Abstract
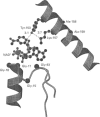
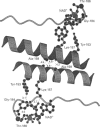
We have determined the nucleotide sequences of eight ethyl methanesulphonate-induced mutants in Drosophila alcohol dehydrogenase (ADH), of which six were previously characterized by Hollocher and Place [(1988) Genetics 116, 253-263 and 265-274]. Four of these ADH mutants contain a single amino acid change: glycine-17 to arginine, glycine-93 to glutamic acid, alanine-159 to threonine, and glycine-184 to aspartic acid. Although these mutants are inactive, three mutants (Gly17Arg, Gly93Glu and Gly184Asp) form stable homodimers, as well as heterodimers with wild-type ADH, in which the wild-type ADH subunit retains full enzyme activity [Hollocher and Place (1988) Genetics 116, 265-274]. Interestingly, the Ala159Thr mutant does not form either stable homodimers or heterodimers with wild-type ADH, suggesting that alanine-159 is important in stabilizing ADH dimers. The mutations were analysed in terms of a three-dimensional model of ADH using bacterial 20 beta-hydroxysteroid dehydrogenase and rat dihydropteridine reductase as templates. The model indicates that mutations in glycine-17 and glycine-93 affect the binding of NAD+. It also shows that alanine-159 is part of a hydrophobic anchor on the dimer interface of ADH. Replacement of alanine-159 with threonine, which has a larger side chain and can hydrogen bond with water, is likely to reduce the strength of the hydrophobic interaction. The three-dimensional model shows that glycine-184 is close to the substrate binding site. Replacement of glycine-184 with aspartic acid is likely to alter the position of threonine-186, which we propose hydrogen bonds to the carboxamide moiety of NAD+. Also, the negative charge on the aspartic acid side chain may interact with the substrate and/or residues in the substrate binding site. These mutations provide information about ADH catalysis and the stability of dimers, which may also be useful in understanding homologous dehydrogenases, which include the human 17 beta-hydroxysteroid, 11 beta-hydroxysteroid and 15-hydroxyprostaglandin dehydrogenases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albalat R., González-Duarte, Atrian S. Protein engineering of Drosophila alcohol dehydrogenase. The hydroxyl group of Tyr152 is involved in the active site of the enzyme. FEBS Lett. 1992 Aug 24;308(3):235–239. doi: 10.1016/0014-5793(92)81282-q. [DOI] [PubMed] [Google Scholar]
- Baker M. E. A common ancestor for human placental 17 beta-hydroxysteroid dehydrogenase, Streptomyces coelicolor actIII protein, and Drosophila melanogaster alcohol dehydrogenase. FASEB J. 1990 Feb 1;4(2):222–226. doi: 10.1096/fasebj.4.2.2153594. [DOI] [PubMed] [Google Scholar]
- Baker M. E. Evolution of enzymatic regulation of prostaglandin action: novel connections to regulation of human sex and adrenal function, antibiotic synthesis and nitrogen fixation. Prostaglandins. 1991 Nov;42(5):391–410. doi: 10.1016/0090-6980(91)90031-a. [DOI] [PubMed] [Google Scholar]
- Baker M. E. Sequence analysis of steroid- and prostaglandin-metabolizing enzymes: application to understanding catalysis. Steroids. 1994 Apr;59(4):248–258. doi: 10.1016/0039-128x(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Bodmer M., Ashburner M. Conservation and change in the DNA sequences coding for alcohol dehydrogenase in sibling species of Drosophila. 1984 May 31-Jun 6Nature. 309(5967):425–430. doi: 10.1038/309425a0. [DOI] [PubMed] [Google Scholar]
