Abstract
Most Plasmodium vivax infections contain genetically distinct parasites, but the consequences of this polyclonality on the development of asexual parasites, their sexual differentiation, and their transmission remain unknown. We describe infections of Saimiri monkeys with two strains of P. vivax and the analyses of 80,024 parasites characterized by single cell RNA sequencing and individually genotyped. In our model, consecutive inoculations fail to establish polyclonal infections. By contrast, simultaneous inoculations of two strains lead to sustained polyclonal infections, although without detectable differences in parasite regulation or sexual commitment. Analyses of sporozoites dissected from mosquitoes fed on coinfected monkeys show that all genotypes are successfully transmitted to mosquitoes. However, after sporozoite inoculation, not all genotypes contribute to the subsequent blood infections, highlighting an important bottleneck during pre-erythrocytic development. Overall, these studies provide new insights on the mechanisms regulating the establishment of polyclonal P. vivax infections and their consequences for disease transmission.
Subject terms: Parasite genetics, Transcriptomics
Single cell analyses of non-human primates infected with one or two strains of Plasmodium vivax provide insights on the origin of polyclonality in patients and reveal an important population bottleneck occurring during pre-erythrocytic development.
Introduction
Despite major progress in the last decades, malaria remains a leading cause of morbidity and mortality around the world, with an estimated 247 million clinical infections in 20221. While Plasmodium falciparum is responsible for the vast majority of malaria cases in Africa (and most deaths worldwide), Plasmodium vivax contributes to a significant proportion of infections outside of Africa and accounts for 40% of the malaria cases in Southeast Asia, 32% in the Pacific region and 71% in the Americas1–3.
P. vivax differs from P. falciparum in several key biological features that negatively impact our ability to control and eliminate vivax malaria. These idiosyncrasies include a greater genetic diversity4–8, a typically lower parasitemia9–11, an earlier appearance of gametocytes during the course of an infection11,12, and a dormant liver-stage, that can cause relapses months or years after an initial infection1–3,13,14. In addition, genetic analyses of infected patients have shown that most P. vivax blood infections are polyclonal and that patients are typically infected with several related or unrelated parasite genotypes5–8,13,15–18. The simultaneous presence of multiple parasites in one infection could complicate efficient control or clearance of the infection by the host immune response (especially if immunity is strain-specific) but may also influence the regulation of the parasites themselves. Distinct Plasmodium parasites present in the same host might alter their respective regulation to compete more efficiently for scarce resources or modify their sexual commitment to improve transmission and outcrossing19–22, similar to the well-known mating type regulation in yeast or the mechanisms limiting self-fertilization in plants. In this regard, it is important to note that sexual commitment is, at least partially, determined by environmental factors23–28, supporting the hypothesis that the parasites are able to sense their environment to modify their asexual development and sexual differentiation.
However, despite its prevalence among natural infections, the consequences of polyclonality on blood-stage infections have not been comprehensively examined, especially for P. vivax that cannot be continuously propagated in in vitro cultures29. Animal models - the infection of non-human primates with patient-derived P. vivax parasites30–32 or the recently developed humanized mouse models33,34 - could enable analyses of polyclonal infections but, so far, have primarily focused on infections with a single, well-characterized, clone (with the exception of one genetic cross study35). In addition to these limitations in generating experimental polyclonal Plasmodium infections with human parasites, it was until recently extremely difficult to differentiate the regulation of different clones present within a single infection18,36. The advent of single cell genomics and the ability to generate gene expression profiles from individual parasite by single-cell RNA sequencing (scRNA-seq) provides a unique opportunity to disentangle this complexity. scRNA-seq has been successfully applied to different Plasmodium species, including P. vivax, to extensively characterize variations in gene expression throughout the life cycle of the parasites26,37–45. Moreover, since scRNA-seq relies on sequencing the mRNA molecules, the data generated can also be leveraged to analyze DNA polymorphisms in these regions and genotype individual parasites46.
Here, we use scRNA-seq to comprehensively investigate the effects of P. vivax polyclonal infections in a non-human primate model. We infected Saimiri boliviensis monkeys with one or two strains of P. vivax and analyzed blood samples from mono-infections, consecutive infections (where animals were first infected with one strain then another) and simultaneous coinfections with both strains, and compared the stage composition and parasites’ transcriptional regulation in each sample. We also evaluated the consequences of polyclonal infections on parasite transmission to mosquitoes, by analyzing salivary gland sporozoites, and assessed, after sporozoite inoculation of new Saimiri monkeys, which of those parasites successfully developed through the liver to establish a blood infection. Overall, our analyses provide novel insights on parameters influencing the regulation of blood-stage P. vivax parasites and their sexual differentiation, and highlight the complexity and critical importance of pre-erythrocytic development.
Results and discussion
Overall experimental design
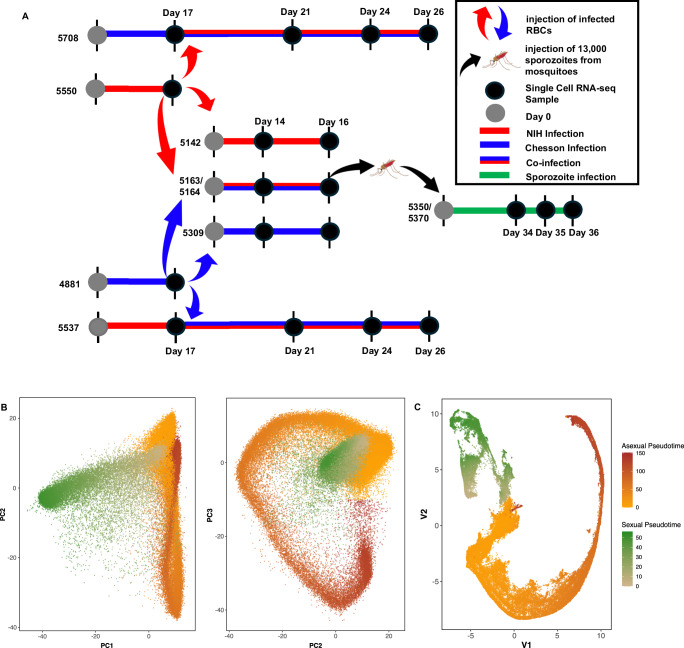
We infected two Saimiri boliviensis monkeys with ~2 million cryopreserved parasitized red blood cells (RBCs) infected with the NIH-1993-F3 strain of P. vivax (a third-generation self-cross of NIH-199347, Supplemental Fig. 1), and two Saimiri monkeys with ~2 million cryopreserved parasitized RBCs infected with the Chesson strain (later referred to as mono-infections). Once the parasitemia reached approximately 0.2% (14–17 days later), we collected ~2 million parasitized RBCs from one of the monkeys infected by one strain to inoculate one of the other monkeys initially infected with the other strain (consecutive infections). We also used parasitized RBCs collected from these initial mono-infections to 1) simultaneously inoculate 0.5 million parasites of the NIH-1993-F3 plus 0.5 million parasites of the Chesson strains to two new Saimiri monkeys (simultaneous infections) and 2) inoculate separately 1 million parasitized RBCs of each strain to two new monkeys (mono-infections) (Fig. 1A).
Fig. 1. Overall study design and summary of the scRNA-seq analysis and cell assignment.
