Abstract
Small-molecule compounds that elicit mRNA-selective translation repression have attracted interest due to their potential for expansion of druggable space. However, only a limited number of examples have been reported to date. Here, we show that desmethyl desamino pateamine A (DMDA-PatA) represses translation in an mRNA-selective manner by clamping eIF4A, a DEAD-box RNA-binding protein, onto GNG motifs. By systematically comparing multiple eIF4A inhibitors by ribosome profiling, we found that DMDA-PatA has unique mRNA selectivity for translation repression. Unbiased Bind-n-Seq reveals that DMDA-PatA-targeted eIF4A exhibits a preference for GNG motifs in an ATP-independent manner. This unusual RNA binding sterically hinders scanning by 40S ribosomes. A combination of classical molecular dynamics simulations and quantum chemical calculations, and the subsequent development of an inactive DMDA-PatA derivative reveals that the positive charge of the tertiary amine on the trienyl arm induces G selectivity. Moreover, we identified that DDX3, another DEAD-box protein, is an alternative DMDA-PatA target with the same effects on eIF4A. Our results provide an example of the sequence-selective anchoring of RNA-binding proteins and the mRNA-selective inhibition of protein synthesis by small-molecule compounds.
Subject terms: Translation, RNA, Small molecules, RNA, Target identification
Here the authors report that DMDA-PatA, an anti-tumor compound, functions as a mRNA-selective translational inhibitor. This drug clamps the target eIF4A and DDX3 RNA-binding proteins on the GNG RNA motif, providing steric hindrance for scanning ribosomes.
Introduction
The production of harmful proteins often leads to deleterious outcomes in cells, causing a wide variety of diseases. Due to the limited druggable proteome1, compounds that modulate the synthesis of unwelcome proteins at the translational level provide attractive therapeutic opportunities2. Although several compounds that suppress translation in an mRNA-selective manner have been identified2,3, the number of such compounds is still limited, warranting further identification of a new class with such activity.
Repurposing natural secondary metabolites for pharmacological use has been a common strategy in drug development4. Indeed, translation inhibitors are not exceptions, as a variety of antibiotics targeting ribosomes have been exploited5. In addition to ribosomes, eukaryotic translation initiation factor (eIF) 4 A has been found to be a common target of a variety of natural products, presenting a vulnerability in cancer6. These compounds include hippuristanol (Hipp) from a soft coral (Isis hippuris)7–11, rocaglates from plants of the Aglaia genus12–20, pateamine A (PatA) from a sponge (Mycale sp.) or its microbiome symbionts21–32, and sanguinarine (San) from poppy plants (Macleaya cordata and Argemone Mexicana)33,34.
eIF4A is an ATP-dependent DEAD-box type RNA-binding protein that forms a complex with the cap-binding protein eIF4E and scaffold protein eIF4G and then facilitates the loading of the 43S preinitiation complex onto the 5′ ends of mRNA and subsequent scanning of the 5′ untranslated region (UTR)35,36. In mammals, this protein is encoded by two genes, EIF4A1 and EIF4A2. Hipp and San have been shown to reduce the RNA-binding ability of eIF4A8–10,34, simply inactivating the function of eIF4A in translation. In contrast, rocaglates have a unique mode of action, clamping eIF4A onto a polypurine (repeats of A and G nucleotides) RNA motif16–19,37. This artificial clamping sterically hinders ribosome scanning, blocks the recruitment of the 43S preinitiation complex at the 5′ ends of mRNAs, and ultimately reduces the amount of eIF4A available for translation initiation (i.e., the bystander effect)16–19,37. It has also been suggested that PatA does not phenocopy the loss of eIF4A function, thus enhancing the interaction between eIF4A and RNA22–25,28,32. However, the molecular mechanism by which PatA blocks protein synthesis remains unclear.
Here, we systematically investigated the mode of action of PatA and found that this compound leads to RNA sequence-selective translation repression. Genome-wide ribosome profiling revealed that PatA induces distinct translational output compared to Hipp, San, and rocaglamide A (RocA)—a potent rocaglate. RNA pulldown-Seq in cells and RNA Bind-n-Seq in vitro showed that PatA clamps to the GNG motif in RNA in an ATP-independent manner. PatA-mediated clamping causes mRNA-selective translation repression, most likely causing steric hindrance to scanning ribosomes. Our classical molecular dynamics (MD) simulations and subsequent fragment molecular orbital (FMO) calculations revealed that the G nucleotide preference of PatA stemmed from the tertiary amine on the trienyl arm. The designed PatA derivative confirmed the importance of the amine in RNA selectivity, translation repression, and cytotoxicity. Our study provides an additional example of a sequence-selective translation inhibitor, expanding the space of exploitable proteomes for drug development.
Results
Differential impacts of multiple eIF4A inhibitors on cellular translation
To understand the variation in translation repression induced by eIF4A inhibitors and the mechanism of the effect evoked by PatA, we systematically compared the translatome alterations with ribosome profiling38,39 (Fig. 1a). Here, instead of the original PatA, we used the simplified derivative desmethyl desamino pateamine A (DMDA-PatA) (Supplementary Fig. 1a) due to its comparable activity40. We conducted the experiments in human embryonic kidney (HEK) 293 cells treated with 2–3 different concentrations of Hipp, San, and DMDA-PatA. To minimize the effect on the transcriptome, we treated cells with the compounds for 15–30 min, limiting the change in mRNA abundance16. We also mined the published ribosome profiling data with RocA16. Normalization by mitochondrial footprints as internal spike-ins16,18,37,41,42 enabled us to monitor global translation changes. Indeed, the calibration of ribosome profiling data allowed us to monitor the dose-dependent changes in translation caused by the compounds (Supplementary Fig. 1b).
Fig. 1. Comparative analysis of translation changes induced by eIF4A-targeting compounds in cells.
a Schematic of ribosome profiling experiments. The chemical structures of the eIF4A-targeting compounds used in the experiments are shown. b Principal component (PC) analysis of the translation changes analyzed by ribosome profiling under the indicated conditions. c Spearman’s correlation coefficients (ρ, two-tailed) for translation changes induced by drug treatments. The color scales for ρ are shown. d MA (M, log ratio; A, mean average) plot of the translation fold change with 0.1 μM DMDA-PatA treatment. Low-sensitivity mRNAs (FDR ≤ 0.01 and log2-fold change ≥1 from the mean) and high-sensitivity mRNAs (FDR ≤ 0.01 and log2-fold change ≤ −1 from the mean) are highlighted. e Schematic of the RNA pulldown-Seq experiments. mRNAs associated with SBP-tagged eIF4A1 in the cells were isolated and subjected to deep sequencing. f Cumulative distribution of the mRNA fold change in RNA pulldown-Seq data for SBP-tagged eIF4A1 upon 0.01 μM DMDA-PatA treatment. DMDA-PatA low-sensitivity and high-sensitivity mRNAs (defined in d) were compared to total mRNAs. The significance was calculated by the Mann‒Whitney U test (two-tailed). Source data are provided as a Source Data file.
This comparative analysis revealed the similarities and differences in translation inhibition by eIF4A-targeting compounds. Principal component analysis (PCA), in which the first component (PC1) explained ~60% of the variance in the data (Supplementary Fig. 1c), revealed the dose-dependent, directional effects of each drug (Fig. 1b and Supplementary Fig. 1d). Strikingly, the eIF4A inhibitors could be categorized into two groups: group 1, San and Hipp; group 2, RocA and DMDA-PatA (Fig. 1c and Supplementary Fig. 1e).
Given that both San and Hipp reduce the binding affinity between eIF4A and RNA8,9,11,34,43, their high correspondence in translation repression was a compelling result (Fig. 1c and Supplementary Fig. 1e, f). On the other hand, the polypurine-selective eIF4A clamping induced by RocA should be distinct from the effects of San and Hipp16–19,37. Our analysis revealed that DMDA-PatA has a similar (but not identical) mode of translational repression to RocA, providing widespread sensitivity in translation across the transcriptome (Fig. 1d).
mRNA-selective clamping of eIF4A1 by DMDA-PatA leads to translational repression
Considering that PatA and its derivatives stabilize the interaction between RNA and eIF4A22–25,32, we reasoned that the biased interaction of eIF4A with mRNAs may be associated with the mRNA selectivity of DMDA-PatA in translation repression. To monitor the mRNAs associated with eIF4A, we conducted RNA pulldown and subsequent RNA sequencing (RNA pulldown-Seq) with streptavidin-binding peptide (SBP)-tagged eIF4A1, a major eIF4A paralog, from a HEK293 cell line (Fig. 1e)16. DMDA-PatA treatment evoked diverse alterations in mRNAs bound to eIF4A1 (Supplementary Fig. 1h).
Through comparison of the changes in the mRNA association with eIF4A and translation repression, we found that the tight association between eIF4A and a subset of mRNAs upon drug treatment confers translation repression. mRNAs highly sensitive to DMDA-PatA in terms of translation repression were more stably associated with eIF4A upon drug treatment, whereas mRNAs with low sensitivity showed the opposite behavior (Fig. 1f and Supplementary Fig. 1i). These data suggested that the DMDA-PatA-mediated mRNA-selective eIF4A interaction determines the efficacy of translational repression.
DMDA-PatA leads to ATP-independent GNG RNA clamping by eIF4A
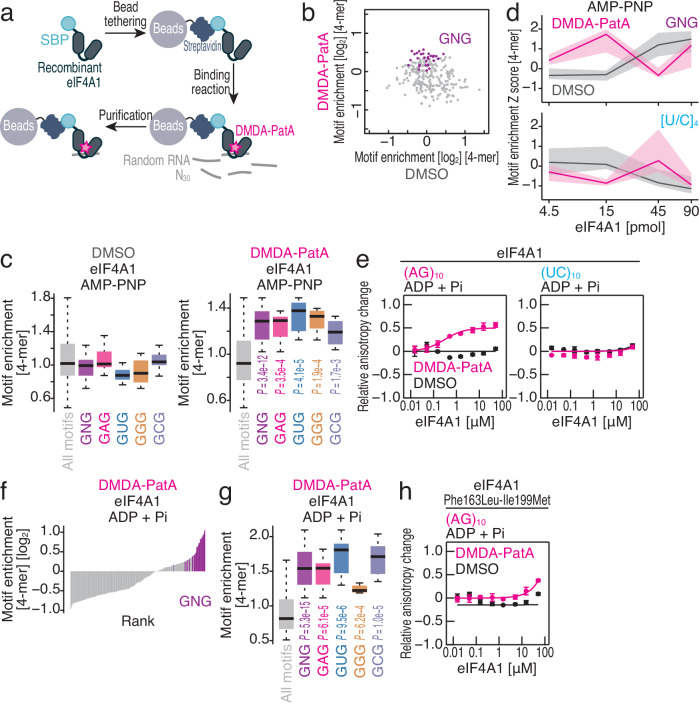
These data led us to investigate whether DMDA-PatA clamps eIF4A on selective RNA motifs. To systematically survey the RNA motif selectivity provided by DMDA-PatA, we conducted RNA Bind-n-Seq44,45 with random 30 nt RNA and recombinant human eIF4A1 in the presence and absence of DMDA-PatA (Fig. 2a). eIF4A requires ATP to interact with RNA but dissociates from RNA upon ATP hydrolysis46,47. To stabilize the ATP-bound ground state, we used a nonhydrolyzable ATP analog (5′-adenylyl-imidodiphosphate or AMP-PNP) for RNA Bind-n-Seq. Our analysis revealed an unexpected nucleotide specificity of DMDA-PatA toward a subset of motifs, which often included GNG sequences in a 4-mer motif survey (Fig. 2b). This same motif preference was found when longer 5-mer or 6-mer motifs were considered (Supplementary Fig. 2a, b). Any nucleotide sandwiched by two Gs appeared to be interchangeable in terms of selectivity (Fig. 2c).
Fig. 2. DMDA-PatA provides GNG motif preference on eIF4A1.
a Schematic of RNA Bind-n-Seq experiments. Randomized RNAs associated with recombinant SBP-tagged eIF4A1 in vitro were isolated and subjected to deep sequencing. b Comparison of the enrichment of 4-mer motifs with DMSO and those with DMDA-PatA. AMP-PNP and 15 pmol of recombinant eIF4A1 were included in the reaction. Motifs containing GNG are highlighted. c Box plots for motif enrichment in RNA Bind-n-Seq (with AMP-PNP) on eIF4A1 with DMSO or DMDA-PatA in the indicated 4-mer species. d Motif enrichment Z score along the titrated recombinant eIF4A1 in RNA Bind-n-Seq (with AMP-PNP) for the indicated 4-mer species with or without DMDA-PatA. The median (line) and upper/lower quartiles (shade) are shown. e, h Fluorescence polarization assay for FAM-labeled RNAs along the titrated recombinant eIF4A1 (wild type or Phe163Leu-Ile199Met mutant) with ADP and Pi. The indicated RNA sequences at 10 nM were used with or without 50 μM DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). f Rank plot for 4-mer motifs enriched in RNA Bind-n-Seq (with ADP and Pi) on eIF4A1 in the presence of DMDA-PatA. Motifs containing GNG are highlighted. g Box plots for motif enrichment in RNA Bind-n-Seq (with ADP and Pi) on eIF4A1 with DMDA-PatA in the indicated 4-mer species. In the box plots, the medians (centerlines), upper/lower quartiles (box limits), and 1.5× interquartile ranges (whiskers) are shown. The significance was calculated by the Mann‒Whitney U test (two-tailed) for all motifs (n = 256), GNG motifs (n = 31), GAG motifs (n = 8), GUG motifs (n = 8), GGG motifs (n = 8), and GCG motifs (n = 8) (c, g). Source data are provided as a Source Data file.
