Abstract
Pythium amaminum sp. nov. was isolated from river and reservoir water on Amami island, Kagoshima Prefecture, Japan. The species can grow at temperatures between 10 °C and 35 °C. At the optimum temperature of 25 °C, the radial growth rate is 22.5 mm per day. Pythium amaminum produces filamentous sporangia consisting of branched, lobulate or digitate elements forming large complexes. Zoospores form inside the vesicle, which is discharged through a long tube at least 320 μm. Globose oogonia are ornamented with conical blunt spines. Oospores are aplerotic and globose. Antheridia twine around the oogonia or stick to them. These features having a both of the long discharge tube from sporangium and oogonia with spines are not observed in any other species of the genus Pythium, and thus we conclude that P. amaminum is a new Pythium species.
Keywords: morphological analysis, oomycete
The genus Pythium comprises approximately 150 recognized species and exists worldwide (Kirk et al., 2008). Many Pythium species are saprophytes, but some are plant pathogens of agricultural importance, such as P. arrhenomanes Drechsler, P. aphanidermatum (Edson) Fitzp., and P. myriotylum Drechsler (van der Plaats-Niterink, 1981).
The genus Pythium was subdivided into 11 clades (A-K) by Lévesque and de Cock (2004), in a phylogeny of 116 species based on the internal transcribed spacer (ITS) region. Uzuhashi et al. (2010) proposed that the genus Pythium be divided into Pythium sensu stricto and four new genera: Pythium characterized by filamentous sporangia which is comparable clade A, B, C, and D, Ovatisporangium characterized by ovoid sporangia which is comparable clade K, Globisporangium characterized by globose sporangia which is comparable clade E, F, G, I, and J, Elongisporangium characterized by elongated clavate sporangia which is comparable clade H, and Pilasporangium, composed of the single species P. apinafurcum (Uzuhashi & Tojo) Uzuhashi, Tojo & Kakish. Bala et al. (2010) retained the genus name Pythium for clades A to J, but re-classified the species in clade K as Phytopythium. Ovatisporangium has been treated as a synonym of the Phytopythium because of nomenclature priority.
Amami is the main island of the subtropical Amami Archipelago in Kagoshima Prefecture, Japan. In this study, we collected 7 water samples from rivers and reservoirs to survey the Pythium and Phytopythium species on Amami island. For sampling, we selected parts of the river where the current was slow and where fallen leaves had accumulated (see Table 1 for coordinates). The water was gathered in 200 mL autoclaved bottles containing 5 mm sections of autoclaved zoysia grass leaf blades (Abdelzaher & Kageyama, 2020). After incubation for 1 wk at 25 °C, the zoysia grass leaf blades were recovered from the water and placed on nystatin +ampicillin +rifampicin +miconazole (NARM) medium (Morita & Tojo, 2007). After incubation on the NARM medium for 24 h at 25 °C, mycelial tip was transferred to corn meal agar (CMA) medium (Abdelzaher & Kageyama, 2020). Total 70 isolates were obtained from 7 samples. Preliminary morphological observation revealed 12 isolates showing unique characteristics such as lobulate filamentous sporangia and ornamented oogonia with conical blunt spines. The isolates were recovered from two rivers and a small reservoir. Our goal for this study was to characterize the isolates using morphological features and a phylogenetic analysis. Two of the 12 isolates (AO.GPS005w2 and AO.GPS015w3) were selected as representatives for further analysis.
