Abstract
mRNA-based vaccines and therapeutics are gaining popularity and usage across a wide range of conditions. One of the critical issues when designing such mRNAs is sequence optimization. Even small proteins or peptides can be encoded by an enormously large number of mRNAs. The actual mRNA sequence can have a large impact on several properties, including expression, stability, immunogenicity, and more. To enable the selection of an optimal sequence, we developed CodonBERT, a large language model (LLM) for mRNAs. Unlike prior models, CodonBERT uses codons as inputs, which enables it to learn better representations. CodonBERT was trained using more than 10 million mRNA sequences from a diverse set of organisms. The resulting model captures important biological concepts. CodonBERT can also be extended to perform prediction tasks for various mRNA properties. CodonBERT outperforms previous mRNA prediction methods, including on a new flu vaccine data set.
mRNA vaccines have emerged as a high-potency, fast-production, low-cost, and safe alternative to traditional vaccines (Pardi et al. 2018, 2020; Zhang et al. 2019; Jackson et al. 2020b). mRNA vaccines are currently being developed for a broad range of human viruses and bacteria, including SARS-CoV-2, influenza, Zika, chlamydia, and more (Maruggi et al. 2017; Pardi et al. 2017; Jackson et al. 2020a; Pilkington et al. 2021). They are also being investigated as potential treatments for several diseases, including lung cancer, breast cancer, and melanoma (Miao et al. 2021; Lorentzen et al. 2022).
The expression level of a vaccine directly affects its potency, ultimate immunogenicity, and efficacy (Schlake et al. 2012). The higher the level of expression of the antigenic protein encoded by the mRNA sequence, the smaller amount of the vaccine is needed to achieve the desired immune response, which can make the vaccine more cost-effective and easier to manufacture (Pardi et al. 2018). Consequently, using a lower dose can help reduce reactogenicity (Ahmad et al. 2022) and maintain the immune response over a longer period (Leppek et al. 2022), leading to better safety and efficacy.
A human protein with an average length of 500 amino acids can be encoded by roughly 3500 different codon sequences. Although only one of those is encoded in the virus or DNA of interest, this is not necessarily the optimal sequence for a vaccine. The classical method to find the optimal mRNA sequence is codon optimization, which selects the most optimal codon for each amino acid using the codon bias in the host organism (Mauro and Chappell 2014). This method has been widely applied, including for optimizing recombinant protein drugs, nucleic acid therapies, gene therapy, mRNA therapy, and DNA/RNA vaccines (Al-Hawash et al. 2017; Webster et al. 2017; Mauro 2018). However, codon optimization alone does not consider several key properties that impact protein expression (Parret et al. 2016). For instance, RNA structural properties (e.g., stem loops and pseudoknots) have been shown to play a major role for noncoding RNAs (such as riboswitches or aptamers) (Groher et al. 2018; Schmidt et al. 2020).
Although mRNA sequence heavily influences cellular RNA stability (Agarwal and Shendure 2020; Agarwal and Kelley 2022), secondary structure can also impact mRNA stability in solution and modulate protein expression (Mauger et al. 2019; Leppek et al. 2022; Nieuwkoop et al. 2023; Zhang et al. 2023). For example, replacing a codon with a synonymous codon can alter the local base-pairing interactions and affect nearby structural motifs (Groher et al. 2019; Li et al. 2021). Thus, optimizing each codon independently is not sufficient to generate highly expressed proteins.
Pretraining a large language model (LLM) based on large-scale unlabeled text, followed by fine-tuning, has been widely adopted for natural language processing (Peters et al. 2018; Radford et al. 2018; Devlin et al. 2019). Recently, this concept has been scaled to biological sequences (protein, DNA, and RNA) (Bepler and Berger 2021; Ji et al. 2021; Rives et al. 2021; Akiyama and Sakakibara 2022; Chen et al. 2022). Such models can be used to embed nucleotides and use these embeddings for downstream supervised learning tasks. However, as we show, such LLMs may not be ideal for predicting protein expression owing to their focus on individual nucleotides and noncoding regions. More recent work, such as cdsBERT (Hallee et al. 2023), addresses the issue of codon awareness for a protein language model.
To address these limitations, we developed CodonBERT, an LLM that extends the BERT model and applies it to the language of mRNAs. CodonBERT uses a multihead attention transformer architecture framework. The pretrained model can also be generalized to a diverse set of supervised learning tasks. We pretrained CodonBERT using 10 million mRNA coding sequences (CDSs) spanning an evolutionarily diverse set of organisms. Next, we used it to perform several mRNA prediction tasks, including protein expression and mRNA degradation prediction. As we show, both the pretrained and the fine-tuned version of the models can learn new biology and improve on current state-of-the-art methods for mRNA vaccine design.
To assess generalization of our CodonBERT model, we collected a novel hemagglutinin flu vaccine data set. Different mRNA candidates that encode the influenza hemagglutinin antigen (i.e., with fixed untranslated regions and a variable coding region) were designed, synthesized, and transfected into cells. The protein expression levels corresponding to these mRNA sequences were measured and used as labels for a supervised learning task. CodonBERT leads to better performance than existing methods.
Results
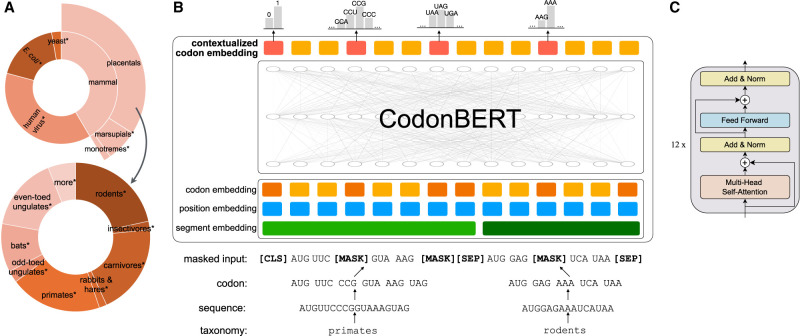
We developed a LLM, CodonBERT, for mRNA analysis and prediction tasks. CodonBERT was pretrained using 10 million mRNA sequences derived from mammals, bacteria, and human viruses. All sequences were hierarchically labeled using 14 categories as shown in Figure 1A. CodonBERT takes the coding region as input, using codons as tokens, and outputs an embedding that provides contextual codon representations. The embeddings provided by CodonBERT can be combined with additional trainable layers to perform various downstream regression and prediction tasks, including the prediction of protein expression and mRNA degradation.