To further monitor the affinity landscape of RNA-eIF4A1 binding, we titrated the amount of recombinant eIF4A1 for RNA Bind-n-Seq. With DMDA-PatA, the frequency of the GNG motif increased at the medium amount (15 pmol) of eIF4A1, whereas the frequency of the GNG motif decreased at higher amounts of the protein (Fig. 2d top). This peak in motif interaction could be expected since at high protein concentrations, the interaction with the strong-affinity motif becomes saturated, and competition is initiated with lower-affinity motifs44. In contrast, the interaction with irrelevant polypyrimidine sequences ([U/C]4) was enhanced with a higher amount of eIF4A1 (45 pmol) by DMDA-PatA (Fig. 2d bottom), indicating that this motif was a less preferred sequence for DMDA-PatA. A conventional fluorescence polarization assay with fluorescein (FAM)-conjugated RNAs confirmed above observations; GNG-possessing (AG)10 RNA associated more tightly with eIF4A1 upon DMDA-PatA treatment than the control (UC)10 RNA did (Supplementary Fig. 2c and Table 1). Notably, DMDA-PatA enhanced the interaction of both RNAs with eIF4A1, as reported in an earlier study32.
Table 1.
Kd (μM) values between proteins and RNAs obtained in this study
| eIF4A1 | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | 0.30 ± 0.061 | 26 ± 17 | 2.0 ± 0.50 | 0.41 ± 0.053 | 12 ± 1.2 | 1.5 ± 0.34 |
| (UC)10 | ND | ND | 28 ± 5.1 | 2.8 ± 0.24 | |||
| eIF4A1 Phe163Leu-Ile199Met | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | ND | |||||
| eIF4A2 | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | 1.6 ± 0.17 | |||||
| DDX3X (helicase core) | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | 0.80 ± 0.12 | 29 ± 61 | 0.71 ± 0.087 | |||
| (UC)10 | ND | ND | ND | ND | |||
| DDX3X Gln360Leu (helicase core) | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | 5.1 ± 2.3 | |||||
| DDX3X Gln360Pro (helicase core) | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | ND | |||||
| DDX6 (helicase core) | |||||||
|---|---|---|---|---|---|---|---|
| ADP + Pi | AMP-PNP | ATP | |||||
| RNA | DMSO | DMDA-PatA | iPr-DMDA-PatA | DMSO | DMDA-PatA | DMSO | DMDA-PatA |
| (AG)10 | ND | ND | 1.8 ± 0.72 | 2.6 ± 0.71 | |||
The fluorescence polarization of FAM-labeled RNAs was determined and fitted to the Hill equation to determine the Kd. ND, not determined.
We found that the GNG-selective clamping of eIF4A1 could occur in an ATP-independent manner. In the presence of ADP and Pi, eIF4A1 per se could not bind to (AG)10 or (UC)10 RNA (Fig. 2e and Table 1), as reported previously16. However, DMDA-PatA enabled the association with (AG)10 (Fig. 2e and Table 1). This ATP-independent clamping did not occur on (UC)10 (Fig. 2e and Table 1). To comprehensively survey ATP-independent RNA selectivity, we again performed RNA Bind-n-Seq with ADP and Pi. As observed in the experiments with AMP-PNP, we detected a strong enrichment of GNG motifs (Fig. 2f, g). This enhanced interaction was maintained in the presence of ATP, which reflects cellular conditions, in the fluorescence polarization assay (Supplementary Fig. 2d and Table 1).
This ATP-independent sequence-selective clamping followed the reported biochemical modes of this compound. We observed that mutations in the binding interface of compound32 (Phe163Leu-Ile199Met substitutions18) attenuated the ATP-independent association of eIF4A1 with (AG)10 RNA in the presence of ADP and Pi (Fig. 2h and Table 1). A PatA derivative was also known to target eIF4A2, a minor paralog of eIF4A32. We found that ATP-independent GNG-selective clamping of this paralog also occurred (Supplementary Fig. 2e and Table 1).
Thus, we conclude that DMDA-PatA provides GNG motif selectivity on eIF4A, evading the need for ATP.
eIF4A clamping on the GNG motif sterically impedes scanning
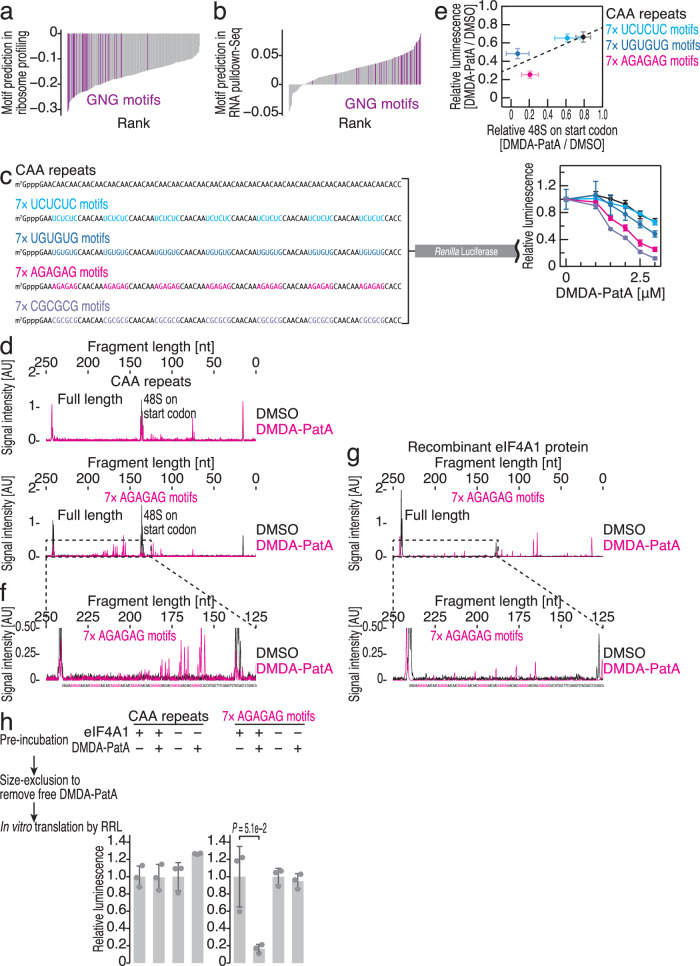
We then investigated whether GNG motif-selective eIF4A clamping by DMDA-PatA could result in mRNA selectivity for translation repression in cells. Here, we calculated the correlation between motif numbers in the 5′ UTR and DMDA-PatA-mediated translational repression via ribosome profiling. Through a survey of all possible 4-mer motifs, we found that more GNG motifs led to stronger repression by DMDA-PatA (Fig. 3a). We noted that in this analysis, all the motifs exhibited a negative correlation in general, probably due to the dependency of DMDA-PatA-mediated translational repression on 5′ UTR length (Supplementary Fig. 3a). Similarly, the number of GNG motifs was associated with tight mRNA interactions with eIF4A1 under DMDA-PatA treatment in RNA pulldown-Seq (Fig. 3b). Thus, the presence of GNG motifs explains the selective repression of translation by DMDA-PatA in cells.
Fig. 3. GNG motif-selective clamping of eIF4A causes the repression of translation initiation.
a Rank plot for motif prediction by ribosome profiling under 0.1 μM DMDA-PatA treatment. Spearman’s correlation coefficients (ρ, two-tailed) between the number of 4-mer motifs found in the 5′ UTRs and translation changes of the mRNAs were calculated. Motifs containing GNG are highlighted. b Rank plot for motif prediction by RNA pulldown-Seq under 0.01 μM DMDA-PatA treatment. Spearman’s correlation coefficients (ρ, two-tailed) between the number of 4-mer motifs found in 5′ UTRs and mRNA changes on SBP-tagged eIF4A1 were calculated. Motifs containing GNG are highlighted. c Schematic of reporter mRNAs with 7× NGNGNG motifs and the control CAA repeats (left). These mRNAs were subjected to in vitro translation with RRL and titration with DMDA-PatA (right). The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). d, f Toeprinting assay to probe the 48S ribosomes assembled on the start codons in the indicated reporter mRNAs with or without 10 μM DMDA-PatA. cDNA synthesized with FAM-labeled reverse transcription primers was analyzed by capillary electrophoresis. A magnified view of the results for reporter mRNA with 7× AGAGAG motifs (the area defined by the dashed line in d) is shown in f. AU, arbitrary unit. e Relationships between translational repression observed during in vitro translation (at 3 μM DMDA-PatA) (c) and the reduction in 48S formation (Supplementary Fig. 3g) for the indicated reporter mRNAs. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). The regression line (dashed line) is shown. g Toeprinting assay with the recombinant eIF4A1 protein on the reporter mRNA with 7× AGAGAG motifs with or without 10 μM DMDA-PatA. cDNA synthesized with FAM-labeled reverse transcription primers was analyzed by capillary electrophoresis. A magnified view of the results is shown at the bottom. h In vitro translation of reporter mRNAs (with 7× AGAGAG motifs or CAA repeats, both at 90.9 nM) preincubated with recombinant eIF4A1 and DMDA-PatA. A size exclusion column was used to eliminate free DMDA-PatA. The data are presented as the mean (bar) and s.d. (error) for replicates (point, n = 3). The significance was calculated by Student’s t test (two-tailed). Source data are provided as a Source Data file.
We recapitulated GNG motif-selective repression with an in vitro translation system using rabbit reticulocyte lysate (RRL). We prepared reporter mRNAs bearing unstructured CAA repeats48 in the 5′ UTR as a control (Fig. 3c). The substitution of a part of the CAA repeats with GNG motifs (7× UGUGUG, 7× AGAGAG, and 7× CGCGCG) (Fig. 3c, left) strongly sensitized the reporter translation to DMDA-PatA (Fig. 3c, right, and Supplementary Fig. 3b). In contrast, the non-GNG motif 7× UCUCUC (Fig. 3c, left) did not affect DMDA-PatA sensitivity (Fig. 3c and Supplementary Fig. 3b). Unmodified PatA also showed essentially the same trends in motif dependency for translational repression (Supplementary Fig. 3c). The presence of many G nucleotides in RNA may increase the likelihood of forming G-quadruplexes, which may enhance the inhibitory effects of eIF4A deficiency49–51. However, we did not observe a significant difference between the G-quadruplex-forming reporter and the sequence-randomized control in DMDA-PatA-mediated translational repression16,49 (Supplementary Fig. 3d), indicating that the target selectivity of this compound is independent of the G-quadruplex structure.
We observed that translational repression depended on the number of motifs (Supplementary Fig. 3e), as a single AGAGAG motif conferred weaker repression than 7 motifs did (Supplementary Fig. 3e). We found limited positional effects of a single motif along the 5′ UTR (Supplementary Fig. 3e).
Using this setup, we further investigated the mechanism of DMDA-PatA-mediated translational repression. Here, we employed a toeprinting assay, which harnesses primer extension by reverse transcriptase and its blocking due to stable 48S formation on the start codon in an in vitro translation system16,48,52,53. cDNAs extended with FAM-labeled reverse transcription primers were analyzed by capillary electrophoresis (Fig. 3d and Supplementary Fig. 3f, g). This assay showed that DMDA-PatA suppresses 48S formation on AUG for GNG motif-containing reporters but not for the control CAA repeats (Fig. 3d and Supplementary Fig. 3f, g). Overall, we found a correlation between translation repression efficiency and the blocking of 48S formation among the reporters (Fig. 3e). These data indicated that DMDA-PatA inhibits a process in translation initiation upstream of 48S formation on the start codon.
In our toeprinting assay on GNG motif-containing reporters, we detected additional cDNA fragments immediately downstream of GNG motifs upon DMDA-PatA treatment (Fig. 3f), suggesting that the stable association of eIF4A in the lysate on these regions becomes a roadblock to reverse transcriptase. Indeed, recombinant eIF4A1 produced cDNA truncated downstream of GNG motifs, similar to that found in the lysate (Fig. 3g).