Table 1 Isolates obtained from river and reservoir water on Amami island.
| Obtained from | Latitude | Longitude | Isolate number | Ex-type strain |
| Oomi River | N28°23'58.1" | E129°35'28.5" | AO.GPS005w2a | NBRC115126 |
| AO.GPS005w4 | ||||
| AO.GPS005w6 | ||||
| AO.GPS005w10 | ||||
| Small reservoir | N28°26'00.4" | E129°35'16.5" | AO.GPS006w1 | |
| AO.GPS006w3 | ||||
| AO.GPS006w7 | ||||
| Koshukuo River | N28°22'40.2" | E129°27'29.5" | AO.GPS015w1 | |
| AO.GPS015w2 | ||||
| AO.GPS015w3a | ||||
| AO.GPS015w6 | ||||
| AO.GPS015w7 |
a The selected isolates as representatives of the sequence and the phylogenetic analysis
For morphological observation, we used grass blade culture (Waterhouse, 1967). Autoclaved zoysia grass leaf blades were placed around each isolate on V8 juice agar (V8A) (Abdelzaher & Kageyama, 2020). After 1-2 d incubation at 25 °C, the colonized blades were transferred to autoclaved pond water (pond water: distilled water = 1: 2) and incubated at 25 °C in the dark. We observed the morphology by a light microscope.
Furthermore, oogonia were observed using a scanning electron microscope, the SEM-4300 (Hitachi, Tokyo, Japan) because a spine of oogonia could not be observed clearly by a light microscope. We observed the spine of oogonia and measured the length of the spine by SEM. Autoclaved grass leaf blades were incubated with an isolate on V8A at 25 °C for more than 1 wk. The colonized blades were then processed using Karnovsky's fixative, prepared as described by Karnovsky (1965). The colonized blades were fixed at room temperature for 20 h. The fixed samples were dehydrated by sequential immersion in 50%, 70%, 80%, 90%, 95%, and 100% ethyl alcohol for 15 min each. The dehydrated samples were then immersed twice in tert-butyl alcohol for 30 min each, then covered with tert-butyl alcohol and frozen at -30 °C for 10 min. The tert-butyl alcohol was sublimated, and the samples were dried at -90 kPa for 3 h in a decompressor. The dried samples were covered with osmium using an osmium coater (Meiwafosis Neoc-Pro, Tokyo, Japan), and then the oogonia were observed using the SEM-4300.
For growth analyses, mycelial growth for each isolate was measured for each temperature on 3 replicate plates of potato carrot agar (PCA) medium (van der Plaats-Niterink, 1981). Each plate was inoculated with a 6-mm mycelial disk taken from the margin of a colony grown on PCA. The plates were incubated in the dark at 5, 10, 15, 20, 25, 30, 35, and 40 °C, and colony diameter were measured daily. Colonies were also grown in the dark on CMA and V8A at 25 °C to evaluate colony pattern.
For DNA extraction, the two representative isolates were grown for 7 d at 25 °C on V8A. A small amount of mycelium was transferred to 100 μL PrepMan Ultra Reagent (Applied Biosystems, Inc., California, USA) and heated to 100 °C for 10 min. After 3 min incubation at room temperature, the sample was centrifuged at 15,000 g for 3 min, and 85 μL of the supernatant was transferred to a new tube.
Phylogenetic analysis was conducted by using the cytochrome c oxidase subunit 1 (cox1) gene and the ribosomal DNA internal transcribed spacer (rDNA-ITS) region. The cox1 were amplified using Oom-COl-Lev-up/FM85-mod primers (Robideau et al., 2011) and rDNA-ITS regions were amplified using ITS5/ITS4 primers (White et al., 1990). Each 25 μL reaction mixture contained 1 μL DNA, 0.125 units rTaq DNA polymerase (TaKaRa Bio Inc., Otsu, Japan), 0.2 mM dNTP mixture, 0.4 μg/μL BSA, PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl and 1.5 mM MgCl2), and either the cox1 primers (0.5 μM each) or the ITS primers (0.2 μM each). The reactions were carried out in a 2700 DNA Thermal Cycler (Applied Biosystems, Inc.). The amplification conditions were as follows: For cox1: 94 °C for 2 min; 35 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min; then 72 °C for 10 min. For the rDNA-ITS region: 94 °C for 3 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; then 72 °C for 10 min. The PCR products were purified using the ExoSAP-IT Express kit (Applied Biosystems, Inc.) following the manufactures' instructions. Each 10 μL sequencing reaction contained 1 μL purified PCR product, 1 μL primer (1.6 μM), 4 μL BigDye Terminator ver.3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Inc.), and 4 μL H2O. Same primers for PCR were used for sequencing. The reaction conditions were: 96 °C for 1 min followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min, with a final incubation at 15 °C. The reaction products were purified by ethanol precipitation and analyzed using the ABI3100 DNA Sequencer (Applied Biosystems, Inc.). The sequences were edited using the ChromasPro ver.1.5 software (Technelysium Pty Ltd., Queensland, Australia), and the consensus sequences were used in alignments. The nucleotide sequence data were deposited in the DNA Data Bank of Japan (DDBJ) sequence database with the accession numbers shown in Fig. 1.
