Abstract
Reduced brain volumes and more prominent white matter hyperintensities on MRI scans are commonly observed among older adults without cognitive impairment. However, it remains unclear whether rates of change in these measures among cognitively normal adults differ as a function of genetic risk for late-onset Alzheimer’s disease, including APOE-ɛ4, APOE-ɛ2 and Alzheimer’s disease polygenic risk scores (AD-PRS), and whether these relationships are influenced by other variables. This longitudinal study examined the trajectories of regional brain volumes and white matter hyperintensities in relationship to APOE genotypes (N = 1541) and AD-PRS (N = 1093) in a harmonized dataset of middle-aged and older individuals with normal cognition at baseline (mean baseline age = 66 years, SD = 9.6) and an average of 5.3 years of MRI follow-up (max = 24 years). Atrophy on volumetric MRI scans was quantified in three ways: (i) a composite score of regions vulnerable to Alzheimer’s disease (SPARE-AD); (ii) hippocampal volume; and (iii) a composite score of regions indexing advanced non-Alzheimer’s disease-related brain aging (SPARE-BA). Global white matter hyperintensity volumes were derived from fluid attenuated inversion recovery (FLAIR) MRI. Using linear mixed effects models, there was an APOE-ɛ4 gene-dose effect on atrophy in the SPARE-AD composite and hippocampus, with greatest atrophy among ɛ4/ɛ4 carriers, followed by ɛ4 heterozygouts, and lowest among ɛ3 homozygouts and ɛ2/ɛ2 and ɛ2/ɛ3 carriers, who did not differ from one another. The negative associations of APOE-ɛ4 with atrophy were reduced among those with higher education (P < 0.04) and younger baseline ages (P < 0.03). Higher AD-PRS were also associated with greater atrophy in SPARE-AD (P = 0.035) and the hippocampus (P = 0.014), independent of APOE-ɛ4 status. APOE-ɛ2 status (ɛ2/ɛ2 and ɛ2/ɛ3 combined) was not related to baseline levels or atrophy in SPARE-AD, SPARE-BA or the hippocampus, but was related to greater increases in white matter hyperintensities (P = 0.014). Additionally, there was an APOE-ɛ4 × AD-PRS interaction in relation to white matter hyperintensities (P = 0.038), with greater increases in white matter hyperintensities among APOE-ɛ4 carriers with higher AD-PRS. APOE and AD-PRS associations with MRI measures did not differ by sex. These results suggest that APOE-ɛ4 and AD-PRS independently and additively influence longitudinal declines in brain volumes sensitive to Alzheimer’s disease and synergistically increase white matter hyperintensity accumulation among cognitively normal individuals. Conversely, APOE-ɛ2 primarily influences white matter hyperintensity accumulation, not brain atrophy. Results are consistent with the view that genetic factors for Alzheimer’s disease influence atrophy in a regionally specific manner, likely reflecting preclinical neurodegeneration, and that Alzheimer’s disease risk genes contribute to white matter hyperintensity formation.
Keywords: Alzheimer’s disease (AD), APOE, polygenic risk score (PRS), magnetic resonance imaging (MRI), white matter hyperintensities
Soldan et al. examined Alzheimer’s disease genetic risk in relation to changes in brain volumes and white matter lesions in cognitively unimpaired adults. The APOE-ɛ4 gene and Alzheimer’s disease polygenic risk scores independently influenced atrophy and synergistically influenced white matter lesions. The APOE-ɛ2 gene increased white matter lesions, not atrophy.
Graphical Abstract
Graphical Abstract.
Introduction
Alzheimer’s disease (AD) pathology and neurodegeneration, as measured by atrophy on magnetic resonance imaging (MRI), are present many years prior to the emergence of clinical symptoms when individuals are cognitively normal.1,2 Older adults without cognitive impairment also frequently have evidence of small vessel cerebrovascular disease, which most commonly manifests as white matter hyperintensities (WMHs) on MRI scans.3 Recent evidence suggests that WMH may also play a role in Alzheimer's disease,4,5 with both vascular and Alzheimer’s disease-specific pathways contributing to WMHs.6,7 Both brain atrophy8,9 and WMH burden10,11 among individuals with normal cognition have been shown to predict subsequent cognitive decline and impairment. It remains unclear, however, whether rates of brain atrophy and WMH accumulation among cognitively normal adults differ as a function of genetic risk for late-onset Alzheimer’s disease and whether this relationship is influenced by other variables, such as age, sex, vascular risk factors and education. This is an important topic for investigation because an examination of non-modifiable and modifiable factors that influence longitudinal changes in atrophy and WMH may help identify ways to reduce brain deterioration and eventual cognitive decline in older persons.
The major genetic risk factor for late-onset Alzheimer’s disease is the apolipoprotein E (APOE) gene, with the ɛ4 allele increasing risk of dementia12,13 and the ɛ2 allele decreasing risk.14,15 Multiple additional genetic loci have been identified in genome-wide association studies (GWAS)16 to increase late-onset Alzheimer’s disease-dementia risk, though to a smaller degree than APOE-ɛ4. To assess the cumulative impact of these other genetic loci on dementia risk, they are often combined into polygenic risk scores for Alzheimer’s disease (AD-PRS).17-20
Many prior studies investigating Alzheimer’s disease-genetic risk in relation to brain atrophy or WMH accumulation have included a mixture of participants across the clinical spectrum [i.e. cognitively normal, mild cognitive impairment (MCI) and dementia] or non-demented cohorts (i.e. cognitively normal and MCI). Taken together, these studies suggest that APOE-ɛ4 genetic status21-27 and higher AD-PRS scores21,24,26,28,29 are both associated with lower volumes or thickness of Alzheimer’s disease-vulnerable regions, with higher rates of atrophy in these regions,18,30-36 and with higher levels of37-41 and greater increases in WMH burden over time27,42,43 (but see Tank et al.,24 Habes et al.,44 Lyall et al.,45 Lane et al.46 and Debette et al.47).
Research among middle-aged and older individuals with normal cognition, however, has primarily included cross-sectional studies that cannot address whether observed differences in brain volumes as a function of Alzheimer’s disease-genetic risk reflect lifelong differences in brain structure, as opposed to differential atrophy that occurs during the preclinical phase of Alzheimer’s disease. Results from cross-sectional studies have been mixed, with some finding higher WMH burden48,49 and lower volumes or thinner cortex23,50-53 among individuals at greater Alzheimer’s disease-genetic risk (i.e. APOE-ɛ4 carriers and/or higher AD-PRS), and others not finding such differences.23,54-59 For APOE-ɛ2 genetic status, results from cross-sectional studies have also been mixed.22,52,54,60,61 Few prior longitudinal studies have been conducted among cognitively unimpaired older individuals. Of these, two reported greater volume loss in Alzheimer’s disease-vulnerable regions among APOE-ɛ4 carriers compared to non-carriers,62,63 whereas two others found no APOE-ɛ4-related differences.64,65 Additionally, a relatively small study reported less hippocampal atrophy among older cognitively normal APOE-ɛ2 carriers relative to ɛ3 homozygouts,66 consistent with a study that included individuals across the Alzheimer’s disease-spectrum.33 To our knowledge, the relationship of AD-PRS and longitudinal atrophy rates or WMH accumulation have not been examined among middle-aged and older cognitively unimpaired individuals. Likewise, although cross-sectional studies across the AD-spectrum have found higher WMH burden in APOE-ɛ2 carriers relative to ɛ3/ɛ3 homozygotes,40 the impact of the APOE-ɛ2 allele on longitudinal changes in WMH burden in cognitively normal individuals remains unclear.
To address these gaps, the current study examined rates of change in regional brain volumes and WMH in a large, harmonized dataset of middle-aged and older individuals with normal cognition at baseline (mean MRI follow-up = 5.3 years, max = 24 years), with both APOE genotypes (N = 1541) and AD-PRS scores (N = 1093) available. The current study expands on prior ones in several ways. First, prior studies have been characterized by either short follow-up periods (i.e. mean follow-up 2–3.5 years),63-66 or by small sample sizes (i.e. N < 110),62,65,66 limiting their ability to draw inferences regarding less frequent alleles, including ɛ2 carrier status and ɛ4 homozygosity. Second, the large sample size allowed us to examine potential interactions between Alzheimer’s disease-genetic risk and other variables in relationship to brain atrophy or WMH accumulation, including age, sex, education, vascular risk and progressor status (i.e. remained cognitively normal versus progressed to MCI or dementia). Third, we examined atrophy rates in three different measures: (i) a composite score of Alzheimer’s disease-vulnerable regions derived from machine learning (SPARE-AD); (ii) hippocampal volume; and (iii) a composite score of regions sensitive to non-Alzheimer’s disease-related aging (SPARE-BA), also derived from machine learning. This allows for a comparison of the influence of Alzheimer’s disease-genetic risk on atrophy in Alzheimer’s disease-vulnerable and non-vulnerable regions. Lastly, we examined the impact of AD-PRS on longitudinal brain atrophy and WMH change, as well as interactions between AD-PRS and APOE genotypes on these measures.