- Briganti F., Fong W. P., Auld D. S., Vallee B. L. In vitro dissociation and reassociation of human alcohol dehydrogenase class I isozymes. Biochemistry. 1989 Jun 27;28(13):5374–5379. doi: 10.1021/bi00439a009. [DOI] [PubMed] [Google Scholar]
- CRICK F. H., ORGEL L. E. THE THEORY OF INTER-ALLELIC COMPLEMENTATION. J Mol Biol. 1964 Jan;8:161–165. doi: 10.1016/s0022-2836(64)80156-x. [DOI] [PubMed] [Google Scholar]
- Chen Z., Jiang J. C., Lin Z. G., Lee W. R., Baker M. E., Chang S. H. Site-specific mutagenesis of Drosophila alcohol dehydrogenase: evidence for involvement of tyrosine-152 and lysine-156 in catalysis. Biochemistry. 1993 Apr 6;32(13):3342–3346. doi: 10.1021/bi00064a017. [DOI] [PubMed] [Google Scholar]
- Chen Z., Lu L., Shirley M., Lee W. R., Chang S. H. Site-directed mutagenesis of glycine-14 and two "critical" cysteinyl residues in Drosophila alcohol dehydrogenase. Biochemistry. 1990 Feb 6;29(5):1112–1118. doi: 10.1021/bi00457a003. [DOI] [PubMed] [Google Scholar]
- Chen Z., Tsigelny I., Lee W. R., Baker M. E., Chang S. H. Adding a positive charge at residue 46 of Drosophila alcohol dehydrogenase increases cofactor specificity for NADP+. FEBS Lett. 1994 Dec 12;356(1):81–85. doi: 10.1016/0014-5793(94)01234-2. [DOI] [PubMed] [Google Scholar]
- Chia W., Savakis C., Karp R., Pelham H., Ashburner M. Mutation of the Adh gene of Drosophila melanogaster containing an internal tandem duplication. J Mol Biol. 1985 Dec 20;186(4):679–688. doi: 10.1016/0022-2836(85)90388-2. [DOI] [PubMed] [Google Scholar]
- Cols N., Marfany G., Atrian S., Gonzàlez-Duarte R. Effect of site-directed mutagenesis on conserved positions of Drosophila alcohol dehydrogenase. FEBS Lett. 1993 Mar 15;319(1-2):90–94. doi: 10.1016/0014-5793(93)80043-t. [DOI] [PubMed] [Google Scholar]
- Ehrig T., Muhoberac B. B., Brems D., Bosron W. F. Monomers of human beta 1 beta 1 alcohol dehydrogenase exhibit activity that differs from the dimer. J Biol Chem. 1993 Jun 5;268(16):11721–11726. [PubMed] [Google Scholar]
- Ensor C. M., Tai H. H. Site-directed mutagenesis of the conserved tyrosine 151 of human placental NAD(+)-dependent 15-hydroxyprostaglandin dehydrogenase yields a catalytically inactive enzyme. Biochem Biophys Res Commun. 1991 Apr 30;176(2):840–845. doi: 10.1016/s0006-291x(05)80262-1. [DOI] [PubMed] [Google Scholar]
- Fossett N. G., Arbour-Reily P., Kilroy G., McDaniel M., Mahmoud J., Tucker A. B., Chang S. H., Lee W. R. Analysis of ENU-induced mutations at the Adh locus in Drosophila melanogaster. Mutat Res. 1990 Jul;231(1):73–85. doi: 10.1016/0027-5107(90)90178-7. [DOI] [PubMed] [Google Scholar]
- Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollocher H., Place A. R. Partial correction of structural defects in alcohol dehydrogenase through interallelic complementation in Drosophila melanogaster. Genetics. 1987 Jun;116(2):265–274. doi: 10.1093/genetics/116.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollocher H., Place A. R. Reexamination of alcohol dehydrogenase structural mutants in Drosophila using protein blotting. Genetics. 1987 Jun;116(2):253–263. doi: 10.1093/genetics/116.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. C., Lee W. R., Chang S. H., Silverman H. Mechanisms for dominance: Adh heterodimer formation in heterozygotes between ENU or X-ray induced null alleles and normal alleles in Drosophila melanogaster. Environ Mol Mutagen. 1992;20(4):260–270. doi: 10.1002/em.2850200404. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983 Aug 4;304(5925):412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Krook M., Ghosh D., Strömberg R., Carlquist M., Jörnvall H. Carboxyethyllysine in a protein: native carbonyl reductase/NADP(+)-dependent prostaglandin dehydrogenase. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):502–506. doi: 10.1073/pnas.90.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krozowski Z. 11 beta-hydroxysteroid dehydrogenase and the short-chain alcohol dehydrogenase (SCAD) superfamily. Mol Cell Endocrinol. 1992 Mar;84(1-2):C25–C31. doi: 10.1016/0303-7207(92)90064-d. [DOI] [PubMed] [Google Scholar]