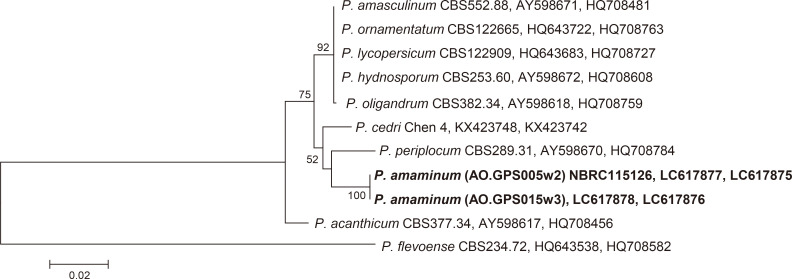
Fig. 1 Phylogenetic tree of Pythium clade D species, based on the combined sequences of the rDNA-ITS region and the cox1 gene and generated by maximum parsimony analysis.
The numbers at the nodes are bootstrap values (only values greater than 50% are shown). Pythium flevoense (CBS 234.72) served as the out-group. The accession (the rDNA-ITS region and the cox1 gene) numbers used for the phylogenetic analysis are shown after the taxon name.
The sequence identities with two representative isolates were 99.2% and 100% in rDNA-ITS and cox1, respectively. A BLAST search of the DDBJ revealed that our isolates show high homology of rDNA-ITS and cox1 with some clade D species, such as P. hydnosporum (Mont.) J. Schröt., P. periplocum Drechsler and P. acanthicum Drechsler, but the value were low for species identification. To determine the relationships between the isolate and closely related species, we constructed a phylogenetic tree based on the combined rDNA-ITS and cox1 sequences using the two Amami isolates, 8 species belonging to clade D of the genus Pythium, and P. flevoense van der Plaäts-Nit. belonging to clade B as an out-group species. Sequences obtained from the NCBI database are shown in Fig. 1. All sequences were first aligned using the Clustal X multiple sequence alignment software (Thompson et al., 1997), and MEGA6 (Tamura et al., 2016) was used to construct the tree. The tree was generated using maximum parsimony, and bootstrap values were calculated from 1,000 replicates. The tree showed that the Amami isolates were closely related to P. periplocum and P. cedri Jia J. Chen & X.B. Zheng (Fig. 1). The isolates formed a monophyletic group with high bootstrap support (100%) in the maximum parsimony analysis. Based on the morphology and phylogeny, we concluded the isolates to be a new species, and named as P. amaminum.
Pythium amaminum H. Kikuchi, A. Hieno & K. Kageyama, sp. nov. Figs. 1, 2, 3.
MycoBank no.: MB 839094.