Materials and methods
Participants
This study used data from the Preclinical Alzheimer’s disease Consortium (PAC), a multi-site collaboration established to investigate the earliest phases of Alzheimer’s disease. The PAC study includes harmonized cognitive, clinical, genetic, MRI and amyloid imaging data from five on-going longitudinal cohort studies: the Adult Children Study (ACS),67 the Australian Imaging, Biomarker, and Lifestyle study (AIBL study),68 the Biomarkers of Cognitive Decline Among Normal Individuals (BIOCARD) study,69 the Neuroimaging Substudy of the Baltimore Longitudinal Study of Aging (BLSA)70 and the Wisconsin Registry for Alzheimer’s Prevention (WRAP).71 To be included in the PAC data files, each participant had to be cognitively normal at baseline and have at least one molecular biomarker (derived from cerebrospinal fluid or positron emission tomography) collected while they were cognitively normal. By design, at least half of the participants in each cohort, except BLSA, had a family history of dementia. Individuals with epilepsy, recent strokes or remote strokes with residual effects were excluded at baseline. Additional details regarding study design and inclusion/exclusion criteria have been published previously for each cohort.67,69-72 Molecular biomarkers were not considered in the present analyses. Participants in all cohorts provided written informed consent according to the Declaration of Helsinki. The study protocols were approved by each site’s local institutional review board.
Clinical and cognitive assessments
Participants in all cohorts undergo longitudinal clinical and cognitive assessments, as well as medical, neurologic and psychiatric evaluations at regular intervals, depending on the protocol for each site (e.g. every 12, 18 or 24 months). The cognitive assessments at each site include a comprehensive neuropsychological battery covering all major cognitive domains (for details, see Gross et al.73 and Pettigrew et al.74). All sites conduct regular consensus diagnoses for all participants using published criteria, e.g. the National Institute on Aging/Alzheimer’s Association criteria for MCI75 and dementia.76
The diagnostic process for each case is handled in a comparable manner at each site: (i) clinical data are examined pertaining to the medical, neurologic and psychiatric status of the subject; (ii) reports of changes in cognition by the subject and by collateral sources are examined, based on the Clinical Dementia Rating scale; and (iii) change in cognitive performance is established. This information is then used to determine whether the subject has become cognitively impaired, and determine the likely aetiology of the impairment. Clinical diagnoses were made without knowledge of the biomarker measures. To be included in the current analyses, participants had to be cognitively normal at the time of their first MRI scan (which is considered the ‘baseline’ in these analyses) and have non-missing APOE genetic and vascular risk score data (see below). Participants with a diagnosis of ‘impaired not MCI’ were included with the cognitively normal participants, as they do not meet criteria for MCI, consistent with prior publications.69
Summary vascular risk scores were calculated using a previously validated method, based on the presence or absence of five vascular risk factors: hypertension, diabetes, obesity (defined as a body mass index > 30 kg/m2), hypercholesterolaemia and smoking within the 30 days prior to data collection.77 This information was obtained from medical history reports or medical records collected at visits coinciding with the MRI visits (±12 months). The risk factors were coded dichotomously (0 if absent and 1 if present or remote) and then summed to calculate summary scores (max = 5) for each visit, consistent with prior publications.78-80
Genetic measures
Participants at each site provided blood that was used for DNA extraction. APOE alleles were determined using standard targeted genotyping, i.e. by direct genotyping (rs7412 and rs429358) in ACS, WRAP and AIBL, or by restriction isotyping (codon 112 and 158) in BIOCARD and BLSA. Dichotomous indicators were created for APOE-ɛ2 carriers (ɛ2/ɛ2 and ɛ2/3 = 1; otherwise 0), APOE-ɛ3/ɛ3 homozygous carriers (ɛ3/ɛ3 = 1, otherwise 0) and APOE-ɛ4 carriers (ɛ2/ɛ4, ɛ3/ɛ4 and ɛ4/ɛ4 = 1; otherwise 0). Participants with ɛ2/ɛ4 alleles were included in the APOE-ɛ4 group given their risk for AD pathology is similar to that of ɛ4 carriers, rather than ɛ2 carriers.81 An additional categorical variable for APOE-ɛ4 carrier status was also created to examine potential differences between ɛ4 homozygous versus heterozygous individuals (i.e. ɛ2/ɛ2 and ɛ2/ɛ3 versus ɛ3/ɛ3 versus ɛ3/ɛ4 versus ɛ4/ɛ4). The ɛ2/ɛ2 was combined with the ɛ2/ɛ3 group due to their sample size (n = 7 across all sites).
Details regarding the generation of the AD-PRS for the PAC dataset have been described previously.74 Briefly, each site generated GWAS data using various genotyping arrays and the raw GWAS data were imputed by chip using a standard pipeline that included variant filtering for genotyping efficiency (95%), minor allele frequency (>1%) and Hardy-Weinberg equilibrium (P > 1 × 10−6). Given the racial and ethnic makeup of the included studies, all GWAS analyses were restricted to those of European ancestry that was confirmed using population principal component analysis. For the purpose of the AD-PRS analysis, we restricted all GWAS datasets to overlapping variants leaving a total of 6 739 456 common variants available in all five datasets for analysis. AD-PRS were generated using imputed GWAS data, leveraging the summary statistics provided by Kunkle et al.16 that were regenerated for us removing PAC participants who were included in the original GWAS analysis (n = 93 220). AD-PRS were computed with PLINK using a previously published method.18 The current analyses only used AD-PRS without the APOE region (i.e. 1 MB upstream and downstream of the APOE gene) to assess the independent associations of APOE and other Alzheimer’s disease risk genes on the MRI measures. AD-PRS were transformed to Z-scores to simplify interpretation, using the mean and standard deviation (SD) across all five datasets (see Pettigrew et al.74 for the distribution of harmonized AD-PRS in the PAC cohorts).
MRI assessments
Image acquisition
All PAC sites have collected structural MRI scans longitudinally, with the majority of scans acquired on 3 T scanners, but a subset on a 1.5 T scanner, since some of the studies began in the mid-1990s. See Supplementary Table 1 for details regarding the types of scanners and acquisition protocols for each site.
Image processing and harmonization
Processing of T1-weighted images included correction of intensity inhomogeneities,82 skull stripping83 and segmentation of the brain into a set of anatomical regions of interest (ROIs) using the Multi-atlas Region Segmentation Utilizing Ensembles (MUSE) software platform.84 This method was specifically designed for longitudinal studies to handle differences in scanners and imaging protocols over time and across sites and employs harmonized acquisition-specific atlases. For a detailed description of these methods, see Erus et al.85 and Habes et al.86 Briefly, MUSE uses a consensus labelling framework that combines an ensemble of labelled atlases in target image space by using multiple atlases reflecting a broad representation of anatomy. Scanner-specific atlases share the same ROI labels, imposing consistency of segmentations, while each atlas set preserves the image intensity characteristics of the specific scanner. The MUSE pipeline has been extensively validated against benchmark methods and applied in various cross-sectional and longitudinal studies.64,84,86,87 In comparison to most commonly used segmentation tools, such as FreeSurfer, MUSE has demonstrated significant improvement in accuracy and more consistent segmentations across scanners, particularly in segmentation of deep brain structures.88 The MUSE software package is freely available: https://www.med.upenn.edu/cbica/sbia/muse.html.
