Abstract
Tepotinib is approved for the treatment of patients with non‐small‐cell lung cancer harboring MET exon 14 skipping alterations. While edema is the most prevalent adverse event (AE) and a known class effect of MET inhibitors including tepotinib, there is still limited understanding about the factors contributing to its occurrence. Herein, we apply machine learning (ML)‐based approaches to predict the likelihood of occurrence of edema in patients undergoing tepotinib treatment, and to identify factors influencing its development over time. Data from 612 patients receiving tepotinib in five Phase I/II studies were modeled with two ML algorithms, Random Forest, and Gradient Boosting Trees, to predict edema AE incidence and severity. Probability calibration was applied to give a realistic estimation of the likelihood of edema AE. Best model was tested on follow‐up data and on data from clinical studies unused while training. Results showed high performances across all the tested settings, with F1 scores up to 0.961 when retraining the model with the most relevant covariates. The use of ML explainability methods identified serum albumin as the most informative longitudinal covariate, and higher age as associated with higher probabilities of more severe edema. The developed methodological framework enables the use of ML algorithms for analyzing clinical safety data and exploiting longitudinal information through various covariate engineering approaches. Probability calibration ensures the accurate estimation of the likelihood of the AE occurrence, while explainability tools can identify factors contributing to model predictions, hence supporting population and individual patient‐level interpretation.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Edema is recognized as the most prevalent AE and a known class effect of MET inhibitors, including tepotinib. Current efforts aim to understand the efficacy of dose modifications in reducing this AE, exploring its relationship with potential prognostic factors.
WHAT QUESTION DID THIS STUDY ADDRESS?
Are there baseline and time‐varying factors to support the identification of higher likelihood of edema occurrence in patients receiving tepotinib treatment?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study assesses 54 covariates as predictors of edema using ML. Explainability tools investigate the relationship between input covariates and predicted outcomes. The identified drivers align with the existing knowledge of the investigated AE behavior.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
This study presents a framework to apply ML and explainability algorithms on longitudinal clinical data, ensuring a correct estimation of the probability of occurrence of the predicted events. Findings from the presented use case showcase the potential of the framework to enhance insights in clinical pharmacology and increase confidence in safety model outcomes.
INTRODUCTION
Drug‐disease models play a pivotal role in quantitative understanding of the trajectory of disease pathophysiology and the effects of drug treatment. These empirical or mechanistic models, often requiring the abstraction of data into dynamical systems, have proven to be valuable tools in drug development and therapeutic research. 1 However, their ability to mathematically describe complex datasets is highly dependent on the availability, quality, and quantity of data, and on the assumptions made by the modeler. 2 In parallel, the recent digital healthcare revolution has significantly expanded the opportunities to collect diverse, multimodal, high‐dimensional data, including clinical information, multi‐omics data, electronic health records, and imaging data, enabling advancements in precision medicine. 3 , 4 , 5 While this presents new opportunities to enhance drug development efficiency and improve patient care, it also poses a new challenge for traditional modeling approaches. Indeed, extracting meaningful insights from such vast volumes of diverse data has become increasingly difficult.
In this context, machine learning (ML) approaches have emerged as promising in advancing understanding in drug and disease in the context of drug development, while complementing and enhancing conventional approaches like pharmacometrics models. 6 ML models are universal, nonlinear, function approximation tools capable of learning patterns from empirical data by exploiting highly dimensional input spaces, that is, potentially incorporating a large number of covariates within the model. 7 , 8 Moreover, no assumptions on data distributions or on the biological process driving the studied phenomena must be a priori taken, as ML models are fully data driven. Such characteristics neatly distinguish ML algorithms from other non‐mechanistic models and traditional statistical approaches. 9 Many ML applications have already proven effective in modeling the occurrence of events or clinical outcomes along with baseline covariate values. However, there is still not a shared unified pipeline for ML application on longitudinal clinical data.
In this study, a framework to apply ML approaches to the prediction of longitudinal clinical end points is proposed. The framework is presented via an application on the case study of tepotinib, a highly selective, potent, orally available, reversible, type Ib adenosine triphosphate (ATP)‐competitive, small molecule inhibitor of the mesenchymal–epithelial transition factor (MET). 10 Tepotinib inhibits hepatocyte growth factor (HGF)‐dependent and ‐independent MET tyrosine kinase signaling by blocking MET phosphorylation, and showed antitumor activity in multiple tumor models derived from diverse cancer types. 11 , 12 The antitumor activity of tepotinib is particularly pronounced in tumors with oncogenic alterations of MET, such as MET exon 14 (METex14) skipping and high‐level MET amplification. Based on results from the Phase II VISION (NCT02864992) clinical trial, 13 , 14 , 15 , 16 tepotinib is approved in multiple regions for the treatment of patients with non‐small‐cell lung cancer (NSCLC) with METex14 skipping alterations, representing ~3–4% of this type of cancer.
Previous clinical studies have revealed edema as the most prevalent adverse event and a known class effect of MET inhibitors including tepotinib. 17 , 18 However, the links between the participants' clinical history, the time and duration of tepotinib treatment, and the occurrence of edema remain unclear, and understanding the effect of dose modifications to mitigate such adverse event is the object of ongoing investigations.
The objectives of this study were twofold. First, ML models were tested to predict the occurrence of edema grade in patients undergoing tepotinib treatment. To this aim, a framework to apply classical ML approaches to the specific task of edema prediction was designed. In doing so, two ML models belonging to the family of classification trees algorithm, namely Random Forest (RF) and Gradient Boosting Trees (GBT), were applied and benchmarked. Classification trees are known to outperform more complex models, including neural networks, when used for prediction tasks on structured tabular data such as those collected for the present study. 19 However, they are not able to intrinsically account for the longitudinal dimension characterizing clinical data. To overcome this limitation, different ways of introducing the temporal dimension into the model via covariate engineering were evaluated. The performances of the best predictive model were verified on longer term follow‐up data, and on data for a different set of patients. Finally, the model was retrained including the entire dataset and only on a reduced set of the most relevant input covariates.