This led us to test whether DMDA-PatA-mediated clamping on the 5′ UTR directly causes translational repression. For this purpose, the mRNA reporter, preincubated with DMDA-PatA and recombinant eIF4A1, was subjected to in vitro translation with RRL16,18,54. Purification through a gel-filtration column ensured the removal of free DMDA-PatA from the reaction. Nevertheless, protein synthesis from the reporter possessing GNG motifs was attenuated (Fig. 3h and Supplementary Fig. 3h). This experiment indicated that the clamped eIF4A by DMDA-PatA suppressed protein synthesis from the mRNA.
Taken together, these results revealed that RNA-selective eIF4A binding caused by DMDA-PatA blocks translation, most likely by sterically hindering ribosome scanning.
The tertiary amine on the trienyl arm confers GNG motif preference
Recent structural determination of the complex of desmethyl PatA (DM-PatA) (Supplementary Fig. 1a), eIF4A1, and polypurine RNA suggested that the compound does not have a clear interaction with RNA to discriminate bases32. Thus, we wondered how PatA derivatives could exhibit sequence selectivity. To address this point, we performed classical MD simulations and ab initio FMO calculations55–58 based on the reported structure of human eIF4A1•DM-PatA•polypurine RNA32. For consistency with our experiments, we replaced DM-PatA with DMDA-PatA for the MD + FMO analysis59. We note that the role of the MD simulations here is to sample the structural fluctuations in a narrow space around the crystal structure for downstream FMO calculations. We found that three independent MD simulations showed a similar range of structural fluctuations in DMDA-PatA and eIF4A1 (Supplementary Fig. 4a, b).
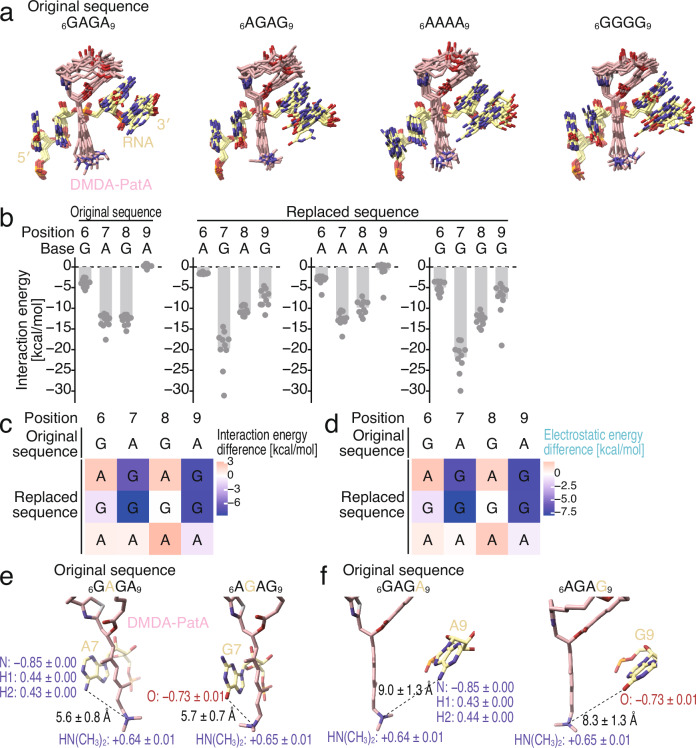
To investigate the origin of the G preference, we replaced the RNA motif (6GAGA9) surrounding DMDA-PatA with 6AGAG9, 6AAAA9, or 6GGGG9 (Fig. 4a) and calculated inter-fragment interaction energy (IFIE) and its pair interaction energy decomposition analysis (PIEDA)60,61 between the nucleotides and DMDA-PatA (Supplementary Fig. 4c). Among the 4 sequences analyzed, G nucleotides at positions 7 and 9 enabled more stable interactions with DMDA-PatA (Fig. 4b, c). This stabilized interaction primarily stemmed from electrostatic energy (Fig. 4d and Supplementary Fig. 4c). These results provided a quantitative energetic explanation for the GNG motif preference conferred on eIF4A by DMDA-PatA.
Fig. 4. MD simulations and FMO calculations elucidated the energetic impact of RNA sequences on the association with DMDA-PatA.
a Overlaid simulated structures (n = 10) of polypurine RNA•DMDA-PatA•eIF4A1 complexes analyzed by MD simulation. The complexes with RNAs possessing the indicated substitutions were investigated. b The interaction energy between DMDA-PatA and the indicated bases along the investigated complexes through FMO calculations. The data are presented as the mean (bar) for 10 simulated complexes (point). c The difference in the interaction energy between DMDA-PatA and the indicated bases from the original sequence (6GAGA9). See b for the source data. The color scale is shown. d The difference in the electrostatic interaction energy between DMDA-PatA and the indicated bases from the original sequence (6GAGA9). See Supplementary Fig. 4c for the source data. The color scale is shown. e, f Representative structures obtained by MD simulations (at 100 ns) with 6GAGA9 (original sequence) or 6AGAG9. The net charge (e) on the indicated groups and the distances are shown (mean ± s.d. for 10 simulated structures). Source data are provided as a Source Data file.
We found that the tertiary amine on the trienyl arm of DMDA-PatA plays a key role in the electrostatic interaction with the G nucleotides at positions 7 and 9. The MD + FMO analysis revealed that the tertiary amine was positively charged regardless of the RNA sequence (Fig. 4e and Supplementary Fig. 4d). This led to an attractive interaction with the negatively charged O6 of the G nucleotide at position 7 (Fig. 4e and Supplementary Fig. 4d). On the other hand, the positively charged hydrogens on the amine at the same position in the A nucleotide repelled the tertiary amine of DMDA-PatA (Fig. 4e and Supplementary Fig. 4d). Essentially, a similar theoretical explanation could be applied to the G and A nucleotides at position 9 (Fig. 4f and Supplementary Fig. 4e). Notably, we found that the distances between the tertiary amine of DMDA-PatA and the corresponding N6/O6 of the A and G nucleotides were highly reproducible in three independent MD simulations (Supplementary Fig. 4f–i).
These analyses provide an understanding of the G preference by DMDA-PatA on eIF4A1 at the atomic level.
The tertiary amine of DMDA-PatA leads to sequence-selective translation repression
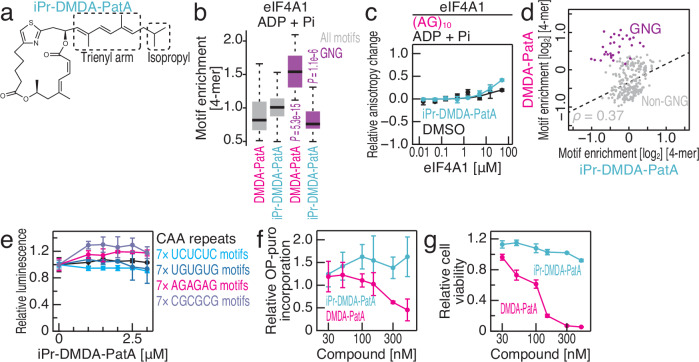
Given the above observation, we directly tested whether a positively charged tertiary amine on the trienyl arm contributes to the G-rich motif preference. For this purpose, we prepared a PatA derivative in which the tertiary amine was replaced with an isopropyl group that is similar in size but not basic (Fig. 5a and Supplementary Fig. 5a, isopropyl-terminated DMDA-PatA or iPr-DMDA-PatA). Strikingly, RNA Bind-n-Seq (in the presence of ADP) with iPr-DMDA-PatA revealed that this compound no longer had a GNG motif preference (Fig. 5b). The fluorescence polarization assay also supported this conclusion (Fig. 5c, compared to Fig. 2e, and Table 1). In contrast, other motifs were similarly recovered for both compounds, suggesting that iPr-DMDA-PatA lost selectivity toward only GNG motifs (Fig. 5d). Due to the loss of sequence-selective eIF4A clamping, iPr-DMDA-PatA could not repress translation of reporter mRNAs with GNG motifs in vitro (Fig. 5e, compared to Fig. 3c). Moreover, global translation repression in cells and associated cell growth retardation were weaker with iPr-DMDA-PatA compared to DMDA-PatA (Fig. 5f, g). This reduced potency of iPr-DMDA-PatA could not be attributed to its lower membrane penetration. Via assessing compound penetration through the membrane by measuring the absorbance at 280 nm (Supplementary Fig. 5b) and evaluating signal linearity (Supplementary Fig. 5c), we found that iPr-DMDA-PatA had better cell permeabililty than the original DMDA-PatA (Supplementary Fig. 5d). We concluded that sequence-selective translational repression and the subsequent cytotoxicity are caused by the tertiary amine on the trienyl arm of DMDA-PatA.
Fig. 5. The tertiary amine on the trienyl arm of DMDA-PatA confers GNG selectivity on eIF4A1.
a Chemical structure of iPr-DMDA-PatA. b Box plots for motif enrichment in RNA Bind-n-Seq (with ADP and Pi) on eIF4A1 with DMDA-PatA or iPr-DMDA-PatA in the indicated 4-mer species. c Fluorescence polarization assay for FAM-labeled RNAs along the titrated recombinant eIF4A1 with ADP and Pi. The indicated RNA sequences at 10 nM were used with or without 50 μM iPr-DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). d Comparison of the enrichment of 4-mer motifs with DMDA-PatA and those with iPr-DMDA-PatA. ADP and Pi were included in the reaction. Motifs containing GNG are highlighted. The regression line (dashed line) for non-GNG motifs (gray points) is shown. ρ, Spearman’s correlation coefficient (two-tailed). e The mRNAs shown in Fig. 3c were subjected to in vitro translation with RRL with the titration of iPr-DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). f The relative amount of OP-Puro incorporation into the nascent peptide in HEK293 cells was analyzed with the titrated DMDA-PatA or iPr-DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). g The relative cell viability was analyzed with titrated DMDA-PatA and iPr-DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). In the box plots, the medians (centerlines), upper/lower quartiles (box limits), and 1.5× interquartile ranges (whiskers) are shown. The significance was calculated by the Mann‒Whitney U test (two-tailed) for all motifs (n = 256), GNG motifs (n = 31), GAG motifs (n = 8), GUG motifs (n = 8), GGG motifs (n = 8), and GCG motifs (n = 8) (b). Source data are provided as a Source Data file.
Sequence selectivity differences between RocA and DMDA-PatA
Our results illuminated the similarity in translation repression mode between RocA and DMDA-PatA; both compounds clamp eIF4A on a subset of RNA motifs, sterically hindering ribosome scanning16–19,37. Although the impacts of these compounds on translation were similar (Fig. 1b, c), we noticed a substantial difference in their effects.
To profile the motif selectivity of the compounds, we conducted RNA Bind-n-Seq experiments for RocA with titrated eIF4A1 recombinant protein in the presence of AMP-PNP. As reported previously16–19,37, RocA enriched polypurine ([A/G]4) sequences on eIF4A1 throughout the protein contents we tested (Supplementary Fig. 6a), rather than a peak in interaction at a specific protein amount. This suggested that competition for less preferred motifs is limited in RocA. Considering the 3-mer motifs defined by polypurine and GNG motifs, we expected that GYG (where Y represents U or C) motifs, which are favorable for DMDA-PatA, would not be selected by RocA (Supplementary Fig. 6b). Consistent with this prediction, RNA Bind-n-Seq with RocA showed weak enrichment in GYG motifs (Supplementary Fig. 6c). These data highlighted the similarities and differences in motif selection between the two compounds.
In addition to its effects on translation initiation16–19,37, RocA induces elongation block by clamping eIF4A within the open reading frames (ORFs)62, as represented by ribosome footprint accumulation upstream of polypurine motifs (Supplementary Fig. 6d). In contrast, DMDA-PatA did not cause ribosome stalling in the vicinity of GNG motifs (Supplementary Fig. 6e), suggesting that the clamped eIF4A by DMDA-PatA does not serve as a strong obstacle for elongating ribosomes.