Additional statistical harmonization was applied to the ROI volumes based on the multivariate ComBAT-GAM method89 to remove cohort-related effects and protocol-specific variability. This method simultaneously models scanner effects (unwanted sources of variation) and covariate associations (e.g. age and sex). This harmonization approach integrates a generalized additive model, with a smoothed non-linear term for age, using thin plate regression splines, and linear terms for sex and intra-cranial volume (ICV), thereby preserving age and sex differences across sites.89
Quantification of WMH volumes from fluid attenuated inversion recovery (FLAIR) images was completed using an automated deep learning based segmentation method90 that is built upon the UNet architecture,91 with the convolutional network layers replaced by an Inception ResNet architecture.92 The network model uses inhomogeneity corrected and co-registered FLAIR and T1-weighted images as input, and has been trained using a multi-site training dataset with human-validated WMH labels, as published previously.86 The algorithm was applied to MRI scans of PAC participants to calculate binary WMH masks, and to extract regional WMH volumes. The current analyses used global WMH volumes.
Volumetric regions of interest and spatial patterns of atrophy
Harmonized volumes of the left and right hippocampus were normalized for head size by regressing the average of the left and right hemispheres on ICV. The standardized residuals (mean = 0, SD = 1) were used in analyses presented below. Hippocampal volumes were examined to enable direct comparison to many prior studies that have specifically focused on this structure.
Atrophy in regions vulnerable to Alzheimer's disease was measured using SPARE-AD scores, which represent an imaging signature of Alzheimer's disease-like neurodegeneration derived from machine learning, as previously described and validated.28,93 For SPARE-AD calculation, a support vector machine classifier with a linear kernel was trained to maximally differentiate between cognitively unimpaired participants and participants with AD-dementia, using a curated dataset of over 10 000 individuals, known as the iSTAGING consortium,86 which includes the PAC sites. More positive SPARE-AD scores imply a more Alzheimer's disease-like brain structure (i.e. more AD-related atrophy).
We also calculated a brain signature of age-related brain atrophy, using SPARE-BA scores, to estimate structural brain changes due to aging. As published previously, this MRI approach uses a multivariate pattern regression method based on support vector regression to calculate brain aging scores for each participant.86,94 The model was trained with the T1-MR scans using harmonized ROI volumes for structures. In the present analyses, SPARE-BA scores were regressed on age at scan and the standardized residuals (referred to in the tables as SPARE-BA-resid) were used in the analyses presented below, with more positive scores indicating greater age-related atrophy compared to normative trends. This is comparable to ‘brain age gap’ scores estimated using related techiques.95,96 The regions contributing to the SPARE-BA scores are weighted optimally for estimating age, whereas the regions contributing to SPARE-AD scores are weighted optimally to distinguish between cognitively normal individuals and individuals with Alzheimer's disease-dementia. See Supplementary Fig. 1 for illustrations of the SPARE-AD and SPARE-BA masks and Supplementary Text 1 for additional information on SPARE-AD and SPARE-BA.
Statistical analyses
We used linear mixed effects models with random intercepts and slopes with an unstructured covariance to evaluate whether the trajectories of the MRI measures of atrophy (i.e. SPARE-AD and SPARE-BA), hippocampal volume or WMH burden differed based on AD-genetic risk. Separate models were run for each MRI outcome measure. The primary models evaluating APOE effects included the following predictors: baseline age, sex, education, dichotomous indicators for APOE-ɛ2 and APOE-ɛ4 (with ɛ3/ɛ3 as the reference group), indicators for site (to control for potential site differences), time and the interaction (cross-product) of each predictor with time. The primary models evaluating AD-PRS were the same as the APOE models, but additionally included the AD-PRS. The years of education variable was standardized to Z-scores separately for cohorts within versus outside the USA, given differences in the number of years of compulsory schooling. Final models examining the trajectories of the atrophy measures and hippocampal volume included a time2 term to account for their statistically significant non-linear change over time. For the WMH models, the time2 was not significant and therefore not included. The primary APOE models were re-run with APOE-ɛ4 status coded categorically (as described above) to evaluate whether the MRI trajectories differed between APOE ɛ3/ɛ4 and ɛ4/ɛ4 carriers. In a sensitivity analysis, the primary models were also re-run excluding APOE ɛ2/ɛ4 carriers.
A second set of linear mixed effects models evaluated whether the results remained the same when additionally covarying both vascular risk summary scores (using all available measures over time) and participants’ follow-up diagnostic status, based on their last (i.e. most recent) consensus diagnosis (coded as 0 = remained normal, or 1 = progressed to MCI or dementia). These models were identical to the primary models, but additionally included the vascular risk summary scores over time and binary indicators for progressor status, as well as their interactions with time.
Lastly, to evaluate whether the relationships between the Alzheimer's disease-genetic factors and the MRI measures were modified by the demographic or clinical variables, a third set of linear mixed effects models were run. These models were the same as the primary models, but additionally included three-way interaction terms for the genetic factors × demographic/clinical variable × time (e.g. APOE-ɛ2 × baseline age × time and ɛ4 × baseline age × time; or AD-PRS × baseline age × time), as well as the corresponding lower-order interaction terms. These models were not adjusted for multiple comparisons because they were exploratory in nature and correcting for multiple comparisons in exploratory analyses can increase the likelihood of Type II errors and potentially obscure meaningful findings.
Data analysis was performed using STATA 17.0 and P-values of <0.05 were considered significant.
Results
Table 1 shows baseline characteristics of participants included in the MRI volumetric and WMH analyses, separately for participants in the APOE and AD-PRS analyses. For baseline characteristics by cohort, see Supplementary Tables 2 and 3. On average, participants were in their mid-60s at baseline, primarily White and highly educated. About one-third of participants were APOE-ɛ4 carriers and approximately two-thirds had one or more vascular risk factor. The mean number of volumetric MRI measures over time was 3 (max = 18), with a mean 5.3 years between the first and last MRI scan (max = 24 years). Out of the 1541 participants with volumetric data, 1348 also had one or more WMH measure (mean number of measures over time = 2, max = 10; mean time between baseline and last WMH measure = 2.9 years, max = 19 years).
Table 1.
Participant characteristics at baseline
| Participants in APOE analyses with volumetric data | Participants in APOE analyses with WMH data | Participants in AD-PRS analyses with volumetric data | Participants in AD-PRS analyses with WMH data | |
|---|---|---|---|---|
| N | 1541 | 1348 | 1093 | 972 |
| Age at baseline MRI scan, M (SD) | 66.2 (9.6) | 68.7 (9.6) | 66.1 (9.4) | 68.6 (9.6) |
| Female sex, N (%) | 929 (60.3%) | 816 (60.6%) | 672 (61.5) | 605 (62.2%) |
| Years of education, M (SD) | 15.2 (3.1) | 15.2 (3.2) | 15.0 (3.2) | 15.1 (3.2) |
| Race, White, N (%) | 1449 (94.0%) | 1273 (94.4%) | 1093 (100%) | 972 (100%) |
| MMSE score, M (SD) | 29.0 (1.2) | 29.0 (1.3) | 29.1 (1.1) | 29.1 (1.3) |
| Progressed to MCI/dementia, N (%) | 94 (6.1%) | 82 (6.1%) | 71 (6.5%) | 64 (6.6%) |
| Vascular risk score, M (SD) | 1.1 (1.0) | 1.1 (1.1) | 1.1 (1.0) | 1.1 (1.0) |
| Vascular risk score ≥ 1, N (%) | 1038 (67.4%) | 904 (67.2%) | 727 (66.5%) | 648 (66.7%) |
| Vascular risk score ≥ 2, N (%) | 481 (31.2%) | 433 (32.2%) | 326 (29.8%) | 307 (31.6%) |
| Vascular risk score ≥ 3, N (%) | 164 (10.5%) | 155 (11.5%) | 98 (9.0%) | 101 (10.4%) |
| Genetic factors | ||||
| APOE ɛ2 carriers, N (%)a | 184 (11.9%) | 164 (12.2%) | 125 (11.5%) | 113 (11.6%) |
| APOE ɛ4 carriers, N (%)b | 495 (32.1%) | 417 (31.0%) | 339 (31.0%) | 293 (30.1%) |
| APOE ɛ3/ɛ3 carriers, N (%) | 862 (55.9%) | 765 (56.8%) | 629 (57.5%) | 566 (58.2%) |
| APOE ɛ4/ɛ4 carriers, N (%) | 63 (4.1%) | 52 (3.9%) | 46 (4.2%) | 37 (3.8%) |
| APOE ɛ3/ɛ4 carriers, N (%) | 385 (25.5%) | 328 (24.4%) | 263 (24.1%) | 232 (23.9%) |
| APOE ɛ2/ɛ4 carriers, N (%) | 46 (3.0%) | 37 (2.7%) | 30 (2.7%) | 24 (2.5%) |
| AD-PRS, M (SD) | −0.02 (0.97) | −0.02 (0.97) | ||
| Baseline MRI measures | ||||
| SPARE_AD, M (SD) | −1.3 (0.8) | −1.2 (0.9) | −1.3 (0.8) | −1.2 (0.9) |
| SPARE_BA, M (SD) | 66.8 (11.2) | 68.6 (11.4) | 66.7 (10.9) | 68.6 (11.3) |
| SPARE_BA residual, M (SD) | −0.3 (7.1) | −0.1 (7.2) | −0.3 (7.1) | −0.0 (7.1) |
| Hippocampal volume, M (SD) in mm3 | 3766 (404) | 3734 (404) | 3768 (402) | 3730 (398) |
| WMH volume (in mm3), M (SD) | 2810 (5408) | 3105 (5837) | 2821 (5540) | 3200 (6096) |
| Number of MRI measures over time, M (SD) [range] | 3.0 (2.5) [1–18] | 2.0 (1.2) [1–10] | 3.1 (2.5) [1–18] | 2.0 (1.2) [1–8] |
| N participants with two or more MRI scans over time, (%) | 998 (64.8%) | 769 (57.1%) | 777 (71.1%) | 599 (61.6%) |
| Years between baseline and last MRI, M (SD) [range] | 5.3 (5.7) [0–24.4] | 2.9 (4.0) [0–18.9] | 5.7 (5.7) [0–24.4] | 3.0 (3.9) [0–18.9] |
aIncludes ɛ2/ɛ2 and ɛ2/ɛ3 carriers.