The second objective of the study was the identification of the factors predicting edema occurrence and evolution over time. The Shapley Additive exPlanations (SHAP) method was used to investigate the role different factors have toward a specific estimation of edema occurrence obtained via the best predictive model, both at population and patient level. The use of this approach overcomes the lack of explainability of ML models, which approximate complex nonlinear functions from data in a not straightforwardly interpretable manner. 20 , 21
METHODS
Clinical data
Data from 612 patients enrolled in five Phase I/II clinical studies with tepotinib were collected (NCT01014936, NCT01832506, NCT01988493, NCT02115373, VISION – NCT02864992). The patients in the dataset received tepotinib monotherapy at doses of 30–1400 mg, once daily, including different patterns of dose modifications and with the recommended clinical dose of 500 mg (equivalent to 450 mg active moiety) once daily administered to 481 of them.
Adverse events were coded according to the Medical Dictionary for Regulatory Activities version 23.0 (MedDRA 23.0). Their severity was graded using the National Cancer Institute Common Terminology Criteria for Adverse Event version 4.0 (NCI‐CTCAE v4.0) toxicity grades. 22 Table 1 shows details of the study designs, together with a summary of the worst edema grade observed in patients during tepotinib treatment within the clinical study.
TABLE 1.
Study design summary. For VISION (NCT02864992), data up to a cutoff date of November 2022 were considered.
| Study number | NCT01014936 40 | NCT01832506 41 | NCT01988493 42 | NCT02115373 43 | VISION; NCT02864992 14 |
|---|---|---|---|---|---|
| Title | A phase I open label, non‐randomized, dose‐escalation first‐in‐man trial to investigate the c‐Met kinase inhibitor tepotinib under three different regimens in patients with advanced solid tumors | A Japanese multicenter, open label, phase I trial of c‐Met inhibitor tepotinib given orally as monotherapy to patients with solid tumors | A multicenter, randomized, phase Ib/II trial to evaluate the efficacy, safety, and pharmacokinetics of tepotinib as monotherapy versus sorafenib in Asian patients with MET+ advanced hepatocellular carcinoma and Child‐Pugh Class A liver function | A multicenter, single arm, Phase Ib/II study to evaluate efficacy, safety, and PK of tepotinib as monotherapy in patients with MET+ advanced hepatocellular carcinoma with Child‐Pugh Class A liver function who have failed sorafenib treatment | A Phase II single‐arm trial to investigate tepotinib in advanced (locally advanced or metastatic) non‐small cell lung cancer with METex14 skipping alterations or MET amplification |
| Tepotinib treatment | Tepotinib doses of 30 mg to 1400 mg/day in three different treatment regimens (QD, 2 weeks on – 1 week of, TIW) | Tepotinib 215, 300, or 500 mg/day QD | Tepotinib 300, 500, 1000 mg/day QD | Tepotinib 300 or 500 mg/day QD | Tepotinib 500 mg/day QD over 21‐day cycle(s) until disease progression or undue toxicity |
| Primary objective | MTD for each of three treatment regimens in patients with advanced solid tumors | Confirm MTD and RP2D in Japanese patients with solid tumors | Confirm RP2D and evaluate efficacy as first‐line treatment in patients with HCC and Child‐Pugh class A liver function | Determine RP2D and evaluate efficacy in patients with advanced HCC and Child‐Pugh class A liver function pretreated with sorafenib | Efficacy in patients with advanced (stage IIIB/IV) NSCLC |
| Number of patients | 149 | 12 | 72 | 66 | 152 Cohort A, 161 Cohort C |
| Median number of days before dropout (min–max) | 48 (7–1166) | 56 (40–393) | 213 (11–1931) | 161 (19–715) | 518 (9–2197) |
| Worst edema grade reached by patients, n (%) | |||||
| Grade 0 | 103 (69.1) | 8 (66.66) | 38 (52.8) | 18 (27.3) | 57 (18.2) |
| Grade 1 | 20 (13.4) | 4 (33.33) | 17 (23.6) | 21 (31.8) | 100 (32.0) |
| Grade 2 | 19 (12.8) | – | 17 (23.6) | 21 (31.8) | 107 (34.2) |
| Grade 3 | 7 (4.7) | – | – | 6 (9.1) | 48 (15.3) |
| Grade 4 | – | – | – | – | 1 (0.3) |
Abbreviations: HCC, hepatocellular carcinoma; MTD, maximum tolerated dose; NSCLC, non‐small‐cell lung cancer; QD, once daily; RP2D, recommended Phase II dose; TIW, three times a week.
Figure S1 shows the proportion of patients reaching each grade as worst case over fixed sized temporal windows of 84 days each. Data from only ~5% of patients were available after 1260 days from the beginning of the treatment. In the first 1260, grade 1 was found in at least 20% of patients available in each window; the same applied to grade 2, except for the first window where such proportion was closer to 15%. The proportion of available patients having grade 3 was stable over time and lower than 10%. These data describe edema as a frequent adverse event, as more than half of all patients experienced it during the entire treatment duration.
However, edema events were measured with limited frequency over time; no edema was reported in 45.15% of available safety visits throughout the entire dataset, and grade 1 was reported in 29.35%. Grade 2 was reported in 21.68% of safety visits, while grade 3 and grade 4 were reported in <4% and 0.01%, respectively. Given the extremely low representativeness of grades 3 and 4 in the dataset, during the modeling phase, grades 2, 3, and 4 were aggregated into a single class representing edema of grade 2 or higher severity. Transitions from the edema grades reported at a safety visit and grades reported at the following one are shown in Figure S2.
A total of 54 covariates, 34 of which are time‐varying, were available as input for the edema prediction models (Table S1). Missing values in time‐invariant covariates have been imputed to the most frequent value in the case of categorical or ordinal variables, and to the median value in case of continuous ones. Time‐varying covariates have been imputed via the last observation carried forward method. Missing baseline values (i.e., most recent measurement of the covariate prior to first dosing for each patient) have been imputed with first observation carried backward.