Figure 1.
Pretraining data distribution and CodonBERT model architecture. (A) Hierarchically classified mRNA sequences for pretraining. All the 14 leaf-level classes (those annotated with an asterisk are numbered). The angle of each segment is proportional to the number of sequences belonging to this group. (B) Model architecture and training scheme deployed for two tasks of CodonBERT. (C) A stack of 12 transformer blocks employed in CodonBERT model.
A schematic representation of CodonBERT's architecture is provided in Figure 1B. We pretrained CodonBERT with two tasks: masked language model (MLM) learning and sequence taxonomy prediction (STP). The MLM task learns the codon representation, interactions between codons, and relationships between codons and sequences. The STP task aims to directly model the sequence representation and understand the evolutionary relationships between mRNA sequences. In short, a pair of mRNA sequences, which is randomly sampled from either the same or different categories, is codon-tokenized, concatenated, and randomly masked. The masked inputs are further encoded with codon-, position-, and segment-based embeddings and fed into a stack of transformer layers using a multihead attention network with residual connections. CodonBERT is self-supervised and relies on masked token prediction and taxonomic sequence prediction for optimizing parameters (Methods).
Pretrained representation model
To assess our pretrained CodonBERT model, we built a held-out data set by randomly leaving out 1% of mRNA sequences for each category and trained the model with the remaining sequences. As illustrated in Supplemental Figure S1, during the pretraining phase, the model performance on two tasks (MLM and STP) substantially improves on both the training and evaluation sets. For example, the entropy loss of the MLM task (ℒMLM) decreased to 2.85 on the evaluation set, which means that the model was able to narrow down the choice from 64 (uniform distribution) to only eight codons for each masked position (Methods).
CodonBERT learns, on its own, the genetic code and evolutionary homology
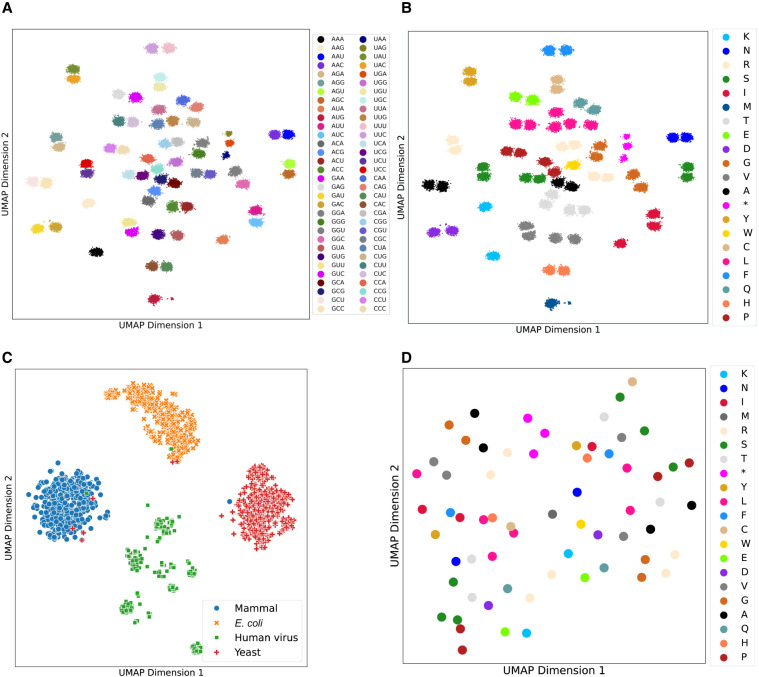
In addition to the quantitative evaluation of model predictions, for example, loss and accuracy, we also performed several qualitative analyses of the embeddings provided by CodonBERT. To decipher what kind of biological information has been learned by the model and encoded in the representation, we randomly sampled 500 sequences for each category from the held-out data set and extracted high-dimensional codon and sequence embeddings from CodonBERT. These were projected onto a two-dimensional space (2D) by UMAP (McInnes et al. 2018).
In Figure 2, A and B, each dot represents a codon and is annotated with different colors based on its type of codon and amino acid. Codons that encode the same amino acid, namely, synonymous codons, are spatially close to each other in Figure 2B, which implies that CodonBERT learns the genetic code from the large-scale training set. For example, the amino acid valine, whose one-letter code is V, can be encoded by four codons: {GUA, GUU, GUG, GUC}. Figure 2B illustrates four separate gray clusters for four possible codons, and four clusters are close to each other. We applied the k-nearest neighbor algorithm (kNN) on the output codon embeddings directly. Five hundred embeddings were sampled for each codon; 99.4% of the 500-nearest neighbors are the same codons. For the remaining misclassified codons, 87.7% are stop codons and are classified to other stop codons.
Figure 2.
Genetic code and evolutionary taxonomy information learned by the pretrained, unsupervised CodonBERT model. High-dimensional embeddings were projected into two-dimensional space using UMAP (McInnes et al. 2018). (A,B) Projected codon embeddings from the pretrained CodonBERT model. Each point represents a codon with different contexts, and its color corresponds to the type of codon (A) or amino acid (B) accordingly. (C) Projected sequence embedding from the pretrained CodonBERT model. Each point is a mRNA sequence, and its color represents the sequence label. (D) Projected codon embedding from the pretrained Codon2vec model. Each point shows a codon, and its color is the corresponding amino acid.