A Four Saimiri boliviensis monkeys were first infected with parasitized RBCs of the NIH-1993-F3 (red) or Chesson (blue) strain. 17 days post inoculation, parasitized RBCs from two of the monkeys were used to i) re-inoculate two of the monkeys previously infected (consecutive infections) and ii) inoculate four additional Saimiri monkeys (two monkeys inoculated with only one strain each - mono-infection, and two monkeys inoculated with both strains together – simultaneous coinfections). Anopheles freeborni mosquitoes were fed on the blood of the simultaneously coinfected monkeys and sporozoites derived from these feedings were used for inoculating two additional Saimiri boliviensis (in green). Black dots indicate the timing of the blood collections used for generating single-cell RNA-seq data. B Principal component analysis of all blood-stage parasites characterized by more than 1000 UMIs mapped within genes. Each dot represents one individual P. vivax parasite and is displayed based on its gene expression and colored according to its pseudotime (Orange to Red - asexual parasites, Green - female gametocytes). C UMAP representation colored by pseudotime (Orange and Red - asexual cells, Green - female gametocytes).
Additionally, we collected blood from the two monkeys simultaneously infected with Chesson and NIH-1993-F3 and fed Anopheles stephensi and An. freeborni mosquitoes by membrane feeding (see Materials and Methods for details). 18–20 days later, we dissected the salivary glands of 20 mosquitoes from each feeding to collect sporozoites and inoculate two additional animals with 13,000 sporozoites intravenously. All animals developed detectable parasitemia within three weeks of the initial inoculation (Supplemental Fig. 2).
To rigorously characterize the genetic differences between the two P. vivax strains, we sequenced at very high coverage (>400 X) their entire genomes using blood collected from two of the initial mono-infections (Supplemental Data 1). Overall, we identified 26,719 single nucleotide differences between the two strains, as well as 210 deleted genes (116 genes deleted in the Chesson strain, 94 in the NIH-1993-F3) (Supplemental Data 2).
Characterization of parasite gene expression profiles by scRNA-seq
Once the parasitemia reached ~0.1%, and at regular intervals afterwards, we collected blood from each animal (Fig. 1A and Supplemental Fig. 2) and prepared 10X Genomics 3’ end scRNA-seq libraries after enrichment of parasitized RBCs using MACS columns. Overall, we prepared 24 single-cell RNA-sequencing libraries from blood samples and eight libraries from dissected salivary glands of infected mosquitoes, and generated 122,578,812 – 311,727,931 sequencing reads from each library (Supplemental Data 1).
From the blood sample libraries, 23,953,801 - 173,969,055 unique reads mapped to the P. vivax genome (37–87%) and 12–82% of those mapped to an annotated P. vivax gene39(Supplemental Data 1). Overall, we obtained a total of 80,024 individual cells, each characterized by at least 1000 unique reads mapped to annotated genes (Table 1).
Table 1.
Single cell data summary
| Condition | No. of animals | No. samples/animal | % Mapped | Total # Cellsa |
|---|---|---|---|---|
| Mono-infection Chesson | 3 | 1–2 | 46.5–74.3% | 22,847 |
| Mono-infection NIH-1993-F3 | 3 | 1–2 | 37.5–79.6% | 16,292 |
| Consecutive infection | 2 | 3 | 66.8–87.3% | 18,286 |
| Simultaneous infection | 2 | 2 | 41.1–55.4% | 11,715 |
| Sporozoite inoculation | 2 | 3 | 39.4–82.9% | 10,884 |
| Sporozoites | na | 8 pools | 0.28–7.1% | 16,527b |
aCells with >1000 unique UMIs in annotated genes
bCells with >250 unique UMIs in annotated genes
To assign each individual cell characterized by scRNA-seq to a specific developmental stage, we clustered the cells using principal component analysis (PCA) and uniform manifold approximation and projection (UMAP), and calculated a pseudotime for asexual and sexual stages, separately, based on each cell PC coordinates (Fig. 1B and Supplemental Fig. 3). Lowering the cutoff for defining individual cells to 250 reads mapped to annotated genes per cell increased the number of cells recovered to 256,935 and enabled recovering many ring stage parasites (Supplemental Fig. 4). However, this lower cutoff hampered accurate genotyping of the cells and differential expression analyses (see below) and we therefore focused for subsequent analyses on the 80,024 cells characterized by more than 1000 unique reads mapped to annotated genes.
Analysis of monoclonal P. vivax infections
We first analyzed the scRNA-seq data generated from the blood of monkeys infected with either the Chesson or the NIH-1993-F3 strain of P. vivax (Fig. 2A). To evaluate our ability to genotype individual parasites using scRNA-seq data, we determined, for each individual cell, the number of reads carrying sequence information at each of the 26,719 nucleotide positions that differentiate the two strains. All but one cell had at least one read spanning one of these differentiating nucleotide positions and 95% of the cells were genotyped at more than 20 positions (Supplemental Fig. 5) allowing for accurate determination of the parasite genotype (Fig. 2B). For the remaining analyses, we focused on the cells that were robustly assigned to one or the other genotype based on 20 positions or more.
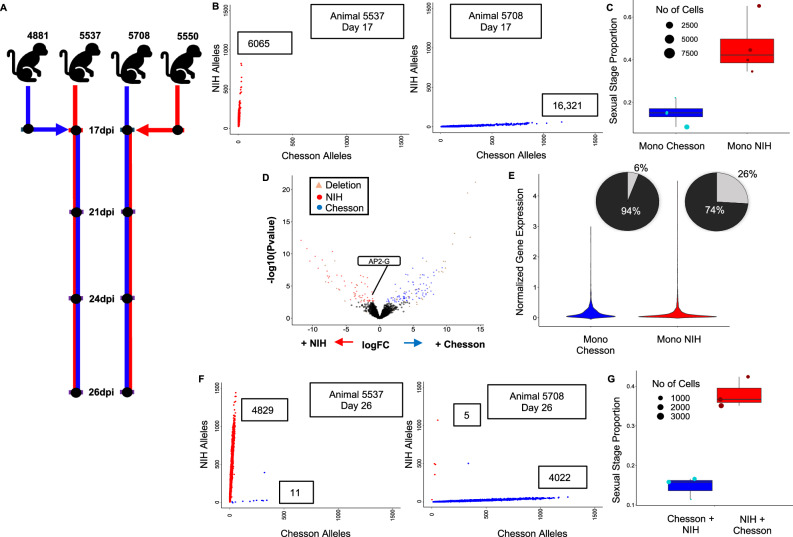
Fig. 2. Analyses of mono- and consecutive infections.
Boxplots show means as center line, boxes represent 75% and 25%, and whiskers minima and maxima. A Experimental design showing the initial monkey infections with the NIH-1993-F3 (red) or Chesson (blue) strain of P. vivax, the second inoculation with the other strain at 17dpi and the timing of the blood draws used for generating scRNA-seq data (black dots). B Genotyping of the parasites in mono-infections. The figures show the number of NIH-1993-F3 alleles (y-axis) and Chesson alleles (x-axis) determined from scRNA-seq reads for each individual parasite from one monkey infected with the NIH-1993-F3 strain (left) or Chesson strain (right). C Proportion of blood-stage parasites determined to be female gametocytes (y-axis) for the NIH-1993-F3 and Chesson mono-infections. Each dot shows the proportion in one blood sample (n = 8) and is sized proportionally to the number of cells. D Volcano plot showing differences in gene expression between Chesson and NIH-1993-F3 early asexual parasites (Group A, complete results provided in Supplemental Data 3). Each dot represents one P. vivax gene and is displayed according to the log fold-change differences in gene expression (x-axis) and statistical significance (-log10 of the p-value). Red dots represent genes more expressed in NIH-1993-F3 parasites; blue dots, genes more expressed in Chesson parasites (FDR = 0.1). Green dots are members of the PIR multigene family; and triangles highlight genes that are deleted in one of the two strains based on whole genome sequence data. E Violin plots showing normalized expression of AP2-G (PVP01_1440800) in Chesson and NIH-1993-F3 parasites with detectable AP2-G expression. Pie charts show the proportion of cells from each strain with detectable expression of AP2-G (in light gray). F Genotyping of parasites in consecutive infections at 26 dpi. The two panels show the number of NIH-1993-F3 alleles and Chesson alleles determined from the scRNA-seq reads for each individual parasite from one monkey infected first with the NIH-1993-F3 and then the Chesson strain (left) or by the Chesson strain first and NIH-1993-F3 second (right). G Proportion of blood-stage parasites determined to be female gametocytes (y-axis) for the NIH-1993-F3 and Chesson parasites in consecutive infections (n = 6).