Covariate engineering of longitudinal data
To identify how to better leverage the longitudinal dimension of the input data, additional inputs were designed by processing the time‐varying covariates. To incorporate the temporal evolution of input data into the models, the following distinct covariate engineering approaches were tested:
Actual values at visit: the time‐varying covariates measured during a safety visit are used without manipulations.
Multiple visits: for each time‐varying covariate, the last three values and their corresponding timestamps are flattened into a vector, eventually padded with the baseline value when the number of available visits is lower than three. The obtained encoded vector is concatenated to the remainder of the covariates.
Multiple windows: average values were computed on three temporal windows – from 1 to 7 days before the visit, 8–21 days, and 22–42 days. Number and size of the windows were determined based on the distribution of the frequency of the safety visits. Averaged values of each of the time‐varying covariate were therefore concatenated to the remainder covariates and used as input for the different models.
Long‐/short‐term windows: mean and standard deviation of time‐varying covariates were estimated in two windows: one with data from 1 to 14 days before the actual visit date, and another with data from start of treatment to 15 days before the visit. The two windows showed short‐ and long‐term covariate variability patterns, respectively.
Baseline plus delta: both the baseline value and the difference from baseline computed at the safety visit day were included.
In addition to the engineered time‐varying covariates, model input also included the collected time‐invariant covariates. The current edema grade of each patient was also included in the input spaces; note that the model target is the edema grade at the following safety visit. Treatment exposure was accounted for by calculating the cumulative tepotinib dose administered in the time interval , that is, from start of treatment to 15 days prior to the visit date, and the one administered in the interval , that is, from 14 days prior to the visit date up to the current visit date. This approach allows for the consideration of long‐term and short‐term cumulative exposure of tepotinib, the latter being also representative of dose modifications. Most of such modifications were made in response to the emergence of adverse events, particularly edema. Hence, treatment exposure serves as both input for the treating physician and the model, while also acting as an effector mechanism. This dual role makes interpreting its influence both highly relevant and complex. Finally, the time elapsed between the current safety visit and the subsequent visit was included as an input, as it can inform the forecasting horizon of the models, given that this period is irregular over time and across patients.
Machine learning modeling framework
A schematization of the workflow used to train and validate the models is presented in Figure 1. Data were divided into train (accounting for data from 80% of patients) and test sets (data from the remaining 20% of patients). Train data collected for each patient at each safety visit, preprocessed with the covariate engineering techniques previously described, were used as input to train the ML model. The aim of such a multiclass classification model was to predict the expected edema by grade at the following safety visit for the given patients. Different ML algorithms belonging to the family of classification trees algorithms were benchmarked. Classification trees build a tree‐like model of decisions and their possible consequences. They linearly partition the covariate space and then fit a simple constant model in each of the partitioned regions. Two tree‐based models were tested – RF and GBT. RF is an ensemble method that combines multiple classification trees to produce a more accurate and stable prediction. 23 A random subset of the input covariates and a separate subset of the training data were used to create each tree. The final prediction was the most frequent among the predictions of all the trees. This reduces the risk of overfitting and stabilizes the model as final output is based on the consensus of several trees. In GBT, classification trees were trained sequentially using a stage‐wise approach. 24 At each stage, the model attempts to reduce the overall prediction error by minimizing the residual error estimated on the preceding trees. GBTs are interdependent, in contrast to RF where trees are trained individually, making the model more prone to overfitting. The XGBoost implementation of the algorithm was used in this study to train the GBT model. 25
FIGURE 1.
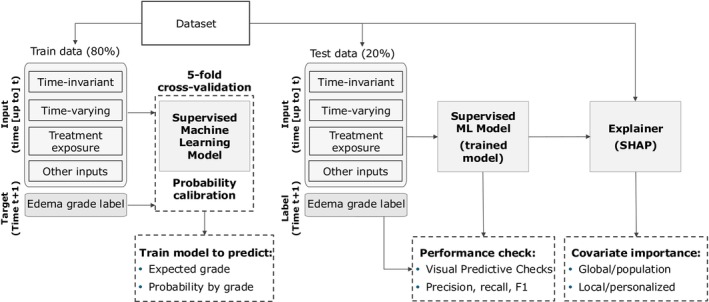
ML modeling framework. Data are divided into training and test sets. On a safety visit at the generic day , new values of the time‐varying covariates are measured for a patient. These values are used together with the time‐invariant covariates and treatment effect features as input for the ML supervised model. Five alternative covariate engineering approaches were adopted to explicitly account for the past measurements of the time‐varying covariates as further inputs for the model. Additional inputs can be generated to account for the time since the first dose administration at and the temporal interval between the visit day and the following safety visit day at time . The supervised model was then trained to forecast the expected edema adverse event grade for the patient at . The probability of having a given severity grade is also estimated for each patient at each safety visit day. The trained model was then applied to the test data to quantify its predictive performance with regard to precision, recall, F1 score, and with visual predictive checks. Finally, SHAP was used to assess the role of the input covariates toward specific predictions obtained. SHAP, Shapley Additive exPlanations.
All models were trained using stratified grouped k‐fold cross‐validation. 26 To account for the imbalance of the edema grades in the classification target, precision, recall, and weighted and macro F1 scores were used as quantitative metrics to evaluate model performances. 27 Probability calibration via Isotonic Regression was used to ensure that confidence scores predicted by the classifier – in this case, one of the RF or GBT models – were matching the true empirical frequencies of edema grades. 28 Practically, the cross‐validation procedure previously described was used to obtain unbiased predictions for all the data. Then, the unbiased predictions within each fold were used to train the Isotonic Regression. Further details on probability calibration and Isotonic Regression are provided in Supplementary Materials S1.
The last step of the methodology deals with model explainability. Here, SHAP 29 values were used to examine the influence of the input covariate on the output obtained at the patient level for each prediction visit. Furthermore, by aggregating the SHAP values computed across all the patient's data, a population‐level evaluation of the relevance of the input covariate in the resulting predictions was generated. 30 Details on computation of SHAP values are provided in Supplementary Materials S1.