Codon2vec, a Word2vec (Mikolov et al. 2013) model trained on the collected mRNA sequences, can also produce codon representations (Methods). However, compared with the codon representation generated by CodonBERT, the embedding of each codon from Codon2vec is fixed regardless of the context surrounding the codon (Fig. 2D). This results in clusters that are often less accurate than the projection of codon representation from CodonBERT.
In addition to codon representation, CodonBERT also optimizes for sequence identification. 2D projections of the sequence embeddings of the held-out data set are presented. Figure 2C illustrates clusters of four high-level sequence categories: (Escherichia coli, human virus, yeast, and mammal). Sequences from the same organism are clustered together with clear boundaries between the taxonomy classes. However, CodonBERT does not reveal a clear separation between different families within mammals as illustrated in Supplemental Figure S2A. This observation could be attributed to the similar codon usage patterns within the taxonomic groups. Codon usage, a critical factor in the translation efficiency of genes, can significantly influence the clustering of genetic sequences in computational analyses. We conducted a statistical analysis on the frequency of codons in different taxonomic groups and computed the Kullback–Leibler divergence of the codon usage between any two organisms (Supplemental Fig. S2B). We found that all taxonomic groups in mammals exhibit significant different codon usage compared with human virus, E. coli, and yeast. However, among mammals, the codon usage is too similar to distinguish.
Evaluating CodonBERT and comparison to prior methods on supervised learning tasks
CodonBERT can be extended to perform supervised learning for specific mRNA prediction tasks. To evaluate the use of our LLM for downstream tasks and to compare it to prior methods, we collected several mRNA prediction data sets. Table 1 presents the data sets and the mRNA properties. As can be seen, these included a diverse set of downstream tasks related to mRNA translation, stability, and regulation. In addition, these data sets represent a range of molecules, including newly published data sets for recombinant protein, bio-computing, and SARS-CoV-2 vaccine design. Finally, we generated a new data set to test CodonBERT in the context of mRNAs encoding the influenza hemagglutinin antigen for flu vaccines.
Table 1.
The collection of the data sets with their corresponding mRNA source and property used for method evaluation
| Data set | Target | Category | No. of mRNAs | Seq length |
|---|---|---|---|---|
| MLOS flu vaccines (Sanofi-Aventis) | Expression | Regression | 543 | 1698–1704 |
| mRFP expression (Nieuwkoop et al. 2023) | Expression | Regression | 1459 | 678–678 |
| Fungal expression (Wint et al. 2022) | Expression | Regression | 7056 | 150–3000 |
| E. coli proteins (Ding et al. 2022) | Expression | Classification | 6348 | 171–3000 |
| Tc-riboswitches (Groher et al. 2019) | Switching factor | Regression | 355 | 67–73 |
| mRNA stability (Diez et al. 2022) | Stability | Regression | 41,123 | 30–1497 |
| SARS-CoV-2 vaccine degradation (Wayment-Steele et al. 2022) | Degradation | Regression | 2400 | 81–81 |
Each data set is split into training, validation, and test with a 0.7, 0.15, and 0.15 ratio. All the methods were optimized on the same data split.
The mRFP expression data set (Nieuwkoop et al. 2023) profiles protein production levels for several gene variants in E. coli. The fungal expression data set (Grigoriev et al. 2014; Wint et al. 2022) includes CDSs >150 bp from a wide range of fungal genomes. The E. coli protein data set (Ding et al. 2022) comprises experimental data for protein expression in E. coli, which are labeled as low, medium, or high expression (2308, 2067, and 1973 mRNA sequences, respectively). The mRNA stability data set (Diez et al. 2022) includes thousands of mRNA stability profiles obtained from human, mouse, frog, and fish. The Tc-riboswitch data set (Groher et al. 2019) consists of a set of tetracycline (Tc) riboswitch dimer sequences upstream of a GFP mRNA. The measured variable in this data set is the switching factor, which refers to the differential effect of the riboswitch in the presence or absence of Tc. The SARS-CoV-2 vaccine degradation data set (Leppek et al. 2022) encompasses a set of mRNA sequences that have been tuned for their structural features, stability, and translation efficiency. The average of the deg_Mg_50C values at each nucleotide is treated as the sequence-level target. Deg_Mg_50C has the highest correlation with other labels, including deg_pH10, deg_Mg_pH10, and deg_50C. The benchmarking data set also included a new data set generated by Sanofi encoding the hemagglutinin antigen for flu vaccines. Briefly, mRNA sequences, encoding the Influenza H3N2 A/Tasmania/503/2020 hemagglutinin protein, were tested for protein expression level in HeLa cells (Methods).
To compare CodonBERT's performance on these tasks, we have also applied several other state-of-the-art methods that have been previously used for mRNA property prediction with different model complexities, including TF-IDF (Rajaraman and Ullman 2011), TextCNN (Kim 2014), Codon2vec, RNABERT (Akiyama and Sakakibara 2022), and RNA-FM (Chen et al. 2022). Table 2 presents the performance of CodonBERT and the other six methods on these downstream tasks. For each task, the first three rows are nucleotide-based methods (plain TextCNN, RNABERT, and RNA-FM), whereas the rest are codon-based methods (TF-IDF, plain TextCNN, Codon2vec, and CodonBERT). Supplemental Table S1 provides complimentary loss values for these comparisons. Overall, we see that codon-based methods outperform nucleotide-based methods on most tasks. This is in part because of the critical role of codons on the protein expression. Moreover, the codon-based variant of TextCNN outperforms the original nucleotide implementation on most tasks.
Table 2.