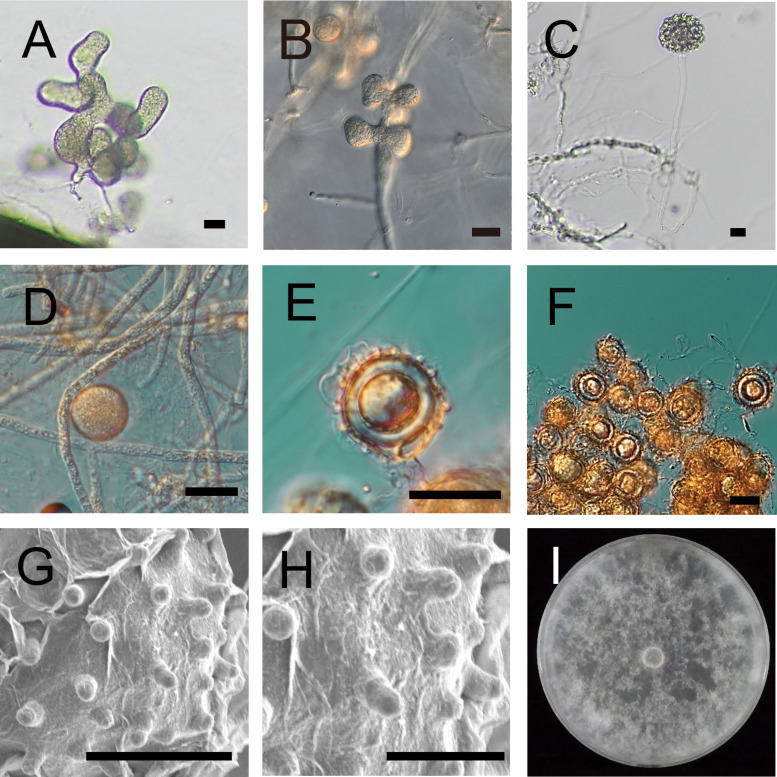
Fig. 2 Morphological characteristics of Pythium amaminum.
A: A sporangium in grass blade culture. B: A sporangium on CMA. C: A discharge tube and a vesicle containing zoospores formed in grass blade culture. D: A hyphal swelling on CMA. E: An oogonium, an antheridium and an aplerotic oospore in grass blade culture. F: Cluster of oogonia in grass blade culture. G, H: Spines of oogonia in grass blade culture. I: Growth pattern on V8A after 1 wk incubation at 25 °C. Bars: A-F 20 μm; G 10 μm; H 5 μm.
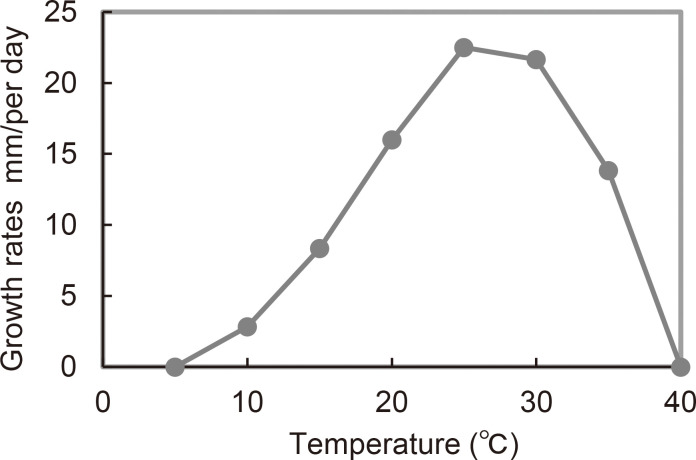
Fig. 3 Mycelial growth rates of Pythium amaminum (NBRC115126) at different temperatures on PCA.
Type: JAPAN, Kagoshima, Amami Island from river water, Nov 21, 2018, Collector K. Kageyama (holotype (frozen dry specimen), NBRC H-13379; ex-type strains, NBRC115126).
Gene sequences ex-holotype: LC617875 (cox1), LC617877 (rDNA-ITS).
Etymology: amaminum refers to the location in Japan where the holotype was isolated.