bIncludes ɛ2/ɛ4, ɛ3/ɛ4 and ɛ4/ɛ4 carriers.
APOE genotypes and MRI trajectories
Results from the primary model examining the binary APOE-ɛ2 and ɛ4 indicators in relationship to trajectories of the MRI measures are shown in Table 2 (with ɛ3/ɛ3 as the reference). APOE-ɛ4 carrier status was not associated with any MRI measure at baseline (all P ≥ 0.14). However, relative to ɛ3/ɛ3 carriers, ɛ4 carriers demonstrated greater increases in SPARE-AD (P < 0.001) scores and greater decreases in hippocampal volume (P ≤ 0.001) over time; they also showed greater increases in SPARE-BA (P = 0.025), though the effect appeared smaller (Z = 2.25 versus Z = 3.45 for SPARE-AD; see Supplementary Text 2 for formal model comparison). APOE-ɛ2 carrier status was also not associated with any baseline MRI measure (all P ≥ 0.12), but ɛ2 carriers showed greater increases in WMH volumes over time (P = 0.014), compared to ɛ3/ɛ3 carriers. There was no association between ɛ2 carrier status and rate of change in the other MRI measures (all P > 0.3).
Table 2.
Mixed effects model results of APOE genetic status in relationship to MRI measures
| SPARE-AD (AD-related atrophy) | SPARE-BA-resid (age-related atrophy) | Hippocampus volume | WMH volume | |||||
|---|---|---|---|---|---|---|---|---|
| Model predictors | Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value |
| Time | −0.184 (0.025) | <0.0001 | −0.037 (0.021) | 0.09 | 0.104 (0.018) | <0.0001 | −0.102 (−0.023) | <0.0001 |
| Time2 | 0.003 (0.0003) | <0.0001 | −0.001 (0.0003) | <0.0001 | −0.0004 (0.0002) | 0.038 | ||
| Age | 0.038 (0.003) | <0.0001 | −0.012 (0.003) | 0.001 | −0.035 (0.003) | <0.0001 | 0.051 (0.003) | <0.0001 |
| Age × time | 0.003 (0.0004) | <0.0001 | 0.001 (0.0003) | <0.0001 | −0.003 (0.0003) | <0.0001 | 0.002 (0.0003) | <0.0001 |
| Sex (female) | −0.002 (0.049) | 0.97 | 0.013 (0.053) | 0.80 | −0.052 (0.048) | 0.29 | 0.022 (0.050) | 0.66 |
| Sex (F) × time | −0.016 (0.006) | 0.009 | −0.028 (0.005) | <0.0001 | 0.007 (0.004) | 0.08 | 0.007 (0.005) | 0.20 |
| Education | 0.008 (0.025) | 0.74 | 0.029 (0.026) | 0.27 | 0.014 (0.024) | 0.56 | −0.001 (0.025) | 0.97 |
| Education × time | 0.008 (0.003) | 0.74 | −0.003 (0.002) | 0.17 | −0.001 (0.002) | 0.75 | −0.001 (0.003) | 0.66 |
| APOE-ɛ2 | −0.042 (0.066) | 0.53 | 0.018 (0.071) | 0.80 | 0.009 (0.065) | 0.89 | 0.008 (0.067) | 0.12 |
| APOE-ɛ2 × time | 0.002 (0.008) | 0.84 | −0.000 (0.006) | 0.99 | 0.005 (0.005) | 0.34 | 0.016 (0.007) | 0.014 |
| APOE-ɛ4 | 0.076 (0.051) | 0.14 | −0.045 (0.055) | 0.41 | −0.010 (0.050) | 0.83 | −0.042 (0.052) | 0.42 |
| APOE-ɛ4 × time | 0.021 (0.006) | 0.001 | 0.011 (0.005) | 0.025 | −0.167 (0.004) | <0.0001 | 0.008 (0.005) | 0.13 |
Additionally, across all models (Table 2), older age was associated with higher SPARE-AD scores, smaller hippocampal volumes and greater WMH volumes at baseline and greater increases in SPARE-AD, SPARE-BA scores and WMH volumes over time, as well as greater decreases in hippocampal volume over time (all P < 0.0001). Participant sex was not associated with any baseline MRI measure, but females had smaller increases in SPARE-AD (P = 0.009) and SPARE-BA (P < 0.0001) scores over time than males. Years of education was not associated with the baseline or rate of change in any MRI measure (all P ≥ 0.17). These results remained the same when excluding APOE-ɛ2/ɛ4 carriers from the analysis (data not shown), except that the association between APOE-ɛ4 carrier status and rate of change in SPARE-BA was no longer significant (estimate = 0.010, SE = 0.005, P = 0.069).
The pattern of results was also the same when additionally adjusting for vascular risk scores and follow-up diagnosis (see Supplementary Table 4). In these models, higher vascular risk scores were associated with higher baseline SPARE-AD scores (P = 0.026), but were not related to the rate of change in any MRI measure (all P ≥ 0.05). Additionally, participants who progressed to MCI/dementia over time had higher baseline WMH volumes (P = 0.012), greater increases in WMH volumes (P = 0.012) and SPARE-AD scores (P < 0.0001) over time, and greater declines in hippocampal volume (P < 0.0001) after also adjusting for vascular risk and follow-up diagnosis.
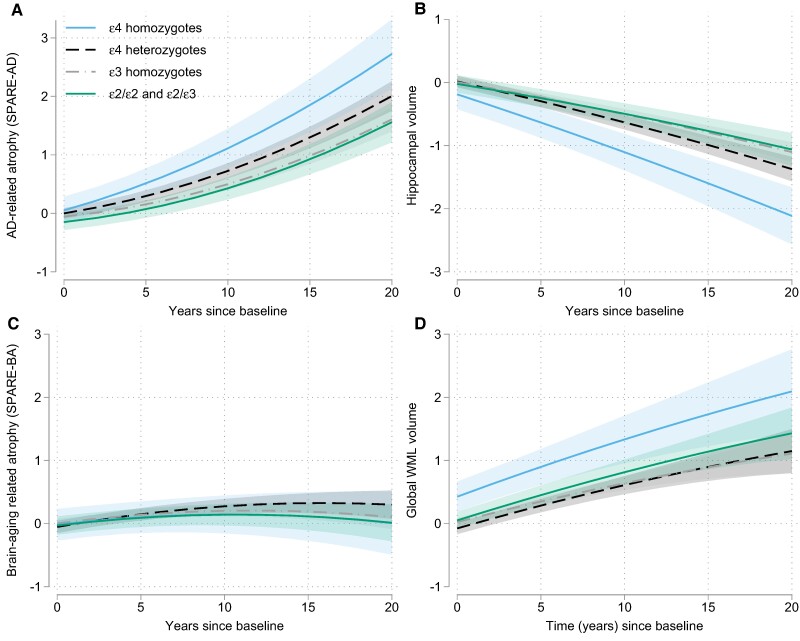
When modelling APOE as a categorical variable, the pattern of results was the same. Additionally, we observed that SPARE-AD scores increased more among APOE ɛ4/ɛ4 carriers relative to ɛ4 heterozygous participants (estimate = 0.033, SE = 0.015, P = 0.028), who had greater increases than ɛ3/ɛ3 carriers (estimate = 0.017, SE = 0.006, P = 0.008). This is illustrated in Fig. 1A. A similar pattern was observed for hippocampal volumes, which showed greater decline over time among ɛ4/ɛ4 carriers compared to ɛ4 heterozygous individuals (estimate = 0.026, SE = 0.011, P = 0.014), who showed greater decline relative to ɛ3/ɛ3 carriers (estimate = 0.013, SE = 0.004, P = 0.003), see Fig. 1B. There was no difference in the SPARE-BA trajectories between individuals with one versus two ɛ4 alleles (P > 0.05; Fig. 1C). For WMH volumes, both ɛ4/ɛ4 carriers (estimate = 0.029, SE = 0.014, P = 0.039) and ɛ2 carriers (ɛ2/ɛ2 and ɛ2/ɛ3 combined, estimate = 0.015, SE = 0.008, P = 0.049) showed greater increases over time than ɛ3/ɛ3 carriers, who did not differ from ɛ4 heterozygous participants (estimate = 0.007, SE = 0.006, P = 0.23), Fig. 2D. Differences between ɛ4 homozygous and heterozygous individuals remained the same when additionally covarying follow-up diagnosis and vascular risk scores (data not shown).