DMDA-PatA also targets DDX3X for selective mRNA clamping and translation repression
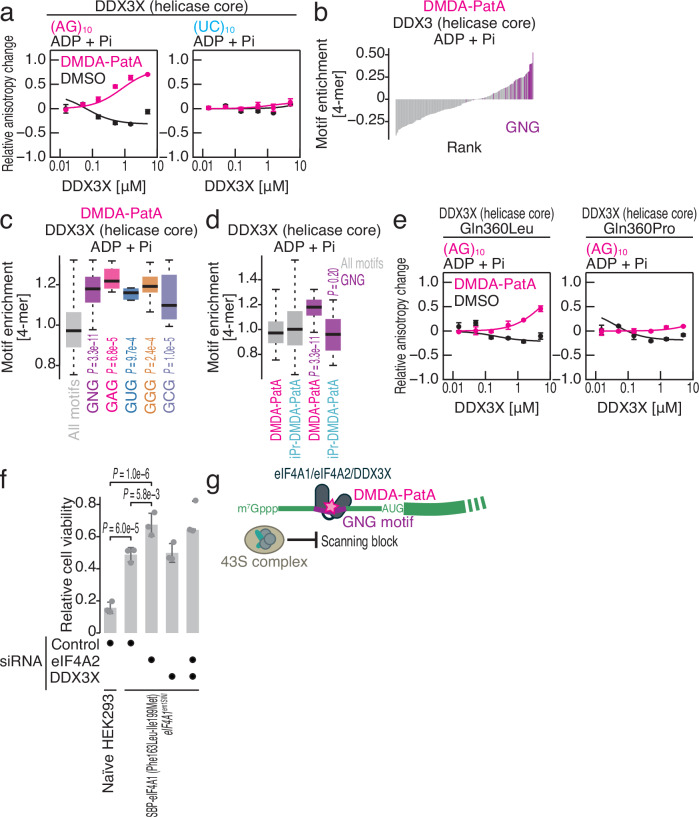
Considering that RocA has been shown to target DDX3X37, we investigated the potential of DMDA-PatA to target DDX3X. We conducted a fluorescence polarization assay with recombinant DDX3X (helicase core) and observed that DMDA-PatA clamped onto GNG motif-containing (AG)10 RNA but not control (UC)10, irrespective of the presence or absence of ATP (Fig. 6a, Supplementary Fig. 7a, and Table 1). More comprehensively, we conducted RNA Bind-n-Seq for recombinant DDX3X with ADP and found that DMDA-PatA allowed DDX3X to bind to GNG motifs (Fig. 6b, c). Again, RocA and DMDA-PatA showed distinct sequence preferences for DDX3X (Supplementary Fig. 7b).
Fig. 6. DMDA-PatA-mediated DDX3X clamping on the GNG motif occurs in an ATP-independent manner.
a, e Fluorescence polarization assay for FAM-labeled RNAs along the titrated recombinant DDX3X (helicase core) (wild type, Gln360Leu mutant, or Gln360Pro mutant) with ADP and Pi. The indicated RNA sequences at 10 nM were used with or without 50 μM DMDA-PatA. The data are presented as the mean (point) and s.d. (error) for replicates (n = 3). b Rank plot for 4-mer motifs enriched in RNA Bind-n-Seq (with ADP and Pi) on DDX3X (helicase core) in the presence of DMDA-PatA. Motifs containing GNG are highlighted. c Box plots for motif enrichment in RNA Bind-n-Seq (with ADP and Pi) on DDX3X (helicase core) with DMDA-PatA in the indicated 4-mer species. d Box plots for motif enrichment in RNA Bind-n-Seq (with ADP and Pi) on DDX3X (helicase core) with DMDA-PatA or iPr-DMDA-PatA in the indicated 4-mer species. f Relative cell viability under the indicated conditions after treatment with 0.1 μM DMDA-PatA for 48 h. The data are presented as the mean (bar) and s.d. (error) of replicates (point, n = 3). g Schematic of DMDA-PatA-mediated repression of mRNA-selective translation. Clamping of eIF4A1/2 and/or DDX3X to the GNG motif in the 5′ UTR provides steric hindrance for ribosome scanning. In the box plots, the medians (centerlines), upper/lower quartiles (box limits), and 1.5× interquartile ranges (whiskers) are shown. The significance was calculated by the Mann‒Whitney U test (two-tailed) for all motifs (n = 256), GNG motifs (n = 31), GAG motifs (n = 8), GUG motifs (n = 8), GGG motifs (n = 8), and GCG motifs (n = 8) (c, d) and by the Tukey‒Kramer test (two-tailed) (f). Source data are provided as a Source Data file.
This led us to test whether the RNA selectivity was also provided by the tertiary amine on the trienyl arm of DMDA-PatA. Indeed, RNA DDX3X Bind-n-Seq confirmed that iPr-DMDA-PatA lost the ability to confer GNG motif preference on the protein (Fig. 6d), suggesting a base preference mechanism similar to that of eIF4A1-bound DMDA-PatA (Fig. 4).
Considering that RocA depends on residues on DDX3X that are slightly different from those on eIF4A1 for binding37, we investigated the role of these residues on DDX3X in DMDA-PatA targeting. As in the case of RocA37, we confirmed the importance of Gln360 for DMDA-PatA-mediated ATP-independent clamping (Fig. 6e and Table 1).
To ensure the specificity for DEAD-box proteins, we repeated the same fluorescence polarization assay with the helicase core of DDX6, which regulates translation and RNA turnover47. DMDA-PatA did not impact the interaction between DDX6 and polypurine RNAs, irrespective of the ATP analog (Supplementary Fig. 7c–e and Table 1), showing that DMDA-PatA is a not a compound for universal DEAD-box proteins.
Finally, we tested the contributions of eIF4A1, eIF4A2, and DDX3X to DMDA-PatA-mediated cytotoxicity. Mutations in the RocA binding pocket (Phe163Leu-Ile199Met)18 desensitized cells to DMDA-PatA in terms of cell viability (Fig. 6f), consistent with its biochemical features (Fig. 2h)32. Moreover, knockdown of eIF4A2 further restored cell viability upon DMDA-PatA treatment (Fig. 6f). In contrast, additional knockdown of DDX3X did not affect cytotoxicity, at least in the HEK293 cell lines that we used (Fig. 6f). Given that DDX3X is overexpressed in a subset of cancer cells, the targeting of DMDA-PatA to this protein may be more significant in other cell types and should be considered for therapeutic purposes.
Overall, we concluded that DMDA-PatA clamps eIF4A1, eIF4A2, and DDX3X on GNG RNA motifs on the 5′ UTR in an ATP-independent manner and sterically hinders ribosome scanning for mRNA-selective translation repression (Fig. 6g).
Discussion
Starting with a comparative study of eIF4A inhibitors, we found that PatA derivatives possessing a tertiary amine on the triene could elicit GNG motif preference by eIF4A1/2 and DDX3X DEAD-box RNA-binding proteins and inhibit protein synthesis from a subset of mRNAs. Our results provide an example of an RNA-selective small molecule that may unlock undruggable targets63,64 but has been identified only in limited cases.
The importance of the tertiary amine on the trienyl arm has been revealed by structure-activity relationship (SAR) analysis of PatA derivatives27. However, the crystal structure of DM-PatA•human eIF4A1•polypurine RNA could not explain the significant contribution of the terminal amine32. Our work accounts for this role in RNA selectivity toward GNG motifs in eIF4A1/eIF4A2/DDX3X clamping, subsequently causing mRNA-selective translation repression and ultimately cytotoxicity. Importantly, our work showed that PatA offers a unique set of small molecules that can be used for RNA motif selection. This work will pave the way for the development of PatA derivatives with improved anticancer22,27,29, anticachexia26, and antiviral therapeutics65–68.
Recent structural analysis has shown that the binding pocket of a PatA analog on eIF4A1 largely overlaps with that of rocaglates32. Although the modes that provide sequence selectivity are different, the sharply bent structure of the RNA, which is a characteristic conformation of eIF4A1-bound RNA, at the compound binding interface18,32 provides a unique context for the sequence-selective clamping of both compounds. Given that rocaglates ultimately reduce the availability of eIF4A or eIF4F (the trimetric complex of eIF4A, eIF4E, and eIF4G) for translation initiation by sequestering them on mRNAs19, a similar bystander effect can be evoked by PatA. Indeed, PatA-mediated widespread clamping of eIF4A on cellular mRNAs has been reported23. Our study exemplifies that two distinct molecules (Fig. 1a) of different origins (microbiome symbionts on marine sponges vs. plants) can converge into the same target, causing similar but not identical mRNA-selective modulation of translation initiation. Given that both RocA13,15,69–79 and PatA27,74,80 show antitumor effects, comparisons of these compounds in their efficacy, cancer-type specificity, and differences in adverse effects on nontumor cells will be an important subject of study.
Limitations
Although long-term incubation with rocaglates in cells has been shown to remodel the translation machinery81, whether similar effects could be induced by PatA was not addressed in this study. Future proteomics studies will address this issue and provide a more comprehensive view of the effects of PatA on cells.
Methods
Compounds
RocA was purchased from Sigma‒Aldrich. Hipp and San were shared by Dr. Junichi Tanaka and Dr. Jun Liu, respectively. DMDA-PatA and Pat-A were synthesized in earlier studies40,82,83. iPr-DMDA-PatA was synthesized as described below. These compounds were dissolved in dimethyl sulfoxide (DMSO).
Chemical synthesis of iPr-DMDA-PatA
(Z)−4,4,5,5-Tetramethyl-2-(5-methylhex-2-en-2-yl)−1,3,2-dioxaborolane: The procedure was adapted from earlier work84. [(ICy)CuCl] (ICy = N,N-dicyclohexylimidazolyl; 87.3 mg, 0.26 mmol)85, NaOt-Bu (42.1 mg, 0.44 mmol) and B2pin2 (2.45 g, 9.63 mmol) were successively added to a solution of 5-methylhexan-2-one (1.00 g, 8.76 mmol), and the resulting mixture was stirred for 24 h at 70 °C (bath temperature). pTsOH•H2O (3.33 g, 17.52 mmol) was then added, and stirring was continued for another 24 h at 65 °C. After reaching ambient temperature, the suspension was filtered through a pad of Celite, which was carefully rinsed with CH2Cl2 in several portions. The combined filtrates were concentrated under reduced pressure, and the crude material was purified by flash chromatography (hexane/tert-butyl methyl ether, 15:1) to give the title compound as a colorless oil (0.57 g, 29%). The analytical and spectroscopic data matched those in the literature86,87.
iPr-DMDA-PatA: Pd(dppf)Cl2 (0.12 mg, 0.2 µmol) and Cs2CO3 (1.6 mg, 4.9 µmol) were successively added to a degassed solution of alkenyl iodide (1.8 mg, 3.2 µmol)83,88 and alkenylpinacolboronate (0.73 mg, 3.2 µmol) in dimethylformamide (DMF) (0.26 ml). The mixture was stirred overnight at ambient temperature, diluted with tert-butyl methyl ether (0.5 ml) and washed with water (3 × 0.5 ml). The aqueous phase was extracted with CHCl3 (2 × 0.5 ml), and the combined organic layers were dried over Na2SO4. The solvent was evaporated under reduced pressure, and the residue was purified by flash chromatography (hexane/EtOAC, 10:1, complemented with 1% Et3N) to give the title compound as a pale yellow solid (1.5 mg, 88%). [α]20D = – 30.0 (c = 0.06, CHCl3). 1H NMR (C6D6, 600 MHz): δ 7.48 (dm, J = 11.6, 1.3 Hz, 1H), 6.73 (dddd, J = 9.6, 9.2, 4.5 Hz, 1H), 6.47 (t, J = 11.6 Hz, 1H), 6.36 (d, J = 15.9 Hz, 1H), 6.22 (d, J = 15.9 Hz, 1H), 6.19 (d, J = 1.0 Hz, 1H), 5.56 (d, J = 11.6 Hz, 1H), 5.51 (d, J = 9.6 Hz, 2H), 5.15 (dqd, J = 10.9, 6.4, 1.7 Hz, 1H), 3.08 (m, 2H), 2.79 (dtd, J = 14.5, 5.7, 4.3, 1.0 Hz, 1H), 2.46 (ddd, J = 16.10, 10.5, 6.4 Hz, 1H), 2.36 (ddd, J = 14.5, 10.5, 4.0, 1H), 2.14 (m, 1H), 2.09 (dd, J = 13.3, 10.9, 1H), 2.06 (m, 1H), 1.97 (d, J = 1.2 Hz, 2H), 1.97 (m, 3H), 1.72 (s, 3H), 1.64 (d, J = 13.3 Hz, 1H), 1.58 (m, 1H), 1.55 (s, 3H), 1.54–1.48 (m, 3H), 0.96 (d, J = 6.4 Hz, 3H), 0.88 (d, J = 6.7 Hz, 3H), 0.87 (d, J = 6.7 Hz, 3H). 13C NMR (C6D6, 151 MHz): δ 172.5, 165.4, 164.9, 157.3, 145.6, 141.2, 138.6, 134.9, 134.6, 133.1, 130.2, 128.7, 124.7, 115.5, 113.1, 69.9, 66.8, 48.4, 39.1, 37.9, 35.0, 31.2, 29.3, 28.5, 23.6, 22.6, 22.6, 21.1, 16.7, 13.4, 12.7. 15N NMR (C6D6, 61 MHz; via 1H-15N HMBC): δ −56.2. IR (film, cm–1): 2952, 2926, 2851, 1732, 1633, 1597, 1523, 1458, 1427, 1379, 1363, 1339, 1265, 1202, 1159, 1122, 1050, 1025, 986, 959, 814, 431. HRMS (ESI) m/z calcd. for C31H43NO4S + Na [M+ + Na]: 548.2807; found 548.2805.