Morphological characteristics: Filamentous sporangia consisting of branched, lobulate, or digitate elements, 25-77 μm (av. 41.6 μm) long and 14-29 μm (av. 19.3 μm) width, forming large complexes (Fig. 2A, B) in water culture at 25 °C. Zoospores inside the vesicle, which discharges through a long tube at least 320 μm from sporangium (Fig. 2C). Globose hyphal swellings observed on CMA and V8A (Fig. 2D), less frequent in grass leaf blade culture. Sexual structure formation after incubation for more than 7 d at 25 °C on V8 or CMA in single culture. Globose oogonia 17-25 μm (av. 20.9±2.1 μm) diam, ornamented with conical blunt spines 2-4 μm (av. 3.0±0.6 μm) long. (Fig. 2E, G, H). Clusters consisting of many oogonia, usually in grass blade culture (Fig. 2F). Antheridia twining around the oogonia or sticking to them, disappearing during long incubations, rarely observed. Oospores aplerotic, globose with 9-17 μm (av. 12.6±2.4 μm) diam (Fig. 2E).
Cultural characteristics: Average radial growth was 22.5 mm/d at the optimum temperature of 25 °C (Fig. 3). The minimum and maximum growth temperatures were 10 °C and 35 °C, respectively. Growth pattern was cottony on V8A (Fig. 2I).
Additional isolate examined: AO.GPS015w3, gene sequences: LC617876 (cox1), LC617878 (rDNA-ITS).
Pythium amaminum shows close relationships with P. cedri and P. periplocum in the phylogenetic tree. These species have similar morphological characteristics with P. amaminum, but there are some differences. The unique morphological characteristics of the new species included long discharge tubes and oogonia ornamented with conical blunt spines (Table 2). Pythium amaminum and P. periplocum produce large complexes of sporangia consisting of branched, lobulate, or digitate elements (van der Plaats-Niterink, 1981), whereas P. cedri produces toruloid sporangia (Chen et al., 2017). Pythium amaminum produces nearly double the size of sporangia compared to P. periplocum, whose sporangia are 10 to 30 μm long and 8 to 20 μm, mostly 10 to 15 μm width (Drechsler, 1930). Pythium amaminum and P. periplocum produce zoospores, but P. cedri does not. All three species form oogonia ornamented with spines, but the oogonia of P. amaminum are smaller than those of the other two species. Pythium cedri forms plerotic oospores whereas P. amaminum and P. periplocum form aplerotic oospores. Pythium amaminum produces smaller oospores than the other two species. The daily growth rate of the new species is intermediate between those of P. cedri and P. periplocum.
Table 2 Comparison of morphological characteristics between Pythium amaminum and the taxonomically related species.
| Organs/characteristics | P. amaminum a | P. cedri b | P. periplocum c |
| Sporangia | Large complexes consisting of branched, lobulate or digitate elements, 25-77 μm (av. 41.6 μm) long and 14-29 μm (av. 19.3 μm) diam | Toruloid | Large complexes consisting of branched, lobulate or digitate elements, 10 to 30 μm long, 8 to 20 μm, mostly 10 to 15 μm |
| Zoospore | Formed | Not formed | Formed |
| Hyphal swelling | Present | Present | No description |
| Oogonia | Ornamented with blunt spines, 17-25 μm (av. 20.9 μm) diam | Ornamented with blunt spines, 17.5-32.5 μm (av. 24.1 μm) diam | Ornamented with blunt spines, 24-27 μm (av. 26 μm) diam |
| Spines | 1.9-4.3 μm long | 2.5-4 μm long | 2-4 μm long |
| Antheridia | Rarely observed | Subclavate, monoclinous, 1-3 per oogonium | Clavate, diclinous, 1-4 per oogonium |
| Oospore | Aplerotic, 9-17 μm (av. 12.6 μm) diam | Plerotic or nearly plerotic, 14.3-26.5 μm (av. 21.3 μm) diam | Aplerotic, 20-24 μm (av. 22 μm) diam |
| Daily growth rate | 22.5 mm/day at 25 °C on PCA | 11 mm/day at 25 °C on PCA | 35 mm/day at 25 °C on PCA |
a Present study, b Chen et al. 2017, c van der Plaats-Niterink 1981
The genus Pythium is mycologically and pathologically important genus because it includes not only saprophytes but also pathogens of plants and animals. In this study, we reported the discovery of a new species from a river unaffected by human activities. The diversity of Pythium spp. in the natural environment has been little studied, and future research will be needed to search for diversity as well as to determine how these species affect human activities.