Interestingly, the proportion of female gametocytes was much higher in the NIH-1993-F3 infections: on average, 47% of all blood-stage parasites were female gametocytes in NIH-1993-F3 infections, while only 10% of all blood-stage parasites were gametocytes in infections with the Chesson strain (Fig. 2C). This observation suggests that sexual commitment is greater in NIH-1993-F3 than Chesson, possibly due to a genetic or epigenetic factor that may have been selected during the serial passages between non-human primates and mosquitoes of the NIH-1993-F3 strain (see e.g.,48,49). We failed to rigorously detect male gametocytes in our experiments while those were certainly present (see the analyses of parasite transmission below). The most likely explanation for the lack of male gametocytes in the scRNA-seq data is that these parasites exflagellated during the MACS column enrichment or the loading of the 10X instrument that were conducted at room temperature.
We then used the scRNA-seq data to evaluate whether the expression levels of specific genes were significantly different between parasites of the two strains. To correct for differences in gene expression among stages, which often confound Plasmodium gene expression analyses, we grouped cells from each infection into four developmental groups (asexual A, B and C, and sexual S) based on their pseudotimes (Supplemental Figure 6). The asexual groups A, B and C roughly correspond to early trophozoites, late trophozoites, schizonts, respectively. Only the early asexual parasite (A) and gametocyte (S) groups had three or more samples with more than 100 cells from each genotype and were further analyzed by pseudobulk RNA-seq, comparing infections with NIH-1993-F3 to infections with Chesson and correcting for multiple testing using false discovery rates50. Overall, 205 genes were differentially expressed between Chesson and NIH-1993-F3 parasites (FDR = 0.1, Supplemental Data 3). Out of the 205 differentially expressed genes, 44 genes were actually deleted in one of the two strains, based on our whole genome sequence data, which validated our differential expression analysis results. Moreover, 109 differentially expressed genes were members of the PIR family, the largest and highly variable Plasmodium multigene family51–55. Interestingly, we observed that AP2-G (PVP01_1440800) had significantly higher expression in asexual parasites of the NIH-1993-F3 strain than in the Chesson parasites (group A, p = 0.00089, Fig. 2D). This gene is the master regulator of gametocyte formation in P. falciparum26,49. Remarkably, while the expression level in parasites with detectable level of AP2-G was similar in NIH-1993-F3 and Chesson parasites (p = 0.663), the proportion of asexual parasites expressing AP2-G differed dramatically between the two strains: only 6% of asexual Chesson parasites has detectable level of AP2-G expression compared to 26% of the asexual NIH-1993-F3 parasites (Fig. 2E). This observation is consistent with the higher proportion of female gametocytes observed in NIH-1993-F3 (Fig. 2C) and could explain the higher sexual conversion in this strain (note that the genetic or epigenetic mechanisms underlying this more frequent expression of AP2-G in NIH-1993-F3 asexual parasites remained undetermined and will need to be further investigated in future studies).
Consecutive infections failed to establish detectable polyclonal infections
In a first attempt to establish P. vivax polyclonal infections, we re-inoculated, two weeks after the first inoculation, animals initially infected with one of the two strains with 1 million fresh RBCs parasitized with the other strain (Fig. 2A). We then collected blood from each animal 4, 7, and 9 days after the second inoculation and generated scRNA-seq data from these samples. Genotyping of the individual parasites from each infection showed, almost exclusively, presence of the initial strain, with very little evidence for the successful establishment of the secondary strain (Fig. 2F). One explanation for the patterns observed is that once one infection is established, with several millions of parasites circulating, subsequent inoculation (or superinfection36) of one million parasites from a different strain is unlikely to lead to a significant presence in the blood unless they have a significant growth advantage: the initial inoculation of one million parasites could lead, four days later, to ~ 1 billion circulating parasites (assuming 32 merozoites produced per schizont every 48 h), which would swamp the one million newly injected parasites from the second strain. Alternatively, the times of sampling might not have been sufficiently afterward to observe successful establishment of the second strain: the latest sampling occurred nine days after the second inoculation ( ~ 4 asexual cycles) and these parasites might not have had enough time to catch up on by the parasites from the first inoculation. These findings could also indicate that the robust establishment of second strain is impeded by the active replication of the first strain that outcompetes it. This hypothesis is especially compelling for P. vivax since this parasite infects reticulocytes that are present in very limited amount9–11 (compared to P. falciparum that can infect RBCs of all ages). Finally, it is possible that an immune response caused by the initial infection slowed the propagation of the second strain and impeded its establishment (similarly to the hepcidin-dependent suppression of superinfection described in56 but with a distinct mechanism since liver development is circumvented in this specific experiment).
Despite this apparent lack of polyclonality, we tested whether introduction of a second strain influenced the regulation of the parasites. Consecutive infection had no effect on sexual stage proportion, with NIH infections still having a greater proportion of sexual stages (Fig. 2G). We also conducted differential gene expression analysis between mono-infected samples and samples consecutively infected but failed to detect any evidence of differential regulation: out of the 6039 genes tested, none were deemed differentially expressed in any of the groups tested (FDR = 0.1). Overall, these results suggested that, under our experimental conditions, the introduction of a second strain had little (if any) impact on the regulation of the parasites already present in the infection.
Simultaneous inoculation of both strains leads to robust polyclonal infections
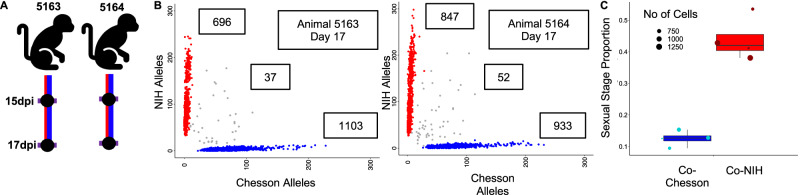
To obtain robust polyclonal infections, we inoculated a new set of malaria-naïve Saimiri monkeys with both strains simultaneously (Fig. 3A). After inoculation of 0.5 million parasitized RBCs from each strain, we observed robust co-infections maintaining polyclonality over several weeks (Fig. 3B).
Fig. 3. Analyses of simultaneous infections.
A Experimental design showing the simultaneous inoculation of two Saimiri monkeys with the NIH-1993-F3 (red) and Chesson (blue) strain of P. vivax and the timing of the blood draws used for generating scRNA-seq data. B Genotyping of the parasites in simultaneous infections. The two panels show the number of NIH-1993-F3 alleles (y-axis) and Chesson alleles (x-axis) determined from the scRNA-seq reads for each individual parasite (red and blue dots) from the two monkeys simultaneous infected with the NIH-1993-F3 and the Chesson strains. Note that the small number of points along the diagonal likely represents doublets containing one cell of each genotype. C The figure shows the proportion of blood-stage parasites determined to be female gametocytes (y-axis) for the NIH-1993-F3 (red dots) and Chesson parasites (blue dots) from the simultaneous infections (see Fig. 2 for details, n = 8).