Library preparation for ribosome profiling
The libraries used in this study are summarized in Supplementary Data 1.
HEK293 Flp-In T-REx cells (Thermo Fisher Scientific, R78007) were treated as follows: DMDA-PatA (0.01 μM or 0.1 μM) for 30 min, San (1 μM or 20 μM) for 30 min, and Hipp (0.1 μM or 1 μM) for 15 min. For the control, cells were incubated with 0.1% DMSO for the same durations as the drug treatments.
Library preparation was conducted following a reported protocol89. Cell lysates containing 10 μg of total RNA were subjected to RNase I (LGC Biosearch Technologies) treatment for 45 min at 25 °C. Ribosomes were collected using sucrose cushion ultracentrifugation. Subsequently, RNA fragments ranging from 26 to 34 nucleotides (nt) for Hipp treatment and from 17 to 34 nt for other samples were selected on a 15% denatured gel (FUJIFILM Wako Pure Chemical Corporation), followed by dephosphorylation and linker ligation. rRNA removal was performed utilizing the Ribo-Zero Gold rRNA Removal Kit (Illumina). The linker-conjugated RNAs were reverse-transcribed, circularized, and PCR-amplified. Single-end, 50-nt sequencing was performed utilizing HiSeq 4000 (Illumina).
Analysis of the ribosome profiling data
The data were analyzed following an approach reported before90. Briefly, the linker sequences were trimmed using fastx_clipper (http://hannonlab.cshl.edu/fastx_toolkit/index.html), followed by alignment of the reads to noncoding RNAs, including rRNAs, tRNAs, snoRNAs, snRNAs, and miRNAs, with Bowtie2 (ver. 2.4.1)91. Unaligned reads were mapped to the hg38 human genome reference and the custom mitochondrial transcript reference using Bowtie2 (ver. 2.4.1). PCR duplicates were removed based on unique molecular identifiers (UMIs) on the linker sequences with a custom script (https://github.com/ingolia-lab/RiboSeq). The distance from the 5′ end to the ribosome A site on the sequenced reads was empirically defined as follows: 15 for 26-30 nt, 16 for 31 nt, and 17 for 32 nt. The read count on each CDS was obtained with a custom script (https://github.com/ingolia-lab/RiboSeq), excluding the first and last 5 codons from the analysis. Regarding mitochondrial footprints, the A-site offset was set to 14 for 26-27 nt, 15 for 28-32 nt, 16 for 33-34 nt, and 17 for 35 nt. RocA-treated ribosome profiling data (0.03, 0.3, or 3 μM for 30 min) were published in an earlier work16.
Changes in ribosome footprint counts were calculated with DESeq92. Subsequently, the data were renormalized to the average values of mitochondrial transcripts to calculate global translation changes.
DMDA-PatA high-sensitivity mRNAs were defined as transcripts showing a log2-fold change of less than −1 from the mean with a false discovery rate (FDR) of less than 0.01 under 0.1 μM DMDA-PatA treatment. Conversely, low-sensitivity mRNAs were characterized as transcripts showing a log2-fold change of more than 1 from the mean with a FDR of less than 0.01.
Principal component analysis was conducted with a built-in function in R. Spearman correlations between the translation changes and the 4-mer numbers in the 5′ UTR were calculated to predict the responsible motifs.
To calculate ribosome occupancy around the motifs, we followed an approach in the previous report62. We first screened mRNAs that had relatively low sensitivity to translation after drug treatment (without mitochondrial footprint normalization) using the threshold of mean + s.d. Then, we focused on the motif sites of these mRNAs, when 16 or more reads were found in the 101-nt region centered on the motif (i.e., 50 nt upstream and 50 nt downstream from the start of the motif). Reads assigned to each codon position were normalized according to the average number of reads on the codon on the transcript. After they were centered on the 4-mer motif, the normalized reads were averaged.
Library preparation for RNA pulldown-Seq
The libraries used in this study are summarized in Supplementary Data 1.
HEK293 Flp-In T-REx cells with an SBP-tagged eIF4A1 integrant16 were seeded in a 10-cm dish and cultured for 3 d in the presence of 1 μg/ml tetracycline. The cells were treated with 0.1% DMSO, 0.01 μM, or 0.1 μM DMDA-PatA for 30 min, washed with 5 ml of ice-cold PBS, and lysed with lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT) containing 1% Triton X-100 and 25 U/ml Turbo DNase (Thermo Fisher Scientific). The cell lysates were clarified by centrifugation at 20,000 × g for 10 min at 4 °C. The supernatants were incubated with 30 μl of Dynabeads M-270 Streptavidin (Invitrogen), preequilibrated with lysis buffer containing 1% Triton X-100, for 30 min at 4 °C. The beads were washed five times with lysis buffer containing 1% Triton X-100 and 1 M NaCl. The SBP-eIF4A1 and the bound RNAs were eluted from the beads with 40 μl of lysis buffer supplemented with 5 mM biotin for 30 min at 4 °C. All buffers used during the above process contained 0.1% DMSO, 0.01 μM, or 0.1 μM DMDA-PatA. RNA was extracted with TRIzol LS reagent (Thermo Fisher Scientific) and the Direct-zol RNA MicroPrep Kit (Zymo Research). Sequencing libraries were generated utilizing a TruSeq Stranded Total RNA kit (Illumina) and sequenced on a HiSeq 4000 (Illumina) with 50-nt single-end reads.
Analysis of the RNA pulldown-Seq data
The data were analyzed as previously reported16. After the linker sequences were removed using fastx_clipper, the reads were aligned to ncRNAs such as rRNAs, tRNAs, snoRNAs, snRNAs, and miRNAs using STAR (ver. 2.7.0a)93. Unaligned reads were mapped to the hg38 human genome reference using STAR (ver. 2.7.0a).
The read counts for each transcript were obtained with the same custom script as described in the ribosome profiling section. The read fold change was calculated with DESeq. Spearman correlations between the mRNA changes and the 4-mer numbers in the 5′ UTR were calculated to predict the responsible motifs.
DNA construction
For His-tagged protein expression, pColdI-eIF4A1 WT18, pColdI-eIF4A1 Phe163Leu-Ile199Met18, pColdI-DDX3X helicase core WT37, pColdI-DDX3X helicase core Gln360Leu37, and pColdI-DDX3X helicase core Gln360Pro37 were used. A DNA fragment encoding the human DDX6 helicase core (amino acids 95-469) was amplified from the cDNAs of HEK293 Flp-In T-REx cells and then inserted downstream of the His tag sequence of pColdI (TaKaRa).
For His- and SBP-tagged protein expression, pColdI-SBP-eIF4A137 and the pColdI-SBP-DDX3X helicase core37 were used.
For reporter mRNA preparation, psiCHECK2-7×AGAGAG motifs, CAA repeats, 1×AGAGAG left, 1×AGAGAG middle, 1×AGAGAG right, G-quadruplex, and randomized control for G-quadruplex16 (gifts from Dr. Nicholas T. Ingolia) were used.
For psiCHECK2-7×UCUCUC, 7×UGUGUG, and 7×CGCGCG, the synthesized DNA fragments listed below were amplified by PCR. The PCR products were inserted between the T7 promoter and the CDS of Renilla luciferase in psiCHECK2 (Promega):
psiCHECK2-7×UCUCUC, 5′-ATACGACTCACTATAGGGAATCTCTCCAACAATCTCTCCAACAATCTCTCCAACAATCTCTCCAACAATCTCTCCAACAATCTCTCCAACAATCTCTCCACCATGGCTTCCAAGGTG-3′;
psiCHECK2-7×UGUGUG, 5′-ATACGACTCACTATAGGGAATGTGTGCAACAATGTGTGCAACAATGTGTGCAACAATGTGTGCAACAATGTGTGCAACAATGTGTGCAACAATGTGTGCACCATGGCTTCCAAGGTG-3′; and
psiCHECK2-7×CGCGCG, 5′-ATACGACTCACTATAGGGAACGCGCGCAACAACGCGCGCAACAACGCGCGCAACAACGCGCGCAACAACGCGCGCAACAACGCGCGCAACAACGCGCGCACCATGGCTTCCAAGGTG-3′.
Recombinant protein purification
E. coli BL21 Star (DE3) cells (Thermo Fisher Scientific) transformed with the pColdI plasmids were cultured in 1 l of LB medium supplemented with ampicillin at 37 °C to an OD600 of 0.6. Then, the cells were chilled for 30 min at 4 °C, followed by overnight cultivation at 15 °C in the presence of 1 mM IPTG. The cells were harvested by centrifugation at 8000 × g for 2 min, flash-frozen in liquid nitrogen, and stored at −80 °C.
The pellet was suspended in bacterial lysis buffer (20 mM HEPES, 500 mM NaCl, 10 mM imidazole, 0.5% NP-40, and 10 mM β-mercaptoethanol, adjusted to pH 7.5 with NaOH) and subsequently sonicated on ice. The lysate was clarified by centrifugation at 10,000 × g for 20 min at 4 °C. The supernatant was then incubated with 3 ml of Ni-NTA Superflow agarose beads (QIAGEN), which were preequilibrated with bacterial lysis buffer, for 1 h at 4 °C in a sealed gravity column (Bio-Rad). The beads on the gravity column were washed with 50 ml of high-salt wash buffer (20 mM HEPES, 1 M NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol, adjusted to pH 7.5 by NaOH) and then with 50 ml of low-salt wash buffer (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol, adjusted to pH 7.5 by NaOH). The His-tagged protein was eluted with 8 ml of elution buffer (20 mM HEPES, 10 mM NaCl, 250 mM imidazole, 10% glycerol, and 10 mM β-mercaptoethanol, adjusted to pH 7.5 by NaOH).
The eluted protein was further purified using an NGC chromatography system (Bio-Rad). Specifically, the protein was loaded on a HiTrap 1 ml Heparin HP column (Cytiva) and fractionated through a gradient of increasing salt concentrations using a mixture of buffer A (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 10% glycerol, and 1 mM DTT) and buffer B (20 mM HEPES-NaOH pH 7.5, 1 M NaCl, 10% glycerol, and 1 mM DTT). The fractions containing the target protein were collected and buffer-exchanged to storage buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 10% glycerol, and 1 mM DTT) with a PD-10 column (Cytiva). The protein was concentrated using an Amicon Ultra4 10 kDa MWCO (Millipore) according to the manufacturer’s instructions. The recombinant protein was flash-frozen with liquid nitrogen and stored at −80 °C. The proteins were separated by SDS‒PAGE and stained with EzStainAQua (ATTO). Then, images were acquired by an ODYSSEY CLx (LI-COR Biosciences) with an infrared 700 nm channel.
Fluorescence polarization assay
The reaction mixtures (10 μl each) were prepared as follows: 0-50 μM recombinant eIF4A1 or 0-5 μM recombinant DDX3X, 10 nM FAM-labeled RNA (5′-FAM-AGAGAGAGAGAGAGAGAGAG-3′ and 5′-FAM-UCUCUCUCUCUCUCUCUCUC-3′, both Hokkaido System Science), 1% DMSO, 50 μM DMDA-PatA or 50 μM iPr-DMDA-PatA, 1 mM AMP-PNP, 1 mM MgCl2, 20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM DTT, and 5% glycerol. The mixtures were incubated at room temperature for 30 min and transferred to black 384-well microplates (Corning). Then, the anisotropy change was measured by an Infinite F-200 PRO (Tecan).
To test the ATP requirement, AMP-PNP was replaced with 1 mM ADP and 1 mM Na2HPO4.
The data were fitted to the Hill equation with Igor Pro 8 (Wavemetrix) to estimate Kd.
Library preparation for RNA Bind-n-Seq
The libraries prepared in this study are summarized in Supplementary Data 1.