Practically, the framework hereby presented was applied in two steps. First, only data from Cohort A of the VISION study (NCT02864992) up until February 2021, along with data from the remainder of the studies, were used. This reduced dataset was employed to determine which combination of ML algorithms and covariate engineering approaches would provide the best performance on the task of edema grade prediction. The resulting model was used to obtain predictions for the test set and on previously excluded data, that is, the follow‐up data for VISION Cohort A from February 2021 to November 2022, as well as data for patients in Cohort C of the same study. The performances obtained on such data were used to evaluate the model's generalization capability; its ability to provide accurate predictions for longer term follow‐up data, and for a different set of patients than that on which the model was trained.
In a second step, the best model was retrained using data from all available patients, divided again into train and test sets. SHAP was then used to identify the 10 most important predictors for model‐based predictions. A final model was subsequently trained using only these 10 predictors, and a comprehensive investigation of its performance and contributing factors was carried out.
RESULTS
Best model identification
To determine the best algorithm and covariate engineering approach to predict edema occurrence and grade, the five different input spaces obtained after covariate engineering were used as input to both RF and GBT, yielding a total of 10 different model settings. Only data up to February 2021 and without VISION Cohort C patients were used to generate the train and the test set. After model training, performances of the different settings have been assessed via the mean cross‐validation error obtained over the fivefold for the best combination of hyperparameters for each model. Table 2 shows the mean F1 score and the corresponding standard deviation for all the settings.
TABLE 2.
Mean weighted F1 scores and the corresponding standard deviations (in parenthesis) computed over the cross‐validation folds obtained via the different covariate engineering approaches tested.
| Covariate engineering | Random Forest | Gradient boosting |
|---|---|---|
| Actual values | 0.890 (0.026) | 0.876 (0.019) |
| Multiple visits | 0.917 (0.017) | 0.880 (0.012) |
| Multiple windows | 0.927 (0.010) | 0.879 (0.017) |
| Long‐/short‐term statistics | 0.929 (0.012) | 0.886 (0.019) |
| Baseline plus delta | 0.924 (0.016) | 0.885 (0.016) |
Although RF performs better than GBT for the specific task of edema prediction, there were no significant differences in the F1 score across the models trained with covariates resulting from different engineering approaches. Therefore, the RF model that utilized the multiple visit approach for time‐varying covariates was deemed the best compromise between model performance, longitudinal data exploitation, and easiness of result interpretation.
The performances of the selected model were further assessed on the VISION Cohort A data collected from February 2021 to November 2022 and on data from VISION Cohort C. No changes were observed in evaluation metrics when the model was used for predicting edema occurrence and grade in the patients from Cohort C, with the weighted F1 score estimated as 0.944. The F1 score reached 0.994 for follow‐up data from Cohort A. Such an increase in the metric is mostly to be attributed to the fact that stable edema conditions were assessed for 74 of the 94 patients, for whom follow‐up data were available.
Final model on extended dataset
The RF model using the actual value at the visit for time‐varying covariates was retrained using data from all available patients. Generalization performances were assessed on the previously unused test set, resulting in a weighted F1 score of 0.959. Effects of calibration via Isotonic Regression on such model are shown in Figures S3, S4. Then, SHAP values were used to determine the 10 most relevant predictors for this model. Finally, a last model was trained using only such predictors, leading to a weighted F1 score of 0.961. Precision and recall values for this model are reported in Table S2, showing consistent results across the different output classes. As increased age was previously found to be associated with increasing risk of edema, 10 the performances of the model have been verified within the different age terciles, showing consistent results across them (weighted F1 score equal to 0.969, 0.975, and 0.938 for the three terciles, respectively).
To characterize the influence of tepotinib exposure on the model, the latter was retrained by excluding the cumulated dose in the interval , the cumulated dose in the interval , or both. Results of this ablation study are reported in Table 3. For models including current edema grade as input to the model, only limited fluctuations of the F1 score were observed. When the current edema grade was not included, the exclusion of both the exposure‐derived features was associated with only a slight reduction of the F1 score, potentially suggesting that exposure effect may have been accounted for by the model based on its (nonlinear) relationship with other input features, such as albumin. 10 Moreover, when accounting only for the dose, only a moderate increase of the F1 score was found with respect to the previous case of excluding both exposure descriptors, while the model including only the dose attained a similar F1 score to that including both exposure‐derived features.
TABLE 3.
Sensitivity analysis to the presence of the exposure‐derived features and to the inclusion of the current edema grade in the inputs of the best model (Random Forest using as input for the time‐varying covariates the actual values at visit engineering approach). Mean values and standard deviations (in parenthesis) of the weighted F1 score computed over the cross‐validation folds are shown.
| Mean cross‐validation F1 score for models including current edema grade (standard deviation) | Mean cross‐validation F1 score for models excluding current edema grade (standard deviation) | |
|---|---|---|
| Both exposure‐derived features | 0.957 (0.007) | 0.596 (0.073) |
| Only cumulated dose since t − 14 | 0.958 (0.009) | 0.561 (0.118) |
| Only cumulated dose until t − 15 | 0.960 (0.007) | 0.604 (0.093) |
| No dose‐derived features | 0.944 (0.026) | 0.550 (0.119) |
Figure 2 displays the 10 most predictive inputs used to train the final model, presented in descending order, and their relationship to the output. Consistently with the above sensitivity analysis, past current edema grade was found to be the most influential input, particularly if a same grade persisted to the following safety visit. The exposure‐derived features were also informative for the model probability predictions. Albumin was found as the most informative time‐varying covariate, especially for predicting edemas of grades 2+.
FIGURE 2.