Comparison of CodonBERT to prior methods on seven downstream tasks
| Model | Flu vaccines | mRFP expression | Fungal expression | E. coli proteins | mRNA stability | Tc-riboswitch | SARS-CoV-2 vaccine degradation |
|---|---|---|---|---|---|---|---|
| Nucleotide-based | |||||||
| Plain TextCNN | 0.72 | 0.62 | 0.53 | 0.39 | 0.01 | 0.41 | 0.55 |
| RNABERT+TextCNN | 0.65 | 0.40 | 0.41 | 0.39 | 0.16 | 0.47 | 0.64 |
| RNA-FM+TextCNN | 0.71 | 0.80 | 0.59 | 0.43 | 0.34 | 0.58 | 0.74 |
| Codon-based | |||||||
| TF-IDF | 0.68 | 0.57 | 0.68 | 0.44 | 0.54 | 0.49 | 0.69 |
| Plain TextCNN | 0.71 | 0.78 | 0.76 | 0.36 | 0.26 | 0.43 | 0.80 |
| Codon2vec+TextCNN | 0.72 | 0.77 | 0.61 | 0.43 | 0.33 | 0.56 | 0.70 |
| CodonBERT | 0.81 | 0.85 | 0.88 | 0.55 | 0.51 | 0.56 | 0.77 |
For regression tasks, the corresponding Spearman's rank correlation values are listed. For the classification task (E. coli protein data set), classification accuracy is calculated. The best values of correlation and accuracy for each task are in bold. The corresponding loss values are listed in Supplemental Table S1.
As for the detailed comparison, we observe that CodonBERT performed best on four of the seven tasks and second best (in most cases with very small difference) on two of the remaining three tasks. Plain codon-based TextCNN produced the best results for SARS-CoV-2 vaccine degradation, whereas it performed poorly on other prediction tasks including riboswitches, flu vaccines, and E. coli. The other two methods that were best performing for one of the data sets, for example, TF-IDF and RNA-FM, did not perform well on the other tasks.
Both secondary structure and codon usage play critical roles in mRNA vaccine expression (Mauger et al. 2019; Agarwal and Kelley 2022). Stable secondary structure increases mRNA stability in solution (Mauger et al. 2019), and optimal codons improve cellular mRNA stability (Agarwal and Kelley 2022). Therefore, mRNA stability, SARS-CoV-2 vaccine degradation, and Tc-riboswitch data sets are strongly affected by local and global secondary structure patterns encoded on top of RNA sequences. Although CodonBERT is a codon-based model, it outperforms RNABERT and RNA-FM, which were demonstrated to capture rich structural information from large-scale noncoding RNAs. This may indicate that CodonBERT also learns coevolutionary information and structural properties from millions of mRNA sequences.
Nucleotide embeddings learned from noncoding RNAs, for example, RNA-FM+TextCNN, leads to significantly better results than plain nucleotide-based TextCNN on most tasks. This may indicate that structural information, even from noncoding RNA sequences, is beneficial to solving mRNA translation and stability problems. Although both RNABERT and RNA-FM are pretrained BERT models from noncoding RNA sequences, their performance differs. This may be attributed to the training data size and model capacity of RNA-FM, which is significantly larger than that of RNABERT.
Discussion
To enable the analysis and prediction of mRNA properties, we utilized 10 million mRNA CDSs from several species to train a LLM (CodonBERT) and to establish a foundational model. Our primary focus is on optimizing mRNA vaccines and drugs, concentrating specifically on sequences pertinent to these applications, including those from host cells and viruses critical for vaccine development.
The model optimizes two self-supervised tasks: codon completion and taxonomic identification. Like other unsupervised LLMs, we expected that such a foundational model will learn to capture aspects of natural selection that favor mRNA sequences with high expression and stable structure. Analysis of the resulting model indicates that it indeed learns several relevant biological properties for codons and sequences.
Projection of codon embedding obtained from CodonBERT produces distinct clusters that adhere to the amino acid types. Besides, the analysis of alanine codons revealed notable clustering patterns: “GCU” and “GCC” cluster separately from “GCA” and “GCG.” Cosine similarity calculations (Supplemental Fig. S3) supported this grouping, showing higher similarities between “GCG” and the nonsynonymous codons “CCG” (proline), “UCG” (serine), and “ACG” (threonine) than with its synonymous counterparts. This unexpected pattern, consistent with our projection plot findings, presents an interesting anomaly in codon behavior. The reasons for these unusual similarities remain unclear, suggesting an area for further exploration that could provide new insights into codon usage and gene expression mechanisms. In-depth analysis of CodonBERT representation of a set of genes from different organisms revealed that CodonBERT autonomously learns the genetic code and principles of evolutionary taxonomy.
We also utilized CodonBERT to perform several supervised prediction tasks for mRNA properties. These include data sets testing for recombinant protein expression, mRNA degradation, mRNA stability, and more. Our results indicate that CodonBERT is the top-performing method overall and ranks first or second in performance for six of the seven tasks. All other methods we compared performed poorly on all, or some of the tasks. Thus, for a new task, the use of CodonBERT is likely to lead to either the best or close to best results. CodonBERT's success in the more structurally related tasks (including mRNA stability and the TC riboswitch data sets) indicates that it can learn coevolutionary and structural concepts using large-scale mRNA sequences.
For the vaccine-related downstream tasks, CodonBERT generally exhibited robust performance. It was 10% better than the second-best method for the new hemagglutinin flu vaccine expression data set and very close (2% difference) to the top-performing model for the SARS-CoV-2 vaccine degradation data set. Although fungal sequences are not included in pretraining sequences, the CodonBERT model shows its generalization to a downstream fungal data set. RNABERT and RNABERT are also pretrained RNA LLMs; however, they are nucleotide based and trained on noncoding RNAs. Because they do not explicitly capture codon usage, these methods are inferior to CodonBERT on the protein expression-related tasks.
The flu vaccine data set uses N1-methylpseudouridine in RNA modification for bypassing the innate immune response and enhancing protein synthesis from mRNA. CodonBERT model's ability to adapt to these modifications without prior exposure to similar natural data exemplifies its robustness and versatility.