For the experiment with AMP-PNP, 4.5, 15, 45, or 90 pmol of SBP-tagged eIF4A1 protein was incubated with 30 μl of Dynabeads M-270 Streptavidin (Thermo Fisher Scientific), which had been preequilibrated with equilibration buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT) containing 1% Triton X-100, for 30 min at 4 °C. The beads were washed 3 times with 60 μl of equilibration buffer containing 1% Triton X-100 and 1 M NaCl and then twice with 60 μl of equilibration buffer containing 0.1% Triton X-100. Subsequently, the beads were incubated with a 1 μM N30 oligonucleotide [5′-ctctttccctacacgacgctcttccgatct-N30-atcgtagatcggaagagcacacgtctgaa-3′ (Gene Design), where the lower cases represent the DNA sequence and N represents a random RNA sequence], for 30 min at 37 °C in 30 μl of equilibration buffer containing 0.1% Triton X-100, 0.33 U/μl SUPERase•In RNase Inhibitor (Thermo Fisher Scientific), 2 mM AMP-PNP, and 3 μM DMDA-PatA. Then, the beads were washed 3 times with equilibration buffer containing 0.1% Triton X-100, 2 mM AMP-PNP, and 3 μM DMDA-PatA. The protein-oligonucleotide complexes bound to the beads were eluted using 30 μl of equilibration buffer containing 0.1% Triton X-100, 5 mM biotin, 2 mM AMP-PNP, and 3 μM DMDA-PatA for 30 min at 4 °C. The oligonucleotides were purified with an Oligo Clean & Concentrator Kit (Zymo Research) and converted into a DNA library as described in the ribosome profiling section89. For the control experiment, DMDA-PatA was substituted with the same volume of DMSO (0.1% in the reaction). For the input experiment, the random oligonucleotides were directly converted to a DNA library. Experiments with RocA were also conducted as described above, but 3 μM DMDA-PatA was replaced with 3 μM RocA.
For the experiments with ADP and Pi, experiments were performed with 2 mM ADP and 2 mM Na2HPO4 instead of 2 mM AMP-PNP. For complex assembly, 50 μM N30 oligonucleotide and 90 pmol of SBP-tagged recombinant proteins (eIF4A1 or DDX3X) were used. iPr-DMDA-PatA was also used at 3 μM throughout the experiments.
DNA libraries were sequenced on HiSeq 4000 (Illumina) with 50-nt single-end read mode or HiSeq X Ten (Illumina) with 150-nt paired-end mode.
Analysis of the RNA Bind-n-Seq data
In the case of paired-end reads, fastp (ver. 0.21.0)94 was employed to correct read errors and read 1 was used for downstream analysis. fastx_clipper was utilized to eliminate the linker sequence, followed by fastx_collapser, which aggregated identical sequences into single sequences.
The frequency of all possible 4-mers was calculated, and the motif enrichment was expressed as the ratio to that in the input library.
RNA Bind-n-Seq data for RocA in the presence of ADP and Pi were obtained from an earlier study37.
Reporter mRNA preparation
The DNA fragments were PCR-amplified from psiCHECK2-based plasmids and used for in vitro transcription with a T7-Scribe Standard RNA IVT Kit (CELLSCRIPT). Then, capping and polyadenylation were performed using a ScriptCap m7G Capping System (CELLSCRIPT), a ScriptCap 2′-O-Methyltransferase (CELLSCRIPT), and an A-Plus Poly(A) Polymerase Tailing Kit (CELLSCRIPT).
In vitro translation
The reaction mixture (10 μl) was prepared with 5 μl of rabbit reticulocyte lysate nuclease-treated (Promega), 2 μl of H2O, 1 μl of DMDA-PatA, PatA, or iPr-DMDA-PatA dissolved in 1% DMSO, 1 μl of 500 nM mRNA reporter, and 1 μl of premix [100 μM amino acid mixture minus methionine (Promega), 100 μM amino acid mixture minus leucine (Promega), and 0.5 U/μl SUPERase•In RNase Inhibitor (Thermo Fisher Scientific)] and incubated for 1 h at 30 °C. After the translation reaction was quenched by adding 30 μl of 1× Passive Lysis Buffer (Promega), 10 μl of the mixture was placed on a 96-well white assay plate (Coster), and the fluorescence signal was measured by the Renilla-Glo Luciferase Assay System (Promega) and GloMax Navigator System (Promega).
To perform in vitro translation of the preformed mRNA reporter complexes, a reaction mixture (27.5 μl) containing 9.1 μM recombinant eIF4A1, 9.1 μM DMDA-PatA (dissolved in 2% DMSO), 90.9 nM reporter mRNA with 7× AGAGAG motifs or CAA repeats, 16.6 mM HEPES-NaOH at pH 7.5, 55.3 mM potassium acetate, 2.8 mM magnesium acetate, 1.8 mM ATP, and 552.7 μM DTT was incubated for 5 min at 30 °C. Subsequently, 2.5 μl of 285 nM magnesium acetate was added to the mixture. The reaction mixture was loaded into a MicroSpin G-25 column (Cytiva) that had been equilibrated with buffer containing 30 mM HEPES-NaOH at pH 7.5, 100 mM potassium acetate, 1 mM magnesium acetate, and 1 mM DTT and centrifuged at 700 × g for 1 min at 4 °C to eliminate free DMDA-PatA. The eluted fraction was mixed with 2.5 μl of storage buffer (20 mM HEPES-NaOH at pH 7.5, 150 mM NaCl, 10% glycerol, and 1 mM DTT). Then, 4 μl of the eluted solution was combined with 5 μl of RRL and 1 μl of premix and incubated for 1 h at 30 °C. In the case of experiments using mRNA reporters with 7× CGCGCG motifs, the concentrations of the mRNA reporters were adjusted to 181.8 nM.
In the control experiments, the recombinant eIF4A1 protein was replaced with storage buffer in the preformation reaction, and the recombinant eIF4A1 protein was added to the G-25 column flowthrough instead of the storage buffer. DMDA-PatA was substituted with 2% DMSO.
Toeprinting assay
A 10-μl reaction mixture consisting of 0.5× RRL, 2 mM GMPPNP, 2.5 mM magnesium acetate, and 10 μM DMDA-PatA (with 0.2% DMSO) was incubated for 5 min at 30 °C. Then, after the addition of 1 μl of 500 nM mRNA reporter, the mixture was further incubated for 5 min at 30 °C. Subsequently, the resulting mixture was combined with 9 μl of RT mix [22.2 mM Tris-HCl pH 7.5, 111.1 mM KCl, 3.2 mM magnesium acetate, 1.1 mM DTT, 1.1 mM each dNTP (New England Biolabs), 27.8 nM 5′ FAM-labeled reverse transcription primer (5′-FAM-ATGCAGAAAAATCACGGC-3′, Eurofins), and 22.2 U/μl ProtoScript II Reverse Transcriptase (New England Biolabs)] and incubated for 15 min at 30 °C. cDNAs were purified using a Direct-zol RNA MicroPrep kit with TRIzol LS reagent (Thermo Fisher Scientific). Subsequently, the cDNAs were subjected to a second purification with AMPure XP beads (Beckman Coulter). In the control experiments, the reaction contained 0.2% DMSO instead of DMDA-PatA. Instead of 0.5× RRL and 2 mM GMPPNP, 2.5 μM recombinant eIF4A1 protein and 2 mM ATP were also used.
The purified cDNAs were analyzed with a GeneScan 400HD ROX dye Size Standard (Thermo Fisher Scientific) on an Applied Biosystems 3130xl Genetic Analyzer (Thermo Fisher Scientific). The data were analyzed using Peak Scanner 2.
Dideoxy-termination sequencing was employed to calibrate the GeneScan 400HD ROX dye Size Standard for the length of cDNA synthesized in the present study. The reaction mixture containing 25 nM mRNA reporter, 12.5 nM FAM-labeled reverse transcription primer, 0.5 mM (each) dNTPs, 0.5 mM ddNTPs (ddATP, ddTTP, ddGTP, or ddCTP), ProtoScript II Reverse Transcriptase, and 1× ProtoScript II RT Reaction Buffer (New England Biolabs) was incubated for 1 h at 30 °C. cDNAs were purified with an Oligo Clean & Concentrator Kit and analyzed as described above.
MD and FMO calculations
The structure of the DM-PatA•eIF4A1•polypurine RNA complex was obtained from the Protein Data Bank (PDB) (6XKI)32. DMDA-PatA was created by removing amines from DM-PatA using the Molecular Operating Environment (MOE, https://www.chemcomp.com/Products.htm)95. Subsequently, hydrogen atoms not determined by X-ray crystallography were added using the “Protonate 3D” function in MOE, considering a protonation state at pH 7.0. Afterward, the atomic coordinates were optimized. Moreover, the DNA/RNA builder of the MOE was utilized to generate complexes in which the 6GAGA9 sequences in the RNA surrounding DMDA-PatA were substituted with 6AGAG9, 6AAAA9, and 6GGGG9. All MOE modeling was performed using the AMBER10:EHT force field96,97.
MD simulations were performed for the four complexes created for 100 ns. A heat process from 0 K to 310 K was performed for 50 ps using the NVT ensemble. Next, an equalization process was performed at 310 K for 50 ps (NPT ensemble). Furthermore, density relaxation was performed for 1 ns (NPT ensemble), and a production run was performed for 100 ns at 310 K (NPT ensemble). Note that the pressure at the NPT was 1013 hPa. The force fields used in this MD simulation were Amberff14SB98 for the protein, OL399 for RNA, and Gaff2100–102 for DMDA-PatA. The TIP3P water model was utilized as the solvent, and Na+ ions were used as the counterions. The bond distances involving hydrogen were not constrained. The time step was 1 fs. This MD simulation was conducted under periodic boundary conditions. Furthermore, the MD simulations in this study were executed using the AMBER16 program (https://ambermd.org/doc12/Amber16.pdf)103.
From the 100-ns trajectories obtained from the MD simulations, we extracted 10 structures at 3 ns intervals starting from 73 ns, resulting in a total of 40 structures. The geometry of each sampled structure was optimized by applying constraints on the heavy atoms. Then, FMO calculations55–58 were performed. The ABINIT-MP program57,58 was used for the FMO calculations; electron correlation effects were incorporated by second-order Møller–Plesset perturbation (MP2) theory104,105, which was efficiently implemented in ABINIT-MP. For the basis functions, we used 6-31 G*, a standard of FMO calculations. Subsequently, the average value and standard deviation of 40 structures of total IFIE with eIF4A and RNA for DMDA-PatA obtained using these FMO calculations were calculated for each pose. In addition, PIEDA divides the IFIE into four energy components: electrostatic (ES), exchange repulsion (EX), charge transfer (CT), and dispersion (DI), which allows the physicochemical properties of molecular interactions to be evaluated60,61.
Cell viability assay
In 24-well plates, 500 μl of 4 × 104 cells/ml HEK293 Flp-In T-REx cells (Thermo Fisher Scientific, R78007) or HEK293 Flp-In T-REx SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1em1SINI cells18 were seeded and incubated overnight. Transfection was performed with 55 nM control siRNA (Dharmacon, D-001810-10-20), DDX3X-specific siRNA (Dharmacon, L-006874-02-0005), and eIF4A2-specific siRNA (Dharmacon, L-013758-01-0005) using the TransIT-X2 Transfection Reagent System (Mirus). After 2 d of incubation, 200 μl of 2 × 104 cells/ml cells were seeded into individual wells of a 96-well plate and incubated for 6 h. siRNA knockdown was repeated once more, following the same protocol described above. After 24 h of incubation, the cells were subjected to treatment with 0.1 μM DMDA-PatA (with 0.1% DMSO) or 0.1% DMSO for 48 h. Cell viability was assessed utilizing the RealTime-Glo MT Cell Viability Assay System (Promega). Luminescence was measured using a GloMax Navigator System (Promega).
For the experiments without transfection, 4000 HEK293 Flp-In T-REx cells were seeded on a 96-well plate and treated with different concentrations of DMDA-PatA or iPr-DMDA-PatA for 24 h. Cell viability was assessed as described above.
Nascent peptide labeling with OP-Puro
On 24-well plates, 500 μl of 5 × 105 cells/ml HEK293 Flp-In T-REx cells were seeded and then incubated for 24 h. Subsequently, 50 μl of 0.22 mM O-propargyl-puromycin (OP-Puro, Jena Bioscience) dissolved in Opti-MEM (Thermo Fisher Scientific) was added to the medium with varying concentrations of DMDA-PatA or iPr-DMDA-PatA. The cells were incubated for 30 min, washed with PBS, and then lysed with OP-Puro lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 1% Triton X-100). The lysate was centrifuged at 20,000 × g for 10 min at 4 °C. The supernatant was subjected to a click reaction with IRdye800CW azide (LI-COR Biosciences) following the protocol provided in the Click-iT Cell Reaction Buffer Kit (Thermo Fisher Scientific). The reaction mixture was loaded onto a MicroSpin G-25 Column (Cytiva), which was equilibrated with OP-Puro lysis buffer containing 1 mM DTT and centrifuged at 700 × g for 2 min. After the proteins were separated via SDS‒PAGE, the infrared 800 nm (IR800) signal on the gel was detected using an Odyssey CLx (LI-COR Biosciences). Subsequently, the SDS‒PAGE gel was stained with Coomassie brilliant blue (CBB) (EzStainAQua, ATTO), and the total protein abundance was monitored by an Odyssey CLx with the IR700 channel. OP-Puro incorporation (IR800 signal) was normalized to the total protein abundance (IR700 signal).