Figure 1.
Longitudinal volumetric atrophy and WMH volumes as a function of APOE genetic status. Figure shows estimates from mixed effects regression model examining APOE genetic status in relation to (A) AD-related atrophy, measured by SPARE-AD scores, (B) hippocampal volumes, and (C) age-related atrophy, and (D) WMH volumes. The rate of change in AD-related atrophy (estimate = 0.033, SE = 0.015, P = 0.028) and hippocampal volumes (estimate = 0.026, SE = 0.011, P = 0.014) was greater for individuals with two ɛ4 alleles compared to those with one ɛ4 allele, who in turn showed more atrophy than ɛ3/ɛ3 carriers (estimate = 0.017, SE = 0.006, P = 0.008 for SPARE = AD and (estimate = 0.013, SE = 0.004, P = 0.003 for hippocampus). Atrophy rates did not differ between ɛ2 carrier and ɛ3/ɛ3 carriers (all P > 0.3). WMH volumes (D) increased more over time among both ɛ4/ɛ4 carriers (estimate = 0.029, SE = 0.014, P = 0.039) and ɛ2 carriers (estimate = 0.015, SE = 0.008, P = 0.049) relative to ɛ3/ɛ3 carriers (see text for details).
Figure 2.
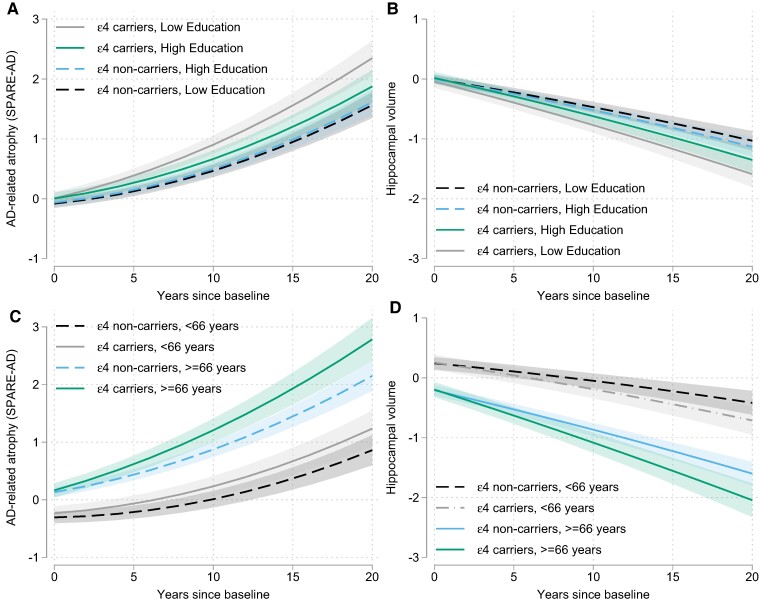
Longitudinal volumetric atrophy based on APOE-ɛ4 genetic status and participant education and age. Estimates from mixed effects regression model showing how years of education (A and B) and baseline age (C and D) modify the association between APOE-ɛ4 genetic status and rate of change of Alzheimer's disease-vulnerable regions, measured by SPARE-AD scores (A and C), and the hippocampus (B and D). The negative effect of APOE-ɛ4 genetic status on rate of atrophy in Alzheimer's disease-vulnerable regions and the hippocampus was greater among older than young participants (C and D), as indicated by significant three-way interactions of ɛ4 × age × time for SPARE-AD (estimate = 0.002, SE = 0.001, P = 0.001) and for the hippocampus (estimate = −0.001, SE = 0.0004, P = 0.027), but was attenuated among individuals with more years of education (A and B), as indicated by significant ɛ4 × education × time interactions for SPARE-AD (estimate = −0.015, SE = 0.006, P = 0.017) and for the hippocampus (estimate = 0.009, SE = 0.004, P = 0.039). For illustration purposes, the 25th and 75th percentiles of the baseline education were used to show trajectories of high versus low education.
Results from models examining whether the demographic and clinical variables modified the relationships between APOE genotypes and the MRI trajectories are shown in Table 3. The associations of APOE-ɛ4 genetic status with rates of change in SPARE-AD scores and hippocampal volume were modified by baseline age and years of education (all P for interaction terms of ɛ4 × (age or education) × time ≤ 0.039). Specifically, ɛ4 related atrophy in SPARE-AD regions and the hippocampus was greater among older participants and weaker among those with more years of education. These three-way interactions remained significant when excluding APOE ɛ2/ɛ4 carriers (all P ≤ 0.012) and are illustrated in Fig. 2. Additionally, the relationship between ɛ4 genetic status and the rate of decline in hippocampal volume was greater among participants with higher compared to lower vascular risk scores (P = 0.038) and among those who progressed to MCI or dementia over time compared to those who remained cognitively unimpaired (P = 0.001), see Table 3. However, the three-way interactions with vascular risk scores or progressor status were not significant when excluding APOE ɛ2/ɛ4 carriers (both P > 0.21). Among APOE-ɛ2 carriers, higher education was unexpectedly associated with greater increases in SPARE-AD over time (P = 0.032); however, this interaction was also not significant after exclusion of APOE ɛ2/ɛ4 carriers (P > 0.15).
Table 3.