The one exception in terms of performance was observed for the mRNA stability task. Stability is known to be structure dependent, and stable structures such as stem-loops or hairpin structures can impede degradation enzymes, protecting the mRNA from rapid decay. A possible reason for the reduction in performance for this data set is that structural properties are highly dependent on nucleotides, whereas CodonBERT is a codon-based model. One possible solution for this is a model that combines codon and nucleotide representation. Similarly, mRNA modification events including capping at the 5′ end and polyadenylation at the 3′ end in eukaryotes are not currently encoded in our model but can also impact mRNA stability. However, extending CodonBERT to include UTRs will be an important direction for future work.
Although the tasks we have focused on are supervised in nature, CodonBERT, as an LLM, can be utilized for generative purposes. Specifically, we envision using this model for codon optimization of various heterologous proteins for vaccines. Other generative tasks can include sampling mRNA for synthetic biology use cases (e.g., creation of optimized mRNA constructs for genome editing).
To conclude, our findings suggest that CodonBERT could serve as a versatile and foundational model for the development of new mRNA-based vaccines and the engineering and recombinant production of industrial and therapeutic proteins.
Methods
Assembly of mRNA sequences for pretraining
We collected mRNA sequences across diverse organisms for pretraining from NCBI (Wheeler et al. 2007). The data sets included mammalian reference sequences (https://www.ncbi.nlm.nih.gov/datasets/taxonomy/40674/), bacteria (E. coli) reference sequences (https://www.ncbi.nlm.nih.gov/datasets/taxonomy/562/), Homo sapiens virus complete nucleotides (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/), and yeast strains (http://sgd-archive.yeastgenome.org/sequence/strains/). Each sequence is with a label representing its taxonomic group.
We preprocessed all the sequences and filtered out some invalid and replicate ones by requiring the mRNA sequences with the sequence length multiples of three, starting with the start codon (“AUG”) and ending with stop codons (“UAA,” “UAG,” or “UGA”), and only including nucleotides from the set {A, U, G, C, N} (replacing T with U). After preprocessing, 10 million mRNA sequences were valid.
To build sequence pairs for the homologous sequence prediction task, 50% of the sequence pairs consisted of two sequences belonging to one of the 14 categories. The remaining 50% of sequence pairs included two sequences that were randomly sampled from two different categories.
Model architecture
A codon is composed of three adjacent nucleotides. There are five different options for each of these three positions {A, U, G, C, N}, leading to a total of 53 (125) possible combinations. Additionally, five special tokens are added to the vocabulary: classifier token ([CLS]), separator token ([SEP]), unknown token ([UNK]), padding token ([PAD]), and masking token ([MASK]). Thus, in total, there are 130 tokens in the vocabulary of CodonBERT.
As shown in Figure 1B, CodonBERT takes a sequence pair as input and concatenates them using a separator token ([SEP]). It then adds a classifier token ([CLS]) and a separator token ([SEP]) at the beginning and end of the combined sequence, respectively. CodonBERT constructs the input embedding by concatenating codon, position, and segment embeddings. Absolute positions are utilized with values initialized from one to n1 + n2 + 3 along the concatenated sequence, where n1 and n2 are the codon-wise length of two sequences plus three specially added tokens ([CLS] and [SPE]). The segment value is either one or two to distinguish two sequences. These three types of embedding matrices are learned across 10 millions of mRNA sequences.
The combined input embedding is fed into the CodonBERT model, which consists of a stack of 12 layers of bidirectional transformer encoders (Vaswani et al. 2017) as shown in Figure 1C. Each transformer layer processes its input using 12 self-attention heads and then outputs a representation for each position with hidden size 768. In each layer, the multihead self-attention mechanism captures the contextual information of the input sequence by considering all the other codons in the sequence. A key benefit of self-attention mechanism is the connection learned between all pairs of positions in an input sequence using parallel computation, which enables CodonBERT to model not only short-range but also long-range interactions, which impact translation efficiency and stability (Aw et al. 2016). Next a feed-forward neural network is added to apply a nonlinear transformation to the output hidden representation from the self-attention network. A residual connection is employed around each of the multihead attention and feed-forward networks. After processing the input sequence with a stack of transformer encoders, CodonBERT produces the final contextualized codon representations, which is followed by a classification layer to produce probability distribution over the vocabulary during pretraining.
Pretraining CodonBERT
Model architecture of CodonBERT and the training for the two tasks are illustrated in Figure 1B. Prior to being fed into the model, the input mRNA sequence is first tokenized into a list of codons. Next, a fraction of the input codons (15%) is randomly selected and replaced by the masking token ([MASK]). The self-training loop optimizes CodonBERT to predict the masked codons based on the remaining ones, taking into account interactions between the missing and unmasked codons. A probability distribution over 64 possible codons is produced by CodonBERT for the masked positions. The average cross entropy loss ℒMLM over the masked positions M is calculated by the optimization function:
where X represents a batch of sequences, x is one sequence, and xi is the original codon for the position i. xM is the masked input with a set of positions M masked. (xi∣xM) indicates the output probability of the real codon xi given all the remaining codons in the masked sequence xM.
For the STP task, the output embedding of the classifier token ([CLS]) is used for prediction about whether these two sequences belong to the same class (binary classification). The average cross entropy loss ℒSTP is computed as
where N represents the number of sequence pairs; yn is the expected value, which is one when two sequences are from the same taxonomic group and zero when they are not; and pn indicates the predicted probability of two sequences belonging to the same category. The total loss is the sum of the losses from both tasks ().
We used a batch size of 128 with a sequence length limit of 1024 and trained the model around seven epochs in 2 weeks. Because the inputs of CodonBERT are sequence pairs, the length of each sequence is limited up to 512 codons; therefore, the length of the combined sequence is less than 1024. Sequences exceeding the length limitation are split into fragments no longer than 512. Pretraining CodonBERT using 10 million mRNA sequences on 4 A10G GPUs with 96 GB GPU memory and 192 GB memory took ∼2 weeks. The model was configured with the following specifications: a sequence length of 1024, as well as 12 layers, each with 12 attention heads. The model's hidden size is set at 768. Overall, the model encompasses about 110 million parameters.