We first evaluated whether the gene expression differences between strains observed from monoinfections (Fig. 2D) held true when the two strains were present in the same animal. Despite the smaller sample size of these analyses, most of the gene expression differences initially observed were recapitulated in polyclonal infections (Supplemental Data S3), indicating that host or batch effect did not dramatically affect these differences.
We then tested whether the polyclonal infections influenced parasite regulation. We observed roughly equal proportions of both strains in the coinfection, with each strain consisting of all identifiable stages in all coinfected samples (Supplemental Fig. 6). In particular, the proportion of sexual stages was similar to that observed in mono-infections, with NIH-1993-F3 displaying, on average, more female gametocytes than Chesson (Fig. 3C). We tested for differential gene expression by comparing parasites, of the same strain, in mono-infections vs. simultaneous coinfections but detected only 33 differentially expressed genes across all groups (again, only early asexuals A and gametocytes S had enough cells characterized across samples to be rigorously assessed) (Supplemental Data 3). Surprisingly, several of the differentially expressed genes observed in asexual parasites were well-known female gametocytes markers (e.g., P25, P28, CPW-WPC or LCCL domain-containing proteins). We hypothesized that these may represent artefacts due to the inclusions of a small number of doublets (i.e., droplets containing more than one cell) containing a gametocyte which may cause spurious gene expression differences. Overall, our findings suggested that, in this Saimiri model, polyclonal infection with two different P. vivax parasites has very little effect (if any) on, either the regulation of gene expression, or sexual commitment.
Coinfected parasites contribute evenly to mosquito infections but some genotypes are lost during pre-erythrocytic development
We then infected Anopheles stephensi and An. freeborni mosquitoes by direct membrane feeding using blood from animals coinfected with NIH-1993-F3 and Chesson. A total of 17–20 days post-infection, we dissected sporozoites from salivary glands pooled from 20 mosquitoes and used these sporozoites for scRNA-seq or for infections of additional monkeys via sporozoite inoculations (see below) (Figs. 1A and 4A).
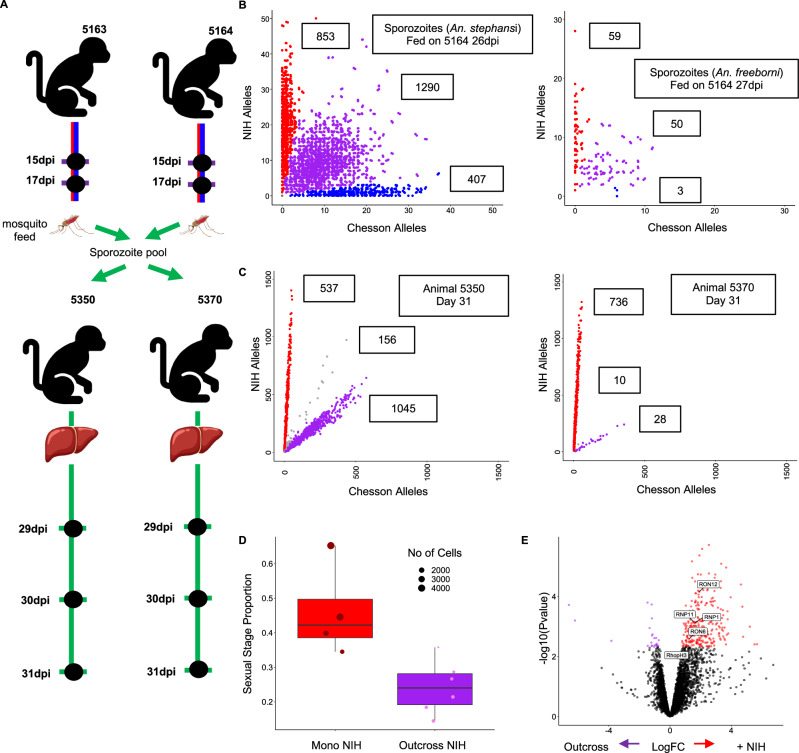
Fig. 4. Analyses of dissected sporozoites and blood-stage infections resulting from sporozoite inoculations.
A Experimental design showing, from top to bottom, mosquitoes fed on the blood of the animals simultaneously infected by NIH-1993-F3 and Chesson (at 17 and 30 dpi), inoculation of two additional animals with 13,000 sporozoites dissected from the salivary glands of 20 infected An. freeborni mosquitoes, and the timing of the blood collected for the generation of scRNA-seq data. B Genotyping of the sporozoites collected from the salivary glands of mosquitoes fed on the blood of simultaneously infected monkeys. The two panels show the number of NIH-1993-F3 alleles (y-axis) and Chesson alleles (x-axis) determined from the scRNA-seq reads for each sporozoite (red, blue and purple dots) from two pools of 20 mosquitoes (left An. stephensi, right An. feeborni, both from feeding using blood from 5164). Note that a large proportion of parasites (in purple) fall along the diagonal and represent sporozoites derived from the mating of a NIH-1993-F3 and Chesson gametocytes (outcrossed). C Genotyping of blood stage parasites from infections initiated by sporozoite inoculation. D The figure shows the proportion of blood-stage parasites determined to be female gametocytes (y-axis) for the NIH-1993-F3 parasites in mono-infections (red dots, animals 5550, 5142, and 5537; n = 4) and NIH-1993-F3 parasites from the infections initiated by sporozoite inoculation (purple dots, animals 5550 and 5570; n = 6). E Volcano plot showing differences in gene expression between NIH-1993-F3 early asexual parasites (Group A) derived from parasitized RBC inoculation (monoinfection) and those derived from inoculation of sporozoites. Blue dots represent genes with higher expression in parasites derived from blood-stage inoculation, red dots genes with higher expression in parasites derived from sporozoite inoculation. Differentially expressed Rhoptry associated genes are highlighted in boxes (FDR = 0.1).
We generated eight scRNA-seq libraries from P. vivax sporozoites dissected from either Anopheles stephensi or An. freeborni mosquitoes (Supplemental Data 1). Despite contamination with mosquito RNAs (comparable to previous studies, see e.g.,37), we obtained 209,323-2,901,437 unique reads mapped to the P. vivax genome (0.28-5.47% mapping) resulting in 102-4,665 single cells characterized by at least 100 reads within genes per library (Supplemental Data 1, Table 1). We genotyped each individual sporozoite as described above and, despite the lower quality of the data, observed three genotype groups (Fig. 4B): some sporozoites only carried NIH-1993-F3 alleles and derived from mating of a NIH-1993-F3 male gametocyte with a NIH-1993-F3 female gametocyte, some sporozoites only carried Chesson alleles and derived from mating of Chesson gametocytes, and some sporozoites carried a mixture of NIH-1993-F3 and Chesson alleles and derived from outcrossed mating of a NIH gametocyte with a Chesson gametocyte. We observed more NIH-1993-F3 than Chesson self-mated sporozoites, probably due to the higher proportion of female gametocytes produced by NIH-1993-F3 parasites compared to Chesson parasites, although the lack of information on male gametocytes precludes drawing definitive conclusions.