Disclosure
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
References
- Abdelzaher, H. M. A., &Kageyama, K.(2020). Diversity of aquatic Pythium and Phytopythium spp. from rivers and a pond of Gifu city, Japan. Novel Research in Microbiology Journal, 4, 1029-1044. https://doi.org/10.21608/NRMJ.2020.130851 [Google Scholar]
- Bala, K., Robideau, G. P., Lévesque, A., de Cock, W. A. M., Abad, Z. G., Lodhi, A. M., Shahzad, S., Ghaffar, A., &Coffey, M. D.(2010). Phytopythium Abad, de Cock, Bala, Robideau, Lodhi & Lévesque, gen. nov. and Phytopythium sindhum Lodhi, Shahzad & Lévesque, sp. nov. Persoonia, 24, 136-137. https://doi.org/10.3767/003158510X512748 [Google Scholar]
- Chen, J. J., Lu, L., Ye, W. W., Wang, Y. C., &Zheng, X. B.(2017). Pythium cedri sp. nov. (Pythiaceae, Pythiales) from southern China based on morphological and molecular characters. Phytotaxa, 309, 135-142. https://doi.org/10.11646/phytotaxa.309.2.4 [Google Scholar]
- Drechsleri, C.(1930) Some new species of Pythium. Journal of the Washington Academy of Sciences, 20, 398-418. [Google Scholar]
- Karnovsky, M. J.(1965). A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. Journal of Cell Biology, 27, 1-149.5857256 [Google Scholar]
- Kirk, P. M., Cannon, P. F., Minter, D. W., &Stalpers, J. A.(2008). Ainsworth & Bisby's Dictionary of the Fungi. CAB International [Google Scholar]
- Lévesque, C. A., &de Cock, W. A. M.(2004). Molecular phylogeny and taxonomy of the genus Pythium. Mycological Research, 108, 1363-1383. https://doi.org/10.1017/S0953756204001431 [DOI] [PubMed] [Google Scholar]
- Morita, Y., &Tojo, M.(2007). Modifications of PARP medium using fluazinam, miconazole, and nystatin for detection of Pythium spp. in soil. Plant Disease, 91, 1591-1599. https://doi.org/10.1094/PDIS-91-12-1591 [DOI] [PubMed] [Google Scholar]
- Robideau, G. P., de Cock, A. W., Coffey, M. D., Voglmarr, H., Brouwer, H., Bala, K., Chitty, D. W., Désaulniers, N., Eggerrtson, Q. A., Gachon, C. M. M., Hu, C-H., Küpper, F. C., Rintoul, T. L., Sarhan, E., Verstappen, E. C. R., Zhang, Y., Bonants, P. J. M., Ristaino, J. B., &Lévesque, C. A.(2011). DNA barcoding of oomycetes with cytochrome c oxidase subunits I and internal transcriber spacer. Molecular Ecology Resources, 11, 1002-1011. https://doi.org/10.1111/j.1755-0998.2011.03041.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., &Kumar, S.(2016). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30, 2725-2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., &Higgins, D. G.(1997). The CLUSTAL_X windows interface: flexible strategies or multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. https://doi.org/10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuhashi, S., Tojo, M., &Kakishima, M.(2010). Phylogeny of the genus Pythium and description of new genera. Mycoscience, 51, 337-365. https://doi.org/10.1007/S10267-010-0046-7 [Google Scholar]
- van der Plaats-Niterink, A. J.(1981). Monograph of the genus Pythium. Studies in Mycology, 21, 1-242. [Google Scholar]
- Waterhouse, G. E.(1967). Key to Pythium Pringsheim. Mycological papers, 109, 1-15. [Google Scholar]
- White, T. J., Bruns, T., Lee, S., &Taylor, J.(1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: A Guide to Methods and Applications (pp. 315-322). Academic Press. [Google Scholar]