CodonBERT was also applied to a wide range of downstream tasks. For this, we can use either a single or a pair of sequences as input (Figs. 1B, 3C). To perform supervised analysis, the output embedding is followed by an output layer that is trained for the specific task (protein expression level prediction, mRNA stability, etc.). We also conducted an ablation study in which we pretrained a model with the same architecture using only the MLM task. Although performance was good, it still did not match performance of the model trained with both types of metrics. See Supplemental Figure S5 and Supplemental Table S2.
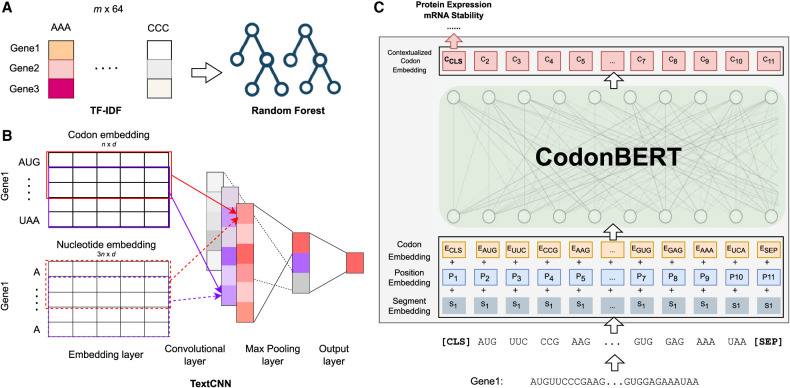
Figure 3.
Comparison to prior methods (TF-IDF, Codon2vec, RNABERT, and RNA-FM) and fine-tuning CodonBERT on downstream data sets. (A) Given an input corpus with m mRNA sequences, TF-IDF is used to construct a feature matrix followed by a random forest regression model. (B) Use a TextCNN model to learn task-specific nucleotide or codon representations. The model is able to fine-tune pretrained representations by initializing the embedding layers with stacked codon or nucleotide embeddings extracted from pretrained language models (Codon2vec, RNABERT, and RNA-FM). n is the number of codons in the input sequence, and d is the dimension of the token embedding. As a baseline, plain TextCNN initializes the embedding layer with a standard normal distribution. (C) Fine-tune the pretrained CodonBERT model on a given downstream task directly by keeping all the parameters trainable.
Pretraining Codon2vec
Word2vec is a popular neural network-based model that is used to learn distributed representations of words in a corpus (Mikolov et al. 2013). Like the LLM mentioned above, Word2vec also learns token representation from a large-scale text corpus, and the embedding from both methods can be utilized as input features for downstream tasks. Unlike LLMs, it only produces a single assignment for each token, which usually correlates with the maximum or most frequent context around the token in the corpus.
An existing work has applied Word2vec to the fungal genomes, studied codon usage bias, and built a predictive model for gene expression (Wint et al. 2022). However, there is no application on a large-scale mRNA sequence data set. Therefore, for comparison, we trained our own Codon2vec model on the collected mRNA sequences. Using the Gensim library (Řehůřek and Sojka 2010), the Codon2vec model opted for a skip-gram architecture, accompanied by a window size of five and a minimum count threshold of 10 codons. The model was trained using hierarchical SoftMax and negative sampling methodologies.
The input sequences for pretraining Codon2vec were processed through a filtration system that selected sequences containing fewer than 1000 nt. This filtration stage was necessitated by the constrained model capacity of the Word2vec neural network. Upon completion of this process, we retrieved a total of around 2 million sequences, which were subsequently subjected to tokenization into k-mers. These k-mers serve as representations of all corresponding codons.
In vitro transcription, cell culture, and transfections
As shown in Supplemental Figure S4, mRNA sequences were designed using to encode the Influenza H3N2 A/Tasmania/503/2020 hemagglutinin protein. Sequences corresponding to these candidates were synthesized as gene fragments and PCR-amplified to generate template DNA for high-throughput in vitro transcription reaction containing N1-methylpseudouridine. The resulting purified precursor mRNA was reacted further via enzymatic addition of a 5′ cap structure (Cap 1) and a 3′ poly(A) tail of ∼200 nt in length as determined by capillary electrophoresis.
HeLa cells were used to evaluate the expression of the protein encoded by different mRNA sequences. Cells were cultured and maintained in MEM (Corning) containing 10% (v/v) heat-inactivated FBS (Gibco). To evaluate the expression of candidate mRNAs, HeLa cells were transiently transfected with mRNAs complexed with Lipofectamine MessengerMax (Thermo Fisher Scientific). Unknown mRNAs were thawed, diluted in Opti-MEM, combined with Lipofectamine for 10 min, and then further diluted in Opti-MEM. To prepare the cells for reverse transfection, HeLa cells were collected from culture flasks using TrypLE and were diluted in complete growth medium such that each well will be seeded with 2E4 live cells. Complexed mRNAs (20 ng/well) were added to triplicate wells of a 96-well poly-D-lysine PhenoPlate (PerkinElmer) and were combined with 2E4 HeLa cells. Plates were rested at RT briefly before incubation in a tissue culture incubator for 20 h + 30 min. At the endpoint, cells were lysed in RIPA (Thermo Fisher Scientific) supplemented with OmniCleave (Lucigen) and HALT protease inhibitor (Thermo Fisher Scientific). The hemagglutinin expression in cell lysates was determined using a quantitative sandwich ELISA, and the expression level of unknown mRNAs was normalized to the value from a known benchmark mRNA sequence.
Comparisons to other methods
We compared CodonBERT to several prior methods that have been used to model and analyze RNA sequences:
Term frequency-inverse document frequency (TF-IDF) (Rajaraman and Ullman 2011) is a numerical statistic that is commonly used as a weighting scheme in information retrieval and natural language processing. In the context of mRNA sequences, TF-IDF is applied to measure the significance of each codon in a sequence. A high TF-IDF value of a codon indicates that the codon is important in a particular mRNA sequence and is rare across all mRNA sequences in the corpus. Figure 3A illustrates the application TF-IDF in the benchmark.