We infected two additional Saimiri boliviensis monkeys by inoculating them with ~13,000 sporozoites dissected from An. freeborni fed with blood from the animals simultaneously coinfected. Both monkeys developed a blood-stage infection detectable by microscopy 29 days post inoculation. We then collected blood from each monkey and prepared scRNA-seq libraries (Fig. 4A, Supplemental Data 1, Table 1). Interestingly, in both monkeys, genotyping of the parasites revealed that the blood-stage infections contained only two of the three genotype groups identified in sporozoites: blood-stage parasites carried either only NIH-1993-F3 alleles (resulting from NIH-1993-F3 self-mating) or an equal proportion of NIH-1993-F3 and Chesson alleles (resulting from outcrossing) but we did not detect any blood-stage parasites carrying only Chesson alleles (Fig. 4C). This observation was unexpected given that the Chesson strain, in monoclonal infections, is able to be transmitted from mosquitoes to Saimiri monkeys and to successfully complete its pre-erythrocytic development to produce blood-stage infections57. Several scenarios could explain this observation. First, the failure of Chesson transmission might simply be due to chance: although we failed to detect Chesson self-mated parasites in two independent monkey infections, it is possible that these parasites (that were less abundant than the NIH-1993-F3 self-mated and the outcrossed sporozoites) were lost stochastically due to a strong bottleneck in population size occurring during the pre-erythrocytic development (58, see also below): for example, if only ~5% of the sporozoites inoculated derived from Chesson self-mating (Fig. 4B) and only 10–15 hepatic schizonts led to the blood infection, it would not be unlikely to see no Chesson blood-stage parasites (p > 0.05). Second, this lack of Chesson parasites detection in the blood might be caused by a delay in their liver-stage development: the consecutive infection experiments (Fig. 2) showed that it was difficult for a second parasite to establish a robust infection once one parasite was already successfully infecting RBCs, and a slightly delayed liver maturation could therefore provide a significant disadvantage in intraerythrocytic development. Alternatively, it is possible that Chesson parasites more readily enter dormancy in the liver while the NIH-1993-F3 parasites disproportionally develop into active liver schizonts. Third, we do not know if all salivary gland sporozoites were fully mature and infectious and it is possible that the Chesson sporozoites detected in mosquitoes were not yet able to be transmitted to mosquitoes (if their maturation was slower than the NIH-1993-F3 or the outcrossed sporozoites). Finally, the sporozoites used for inoculation were different from those used for scRNA-seq and it is possible that the latter did not include Chesson self-mated sporozoites (although the animals used for blood feeding were the same, and parasitemia and strain proportion was roughly similar at the time of collection (Supplemental Figs. 2, 6). A repeated infection and follow up studies, for example a back-cross between Chesson and the outcrossed parasites, could help differentiating among these hypotheses and could potentially provide insights on the genetic loci underlying this observation (if any).
We then compared the NIH-1993-F3 blood-stage parasites resulting from sporozoite infections with the NIH-1993-F3 parasites resulting from infection by blood stage inoculation to determine if the route of infection had any effect on the parasite regulation and development. The proportion of sexual parasites was lower in infections initiated by sporozoite inoculation than in infections initiated by parasitized RBC (27% vs 47%) (Fig. 4D), which could reflect a delay in the initiation of the sexual stage commitment. Infected red blood cell inoculations contain all blood stages from the previous infection, including long-lived gametocytes and already-committed asexual parasites, while infections derived from liver schizonts need to initiate and progress through the entire gametocytogenesis process and might therefore need longer to reach the same gametocytemia (note however that in contrast to P. falciparum59, gametocytogenesis might start during liver development in P. vivax60). Finally, we compared the gene expression profiles of parasites from infections initiated with parasitized RBCs vs. sporozoites. In early asexual parasites (group A) 439 genes were differentially expressed, and 99 genes were differentially regulated in gametocytes (Supplemental Data 3). The results revealed many female gametocyte genes differentially expressed in asexual parasites, again likely reflecting the spurious effect of rare doublets including female gametocytes in the infections initiated by blood inoculations where they are more abundant. However, we also noted a unique transcriptional signature with at least eight rhoptry-associated proteins (RhopH2, RhopH3, RAMA and the rhoptry neck proteins 1, 3, 6, 11 and 12) showing higher expression in blood-initiated infections than in the sporozoite-initiated infections (Fig. 4E). This specific pattern of overexpression of genes involved in RBC invasion could reflect a gradual strengthening of the mechanisms of RBC invasion over time to optimize the parasite growth and intraerythrocytic development61.
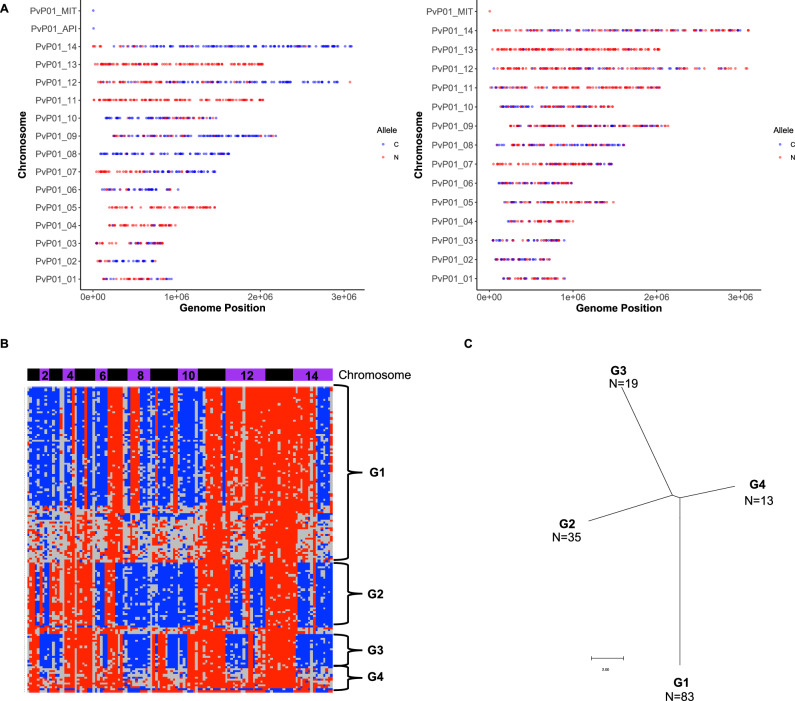
Finally, the observation of blood-stage parasites derived from the outcrossing of Chesson and NIH-1993-F3 gametocytes provides a unique opportunity to estimate how many hepatic schizonts contributed to the blood-stage infections and thus to evaluate the magnitude of the liver bottleneck. We used the genotypes determined by scRNA-seq to infer genome-wide haplotypes for the subset of the outcrossed blood-stage parasites best characterized (see Materials and Methods for details). As expected, these haplotypes showed clear patterns of recombination between NIH-1993-F3 and Chesson, but also a high degree of redundancy: we only identified four distinct genotypes (in roughly equal proportion) in one animal and two distinct genotypes in the second animal (Fig. 5, Supplemental Figure 7). Importantly, these haplotypes remained detectable, and in similar proportion, during the course of the infection and no additional haplotype emerged later (Supplemental Figure 7). These results indicate that the parasites present in the blood of the animals infected by sporozoites derived from a very small number of hepatic schizonts (probably less than 10, when accounting for the unknown number of schizonts leading to all NIH-1993-F3 blood-stage parasites), and that a major bottleneck indeed restricts the development and diversity of Plasmodium pre-erythrocytic stages.
Fig. 5. Analysis of outcrossed blood-stage parasites.