Results from mixed effects models testing whether associations between genetic risk factors and rate of change in MRI measures differ by demographic and clinical variables
| SPARE-AD (AD-related atrophy) | SPARE-BA-resid (age-related atrophy) | Hippocampus volume | WMH volume | |||||
|---|---|---|---|---|---|---|---|---|
| Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value | |
| Age | ||||||||
| APOE-ɛ2 × age × time | −0.001 (0.001) | 0.36 | −0.001 (0.001) | 0.07 | 0.001 (0.001) | 0.10 | −0.000 (0.001) | 0.96 |
| APOE-4 × age × time | 0.002 (0.001) | 0.001 | 0.000 (0.001) | 0.69 | −0.001 (0.000) | 0.027 | −0.000 (0.001) | 0.67 |
| AD-PRS × age × time | −0.000 (0.001) | 0.63 | −0.000 (0.000) | 0.15 | 0.000 (0.000) | 0.45 | 0.000 (0.000) | 0.11 |
| Sex | ||||||||
| APOE-ɛ2 × sex × time | −0.005 (0.016) | 0.78 | −0.008 (0.014) | 0.57 | −0.004 (0.011) | 0.69 | −0.016 (0.014) | 0.27 |
| APOE-4 × sex × time | −0.015 (0.013) | 0.26 | −0.005 (0.011) | 0.63 | −0.000 (0.009) | 0.97 | −0.002 (0.012) | 0.89 |
| AD-PRS × sex × time | 0.008 (0.007) | 0.24 | −0.002 (0.005) | 0.78 | −0.005 (0.005) | 0.26 | −0.001 (0.006) | 0.88 |
| Education | ||||||||
| APOE-ɛ2 × Educ × time | 0.018 (0.008) | 0.032 | 0.005 (0.007) | 0.49 | −0.010 (0.006) | 0.08 | −0.005 (0.008) | 0.48 |
| APOE-ɛ4 × Educ × time | −0.015 (0.006) | 0.017 | −0.007 (0.006) | 0.20 | 0.009 (0.004) | 0.039 | −0.006 (0.006) | 0.28 |
| AD-PRS × Educ × time | 0.007 (0.004) | 0.058 | 0.002 (0.003) | 0.51 | −0.004 (0.002) | 0.10 | −0.000 (0.003) | 0.99 |
| Vascular risk scores (VRS) | ||||||||
| APOE-ɛ2 × VRS × time | 0.005 (0.005) | 0.29 | −0.002 (0.005) | 0.69 | 0.001 (0.004) | 0.77 | 0.006 (0.006) | 0.33 |
| APOE ɛ4 × VRS × time | 0.002 (0.004) | 0.59 | 0.002 (0.004) | 0.58 | −0.006 (0.003) | 0.038 | −0.042 (0.025) | 0.09 |
| AD-PRS × VRS × time | 0.003 (0.002) | 0.20 | −0.002 (0.002) | 0.29 | −0.002 (0.002) | 0.23 | 0.003 (0.003) | 0.30 |
| Progressed | ||||||||
| APOE-ɛ2 × Progr × time | 0.028 (0.026) | 0.29 | −0.019 (0.023) | 0.42 | 0.011 (0.018) | 0.54 | 0.004 (0.030) | 0.89 |
| APOE-ɛ4 × Progr × time | 0.022 (0.019) | 0.25 | −0.008 (0.017) | 0.65 | −0.046 (0.013) | 0.001 | −0.011 (0.019) | 0.55 |
| AD-PRS × Progr × time | −0.004 (0.010) | 0.66 | −0.005 (0.008) | 0.51 | 0.012 (0.007) | 0.09 | −0.002 (0.009) | 0.80 |
Three-way interaction terms for each demographic variable (i.e. baseline age, sex and years of education) or clinical variable (i.e. VRS score and progressed status) with APOE-ɛ4 and APOE-ɛ2 (or AD-PRS scores) and time were tested in separate models, adjusting for baseline age, sex, years of education, cohort indicators and interactions of all predictors with time. All lower-order interaction terms for the three-way interactions were also included. Bold values represent significant effects at P < 0.05.
Alzheimer's disease-polygenic risk score and MRI trajectories
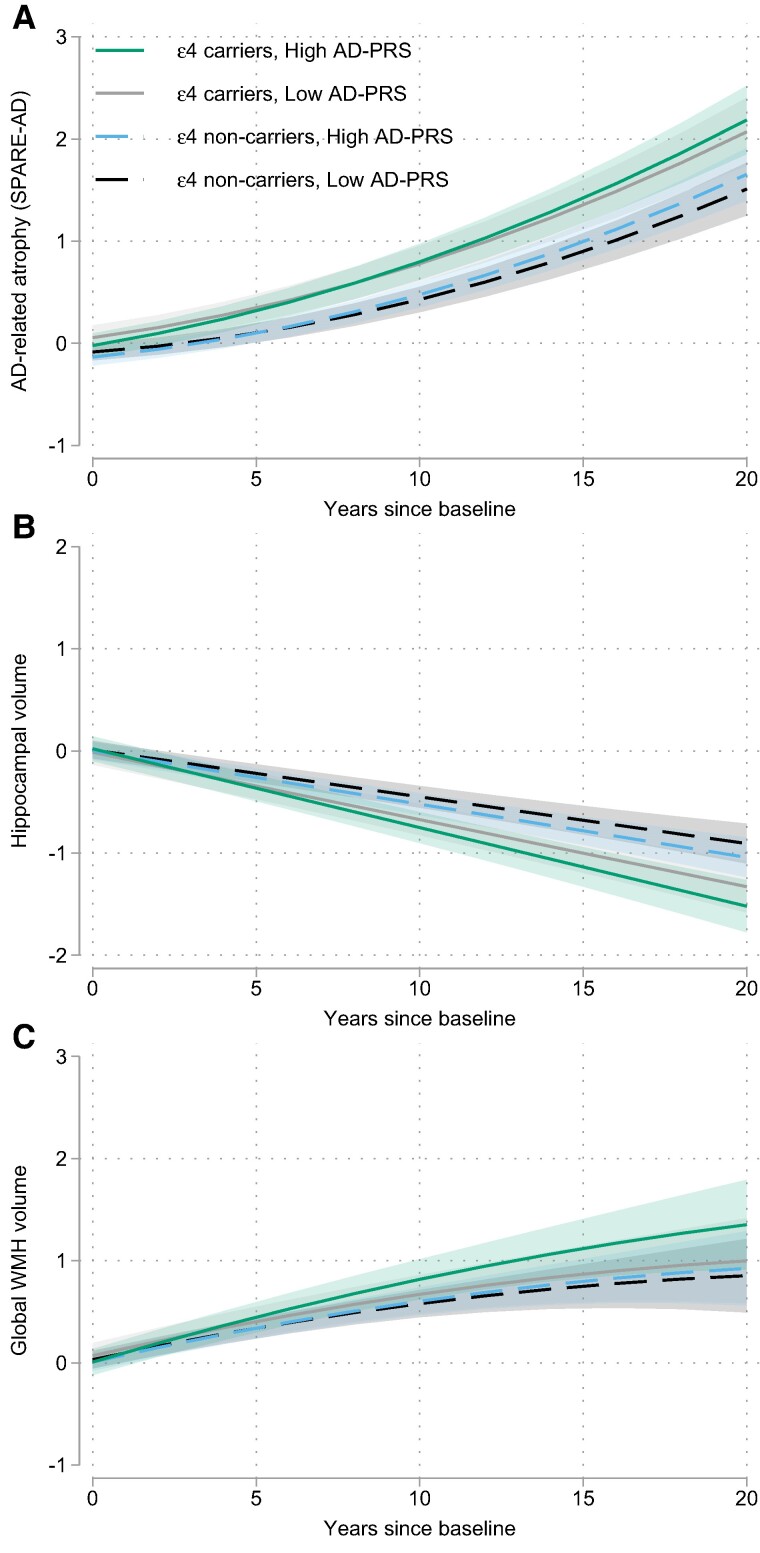
In the primary models, there was no association between AD-PRS scores and baseline levels of the MRI measures (all P ≥ 0.12). By contrast, higher AD-PRS scores were associated with greater increases over time in SPARE-AD scores, greater decreases in hippocampal volume and greater increases in WMH volumes (all P ≤ 0.035, see Table 4 and Fig. 3). Results were similar using a dichotomous AD-PRS (see Supplementary Table 5). The associations between the AD-PRS score and rate of change in the MRI measures were independent of APOE genetic status and were not modified by age, sex, years of education, vascular risk scores or progressor status (see Table 3).
Table 4.
Mixed effects model results of AD-polygenic risk score and APOE genetic status in relationship to MRI measures
| SPARE-AD (AD-related atrophy) | SPARE-BA-resid (age-related atrophy) | Hippocampus volume | WMH volume | |||||
|---|---|---|---|---|---|---|---|---|
| Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value | Estimate (SE) | P-value | |
| AD-PRS | −0.045 (0.029) | 0.12 | −0.001 (0.032) | 0.97 | 0.006 (0.030) | 0.85 | −0.028 (0.031) | 0.38 |
| AD-PRS × time | 0.007 (0.003) | 0.035 | 0.004 (0.003) | 0.15 | −0.006 (0.003) | 0.014 | 0.008 (0.003) | 0.009 |
| APOE-ɛ2 | 0.061 (0.079) | 0.44 | 0.061 (0.086) | 0.48 | −0.093 (0.081) | 0.25 | 0.069 (0.085) | 0.42 |
| APOE-ɛ2 × time | −0.000 (0.010) | 0.99 | −0.004 (0.008) | 0.65 | 0.006 (0.007) | 0.36 | 0.006 (0.008) | 0.43 |
| APOE-ɛ4 | 0.131 (0.061) | 0.032 | −0.049 (0.066) | 0.46 | −0.009 (0.062) | 0.89 | 0.025 (0.065) | 0.70 |
| APOE-ɛ4 × time | 0.021 (0.007) | 0.003 | 0.010 (0.006) | 0.09 | −0.022 (0.005) | <0.0001 | 0.014 (0.006) | 0.027 |
All models were adjusted by baseline age, sex, years of education, indicators for each cohort and included interactions of each predictor with time (e.g. terms for all genetic predictors × time and covariates × time).