Convolutional neural network (CNN) was first proposed and has been commonly used in image recognition and was later also applied for text analysis TextCNN (Kim 2014). TextCNN consists of multiple types of layers, including an embedding layer, convolutional layer, pooling layer, and fully connected layer, as shown in Figure 3B. Each row of the embedding layer represents a token.
RNABERT (Akiyama and Sakakibara 2022) and RNA-FM (Chen et al. 2022) are RNA LLMs. However, they are pretrained on noncoding RNAs to learn and encode structural and functional properties in the output nucleotide embedding.
Software availability
Software and data are available in the Sanofi GitHub (https://github.com/Sanofi-Public/CodonBert) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Eleni Litsa for support in preparing mRNA designs.
Author contributions: Z.B-J. and S.J. proposed this idea. S.L. and S.M. implemented the algorithm and performed data collection and the in silico experiments. S.L., S.M., S.R., and S.J. wrote the paper. R.L., M.B., M.M., J.M., D.Z., J.W., A.B., F.U.M., V.A., and Z.B-J. revised the manuscript. L.K-A. developed the public code repository. M.M, J.M., F.P., D.Z., J.W., A.B., K.T., M.Z., M.W., X.G., R.C., C.A., J.S., L.B., S.C., A.D., T.S., F.U.M., and V.A. built the flu vaccine data set.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278870.123.
Competing interest statement
All authors are Sanofi employees and may hold shares and/or stock options in the company.
References
- Agarwal V, Kelley DR. 2022. The genetic and biochemical determinants of mRNA degradation rates in mammals. Genome Biol 23: 245. 10.1186/s13059-022-02811-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Shendure J. 2020. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks. Cell Rep 31: 107663. 10.1016/j.celrep.2020.107663 [DOI] [PubMed] [Google Scholar]
- Ahmad HI, Jabbar A, Mushtaq N, Javed Z, Hayyat MU, Bashir J, Naseeb I, Abideen ZU, Ahmad N, Chen J. 2022. Immune tolerance vs. immune resistance: the interaction between host and pathogens in infectious diseases. Front Vet Sci 9: 827407. 10.3389/fvets.2022.827407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Sakakibara Y. 2022. Informative RNA base embedding for RNA structural alignment and clustering by deep representation learning. NAR Genom Bioinform 4: lqac012. 10.1093/nargab/lqac012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hawash AB, Zhang X, Ma F. 2017. Strategies of codon optimization for high-level heterologous protein expression in microbial expression systems. Gene Rep 9: 46–53. 10.1016/j.genrep.2017.08.006 [DOI] [Google Scholar]
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AY, et al. 2016. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol Cell 62: 603–617. 10.1016/j.molcel.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Bepler T, Berger B. 2021. Learning the protein language: evolution, structure, and function. Cell Syst 12: 654–669.e3. 10.1016/j.cels.2021.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hu Z, Sun S, Tan Q, Wang Y, Yu Q, Zong L, Hong L, Xiao J, Shen T, et al. 2022. Interpretable RNA foundation model from unannotated data for highly accurate RNA structure and function predictions. bioRxiv 10.1101/2022.08.06.503062 [DOI]
- Devlin J, Chang M-W, Lee K, Toutanova K. 2019. BERT: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) (ed. Burstein J, et al.), pp. 4171–4186. Association for Computational Linguistics, Minneapolis. https://aclanthology.org/N19-1423. 10.18653/v1/N19-1423 [DOI]
- Diez M, Medina-Muñoz SG, Castellano LA, da Silva Pescador G, Wu Q, Bazzini AA. 2022. iCodon customizes gene expression based on the codon composition. Sci Rep 12: 12126. 10.1038/s41598-022-15526-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Guan F, Xu G, Wang Y, Yan Y, Zhang W, Wu N, Yao B, Huang H, Tuller T, et al. 2022. MPEPE, a predictive approach to improve protein expression in E. coli based on deep learning. Comput Struct Biotechnol J 20: 1142–1153. 10.1016/j.csbj.2022.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42: D699–D704. 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher F, Bofill-Bosch C, Schneider C, Braun J, Jager S, Geißler K, Hamacher K, Suess B. 2018. Riboswitching with ciprofloxacin: development and characterization of a novel RNA regulator. Nucleic Acids Res 46: 2121–2132. 10.1093/nar/gkx1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher A-C, Jager S, Schneider C, Groher F, Hamacher K, Suess B. 2019. Tuning the performance of synthetic riboswitches using machine learning. ACS Synth Biol 8: 34–44. 10.1021/acssynbio.8b00207 [DOI] [PubMed] [Google Scholar]
- Hallee L, Rafailidis N, Gleghorn JP. 2023. cdsBERT: extending protein language models with codon awareness. bioRxiv 10.1101/2023.09.15.558027 [DOI]
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. 2020a. An mRNA vaccine against SARS-CoV-2: preliminary report. N Engl J Med 383: 1920–1931. 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson NA, Kester KE, Casimiro D, Gurunathan S, DeRosa F. 2020b. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines 5: 11. 10.1038/s41541-020-0159-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Zhou Z, Liu H, Davuluri RV. 2021. DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome. Bioinformatics 37: 2112–2120. 10.1093/bioinformatics/btab083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. 2014. Convolutional neural networks for sentence classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (ed. Moschitti A, et al. ), pp. 1746–1751. Association for Computational Linguistics, Doha, Qatar. https://aclanthology.org/D14-1181. 10.3115/v1/D14-1181 [DOI] [Google Scholar]
- Leppek K, Byeon GW, Kladwang W, Wayment-Steele HK, Kerr CH, Xu AF, Kim DS, Topkar VV, Choe C, Rothschild D, et al. 2022. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat Commun 13: 1536. 10.1038/s41467-022-28776-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang H, Zhang L, Liu K, Liu B, Mathews DH, Huang L. 2021. LinearTurboFold: linear-time global prediction of conserved structures for RNA homologs with applications to SARS-CoV-2. Proc Natl Acad Sci 118: e2116269118. 10.1073/pnas.2116269118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen CL, Haanen JB, Met Ö, Svane IM. 2022. Clinical advances and ongoing trials of mRNA vaccines for cancer treatment. Lancet Oncol 23: e450–e458. 10.1016/S1470-2045(22)00372-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G, Chiarot E, Giovani C, Buccato S, Bonacci S, Frigimelica E, Margarit I, Geall A, Bensi G, Maione D. 2017. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine 35: 361–368. 10.1016/j.vaccine.2016.11.040 [DOI] [PubMed] [Google Scholar]
- Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ, et al. 2019. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci 116: 24075–24083. 10.1073/pnas.1908052116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP. 2018. Codon optimization in the production of recombinant biotherapeutics: potential risks and considerations. BioDrugs 32: 69–81. 10.1007/s40259-018-0261-x [DOI] [PubMed] [Google Scholar]
- Mauro VP, Chappell SA. 2014. A critical analysis of codon optimization in human therapeutics. Trends Mol Med 20: 604–613. 10.1016/j.molmed.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, Grossberger L. 2018. UMAP: Uniform Manifold Approximation and Projection. J Open Source Softw 3: 861. 10.21105/joss.00861 [DOI] [Google Scholar]
- Miao L, Zhang Y, Huang L. 2021. mRNA vaccine for cancer immunotherapy. Mol Cancer 20: 41. 10.1186/s12943-021-01335-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolov T, Chen K, Corrado G, Dean J. 2013. Efficient estimation of word representations in vector space. arXiv: 1301.3781 [cs.CL].