- Lee W. R. Addition of molecular methods to mutation studies with Drosophila melanogaster. Environ Mol Mutagen. 1989;14 (Suppl 16):99–104. doi: 10.1002/em.2850140618. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C. Population genetics. Annu Rev Genet. 1985;19:81–102. doi: 10.1146/annurev.ge.19.120185.000501. [DOI] [PubMed] [Google Scholar]
- LoMonaco M. B., Lee W. R., Chang S. H. Identification of an X-ray induced deletion mutant flanked by direct repeats. Nucleic Acids Res. 1987 Sep 25;15(18):7641–7641. doi: 10.1093/nar/15.18.7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud J., Fossett N. G., Arbour-Reily P., McDaniel M., Tucker A., Chang S. H., Lee W. R. DNA sequence analysis of X-ray induced Adh null mutations in Drosophila melanogaster. Environ Mol Mutagen. 1991;18(3):157–160. doi: 10.1002/em.2850180303. [DOI] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991 Jun 20;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Obeid J., White P. C. Tyr-179 and Lys-183 are essential for enzymatic activity of 11 beta-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun. 1992 Oct 15;188(1):222–227. doi: 10.1016/0006-291x(92)92373-6. [DOI] [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991 Sep 1;200(2):537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L., Fothergill-Gilmore L. A. The active site architecture of a short-chain dehydrogenase defined by site-directed mutagenesis and structure modeling. Biochemistry. 1994 Jun 14;33(23):7047–7055. doi: 10.1021/bi00189a005. [DOI] [PubMed] [Google Scholar]
- Schwartz D. The molecular basis for allelic complementation of alcohol dehydrogenase mutants of maize. Genetics. 1975 Feb;79(2):207–212. doi: 10.1093/genetics/79.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Sofer W., Martin P. F. Analysis of alcohol dehydrogenase gene expression in Drosophila. Annu Rev Genet. 1987;21:203–225. doi: 10.1146/annurev.ge.21.120187.001223. [DOI] [PubMed] [Google Scholar]
- Tannin G. M., Agarwal A. K., Monder C., New M. I., White P. C. The human gene for 11 beta-hydroxysteroid dehydrogenase. Structure, tissue distribution, and chromosomal localization. J Biol Chem. 1991 Sep 5;266(25):16653–16658. [PubMed] [Google Scholar]
- Thatcher D. R., Sawyer L. Secondary-structure prediction from the sequence of Drosophila melanogaster (fruitfly) alcohol dehydrogenase. Biochem J. 1980 Jun 1;187(3):884–886. doi: 10.1042/bj1870884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Varughese K. I., Skinner M. M., Whiteley J. M., Matthews D. A., Xuong N. H. Crystal structure of rat liver dihydropteridine reductase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6080–6084. doi: 10.1073/pnas.89.13.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varughese K. I., Xuong N. H., Kiefer P. M., Matthews D. A., Whiteley J. M. Structural and mechanistic characteristics of dihydropteridine reductase: a member of the Tyr-(Xaa)3-Lys-containing family of reductases and dehydrogenases. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5582–5586. doi: 10.1073/pnas.91.12.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]