A Genotypes of two individual outcrossed parasites across the genome. The plots show the alleles successfully characterized by scRNA-seq based on their genomic position (x-axis: chromosome, y-axis: nucleotide position) and colored by the genotype (red: NIH-1993-F3, blue: Chesson). B Reconstruction of the haplotypes across the entire genome for 150 outcrossed parasites (rows). Each column represents a 200,000 bp window of the P. vivax genome organized by chromosomes (blue and purple horizontal bars) and colored according to its genotype (red: NIH-1993-F3, blue: Chesson, gray: no data). The data is shown for 150 cells from one animal 29 dpi (the other time points and the data from the second animal are shown in Supplemental Fig. 6). C Phylogenetic tree reconstructed from representative haplotypes shown in (B).
Overall, we showed here that we can use the Saimiri model, in combination with scRNA-seq to accurately genotype individual parasites, to study P. vivax polyclonal infections. While this study is relatively limited in sample size and the reliance on specific P. vivax strains cautions on overinterpreting the results, these analyses revealed interesting observations that may have important biological implications. First, inoculation of a second strain, after the establishment of a blood stage infection, failed to generate robust polyclonal infections, which could suggest that polyclonality in patients primarily derives from co-transmission of parasites by a mosquito bite rather than consecutive infections. This finding is consistent with previous studies36,40,62 but will need to be further evaluated, in particular in the context of non-naïve hosts and over longer infection periods. (Note also that this finding might be specific to P. vivax that is much more limited than P. falciparum in the number of RBCs available for infection4,63). Second, our results suggest that coinfection with multiple strains of P. vivax does not dramatically impact parasite regulation. We did not observe significant changes in asexual stage proportion nor in sexual differentiation, and the gene expression profiles appeared unaffected by the presence of other genotypes. Along the same lines, we observed even transmission of the genotypes to the mosquitoes. Third, the analyses of Saimiri monkeys infected through sporozoite inoculation were consistent with a strong bottleneck during pre-erythrocytic development and suggested that some strains (such as Chesson in our experiments) were less fit and outcompeted by others, or that they matured more slowly in the mosquitoes or in the liver (or that they more readily enter a dormant stage). Overall, these studies demonstrate the potential of animal models combined with modern genomic approaches for studying Plasmodium biology and rigorously evaluating variability (and possibly interactions) among parasite strains during their development, and its consequences for malaria control.
Methods
Ethics statement
All animal procedures were conducted in accordance with the National Institutes of Health (NIH) guidelines and regulations64, under approved protocols by the National Institute of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee (ACUC) (Animal study NIAID ASPLMVR15). Animals were purchased from NIH-approved sources and transported and housed according to Guide for the Care and Use of Laboratory Animals64.
Animal experiments
10 female Saimiri boliviensis monkeys were infected with either the NIH-1993-F3 and/or the Chesson strains of P. vivax. Eight animals were infected via intravenous inoculation of cryopreserved (n = 4) or fresh (n = 4) parasitized RBCs. We examined blood via blood smears following inoculation until parasitemia was microscopically detected (approximately 0.1%).
Three animals were inoculated with NIH-1993-F3 strain, with one animal receiving a second inoculation with the Chesson strain approximately two weeks after the initial inoculation (consecutive infection). Another three animals were inoculated with the Chesson strain, with one of these receiving a second inoculation with the NIH-1993-F3 strain approximately two weeks after initial inoculation (consecutive infection). The other two animals were infected simultaneously with equal numbers of parasites from each strain.
The blood of simultaneously coinfected animals was used for membrane feeding of Anopheles stephensi and Anopheles freeborni mosquitoes. After 17-20 days, 20 female mosquitoes were anesthetized on ice and their salivary glands dissected in PBS under a stereomicroscope. The salivary glands were transferred to a low-retention tube (Protein LoBind Tube; Eppendorf) containing PBS, homogenized with a disposable pestle, spun down, washed and resuspended37. This pool of sporozoites was used to infect two additional monkeys by intra-venous inoculation. One of these monkeys was splenectomized prior to infection and the other was splenectomized 6 days post infection. The overall design of the animal experiments is summarized in Fig. 1A.
Whole genome sequencing
Blood was collected from two of the monkeys monoinfected by Chesson and NIH-1993-F3 and processed through MACS column to enrich for parasitized RBCs. Parasite DNA was extracted by Qiagen DNeasy column [Cat. No. / ID: 69504] and approximately 500 ng of DNA used to prepare Nextera XT DNA libraries [Cat. No./ID FC-131-1024]. Each library was then sequenced on an Illumina NovaSeq to generate 200 million paired-end reads of 150 bp per sample.
All sequencing reads were mapped to P. vivax P01 v54 ref. 65 using Hisat2 with default paired-end parameters66. We then used samtools mpileup and custom scripts to identify all nucleotide differences between the two strains, only considering regions sequenced by at least 50 reads.
10X Genomics scRNA-seq library preparation
Once the parasitemia became measurable, and at regular intervals throughout the course of the infection, we collected 1 ml of blood from each animal to prepare 10X Genomics 3’ end scRNA-seq libraries (for a total of 3-4 samples per animal) [Cat. No./ID 1000268]. 50 µl of whole blood was diluted 1:10 in trizol and stored for later analyses. The remaining blood cells were transferred to 50 mL tubes containing RPMI 1640 to a final 0.5% to 1.0% hematocrit solution. Parasitized cells were enriched using MACS LS separation columns. Enriched cells were quantified by Millipore Scepter 3.0 Handheld Automated Cell Counter. From each sample, we then loaded an estimated 5000 cells on a 10X Chromium controller. Libraries were completed according to the manufacturer’s instructions and sequenced on an Illumina NovaSeq to generate 200 million read pairs per sample.
Two blood samples from two additional Saimiri boliviensis monkeys coinfected with one NIH-1993-F338 and the Chesson P. vivax clones were used for membrane feeding of Anopheles stephensi and Anopheles freeborni. Salivary glands sporozoites were collected from each feeding at 17–21 days post-feed: eight groups of 20 female mosquitoes, each from a separate feed, were anesthetized on ice and their salivary glands dissected in PBS under a stereomicroscope. The salivary glands were transferred to a low-retention tube (Protein LoBind Tube; Eppendorf) containing PBS, homogenized with a disposable pestle, spun down, washed, resuspended, and quantified.
Analysis of 10X scRNA-seq data
10X scRNA-seq data were first demultiplexed using custom scripts and all reads mapped to P. vivax P01 v54 reference65 using Hisat266 with the maximum intron length set to 5000 bp. We then used custom scripts to count the number of reads per cell, only considering reads that mapped within genes, using the annotations generated in39. For the blood-stage parasites, we only considered single cell transcriptomes characterized by at least 1000 unique reads mapped within annotated genes, and 100 unique reads mapped within annotated genes for the sporozoites to account for host RNA contamination (the data for blood-stage parasites using the 250 reads cutoff is presented in supplemental information). Count tables were generated via custom scripts and exported to SCRAN for further analysis67. Asexual and female gametocytes were identified based on UMAP clusters and expression of selected marker genes. We then calculated pseudotime for each cell, separately for the asexual parasites and female gametocytes, by calculating the Euclidean distance in the principal component space from an arbitrarily chosen origin.