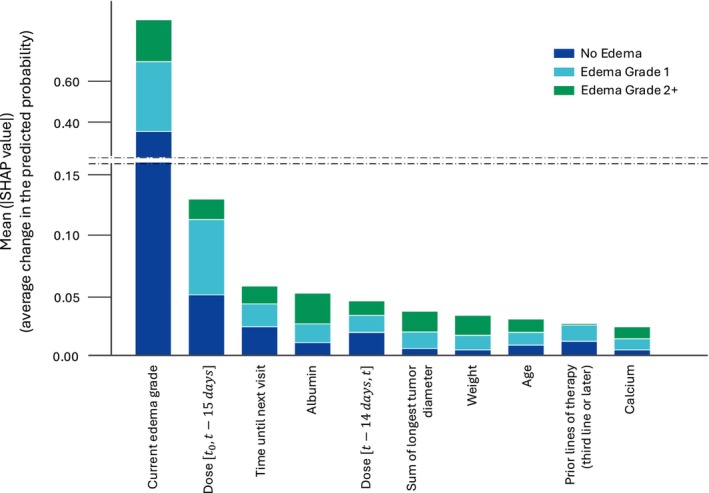
Global input importance via mean SHAP values. Ranking of the model input for the most influential to the less influential for the model. The y‐axis indicates the average change in the predicted probability of edema by grade, on average across the entire test set. SHAP, Shapley Additive exPlanations.
Figure 3 illustrates the contribution of the input variables toward the predicted probability of edemas of grades 2+. The analysis reveals that the current edema grade is the most informative input, as patients with a history of edemas of grades 2+ are considered highly likely to experience the same grade in the future. Interestingly, albumin once again emerges as the most informative among the longitudinal covariates, with lower levels associated with an increase in the predicted probability of edemas of grades 2+. Moreover, age impact on predictions appears to have an evident pattern, with older subjects associated with an increased probability of edemas of grade 2+.
FIGURE 3.
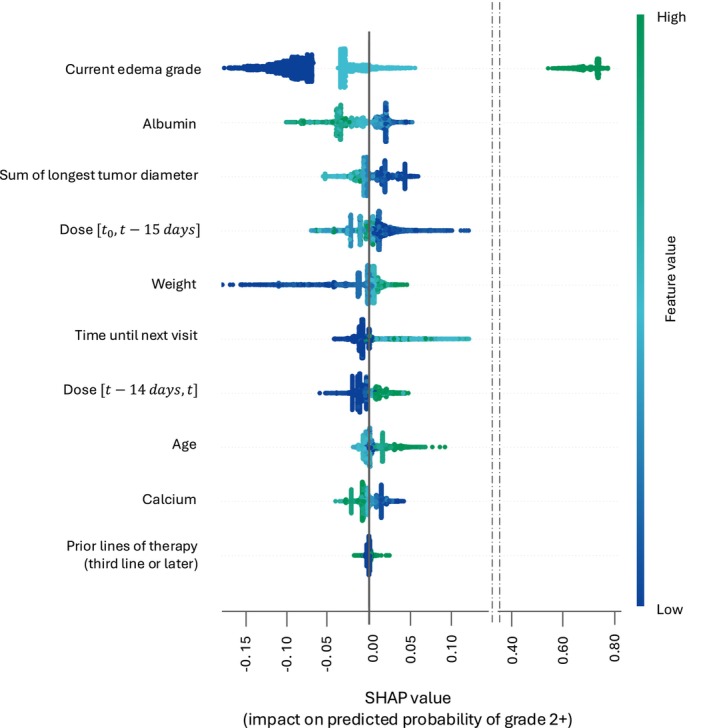
SHAP values – contribution of the inputs toward the predicted probabilities of edemas of grade 2+. List of the eight most influential inputs with respect to the predicted probabilities of edemas of grades 2+. Each point on the plot is a SHAP value for a covariate at a specific patient visit. The position on the y‐axis indicates the covariate importance and on the x‐axis the impact on the predicted probability. Color represents the value of the covariate. SHAP, Shapley Additive exPlanations.
Figure 4 illustrates the relationship between albumin, age, the cumulated dose normalized over 14 days, and their corresponding SHAP values for predicting the likelihood of edemas of grades 2+. For lower albumin levels, positive SHAP contributions between 0 and 0.5 are consistently assigned, signifying an increased risk of developing edema of grade 2+. Notably, very low albumin values are predominantly associated with higher grades of edema, particularly grade 2+. On the other hand, for higher albumin levels the corresponding SHAP values are mostly negative and ranging from 0 to −0.5, suggesting a reduced risk of edema of grade 2+. The association between age greater than 70 years and an increased likelihood of edemas of grades 2+ was also confirmed. Additionally, for all ages, higher SHAP values were assigned to patients who experienced edemas, particularly of grade 2+. Finally, within low ranges of cumulated dose in the interval normalized over 14 days, higher SHAP values were assigned to samples corresponding to edemas of grades 2+. This could reflect the tendency to adjust administered doses in those cases where the risk of edema was identified.
FIGURE 4.
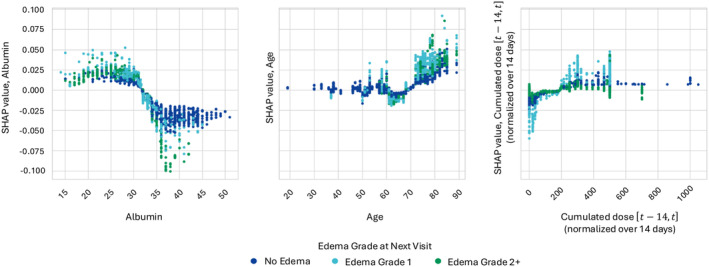
Interactions between covariate values and corresponding SHAP values. Scatterplot of the covariate value against its corresponding SHAP value for albumin, age, and cumulated dose over 2 weeks prior to the time at which prediction is performed. Each point corresponds to a specific patient visit. Points are colored based on the edema grade at the following safety visit. SHAP, Shapley Additive exPlanations.