Figure 3.
Longitudinal volumetric atrophy and WMH volumes as a function of AD-polygenic risk and APOE-ɛ4. Estimates from mixed effects regression model showing independent associations between AD-polygenic risk scores and APOE-ɛ4 genetic status with longitudinal atrophy in AD-vulnerable regions (i.e. SPARE-AD scores, A, estimate = 0.007, SE = 0.003, P = 0.035) and the hippocampus (B, estimate = −0.006, SE = 0.003, P = 0.014). For global WMH volumes (C), APOE-ɛ4 genetic status was more strongly associated with increases in WMH volumes among those with higher compared to lower AD-polygenic risk scores (AD-PRS × APOE-ɛ4 × time interaction, estimate = 0.014, SE = 0.007, P = 0.038). For illustration purposes, the 25th and 75th percentiles of the baseline AD-PRS scores were used to show trajectories of high versus low PRS.
Lastly, we explored potential interactions between the AD-PRS scores and APOE-ɛ4 and ɛ2 genetic status in relationship to change in the MRI measures. To simplify interpretation, APOE-ɛ2/ɛ4 carriers were excluded from these analyses. There were no interactions between the AD-PRS score and APOE-ɛ4 or APOE-ɛ2 genetic status with respect to the rate of change in SPARE-AD, SPARE-BA and hippocampal volume (all P > 0.47). However, for WMH volumes, there was an AD-PRS × APOE-ɛ4 × time interaction (P = 0.038, see Supplementary Table 6), suggesting a stronger association between APOE-ɛ4 genetic status and rate of increase in WMH among participants with higher compared to lower AD-PRS scores (Fig. 3C).
Discussion
The large sample size and substantial follow-up period of the current study provide the basis for several new insights on the relationship between genetic risk factors for late-onset Alzheimer's disease among cognitively normal individuals and changes in MRI measures of brain atrophy and WMH. First, the atrophy rates in a composite volume measure of Alzheimer's disease-vulnerable regions (SPARE-AD) and the hippocampus demonstrated an APOE-ɛ4 gene-dose effect, with greatest atrophy among ɛ4 homozygous participants, followed by ɛ4 heterozygous participants, and least among ɛ4 non-carriers. Second, both APOE-ɛ4 status and AD-PRS scores independently influenced rates of change in AD-vulnerable regions and the hippocampus, suggesting additive effects. Third, the negative impact of APOE-ɛ4 on atrophy in AD-vulnerable regions and the hippocampus was reduced among individuals with higher education and younger baseline ages. Fourth, ɛ4 carrier status was associated with greater increases in global WMH volumes over time, particularly among ɛ4 homozygous participants and those with high Alzheimer's diesease-polygenic risk scores. Fifth, ɛ2 carrier status did not influence atrophy rates in regions sensitive to aging or Alzheimer's disease, but was associated with greater increases in WMH volumes over time. In contrast, neither APOE nor AD-PRS scores showed robust associations with atrophy in a composite measure of regions sensitive advanced non-Alzheimer's disease-related brain aging (SPARE-BA), supporting prior evidence that this measure largely reflects age but not disease-related atrophy. Taken together, these results underscore the impact of AD-genetic risk factors on rates of change in MRI measures of neurodegeneration among middle-aged and older adults with normal cognition and point to potential interactions and synergistic effects between APOE and other Alzheimer's disease risk genes on WMH burden.
APOE-e4 genetic status and brain atrophy
The current results are consistent with prior longitudinal studies among participants across the clinical spectrum of Alzheimer's disease33 and non-demented cohorts30-32 that have reported elevated longitudinal atrophy in Alzheimer's disease-vulnerable regions among ɛ4 carriers compared to non-carriers. Our findings are also in line with, and extend prior work, among individuals with normal cognition62,63,97 by documenting an APOE-ɛ4 gene-dose effect on atrophy rates in the hippocampus and a composite of Alzheimer's disease-vulnerable regions. This gene-dose effect likely reflects the fact that Alzheimer's disease pathology (i.e. amyloid and tau) begins to accumulate at an earlier age among ɛ4 carriers compared to non-carriers, with ɛ4 homozygous individuals showing the youngest age of onset of amyloid accumulation and amyloid positivity, followed ɛ4 heterozygous individuals, and then ɛ4 non-carriers.98-100 It is hypothesized that this earlier age of amyloid accumulation likely initiates an earlier onset of AD-related atrophy in selected brain regions. This is consistent with the view that subtle Alzheimer's disease-related atrophy begins during the preclinical phase of the disease, when individuals are cognitively normal.8,9,101 The lack of volumetric differences between ɛ4 carriers and non-carriers at baseline supports the view that APOE-ɛ4 primarily influences brain volumes during the preclinical phase of Alzheimer's disease, but has limited impact on volumes prior to midlife, though such differences have been demonstrated.26,102 Our results also demonstrate that APOE-ɛ4 related differences in hippocampal atrophy appear to be particularly evident among individuals who progress to MCI or dementia over time, in line with a prior study.97 The finding that ɛ4 was only weakly associated with atrophy in non-Alzheimer's disease regions that are sensitive to aging (SPARE-BA) underscores the specificity of ɛ4 to Alzheimer's disease-related atrophy and might reflect the fact that in some individuals, Alzheimer's disease pathology begins in more atypical regions.103
Our results also showed that the association between APOE-ɛ4 genetic status and rate of atrophy in the hippocampus and the Alzheimer's disease-vulnerable regions increases with advancing age (Fig. 2C and D), in line with the age-related increase in Alzheimer's disease pathology accumulation98 and the age-related increase in Alzheimer's disease-related cognitive impairment. This finding might also explain why some prior studies among middle-aged cohorts have failed to find cross-sectional volumetric differences by ɛ4-status.56-58 We found no evidence that the association between APOE-ɛ4 and atrophy in AD-vulnerable regions or regions sensitive to aging (SPARE-BA) was influenced by overall levels of vascular risk, though for the hippocampus, higher vascular risk scores were associated with a stronger relationship between APOE-ɛ4 genetic status and atrophy over time (but this was not significant when excluding ɛ2/ɛ4 carriers). Although vascular risk factors have been consistently linked with smaller regional brain volumes28,104,105 and atrophy rates,47,97 little is known about whether APOE variants moderate this relationship. Additionally, the relationship between vascular risk and brain atrophy may differ by other factors, such as amyloid burden106 and by type of vascular risk factor (e.g. hypertension versus obesity).64 Thus, summary scores using different risk factors, or differentially weighted risk factors, may potentially show stronger associations with brain atrophy than was observed here.
In this study, we also observed that years of education modified the association between APOE-ɛ4 genetic status and atrophy in the Alzheimer's disease-vulnerable regions composite and the hippocampus, such that participants with more years of education had less ɛ4-related atrophy than those with less education. These findings are in line with a recent study also using data from the PAC cohort (N = 1819), which reported that higher scores on a composite measure of years education and literacy attenuated the negative effect of APOE-ɛ4 genotype on the rate of decline in episodic memory and a global cognitive score.74 The present results suggest that this reduction in APOE-ɛ4 related cognitive decline among participants with more education may be mediated by reduced atrophy in Alzheimer's disease-vulnerable regions. Previous cross-sectional studies have produced mixed results regarding the association of years of education and regional brain volumes among middle-aged and older cognitively unimpaired individuals (e.g. Launer et al.,107 Arenaza-Urquijo et al.,108 Liu et al.109 and Vemuri et al.110). Among the few prior longitudinal studies, most found no association between education or related measures of literacy and change in brain volumes or cortical thickness over time.8,111-113 The present results suggest that some inconsistencies across prior studies may be attributable to the fact that the education-related reduction in atrophy is relatively small and primarily evident among APOE-ɛ4 carriers, making it difficult to detect in studies with smaller samples.