Single cell genotyping based on the scRNA-seq data
For each 10X scRNA-seq library, we used samtools mpileup to identify the reads (and alleles) overlapping each of the 26,719 single nucleotide positions differentiating Chesson and NIH-1993-F3. We then use custom scripts to count the number of Chesson alleles and NIH alleles present in each cell, considering, for blood-stage parasites, only positions sequenced by at least five reads, with 75% of the reads carrying the same allele (after removing potential PCR duplicates). For sporozoites, given the lower sequencing depth, we considered the major allele using positions sequenced by at least three reads. For all analyses, we only considered single cells genotyped by at least 20 positions and for which more than 95% of the genotypes were concordant (e.g., >95% of the genotypes had to be NIH-1993-F3).
For the subset of blood-stage parasites resulting from outcrossed fertilization of Chesson and NIH-1993-F3 gametocytes, we used custom scripts to infer genome-wide haplotypes for each cell by determining, for each non-overlapping 200,000 bp window of the genome, the genotype that was most common among the differentiating positions.
Differential gene expression by pseudobulk
Differential gene expression analysis was conducted by pseudobulk analysis using EdgeR68 after assigning cells from each sample into one of four groups based on their pseudotime (Group A: pseudotime = 0-50, Group B = 50–100, Group C = 100–150, Sexual = female gametocytes) and separating the cells according to their genotype (if applicable). Only samples with at least 100 cells in a given group were considered for differential expression analysis, and only genes detected in at least 100 cells (across all samples) for a given group were tested. A FDR of 0.1 was used to determine statistical significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank S. Ott, H. Bowen, L. Sadzewicz, and L. Tallon in Maryland Genomics at the University of Maryland School of Medicine for their support with Illumina sequencing, and A. Laughinghouse and K. Lee for insectary support, and A. Bruce, A. Roessler, M. Herrera, K. Dicken, B. Lewis, and Y. Littman for technical assistance with non-human primates. This work was supported by an award from the NIH to the University of Maryland School of Medicine (U19 AI110820 to DS), by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH (to TEW) and a grant from the Bill and Melinda Gates Foundation (OPP#1023643 to JHA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, J.M.S., T.E.W. and D.S.; Methodology, J.M.S., T.E.W. and D.S.; Investigation, B.H., J.M.S., H.N.B., T.V.P., A.C.E., S.A., J.S.A. and D.S.; Writing – Original Draft, B.H. and D.S.; Writing – Review & Editing, B.H., J.M.S, T.E.W., and D.S.; Funding Acquisition, J.H.A., T.E.W. and D.S.; Supervision, J.M.S, T.E.W. and D.S.
Peer review
Peer review information
Nature Communications thanks Mara Lawniczak, Sunil Dogga, Jesse Rop and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All sequence data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the BioProject ID PRJNA1047651 (https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1047651). Source Data are provided with this paper, including single cell count table and processed data generated, available through Zenodo (https://zenodo.org/records/12775216).
Code availability
Custom scripts are available at https://github.com/bhazzard11/Coinfection_Analysis and https://zenodo.org/records/12775129.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51949-8.
References
- 1.World Health Organization. World Malaria Report 2022. (2022).
- 2.Howes, R. E. et al. Global Epidemiology of Plasmodium vivax. Am. J. Trop. Med Hyg.95, 15–34 (2016). 10.4269/ajtmh.16-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price, R. N. et al. Vivax malaria: neglected and not benign. Am. J. Trop. Med Hyg.77, 79–87 (2007). 10.4269/ajtmh.2007.77.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgard, C., Albrecht, L., Kayano, A., Sunnerhagen, P. & Costa, F. T. M. Plasmodium vivax Biology: Insights Provided by Genomics, Transcriptomics and Proteomics. Front Cell Infect. Microbiol8, 34 (2018). 10.3389/fcimb.2018.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fola, A. A. et al. Higher Complexity of Infection and Genetic Diversity of Plasmodium vivax Than Plasmodium falciparum Across All Malaria Transmission Zones of Papua New Guinea. Am. J. Trop. Med Hyg.96, 630–641 (2017). 10.4269/ajtmh.16-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich, L. R. et al. Complexity of Infection and Genetic Diversity in Cambodian Plasmodium vivax. PLoS Negl. Trop. Dis.10, e0004526 (2016). 10.1371/journal.pntd.0004526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neafsey, D. E. et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat. Genet44, 1046–1050 (2012). 10.1038/ng.2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson, R. D. et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet48, 959–964 (2016). 10.1038/ng.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. Malaria.
- 10.Andrew, G. & Evans, T. E. W. Coevolutionary Genetics of Plasmodium Malaria Parasites and Their Human Hosts. Integr. Comp. Biol.42, 401–407 (2002). 10.1093/icb/42.2.401 [DOI] [PubMed] [Google Scholar]
- 11.Venugopal, K., Hentzschel, F., Valkiunas, G. & Marti, M. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat. Rev. Microbiol18, 177–189 (2020). 10.1038/s41579-019-0306-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bantuchai, S., Imad, H. & Nguitragool, W. Plasmodium vivax gametocytes and transmission. Parasitology International87(2022). [DOI] [PubMed]
- 13.Genton, B. et al. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med5, e127 (2008). 10.1371/journal.pmed.0050127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, D. K. et al. The Plasmodium liver-specific protein 2 (LISP2) is an early marker of liver stage development. Elife8, e43362 (2019). [DOI] [PMC free article] [PubMed]
- 15.Bozdech, Z. et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl Acad. Sci. USA105, 16290–16295 (2008). 10.1073/pnas.0807404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popovici, J. et al. Genomic analyses reveal the common occurrence and complexity of plasmodium vivax relapses in Cambodia. mBio9, e01888-17 (2018). [DOI] [PMC free article] [PubMed]
- 17.Steenkeste, N. et al. Sub-microscopic malaria cases and mixed malaria infection in a remote area of high malaria endemicity in Rattanakiri province, Cambodia: implication for malaria elimination. Malar. J.9, 108 (2010). 10.1186/1475-2875-9-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dia, A. et al. Single-genome sequencing reveals within-host evolution of human malaria parasites. Cell Host Microbe29, 1496–1506 e3 (2021). 10.1016/j.chom.2021.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollitt, L. C. et al. Competition and the evolution of reproductive restraint in malaria parasites. Am. Nat.177, 358–367 (2011). 10.1086/658175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mideo, N. & Day, T. On the evolution of reproductive restraint in malaria. Proc. Biol. Sci.275, 1217–1224 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor, L. H., Walliker, D. & Read, A. F. Mixed-genotype infections of malaria parasites: within-host dynamics and transmission success of competing clones. Proc. Biol. Sci.264, 927–935 (1997). 10.1098/rspb.1997.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tirrell, A. R. et al. Pairwise growth competitions identify relative fitness relationships among artemisinin resistant Plasmodium falciparum field isolates. Malar. J.18, 295 (2019). 10.1186/s12936-019-2934-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell, A. J. C. et al. Regulators of male and female sexual development are critical for the transmission of a malaria parasite. Cell Host Microbe31, 305–319 e10 (2023). 10.1016/j.chom.2022.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentzschel, F. et al. Host cell maturation modulates parasite invasion and sexual differentiation in Plasmodium berghei. Sci. Adv.8, eabm7348 (2022). 10.1126/sciadv.abm7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, A., Popovici, J., Menard, D. & Serre, D. Plasmodium vivax transcriptomes reveal stage-specific chloroquine response and differential regulation of male and female gametocytes. Nat. Commun.10, 371 (2019). 10.1038/s41467-019-08312-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poran, A. et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature551, 95–99 (2017). 