DISCUSSION
Edema is known to be the most prevalent adverse event, and a known class effect of MET inhibitors 17 , 18 , 31 indicating that the underlying pathology may be related to a target‐mediated effect. 10 The MET ligand, HGF, and the MET/PI3k/Akt pathway could play a role in regulating endothelial permeability. 32 , 33 Blocking the HGF/MET signaling axis may result in compromised endothelial barrier integrity, leading to fluid retention and edema.
A wider understanding of the relationships between clinical covariates and the occurrence of edema is of practical importance in the evaluation and management of risk for treatment‐emergent edema during pharmacotherapy with MET inhibitors.
This study was aimed at formalizing a framework to apply ML algorithms on longitudinal clinical data and testing it on the case of edema occurrence in patients from five Phase I/II clinical studies receiving tepotinib monotherapy at doses of 30–1400 mg, once daily. However, some ML algorithms, including those benchmarked in the present study, are not explicitly designed to handle longitudinal time‐varying covariates. To overcome this limitation, five different covariate engineering approaches were evaluated to embed temporal dimensions into model input. The analysis framework was further completed by applying ML explainability tools to enable an understanding of the relationship between the input covariates and predicted outcomes.
The results indicated high predictive performances, with the best model correctly predicting edema grades in 94–98% of patients' visits. The high performances are mostly driven by the current edema status, the inclusion of which as model input was motivated by the underlying premise that in clinical practice, the presence of adverse events is known, and serves as a foundation for any decision, such as dose modification. The presence of such input is ensuring high model performances when predicting persistence of a given edema grade. The sensitivity analysis to the inclusion of this input revealed a decline in the mean cross‐validation F1 score of ~0.350 when it was removed as a candidate predictor. However, the use of Isotonic Regressions ensures the estimations of correctly calibrated probabilities. For instance, while low predicted probabilities, for example, for edemas of grade 2+, might lead to a classification error, they can still provide valuable details on existing risk of occurrence of an adverse event (Figure S5). Moreover, sensitivity analysis together with the SHAP importance highlighted interesting patterns with respect to the exposure‐related features. Referencing Figure 3, lower values of the dose are associated with a decreased probability of edema of grades 2+ at subsequent visits. As a decrease in such covariate would be a consequence of a dose reduction or of a temporary treatment interruption, this suggest that the latter are an effective approach to mitigate edema. Conversely, dose is influenced by the fact that the longer a patient remains on treatment, the higher the cumulative long‐term dose. Figure 3 indicates that patients with higher cumulative doses have a lower probability of experiencing edema of grade 2+, suggesting that these patients are undergoing a longer duration of treatment without severe adverse effects. The exclusion of the exposure‐related features seems to affect the model classification performances only marginally, as shown in Table 3. However, they were identified as among the most important inputs in the SHAP analyses. The combination of the two results suggests that other covariate(s) included in the model might act as surrogate(s) of the exposure, informing the model about its role even in the case in which dose‐related variables are excluded from its input. This might, for example, be true for serum albumin as treatment‐emergent hypoalbuminemia was already reported for several MET inhibitors, including tepotinib, and a relationship to tepotinib plasma concentrations has been previously described. 10 , 34 , 35 Furthermore, SHAP analysis revealed an association between lower levels of albumin and an increased predicted probability of edemas of grades 2+. This is consistent with previous findings which highlighted a trend indicating a positive relationship between the magnitude of decrease in serum albumin and the maximum severity of edema. 10 Indeed, albumin has a physiological role of maintaining oncotic pressure, hence is potentially a factor for edema pathogenesis. Advanced age was also found as predictive of edemas of grade 2+, in agreement with current knowledge of the investigated adverse event behavior, for which age is known to be a risk factor independently from drug exposure. 36 , 37 Finally, the time until the next visit, used to inform the model about the forecasting horizon, was also informative. However, this input should be seen as a factor reflecting the deteriorating status of patients, as changes in medical condition could prompt clinicians to schedule short‐term (re‐)assessment visits.
One of the primary challenges encountered in this study's classification setting was the unbalanced representation of different edema grades in the data. To overcome this issue, the model was set up to produce a probability for each edema grade of any new patient instance. When dealing with unbalanced data, such probabilities can be small, which is not a problem as long as they are accurate. Given that ML models are prone to poor estimation of class probabilities, probability calibration through Isotonic Regression was employed to obtain reliable probability estimators. Probability calibration has recently been applied to other case studies in the biomedical and clinical research settings, for example, to identify optimal dosing in phase I/II 38 or to predict mortality rates in patients with diffuse large B‐cell lymphoma. 39 Furthermore, the evaluation of model performance through metrics, such as precision or recall, can be viewed not as a part of the ML modeling itself, but rather as part of the decision‐making component. The cost associated with type I and II errors might indeed be defined in different ways by decision makers and clinical practitioners, without requiring any change to the statistical setting of the ML model.
In conclusion, even in cases where the reduced number of data points might preclude the use of complex Deep Learning models like, for example, Recurrent Neural Networks, the methodology hereby presented enables the exploitation of longitudinal data within ML models, furthering progress in model‐informed precision medicine 5 by complementing analyses conducted with other mechanism‐informed and non‐mechanistic models in pharmacometrics and traditional statistical approaches. Future research may focus on the use of models explicitly able to exploit the longitudinal dimension of clinical data, such as recurrent neural networks or Neural Ordinary Differential Equations, for which however a larger set of input observations would be needed to ensure proper model training.
AUTHOR CONTRIBUTIONS
F.A, N.T., R.S., R.C., K.V, R.B., V.G., A.J., and K.B. wrote the manuscript. F.A, N.T., R.S., R.C., K.V, R.B., V.G., A.J., and K.B. designed the research. F.A. and N.T. performed the research. F.A. and N.T. analyzed the data. F.A., N.T., and R.C. contributed new reagents/analytical tools.
FUNDING INFORMATION
This study was sponsored by the healthcare business of Merck KGaA, Darmstadt, Germany (CrossRef Funder ID:10.13039/100009945). Editorial support was provided by Syneos Health, London, UK, and funded by the healthcare business of Merck KGaA, Darmstadt, Germany.