Membrane permeability assay
A Parallel Artifical Membrane Permeability Assay Kit (gastrointestinal, BioAssay Systems) was used with some modifications. Five microliters of a dodecane solution containing 4% lecithin were added to the membranes of the donor wells. The donor wells were placed in acceptor wells containing 300 μl of PBS. Two hundred microliters of 50 μM DMDA-PatA or iPr-DMDA-PatA in PBS were loaded into the donor wells. The assemblies were incubated at 37 °C for 6, 12, or 18 h. After incubation, the absorbance of the solution in the donor well was measured from 220 to 750 nm with a DS-11 Spectrophotometer (DeNovix).
To construct calibration curves, DMDA-PatA solutions at concentrations of 20, 60, and 200 μM or iPr-DMDA-PatA solutions at concentrations of 6, 20, 60, and 200 μM were prepared in PBS. The absorbance at 280 nm was used to establish the calibration curve and calculate the concentration of the compound.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank all the members of the Iwasaki laboratory for constructive discussions and technical help. We are grateful to Dr. K. Dodo, Dr. K. Okuwaki, Dr. K. Kato, Dr. C. Watanabe, and Dr. T. Honma for their helpful advice. Hipp was a kind gift from Dr. J. Tanaka. San was a kind gift from Dr. J. Liu. We thank Dr. C.-X. Zhuo and S. Schulthoff for preparing the pateamine derivatives and C. Wirtz for excellent NMR support (and all at the MPI Mülheim). We are grateful to Dr. N.T. Ingolia for sharing the plasmids. This study used facilities of the HOKUSAI SailingShip supercomputer facility at RIKEN; Sanger sequencing at the Support Unit for Bio-Material Analysis, RIKEN CBS Research Resources Division; and deep sequencing via HiSeq 4000, supported by the National Institutes for Health (NIH) Instrumentation Grant (S10 OD018174), in QB3 Genomics, UC Berkeley, Berkeley, CA, (RRID:SCR_022170). MD simulations and FMO calculations were performed using the Fugaku supercomputer (project ID: hp220143) and the TSUBAME 3.0 supercomputer (Tokyo Institute of Technology, Japan). This work was supported by the Japan Society for the Promotion of Science (JSPS) (JP23H02415 and JP23H00095 to S.I.; JP23H05473 to M.Y.; JP23K05648 to Y.S.), the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (JP20H05784 and JP24H02307 to S.I.; JP21H05281 to T.I.; JP21H05734 and JP23H04268 to Y.S.; JP23H04882 to M.Y.), the Japan Agency for Medical Research and Development (AMED) (JP23gm1410001 to S.I. and T.I.), and RIKEN (Pioneering Projects “Biology of Intracellular Environments” to S.I., T.I., and Y.S.; Incentive Research Projects to T.S.-P.). This work was also supported by the Research Support Project for Life Science and Drug Discovery [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED (JP23ama121030). H.S. was a RIKEN Junior Research Associate. A part of the in silico study was conducted in an activity of FMO drug design consortium (FMODD) (to Y.H. and K.F.). Support from NIH NIGMS (R35 GM052964) to D.R. is gratefully acknowledged.
Author contributions
Conceptualization: H.S. and S.I.; Methodology: H.S., Y.H., M.C., T.S.-P., Y.S., M.T., and A.F.; Formal analysis: H.S., Y.H., M.C., Y.S., M.T., and A.F.; Investigation: H.S., H.S., Y.H., M.C., Y.S., M.T., and A.F.; Resources: D.R. and A.F.; Writing – Original Draft: S.I.; Writing – Review & Editing: H.S., Y.H., M.C., T.S.-P., Y.S., M.T., D.R., M.Y., A.F., T.I., K.F., and S.I.; Visualization: H.S. and S.I.; Supervision: Y.S., M.Y., A.F., T.I., K.F., and S.I.; Project administration: S.I.; and Funding Acquisition: T.S.-P., Y.S., D.R., M.Y., T.I., and S.I.
Peer review
Peer review information
Nature Communications thanks Lars Bock and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The results of ribosome profiling, RNA pulldown-Seq, and RNA Bind-n-Seq (GEO: GSE243312) obtained in this study have been deposited in the National Center for Biotechnology Information (NCBI) database. All the input and result files for the FMO calculations are available at the FMODB [https://drugdesign.riken.jp/FMODB/detail.php?FMODBID = “ID in the list”]106 (see Supplementary Table 1 for the ID list). Source data are provided with this paper.
Code availability
For the data analysis for ribosome profiling, RNA pulldown-Seq, and RNA Bind-n-Seq, we deposited key codes in Zenodo (10.5281/zenodo.11064746)107, which used reported custom script (https://github.com/ingolia-lab/RiboSeq). For the MD and FMO calculations, we used the abmptools on GitHub (https://github.com/kojioku/abmptools).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51635-9.
References
- 1.Valeur, E. & Jimonet, P. New modalities, technologies, and partnerships in probe and lead generation: enabling a mode-of-action centric paradigm. J. Med. Chem.61, 9004–9029 (2018). 10.1021/acs.jmedchem.8b00378 [DOI] [PubMed] [Google Scholar]
- 2.Shichino, Y. & Iwasaki, S. Compounds for selective translational inhibition. Curr. Opin. Chem. Biol.69, 102158 (2022). 10.1016/j.cbpa.2022.102158 [DOI] [PubMed] [Google Scholar]
- 3.Vázquez-Laslop, N. & Mankin, A. S. Context-specific action of ribosomal antibiotics. Annu. Rev. Microbiol.72, 185–207 (2018). 10.1146/annurev-micro-090817-062329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., International Natural Product Sciences Taskforce & Supuran, C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov.20, 200–216 (2021). 10.1038/s41573-020-00114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin, J., Zhou, D., Steitz, T. A., Polikanov, Y. S. & Gagnon, M. G. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem.87, 451–478 (2018). 10.1146/annurev-biochem-062917-011942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen, L. & Pelletier, J. Selective targeting of the DEAD-box RNA helicase eukaryotic initiation factor (eIF) 4A by natural products. Nat. Prod. Rep.37, 609–616 (2020). 10.1039/C9NP00052F [DOI] [PubMed] [Google Scholar]
- 7.Higa, T., Tanaka, J.-I., Tsukitani, Y. & Kikuchi, H. Hippuristanols, cytotoxic polyoxygenated steroids from the gorgonian Isis hippuris. Chem. Lett.10, 1647–1650 (1981). 10.1246/cl.1981.1647 [DOI] [Google Scholar]
- 8.Bordeleau, M. E. et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol.2, 213–220 (2006). 10.1038/nchembio776 [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist, L. et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One3, e1583 (2008). 10.1371/journal.pone.0001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun, Y. et al. Single-molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure22, 941–948 (2014). 10.1016/j.str.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 11.Steinberger, J. et al. Identification and characterization of hippuristanol-resistant mutants reveals eIF4A1 dependencies within mRNA 5′ leader regions. Nucleic Acids Res.48, 9521–9537 (2020). 10.1093/nar/gkaa662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, M. L. et al. X-Ray crystal structure of rocaglamide, a novel antileulemic 1H-cyclopenta[b]benzofuran from Aglaia elliptifolia. J. Chem. Soc. Chem. Commun.1, 1150–1151 (1982). 10.1039/c39820001150 [DOI] [Google Scholar]
- 13.Bordeleau, M. E. et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest.118, 2651–2660 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadlish, H. et al. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem. Biol.8, 1519–1527 (2013). 10.1021/cb400158t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santagata, S. et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science341, 1238303 (2013). 10.1126/science.1238303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki, S., Floor, S. N. & Ingolia, N. T. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature534, 558–561 (2016). 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu, J. et al. Amidino-rocaglates: a potent class of eIF4A inhibitors. Cell Chem. Biol.26, 1586–1593.e3 (2019). 10.1016/j.chembiol.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki, S. et al. The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol. Cell73, 738–748.e9 (2019). 10.1016/j.molcel.2018.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu, J. et al. Rocaglates induce gain-of-function alterations to eIF4A and eIF4F. Cell Rep.30, 2481–2488.e5 (2020). 10.1016/j.celrep.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cencic, R. et al. A second-generation eIF4A RNA helicase inhibitor exploits translational reprogramming as a vulnerability in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA.121, e2318093121 (2024). 10.1073/pnas.2318093121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Northcote, P. T., Blunt, J. W. & Munro, M. H. G. Pateamine: a potent cytotoxin from the New Zealand Marine sponge, mycale sp. Tetrahedron Lett.32, 6411–6414 (1991). 10.1016/0040-4039(91)80182-6 [DOI] [Google Scholar]
- 22.Bordeleau, M. E. et al. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA.102, 10460–10465 (2005). 10.1073/pnas.0504249102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Low, W. K. et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell20, 709–722 (2005). 10.1016/j.molcel.2005.10.008 [DOI] [PubMed] [Google Scholar]
- 24.Bordeleau, M. E. et al. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol.13, 1287–1295 (2006). 10.1016/j.chembiol.2006.10.005 [DOI] [PubMed] [Google Scholar]
- 25.Low, W. K., Dang, Y., Bhat, S., Romo, D. & Liu, J. O. Substrate-dependent targeting of eukaryotic translation initiation factor 4A by pateamine A: negation of domain-linker regulation of activity. Chem. Biol.14, 715–727 (2007). 10.1016/j.chembiol.2007.05.012 [DOI] [PubMed] [Google Scholar]
- 26.Di Marco, S. et al. The translation inhibitor pateamine A prevents cachexia-induced muscle wasting in mice. Nat. Commun.3, 896 (2012). 10.1038/ncomms1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Low, W.-K. et al. Second-generation derivatives of the eukaryotic translation initiation inhibitor pateamine A targeting eIF4A as potential anticancer agents. Bioorg. Med. Chem.22, 116–125 (2014). 10.1016/j.bmc.2013.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popa, A., Lebrigand, K., Barbry, P. & Waldmann, R. Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics17, 52 (2016). 10.1186/s12864-016-2384-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, R. et al. Creating novel translation inhibitors to target pro-survival proteins in chronic lymphocytic leukemia. Leukemia33, 1663–1674 (2019). 10.1038/s41375-018-0364-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rust, M. et al. A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli. Proc. Natl. Acad. Sci. USA.117, 9508–9518 (2020). 10.1073/pnas.1919245117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storey, M. A. et al. Metagenomic exploration of the marine sponge Mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. MBio11, e02997–19 (2020). 10.1128/mBio.02997-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naineni, S. K. et al. Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A. Cell Chem. Biol.28, 825–834.e6 (2021). [DOI] [PMC free article] [PubMed]
- 33.Santos, A. C. & Adkilen, P. The alkaloids of Argemone mexicana. J. Am. Chem. Soc.54, 2923–2924 (1932). 10.1021/ja01346a037 [DOI] [Google Scholar]
- 34.Jiang, C. et al. Targeting the N terminus of eIF4AI for inhibition of its catalytic recycling. Cell Chem. Biol.26, 1417–1426.e5 (2019). 10.1016/j.chembiol.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 35.Hinnebusch, A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem.83, 779–812 (2014). 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- 36.Brito Querido, J. et al. Structure of a human 48S translational initiation complex. Science369, 1220–1227 (2020). 10.1126/science.aba4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, M. et al. Dual targeting of DDX3 and eIF4A by the translation inhibitor rocaglamide A. Cell Chem. Biol.28, 475–486.e8 (2021). 10.1016/j.chembiol.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science324, 218–223 (2009). 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasaki, S. & Ingolia, N. T. The growing toolbox for protein synthesis studies. Trends Biochem. Sci.42, 612–624 (2017). 10.1016/j.tibs.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romo, D. et al. Evidence for separate binding and scaffolding domains in the immunosuppressive and antitumor marine natural product, pateamine a: design, synthesis, and activity studies leading to a potent simplified derivative. J. Am. Chem. Soc.126, 10582–10588 (2004). 10.1021/ja040065s [DOI] [PubMed] [Google Scholar]
- 41.Liu, T. Y. et al. Time-resolved proteomics extends ribosome profiling-based measurements of protein synthesis dynamics. Cell Syst.4, 636–644.e9 (2017). 10.1016/j.cels.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chhipi-Shrestha, J. K. et al. Splicing modulators elicit global translational repression by condensate-prone proteins translated from introns. Cell Chem. Biol.29, 259–275.e10 (2022). 10.1016/j.chembiol.2021.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naineni, S. K. et al. A comparative study of small molecules targeting eIF4A. RNA26, 541–549 (2020). 10.1261/rna.072884.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambert, N. et al. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell54, 887–900 (2014). 10.1016/j.molcel.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert, N. J., Robertson, A. D. & Burge, C. B. RNA Bind-n-Seq: measuring the binding affinity landscape of RNA-binding proteins. Methods Enzymol.558, 465–493 (2015). 10.1016/bs.mie.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linder, P. & Jankowsky, E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol.12, 505–516 (2011). 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- 47.Weis, K. & Hondele, M. The role of DEAD-Box ATPases in gene expression and the regulation of RNA-protein condensates. Annu. Rev. Biochem.91, 197–219 (2022). 10.1146/annurev-biochem-032620-105429 [DOI] [PubMed] [Google Scholar]
- 48.Pestova, T. V. & Kolupaeva, V. G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev.16, 2906–2922 (2002). 10.1101/gad.1020902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe, A. L. et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature513, 65–70 (2014). 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldron, J. A., Raza, F. & Le Quesne, J. eIF4A alleviates the translational repression mediated by classical secondary structures more than by G-quadruplexes. Nucleic Acids Res.46, 3075–3087 (2018). 10.1093/nar/gky108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldron, J. A. et al. mRNA structural elements immediately upstream of the start codon dictate dependence upon eIF4A helicase activity. Genome Biol.20, 300 (2019). 10.1186/s13059-019-1901-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dmitriev, S. E., Pisarev, A. V., Rubtsova, M. P., Dunaevsky, Y. E. & Shatsky, I. N. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett.533, 99–104 (2003). 10.1016/S0014-5793(02)03776-6 [DOI] [PubMed] [Google Scholar]
- 53.Shirokikh, N. E. et al. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res.38, e15 (2010). 10.1093/nar/gkp1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen, M. et al. A parasitic fungus employs mutated eIF4A to survive on rocaglate-synthesizing Aglaia plants. Elife12, e81302 (2023). 10.7554/eLife.81302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitaura, K., Ikeo, E., Asada, T., Nakano, T. & Uebayasi, M. Fragment molecular orbital method: an approximate computational method for large molecules. Chem. Phys. Lett.313, 701–706 (1999). 10.1016/S0009-2614(99)00874-X [DOI] [Google Scholar]
- 56.Fedorov, D. G., Nagata, T. & Kitaura, K. Exploring chemistry with the fragment molecular orbital method. Phys. Chem. Chem. Phys.14, 7562–7577 (2012). 10.1039/c2cp23784a [DOI] [PubMed] [Google Scholar]
- 57.Tanaka, S., Mochizuki, Y., Komeiji, Y., Okiyama, Y. & Fukuzawa, K. Electron-correlated fragment-molecular-orbital calculations for biomolecular and nano systems. Phys. Chem. Chem. Phys.16, 10310–10344 (2014). 10.1039/C4CP00316K [DOI] [PubMed] [Google Scholar]
- 58.Mochizuki, Y., Tanaka, S. & Fukuzawa, K. Recent Advances of the Fragment Molecular Orbital Method: Enhanced Performance and Applicability (Springer Nature Singapore, 2021).