10.1038/nature24280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brancucci, N. M. B. et al. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite plasmodium falciparum. Cell171, 1532–1544 e15 (2017). 10.1016/j.cell.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, C. T. et al. Sexual differentiation in human malaria parasites is regulated by competition between phospholipid metabolism and histone methylation. Nat. Microbiol8, 1280–1292 (2023). 10.1038/s41564-023-01396-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bermudez, M., Moreno-Perez, D. A., Arevalo-Pinzon, G., Curtidor, H. & Patarroyo, M. A. Plasmodium vivax in vitro continuous culture: the spoke in the wheel. Malar. J.17, 301 (2018). 10.1186/s12936-018-2456-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beignon, A. S., Le Grand, R. & Chapon, C. In vivo imaging in NHP models of malaria: challenges, progress and outlooks. Parasitol. Int63, 206–215 (2014). 10.1016/j.parint.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan, E. R., Barnwell, J. W., Zimmerman, P. A. & Serre, D. Comparative analysis of field-isolate and monkey-adapted Plasmodium vivax genomes. PLoS Negl. Trop. Dis.9, e0003566 (2015). 10.1371/journal.pntd.0003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins, W. E. Nonhuman primate models. II. Infection of saimiri and aotus monkeys with plasmodium vivax. Methods Mol. Med72, 85–92 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Minkah, N. K., Schafer, C. & Kappe, S. H. I. Humanized mouse models for the study of human malaria parasite biology, pathogenesis, and immunity. Front Immunol.9, 807 (2018). 10.3389/fimmu.2018.00807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luiza-Batista, C. et al. Humanized mice for investigating sustained Plasmodium vivax blood-stage infections and transmission. Nat. Commun.13, 4123 (2022). 10.1038/s41467-022-31864-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walliker, D., Carter, R. & Morgan, S. Genetic recombination in Plasmodium berghei. Parasitology66, 309–320 (1973). 10.1017/S0031182000045248 [DOI] [PubMed] [Google Scholar]
- 36.Nkhoma, S. C. et al. Co-transmission of related malaria parasite lineages shapes within-host parasite diversity. Cell Host Microbe27, 93–103 e4 (2020). 10.1016/j.chom.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogale, H. N. et al. Transcriptional heterogeneity and tightly regulated changes in gene expression during Plasmodium berghei sporozoite development. Proc. Natl Acad. Sci. USA118(2021). [DOI] [PMC free article] [PubMed]
- 38.Sa, J. M., Cannon, M. V., Caleon, R. L., Wellems, T. E. & Serre, D. Single-cell transcription analysis of Plasmodium vivax blood-stage parasites identifies stage- and species-specific profiles of expression. PLoS Biol.18, e3000711 (2020). 10.1371/journal.pbio.3000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazzard, B. et al. Long read single cell RNA sequencing reveals the isoform diversity of Plasmodium vivax transcripts. PLoS Negl. Trop. Dis.16, e0010991 (2022). 10.1371/journal.pntd.0010991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nair, S. et al. Single-cell genomics for dissection of complex malaria infections. Genome Res24, 1028–1038 (2014). 10.1101/gr.168286.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruberto, A. A. et al. Single-cell RNA sequencing of Plasmodium vivax sporozoites reveals stage- and species-specific transcriptomic signatures. PLoS Negl. Trop. Dis.16, e0010633 (2022). 10.1371/journal.pntd.0010633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Real, E. et al. A single-cell atlas of Plasmodium falciparum transmission through the mosquito. Nat. Commun.12, 3196 (2021). 10.1038/s41467-021-23434-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howick, V. M. et al. The Malaria Cell Atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science365(2019). [DOI] [PMC free article] [PubMed]
- 44.Reid, A. J. et al. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife7, e33105 (2018). [DOI] [PMC free article] [PubMed]
- 45.Brancucci, N. M. B. et al. Probing Plasmodium falciparum sexual commitment at the single-cell level. Wellcome Open Res3, 70 (2018). 10.12688/wellcomeopenres.14645.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods17, 615–620 (2020). 10.1038/s41592-020-0820-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sa, J. M. et al. Plasmodium vivax chloroquine resistance links to pvcrt transcription in a genetic cross. Nat. Commun.10, 4300 (2019). 10.1038/s41467-019-12256-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinha, A. et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature507, 253–257 (2014). 10.1038/nature12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kafsack, B. F. et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature507, 248–252 (2014). 10.1038/nature12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA100, 9440–9445 (2003). 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auburn, S. et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res1, 4 (2016). 10.12688/wellcomeopenres.9876.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlton, J. M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature455, 757–763 (2008). 10.1038/nature07327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hester, J. et al. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl. Trop. Dis.7, e2569 (2013). 10.1371/journal.pntd.0002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.del Portillo, H. A. et al. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature410, 839–842 (2001). 10.1038/35071118 [DOI] [PubMed] [Google Scholar]
- 55.Little, T. S. et al. Analysis of pir gene expression across the Plasmodium life cycle. Malar. J.20, 445 (2021). 10.1186/s12936-021-03979-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Portugal, S. et al. Host-mediated regulation of superinfection in malaria. Nat. Med17, 732–737 (2011). 10.1038/nm.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins, W. E. et al. Chesson strain Plasmodium vivax in Saimiri sciureus boliviensis monkeys. J. Parasitol.73, 929–934 (1987). 10.2307/3282513 [DOI] [PubMed] [Google Scholar]
- 58.Aleshnick, M., Ganusov, V. V., Nasir, G., Yenokyan, G. & Sinnis, P. Experimental determination of the force of malaria infection reveals a non-linear relationship to mosquito sporozoite loads. PLoS Pathog.16, e1008181 (2020). 10.1371/journal.ppat.1008181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Day, K. P., Hayward, R. E. & Dyer, M. The biology of Plasmodium falciparum transmission stages. Parasitology116, S95–S109 (1998). 10.1017/S0031182000084985 [DOI] [PubMed] [Google Scholar]
- 60.Mancio-Silva, L. et al. A single-cell liver atlas of Plasmodium vivax infection. Cell Host Microbe30, 1048–1060 e5 (2022). 10.1016/j.chom.2022.03.034 [DOI] [PubMed] [Google Scholar]
- 61.Topolska, A. E., Black, C. G. & Coppel, R. L. Identification and characterisation of RAMA homologues in rodent, simian and human malaria species. Mol. Biochem Parasitol.138, 237–241 (2004). 10.1016/j.molbiopara.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 62.Nkhoma, S. C. et al. Close kinship within multiple-genotype malaria parasite infections. Proc. Biol. Sci.279, 2589–2598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mueller, I. et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis.9, 555–566 (2009). 10.1016/S1473-3099(09)70177-X [DOI] [PubMed] [Google Scholar]
- 64.Hawkins, P. et al. A guide to defining and implementing protocols for the welfare assessment of laboratory animals: eleventh report of the BVAAWF/FRAME/RSPCA/UFAW Joint Working Group on Refinement. Lab Anim.45, 1–13 (2011). 10.1258/la.2010.010031 [DOI] [PubMed] [Google Scholar]
- 65.Siegel, S. V. et al. Analysis of Plasmodium vivax schizont transcriptomes from field isolates reveals heterogeneity of expression of genes involved in host-parasite interactions. Sci. Rep.10, 16667 (2020). 10.1038/s41598-020-73562-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All sequence data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the BioProject ID PRJNA1047651 (https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1047651). Source Data are provided with this paper, including single cell count table and processed data generated, available through Zenodo (https://zenodo.org/records/12775216).
Custom scripts are available at https://github.com/bhazzard11/Coinfection_Analysis and https://zenodo.org/records/12775129.