CONFLICT OF INTEREST STATEMENT
N.T. is an employee of Ares Trading S.A., Lausanne, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany. R.S., R.B. and A.J. are employees of the healthcare business of Merck KGaA, Darmstadt, Germany. K.B. was an employee of the healthcare business of Merck KGaA, Darmstadt, Germany at the time of the study. V.G. is an employee of Ares Trading S.A. Eysins, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany. F.A. and R.C. are employees of the Swiss Data Science Center – EPFL, Lausanne, Switzerland, formally engaged in a research collaboration with Quantitative Pharmacology, Ares Trading S.A., Lausanne, Switzerland, an affiliate of Merck KGaA, Darmstadt, Germany at the time of the publishing of this study. K.V. is an employee of EMD Serono.
Supporting information
Data S1:
Data S2:
Amato F, Strotmann R, Castello R, et al. Explainable machine learning prediction of edema adverse events in patients treated with tepotinib. Clin Transl Sci. 2024;17:e70010. doi: 10.1111/cts.70010
Karin Berghoff was an employee at the time of the study.
DATA AVAILABILITY STATEMENT
Any requests for data by qualified scientific and medical researchers for legitimate research purposes will be subject to the healthcare business of Merck KGaA, Darmstadt, Germany's (CrossRef Funder ID: 10.13039/100009945) Data Sharing Policy. All requests should be submitted in writing to the healthcare business of Merck KGaA, Darmstadt, Germany's data sharing portal (https://www.emdgroup.com/en/research/our‐approach‐to‐research‐and‐development/healthcare/clinical‐trials/commitment‐responsible‐data‐sharing.html). When the healthcare business of Merck KGaA, Darmstadt, Germany has a co‐research, co‐development, or co‐marketing or co‐promotion agreement, or when the product has been out‐licensed, the responsibility for disclosure might be dependent on the agreement between parties. Under these circumstances, the healthcare business of Merck KGaA, Darmstadt, Germany will endeavor to gain agreement to share data in response to requests.
REFERENCES
- 1. Helmlinger G, Al‐Huniti N, Aksenov S, et al. Drug‐disease modeling in the pharmaceutical industry – where mechanistic systems pharmacology and statistical pharmacometrics meet. Eur J Pharm Sci. 2017;109S:S39‐S46. [DOI] [PubMed] [Google Scholar]
- 2. Meibohm B, Dorendorf H. Basic concepts of pharmacokinetic/pharmacodynamic (PK/PD) modelling. Int J Clin Pharmacol Ther. 1997;35:401‐413. [PubMed] [Google Scholar]
- 3. Terranova N, Venkatakrishnan K. Machine learning in modeling disease trajectory and treatment outcomes: an emerging enabler for model‐informed precision medicine. Clin Pharmacol Ther. 2023;115:720‐726. [DOI] [PubMed] [Google Scholar]
- 4. Terranova N, Venkatakrishnan K, Benincosa LJ. Application of machine learning in translational medicine: current status and future opportunities. AAPS Journal. 2021;23:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venkatakrishnan K, Benincosa LJ. Diversity and inclusion in drug development: rethinking intrinsic and extrinsic factors with patient centricity. Clin Pharmacol Ther. 2022;112:204‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Naik K, Goyal RK, Foschini L, et al. Current status and future directions: the application of artificial intelligence/machine learning for precision medicine. Clin Pharmacol Ther. 2023;115:673‐686. [DOI] [PubMed] [Google Scholar]
- 7. Murphy KP. Machine Learning – A Probabilistic Perspective. The MIT Press; 2012. [Google Scholar]
- 8. Sibieude E, Khandelwal A, Girard P, Hesthaven JS, Terranova N. Population pharmacokinetic model selection assisted by machine learning. J Pharmacokinet Pharmacodyn. 2022;49:257‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker RE, Peña JM, Jayamohan J, Jérusalem A. Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol Lett. 2018;14:20170660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiong W, Hietala SF, Nyberg J, et al. Exposure‐response analyses for the MET inhibitor tepotinib including patients in the pivotal VISION trial: support for dosage recommendations. Cancer Chemother Pharmacol. 2022;90:53‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao CF, Woude GF. Vande HGF/SF‐met signaling in tumor progression. Cell Res. 2005;15:49‐51. [DOI] [PubMed] [Google Scholar]
- 12. Birchmeier C, Birchmeier W, Gherardi E, Woude GF. Vande met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915‐925. [DOI] [PubMed] [Google Scholar]
- 13. Mazieres J, Paik PK, Garassino MC, et al. Tepotinib treatment in patients With MET Exon 14–skipping non–small cell lung cancer: long‐term follow‐up of the VISION phase 2 nonrandomized clinical trial. JAMA Oncol. 2023;9:1260‐1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paik P, Felip E, Veillon R, et al. Tepotinib in non‐small‐cell lung cancer with MET exon 14 skipping mutations. N Engl J Med. 2020;383:931‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Veillon R, Sakai H, Le X, et al. Safety of tepotinib in patients with MET exon 14 skipping NSCLC and recommendations for management. Clin Lung Cancer. 2022;23:320‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le X, Sakai H, Felip E, et al. Tepotinib efficacy and safety in patients with MET exon 14 skipping NSCLC: outcomes in patient subgroups from the VISION study with relevance for clinical practice. Clin Cancer Res. 2022;28:1117‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choueiri TK, Heng DYC, Lee JL, et al. Efficacy of savolitinib vs sunitinib in patients with MET‐driven papillary renal cell carcinoma: the SAVOIR phase 3 randomized clinical trial. JAMA Oncol. 2020;6:1247‐1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heigener DF, Reck M. Crizotinib. Recent Results Cancer Res. 2018;211:57‐65. [DOI] [PubMed] [Google Scholar]
- 19. Grinsztajn L, Oyallon E, Varoquaux G. Why do tree‐based models still outperform deep learning on typical tabular data? Adv Neural Inf Proces Syst. 2022;35:507‐520. [Google Scholar]
- 20. Terranova N, French J, Dai H, et al. Pharmacometric modeling and machine learning analyses of prognostic and predictive factors in the JAVELIN gastric 100 phase III trial of avelumab. CPT Pharmacometrics Syst Pharmacol. 2022;11:333‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Basu S, Munafo A, Ben‐Amor AF, Roy S, Girard P, Terranova N. Predicting disease activity in patients with multiple sclerosis: an explainable machine‐learning approach in the Mavenclad trials. CPT Pharmacometrics Syst Pharmacol. 2022;11:843‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. National Cancer Institute . Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. 2009. https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03/CTCAE_4.03_2010‐06‐14_QuickReference_5x7.pdf
- 23. Breiman L. Random forests. Mach Learn. 2001;45:5‐32. [Google Scholar]
- 24. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189‐1232. [Google Scholar]
- 25. Chen T, He T, Benesty M, et al. Xgboost: extreme gradient boosting. R Package Version 0.4‐2. 2015;1:2‐4. [Google Scholar]
- 26. Hastie T, Tibshirani R, Friedman J. The Element of Statistical Learning: Data Mining, Interference, and Prediction. Springer; 2009. [Google Scholar]
- 27. Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inf Process Manag. 2009;45:427‐437. [Google Scholar]
- 28. Zadrozny B, Elkan C. Transforming classifier scores into accurate multiclass probability estimates. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining . 2002. doi: 10.1145/775047.775151 [DOI]
- 29. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Adv Neural Inf Process Syst . 2017.