- 59.Handa, Y. et al. Prediction of binding pose and affinity of Nelfinavir, a SARS-CoV-2 main protease repositioned drug, by combining docking, molecular dynamics, and fragment molecular orbital calculations. J. Phys. Chem. B128, 2249–2265 (2024). 10.1021/acs.jpcb.3c05564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fedorov, D. G. & Kitaura, K. Pair interaction energy decomposition analysis. J. Comput. Chem.28, 222–237 (2007). 10.1002/jcc.20496 [DOI] [PubMed] [Google Scholar]
- 61.Tsukamoto, T. et al. Implementation of pair interaction energy decomposition analysis and its applications to protein-ligand systems. J. Comput. Chem. Jpn.14, 1–9 (2015). 10.2477/jccj.2014-0039 [DOI] [Google Scholar]
- 62.Li, F. et al. Reanalysis of ribosome profiling datasets reveals a function of rocaglamide A in perturbing the dynamics of translation elongation via eIF4A. Nat. Commun.14, 553 (2023). 10.1038/s41467-023-36290-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mullard, A. Small molecules against RNA targets attract big backers. Nat. Rev. Drug Discov.16, 813–815 (2017). 10.1038/nrd.2017.239 [DOI] [PubMed] [Google Scholar]
- 64.Garber, K. Drugging RNA. Nat. Biotechnol.41, 745–749 (2023). 10.1038/s41587-023-01790-z [DOI] [PubMed] [Google Scholar]
- 65.Khaperskyy, D. A. et al. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog.10, e1004217 (2014). 10.1371/journal.ppat.1004217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.González-Almela, E. et al. Differential action of pateamine A on translation of genomic and subgenomic mRNAs from Sindbis virus. Virology484, 41–50 (2015). 10.1016/j.virol.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 67.Ziehr, B., Lenarcic, E., Cecil, C. & Moorman, N. J. The eIF4AIII RNA helicase is a critical determinant of human cytomegalovirus replication. Virology489, 194–201 (2016). 10.1016/j.virol.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slaine, P. D., Kleer, M., Smith, N. K., Khaperskyy, D. A. & McCormick, C. Stress granule-inducing eukaryotic translation initiation factor 4A inhibitors block influenza A virus replication. Viruses9, 388 (2017). 10.3390/v9120388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lucas, D. M. et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood113, 4656–4666 (2009). 10.1182/blood-2008-09-175430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alachkar, H. et al. Silvestrol exhibits significant in vivo and in vitro antileukemic activities and inhibits FLT3 and miR-155 expressions in acute myeloid leukemia. J. Hematol. Oncol.6, 21 (2013). 10.1186/1756-8722-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boussemart, L. et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature513, 105–109 (2014). 10.1038/nature13572 [DOI] [PubMed] [Google Scholar]
- 72.Wiegering, A. et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov.5, 768–781 (2015). 10.1158/2159-8290.CD-14-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manier, S. et al. Inhibiting the oncogenic translation program is an effective therapeutic strategy in multiple myeloma. Sci. Transl. Med.9, eaal2668 (2017). 10.1126/scitranslmed.aal2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerezo, M. et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat. Med.24, 1877–1886 (2018). 10.1038/s41591-018-0217-1 [DOI] [PubMed] [Google Scholar]
- 75.Chan, K. et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat. Commun.10, 5151 (2019). 10.1038/s41467-019-13086-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishida, Y. et al. Inhibition of translation initiation factor eIF4a inactivates heat shock factor 1 (HSF1) and exerts anti-leukemia activity in AML. Leukemia35, 2469–2481 (2021). 10.1038/s41375-021-01308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skofler, C. et al. Eukaryotic translation initiation factor 4AI: a potential novel target in neuroblastoma. Cells10, 301 (2021). 10.3390/cells10020301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson, P. A. et al. Targeting oncogene mRNA translation in B-cell malignancies with eFT226, a potent and selective inhibitor of eIF4A. Mol. Cancer Ther.20, 26–36 (2021). 10.1158/1535-7163.MCT-19-0973 [DOI] [PubMed] [Google Scholar]
- 79.Wilmore, S. et al. Targeted inhibition of eIF4A suppresses B-cell receptor-induced translation and expression of MYC and MCL1 in chronic lymphocytic leukemia cells. Cell. Mol. Life Sci.78, 6337–6349 (2021). 10.1007/s00018-021-03910-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuznetsov, G. et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol. Cancer Ther.8, 1250–1260 (2009). 10.1158/1535-7163.MCT-08-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho, J. J. D. et al. Proteomics reveal cap-dependent translation inhibitors remodel the translation machinery and translatome. Cell Rep37, 109806 (2021). 10.1016/j.celrep.2021.109806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Romo, D. et al. Total synthesis and immunosuppressive activity of (−)-pateamine A and related compounds: implementation of a β-lactam-based macrocyclization. J. Am. Chem. Soc.120, 12237–12254 (1998). 10.1021/ja981846u [DOI] [Google Scholar]
- 83.Zhuo, C.-X. & Fürstner, A. Catalysis-based total syntheses of pateamine A and DMDA-Pat A. J. Am. Chem. Soc.140, 10514–10523 (2018). 10.1021/jacs.8b05094 [DOI] [PubMed] [Google Scholar]
- 84.Guan, W. et al. Stereoselective formation of trisubstituted vinyl boronate esters by the acid-mediated elimination of α-hydroxyboronate esters. J. Org. Chem.79, 7199–7204 (2014). 10.1021/jo500773t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McIntosh, M. L., Moore, C. M. & Clark, T. B. Copper-catalyzed diboration of ketones: facile synthesis of tertiary alpha-hydroxyboronate esters. Org. Lett.12, 1996–1999 (2010). 10.1021/ol100468f [DOI] [PubMed] [Google Scholar]
- 86.Xu, S. et al. Pincer iron hydride complexes for alkene isomerization: catalytic approach to trisubstituted (Z)-alkenyl boronates. ACS Catal.11, 10138–10147 (2021). 10.1021/acscatal.1c02432 [DOI] [Google Scholar]
- 87.Sanchez, A. & Maimone, T. J. Taming shapeshifting anions: total synthesis of ocellatusone C. J. Am. Chem. Soc.144, 7594–7599 (2022). 10.1021/jacs.2c02627 [DOI] [PubMed] [Google Scholar]
- 88.Zhuo, C.-X. & Fürstner, A. Concise synthesis of a pateamine A analogue with in vivo anticancer activity based on an iron-catalyzed pyrone ring opening/cross-coupling. Angew. Chem. Int. Ed Engl.55, 6051–6056 (2016). 10.1002/anie.201602125 [DOI] [PubMed] [Google Scholar]
- 89.Mito, M., Mishima, Y. & Iwasaki, S. Protocol for disome profiling to survey ribosome collision in humans and zebrafish. STAR Protoc.1, 100168 (2020). 10.1016/j.xpro.2020.100168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kashiwagi, K. et al. eIF2B-capturing viral protein NSs suppresses the integrated stress response. Nat. Commun.12, 1–12 (2021). 10.1038/s41467-021-27337-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890 (2018). 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molecular Operating Environment (MOE). (2022.02 Chemical Computing Group ULC, 910-1010 Sherbrooke St. W., Montreal, QC H3A 2R7, Canada, 2024).
- 96.Gerber, P. R. & Müller, K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided Mol. Des.9, 251–268 (1995). 10.1007/BF00124456 [DOI] [PubMed] [Google Scholar]
- 97.Case, D. A. et al. Amber 10. (University of California, 2008). [Google Scholar]
- 98.Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput.11, 3696–3713 (2015). 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zgarbová, M. et al. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput.7, 2886–2902 (2011). 10.1021/ct200162x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem.25, 1157–1174 (2004). 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- 101.Wang, J., Wolf, R. M., Caldwell, J. W. & Kollman, P. A. & Case, D. A. Junmei Wang, Romain M. Wolf, James W. Caldwell, Peter A. Kollman, and David A. Case, “Development and testing of a general amber force field”Journal of Computational Chemistry (2004) 25(9) 1157–1174. J. Comput. Chem.26, 114–114 (2005). 10.1002/jcc.20145 [DOI] [PubMed] [Google Scholar]
- 102.He, X., Man, V. H., Yang, W., Lee, T.-S. & Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys.153, 114502 (2020). 10.1063/5.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Case, D. A. et al. Amber 16 (University of California, 2016).
- 104.Mochizuki, Y. et al. A parallelized integral-direct second-order Møller–Plesset perturbation theory method with a fragment molecular orbital scheme. Theor. Chem. Acc.112, 442–452 (2004). 10.1007/s00214-004-0602-3 [DOI] [Google Scholar]
- 105.Mochizuki, Y., Koikegami, S., Nakano, T., Amari, S. & Kitaura, K. Large scale MP2 calculations with fragment molecular orbital scheme. Chem. Phys. Lett.396, 473–479 (2004). 10.1016/j.cplett.2004.08.082 [DOI] [Google Scholar]
- 106.Takaya, D. et al. FMODB: the world’s first database of quantum mechanical calculations for biomacromolecules based on the fragment molecular orbital method. J. Chem. Inf. Model.61, 777–794 (2021). 10.1021/acs.jcim.0c01062 [DOI] [PubMed] [Google Scholar]
- 107.Saito, H. Custom scripts for DMDA-PatA mediates RNA sequence-selective translation repression by anchoring eIF4A and DDX3 to GNG motifs. Zenodo. 10.5281/zenodo.11064746 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon request. The results of ribosome profiling, RNA pulldown-Seq, and RNA Bind-n-Seq (GEO: GSE243312) obtained in this study have been deposited in the National Center for Biotechnology Information (NCBI) database. All the input and result files for the FMO calculations are available at the FMODB [https://drugdesign.riken.jp/FMODB/detail.php?FMODBID = “ID in the list”]106 (see Supplementary Table 1 for the ID list). Source data are provided with this paper.
For the data analysis for ribosome profiling, RNA pulldown-Seq, and RNA Bind-n-Seq, we deposited key codes in Zenodo (10.5281/zenodo.11064746)107, which used reported custom script (https://github.com/ingolia-lab/RiboSeq). For the MD and FMO calculations, we used the abmptools on GitHub (https://github.com/kojioku/abmptools).