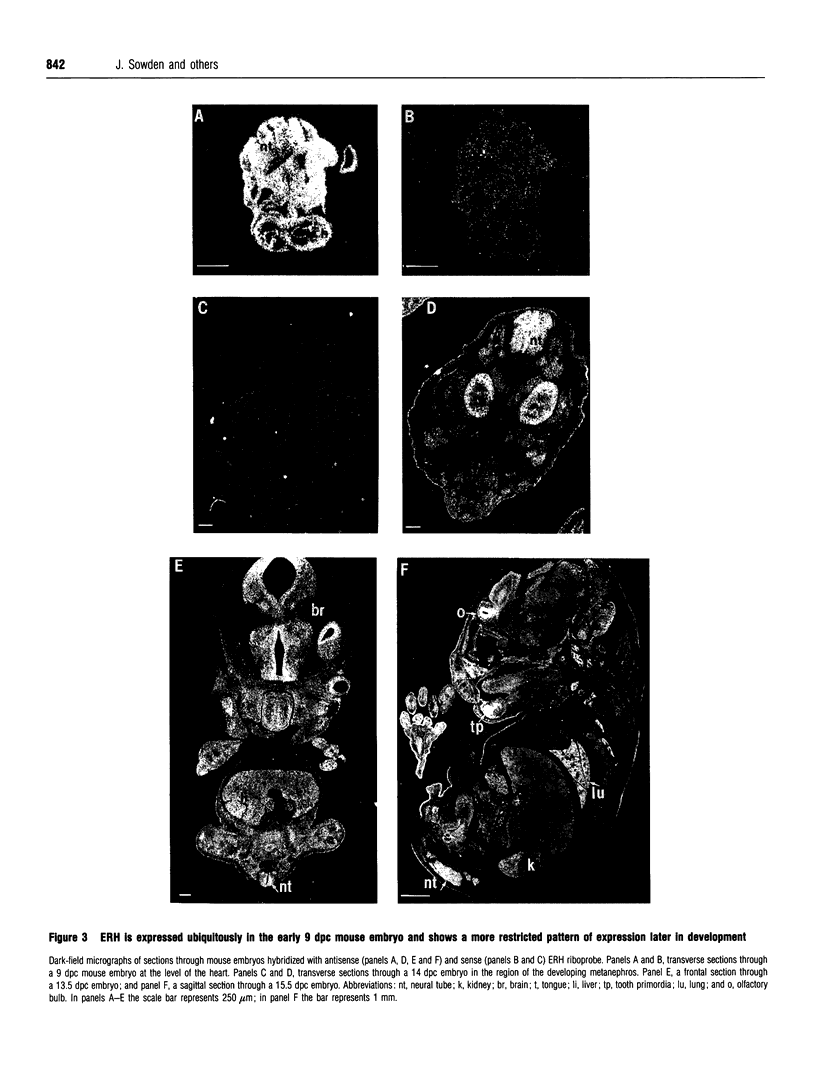
DEAD box proteins share several highly conserved motifs including the characteristic Asp-Glu-Ala-Asp (D-E-A-D in the amino acid single-letter code) motif and have established or putative ATP-dependent RNA helicase activity. These proteins are implicated in a range of cellular processes that involve regulation of RNA function, including translation initiation, RNA splicing and ribosome assembly. Here we describe the isolation and characterization of an embryonic RNA helicase gene, ERH, which maps to mouse chromosome 1 and encodes a new member of the DEAD box family of proteins. The predicted ERH protein shows high sequence similarity to the testes-specific mouse PL10 and to the maternally acting Xenopus An3 helicase proteins. The ERH expression profile is similar, to that of An3, which localizes to the animal hemisphere of oocytes and is abundantly expressed in the embryo. ERH is expressed in oocytes and is a ubiquitous mRNA in the 9 days-post-conception embryo, and at later stages of development shows a more restricted pattern of expression in brain and kidney. The similarities in sequence and in expression profile suggest that ERH is the murine equivalent of the Xenopus An3 gene, and we propose that ERH plays a role in translational activation of mRNA in the oocyte and early embryo.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Ford M. J., Anton I. A., Lane D. P. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988 Apr 21;332(6166):736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Nicol S. M., Reid A. D., Lane D. P. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993 Sep;12(9):3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994 Aug;4(8):271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Gee S. L., Conboy J. G. Mouse erythroid cells express multiple putative RNA helicase genes exhibiting high sequence conservation from yeast to mammals. Gene. 1994 Mar 25;140(2):171–177. doi: 10.1016/0378-1119(94)90541-x. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan R., Perry-O'Keefe H., Melton D. A., Weeks D. L. The Xenopus localized messenger RNA An3 may encode an ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):717–719. doi: 10.1038/349717a0. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T. R., Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990 Feb 15;343(6259):617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Iost I., Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994 Nov 10;372(6502):193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Beggs J. D. A suppressor of yeast spp81/ded1 mutations encodes a very similar putative ATP-dependent RNA helicase. Mol Microbiol. 1991 Apr;5(4):805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988 Oct 13;335(6191):611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Leroy P., Alzari P., Sassoon D., Wolgemuth D., Fellous M. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell. 1989 May 19;57(4):549–559. doi: 10.1016/0092-8674(89)90125-6. [DOI] [PubMed] [Google Scholar]
- Liang L., Diehl-Jones W., Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994 May;120(5):1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Lyons G. E., Buckingham M. E., Tweedie S., Edwards Y. H. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Development. 1991 Jan;111(1):233–244. doi: 10.1242/dev.111.1.233. [DOI] [PubMed] [Google Scholar]
- McGrew L. L., Dworkin-Rastl E., Dworkin M. B., Richter J. D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989 Jun;3(6):803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- Nielsen P. J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988 Jul;7(7):2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K., Morel-Deville F., Hershey J. W., Leighton T., Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988 Dec 1;336(6198):496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- Pause A., Méthot N., Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993 Nov;13(11):6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992 Jul;11(7):2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Rebagliati M. R., Weeks D. L., Harvey R. P., Melton D. A. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985 Oct;42(3):769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- Richter J. D. Translational control in development: a perspective. Dev Genet. 1993;14(6):407–411. doi: 10.1002/dvg.1020140602. [DOI] [PubMed] [Google Scholar]
- Roussell D. L., Bennett K. L. glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9300–9304. doi: 10.1073/pnas.90.20.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallés F. J., Darrow A. L., O'Connell M. L., Strickland S. Isolation of novel murine maternal mRNAs regulated by cytoplasmic polyadenylation. Genes Dev. 1992 Jul;6(7):1202–1212. doi: 10.1101/gad.6.7.1202. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S. R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992 Feb;6(3):283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Thach R. E. Cap recap: the involvement of eIF-4F in regulating gene expression. Cell. 1992 Jan 24;68(2):177–180. doi: 10.1016/0092-8674(92)90461-k. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks D. L., Melton D. A. A maternal mRNA localized to the animal pole of Xenopus eggs encodes a subunit of mitochondrial ATPase. Proc Natl Acad Sci U S A. 1987 May;84(9):2798–2802. doi: 10.1073/pnas.84.9.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse D. B., Putt W., Lovegrove J. U., Morrison K., Hollyoake M., Fox M. F., Hopkinson D. A., Edwards Y. H. Phosphoglucomutase 1: complete human and rabbit mRNA sequences and direct mapping of this highly polymorphic marker on human chromosome 1. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):411–415. doi: 10.1073/pnas.89.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S., Herrmann B. G. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990 Feb 15;343(6259):657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]