- 30. Janssen A, Hoogendoorn M, Cnossen MH, Mathôt RAA, OPTI‐CLOT Study Group and SYMPHONY Consortium . Application of SHAP values for inferring the optimal functional form of covariates in pharmacokinetic modeling. CPT Pharmacometrics Syst Pharmacol. 2022;11:1100‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolf J, Seto T, Han J‐Y, et al. Capmatinib in MET exon 14–mutated or MET‐amplified non–small‐cell lung cancer. N Engl J Med. 2020;383:944‐957. [DOI] [PubMed] [Google Scholar]
- 32. Usatyuk PV, Fu P, Mohan V, et al. Role of c‐met/phosphatidylinositol 3‐kinase (PI3k)/Akt signaling in hepatocyte growth factor (HGF)‐mediated Lamellipodia formation, reactive oxygen species (ROS) generation, and motility of lung endothelial cells*. J Biol Chem. 2014;289:13476‐13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamada N, Nakagawa S, Horai S, et al. Hepatocyte growth factor enhances the barrier function in primary cultures of rat brain microvascular endothelial cells. Microvasc Res. 2014;92:41‐49. [DOI] [PubMed] [Google Scholar]
- 34. Morley R, Cardenas A, Hawkins P, et al. Safety of onartuzumab in patients with solid tumors: experience to date from the onartuzumab clinical trial program. PLoS One. 2015;10:e0139679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tabernero J, Elez ME, Herranz M, et al. A Pharmacodynamic/pharmacokinetic study of ficlatuzumab in patients with advanced solid tumors and liver metastases. Clin Cancer Res. 2014;20:2793‐2804. [DOI] [PubMed] [Google Scholar]
- 36. Paik P, Xiong W, Hietala SF, et al. 584P Tepotinib exposure‐response analyses of safety and efficacy in patients with solid tumours. Ann Oncol. 2020;31:S494‐S495. [Google Scholar]
- 37. Ahn L, Alexander T, Vlassak S, Berghoff K, Lemmens L. Tepotinib: guidance for oncology nurses on management of adverse events in patients with MET exon 14 skipping non‐small‐cell lung cancer. Clin J Oncol Nurs. 2022;26:543‐551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiu Y, Zhao Y, Liu H, Cao S, Zhang C, Zang Y. Modified isotonic regression based phase I/II clinical trial design identifying optimal biological dose. Contemp Clin Trials. 2023;127:107139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan S, Zhao Z, Yu H, et al. Applying probability calibration to ensemble methods to predict 2‐year mortality in patients with DLBCL. BMC Med Inform Decis Mak. 2021;21:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Falchook GS, Kurzrock R, Amin HM, et al. First‐in‐man phase I trial of the selective MET inhibitor tepotinib in patients with advanced solid tumors. Clin Cancer Res. 2020;26:1237‐1246. [DOI] [PubMed] [Google Scholar]
- 41. Shitara K, Yamazaki K, Tsushima T, et al. Phase I trial of the MET inhibitor tepotinib in Japanese patients with solid tumors. Jpn J Clin Oncol. 2020;50:859‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Decaens T, Barone C, Assenat E, et al. Phase 1b/2 trial of tepotinib in sorafenib pretreated advanced hepatocellular carcinoma with MET overexpression. Br J Cancer. 2021;125:190‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ryoo B‐Y, Cheng A‐L, Ren Z, et al. Randomised phase 1b/2 trial of tepotinib vs sorafenib in Asian patients with advanced hepatocellular carcinoma with MET overexpression. Br J Cancer. 2021;125:200‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1:
Data S2:
Data Availability Statement
Any requests for data by qualified scientific and medical researchers for legitimate research purposes will be subject to the healthcare business of Merck KGaA, Darmstadt, Germany's (CrossRef Funder ID: 10.13039/100009945) Data Sharing Policy. All requests should be submitted in writing to the healthcare business of Merck KGaA, Darmstadt, Germany's data sharing portal (https://www.emdgroup.com/en/research/our‐approach‐to‐research‐and‐development/healthcare/clinical‐trials/commitment‐responsible‐data‐sharing.html). When the healthcare business of Merck KGaA, Darmstadt, Germany has a co‐research, co‐development, or co‐marketing or co‐promotion agreement, or when the product has been out‐licensed, the responsibility for disclosure might be dependent on the agreement between parties. Under these circumstances, the healthcare business of Merck KGaA, Darmstadt, Germany will endeavor to gain agreement to share data in response to requests.