- Nieuwkoop T, Terlouw BR, Stevens KG, Scheltema R, de Ridder D, van der Oost J, Claassens N. 2023. Revealing determinants of translation efficiency via whole-gene codon randomization and machine learning. Nucleic Acids Res 51: 2363–2376. 10.1093/nar/gkad035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, Dowd KA, Sutherland LL, Scearce RM, Parks R, et al. 2017. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543: 248–251. 10.1038/nature21428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, Weissman D. 2018. mRNA vaccines: a new era in vaccinology. Nat Rev Drug Discov 17: 261–279. 10.1038/nrd.2017.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Weissman D. 2020. Recent advances in mRNA vaccine technology. Curr Opin Immunol 65: 14–20. 10.1016/j.coi.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Parret AH, Besir H, Meijers R. 2016. Critical reflections on synthetic gene design for recombinant protein expression. Curr Opin Struct Biol 38: 155–162. 10.1016/j.sbi.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L. 2018. Deep contextualized word representations. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers) (ed. Walker M, et al.), pp. 2227–2237. Association for Computational Linguistics, New Orleans. https://aclanthology.org/N18-1202. 10.18653/v1/N18-1202 [DOI] [Google Scholar]
- Pilkington EH, Suys EJ, Trevaskis NL, Wheatley AK, Zukancic D, Algarni A, Al-Wassiti H, Davis TP, Pouton CW, Kent SJ, et al. 2021. From influenza to COVID-19: lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater 131: 16–40. 10.1016/j.actbio.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A, Narasimhan K, Salimans T, Sutskever I. 2018. Improving language understanding by generative pre-training. OpenAI. https://cdn.openai.com/research-covers/language-unsupervised/language_understanding_paper.pdf.
- Rajaraman A, Ullman JD. 2011. Mining of massive datasets. Cambridge University Press, Cambridge. [Google Scholar]
- Řehůřek R, Sojka P. 2010. Software framework for topic modelling with large corpora. In Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks, pp. 45–50. ELRA, Valletta, Malta. http://is.muni.cz/publication/884893/en. [Google Scholar]
- Rives A, Meier J, Sercu T, Goyal S, Lin Z, Liu J, Guo D, Ott M, Zitnick CL, Ma J, et al. 2021. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci 118: e2016239118. 10.1073/pnas.2016239118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake T, Thess A, Fotin-Mleczek M, Kallen K-J. 2012. Developing mRNA-vaccine technologies. RNA Biol 9: 1319–1330. 10.4161/rna.22269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Hamacher K, Reinhardt F, Lotz TS, Groher F, Suess B, Jager S. 2020. SICOR: subgraph isomorphism comparison of RNA secondary structures. IEEE/ACM Trans Comput Biol Bioinform 17: 2189–2195. 10.1109/TCBB.2019.2926711 [DOI] [PubMed] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. 2017. Attention is all you need. In 31st Conference on Neutral Information Processing Systems (NIPS 2017), Long Beach, CA, pp. 6000–6010. [Google Scholar]
- Wayment-Steele HK, Kladwang W, Watkins AM, Kim DS, Tunguz B, Reade W, Demkin M, Romano J, Wellington-Oguri R, Nicol JJ, et al. 2022. Deep learning models for predicting RNA degradation via dual crowdsourcing. Nat Mach Intell 4: 1174–1184. 10.1038/s42256-022-00571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster GR, Teh AY-H, Ma JK-C. 2017. Synthetic gene design: the rationale for codon optimization and implications for molecular pharming in plants. Biotechnol Bioeng 114: 492–502. 10.1002/bit.26183 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. 2007. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 35: D5–D12. 10.1093/nar/gkl1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wint R, Salamov A, Grigoriev IV. 2022. Kingdom-wide analysis of fungal protein-coding and tRNA genes reveals conserved patterns of adaptive evolution. Mol Biol Evol 39: msab372. 10.1093/molbev/msab372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Maruggi G, Shan H, Li J. 2019. Advances in mRNA vaccines for infectious diseases. Front Immunol 10: 594. 10.3389/fimmu.2019.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang L, Lin A, Xu C, Li Z, Liu K, Liu B, Ma X, Zhao F, Jiang H, et al. 2023. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 621: 396–403. 10.1038/s41586-023-06127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.