APOE-ɛ2 genetic status and brain atrophy
Another important finding is that APOE-ɛ2 carriers demonstrated similar rates of brain atrophy over time as ɛ3 homozygous individuals and did not differ in terms of brain volumes at baseline. Given the relatively low prevalence of the ɛ2 allele, prior work on this subject has been limited and largely comprised of cross-sectional studies with small numbers of ɛ2 carriers (ranging from ∼12 to 85 compared to 184 in this study). Our results are consistent with two prior cross-sectional studies among cognitively normal individuals that also found no difference in volumetric measures as a function of ɛ2 carrier status among middle-aged and older adults.54,61 By comparison, a small-scale longitudinal study66 and a few other cross-sectional studies52,60,114 reported reduced 2-year atrophy and greater cortical thickness and volumes among older cognitively normal ɛ2 carriers compared to ɛ3 homozygotes in regions sensitive to Alzheimer's disease. A likely explanation for these discrepancies across studies is that ɛ2 carriers are less likely to harbour preclinical Alzheimer's disease pathology (due to a later age of amyloid accumulation). Consequently, they are less likely to have atrophy in Alzheimer's disease-sensitive regions during middle- and old age compared to ɛ3/ɛ3 carriers, which can appear as reduced atrophy or greater volume. Future studies will be able to test this possibility by covarying amyloid and tau burden when evaluating associations between APOE-ɛ2 status and atrophy.
APOE-ɛ2 and ɛ4 genetic status and white matter hyperintensities
Prior studies among non-demented participants as well as samples spanning the Alzheimer's disease-spectrum have reported higher WMH burden37-40 and greater longitudinal increases in WMH burden42,43 among APOE-ɛ4 carriers relative to carriers, with stronger associations for homozygous than heterozygous participants38 (but see Habes et al.,44 Lyall et al.,45 Lane et al.46 and Debette et al.47 for negative results). Few studies, however, have examined ɛ4-related differences in WMH volumes among individuals with normal cognition.48,49,115 The present study found greater longitudinal increases in global WMH volumes among ɛ4 homozygous compared to ɛ4 heterozygous participants and ɛ3/ɛ3 carriers, who did not differ from one another. These findings are consistent with a cross-sectional study among cognitively normal middle-aged participants.49 Given that cognitively unimpaired middle-aged and older ɛ4/ɛ4 carriers likely harbour the highest level of brain amyloid,98 these findings support the view that Alzheimer's disease-specific pathways contribute to the formation of WMH among individuals with normal cognition.4,5,116 This contribution may be subtle during the preclinical phase of AD, when pathology levels are low, and increase as the disease progresses, as evidenced by more robust associations of WMH burden with ɛ4 genetic status37,43 or Alzheimer's disease-biomarker levels116-118 among symptomatic individuals. Our finding of an interaction between APOE-ɛ4 and AD-PRS scores in relation to WMH trajectories further suggests that associations between APOE-ɛ4 and WMH load may be more evident among those with additional Alzheimer's disease risk genes, beyond APOE.
The current study is the first, to our knowledge, to demonstrate greater longitudinal increases in WMH burden among cognitively unimpaired APOE-ɛ2 carriers relative to ɛ3/ɛ3 carriers. This finding is consistent with and expands prior cross-sectional studies among non-demented and cognitively impaired cohorts that have also reported ɛ2-related elevations in WMH burden.40,119 Two cross-sectional studies among cognitively normal middle-aged participants found no ɛ2-related WMH differences,49,120 consistent with the absence of a baseline difference in WMH volumes by ɛ2 genetic status in this study. Altogether, these results support the view that the APOE-ɛ2 allele promotes cerebrovascular disease, though the mechanisms remain poorly understood.121
Alzheimer's disease-polygenic risk, brain atrophy and WMH burden
Another important finding of this study was that higher AD-PRS scores were associated with greater atrophy over time in Alzheimer's disease-vulnerable regions, including the hippocampus, independent of APOE-ɛ4 status. This extends prior findings from longitudinal studies with participants across the Alzheimer's disease-spectrum18,34,35 and from cross-sectional studies among non-demented corhorts21,23,24,28,29 and suggest that higher Alzheimer's disease-polygenic risk scores increase the risk of neurodegeneration not only during the symptomatic phase of the disease but also among participants with normal cognition. Furthermore, AD-PRS-related atrophy was independent of age, sex, education and vascular risk scores, and not evident for regions that show non-Alzheimer's disease-related atrophy with age, consistent with a cross-sectional study.28 This suggests that APOE-ɛ4 and other AD risk genes influence atrophy in common Alzheimer's disease-susceptible brain regions, including the hippocampus, as early as midlife.
Higher AD-PRS were also associated with greater increases in global WMHs over time, particularly among APOE-ɛ4 carriers. This suggests that other Alzheimer's disease risk genes may exert some of their effects via cerebrovascular mechanisms, in addition to Alzheimer's disease-specific pathways. This interpretation is consistent with evidence linking higher AD-PRS scores to reductions in cerebral blood flow122 and neuropathological markers of cerebrovascular disease,34 though results might differ for PRS scores computed using other methodologies. Future studies that also include AD-biomarker assessments of amyloid and tau are needed to clarify how AD-PRS influence neurodegeneration.
Conclusion
The generalizability of findings from the current study to the broader population is limited because participants were primarily White, well-educated and enriched for a family history of Alzheimer's disease-dementia. For example, atrophy is more likely due to Alzheimer's disease in the current study than in the general population, and we may have overestimated associations between Alzheimer's disease-genetic risk with atrophy and underestimated other factors, like vascular risk. Additionally, these analyses do not include biomarkers of Alzheimer's disease pathology or other measures of brain structure and function, which precludes inferences regarding the precise mechanisms by which Alzheimer's disease-genetic risk influences brain atrophy among unimpaired individuals. Also, the power to detect significant three-way interactions involving Alzheimer's disease-genetic variables for the observed effects was only low to moderate. Nonetheless, the study provides compelling evidence that APOE-ɛ4 and AD-PRS independently and additively influence longitudinal trajectories of neurodegeneration in Alzheimer's disease-sensitive regions and synergistically increase WMH accumulation among cognitively normal individuals. Conversely, APOE-ɛ2 primarily influences WMH accumulation, but not atrophy. These AD-genetic associations did not differ by participant sex, but in some cases were influenced by participant age, years of education and vascular risk, providing potential avenues for reducing the negative impact of AD risk genes on neurodegeneration prior to the development of cognitive impairment. Future studies are needed to examine the degree to which these Alzheimer's disease-genetic-related brain changes mediate changes in cognition.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the participants and staff from each study for their contributions to the research programme.
Contributor Information
Anja Soldan, Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Jiangxia Wang, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, USA.
Corinne Pettigrew, Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Christos Davatzikos, Centre for Biomedical Image Computing and Analytics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Guray Erus, Centre for Biomedical Image Computing and Analytics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Timothy J Hohman, Department of Neurology, Vanderbilt University Medical Center, Nashville, TN 37212, USA.
Logan Dumitrescu, Department of Neurology, Vanderbilt University Medical Center, Nashville, TN 37212, USA.
Murat Bilgel, Laboratory of Behavioral Neuroscience, National Institute on Aging Intramural Research Program, Baltimore, MD 21224, USA.
Susan M Resnick, Laboratory of Behavioral Neuroscience, National Institute on Aging Intramural Research Program, Baltimore, MD 21224, USA.
Leonardo A Rivera-Rivera, Wisconsin Alzheimer's Disease Research Center, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI 53726, USA.
Rebecca Langhough, Wisconsin Alzheimer's Disease Research Center, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI 53726, USA.
Sterling C Johnson, Wisconsin Alzheimer's Disease Research Center, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI 53726, USA.
Tammie Benzinger, Knight Alzheimer Disease Research Center, Washington University School of Medicine, St. Louis, MO 63110, USA.
John C Morris, Knight Alzheimer Disease Research Center, Washington University School of Medicine, St. Louis, MO 63110, USA.
Simon M Laws, Centre for Precision Health, Edith Cowan University, Joondalup, WA 6027, Australia.
Jurgen Fripp, Australian E-Health Research Centre, CSIRO Health & Biosecurity, Herston, QLD 4029, Australia.
Colin L Masters, The Florey Institute, University of Melbourne, Parkville, VIC 3052, Australia.
Marilyn S Albert, Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
The Preclinical AD Consortium is supported by the National Institutes of Health (NIH), USA [grant number RF1-AG059869]. The individual studies in the consortium are funded, in part, by the following grants: U19-AG033655, P01-AG026276, RF1-AG027161 and the Australian Commonwealth Scientific Industrial Research Organization (CSIRO); as well as the National Institute on Aging Intramural Program. M.S.A., C.P. and A.S. are supported by NIH grant P30-AG066507. J.C.M. is supported by NIH grants P30-AG066444, P01-AG003991, U19-AG032438 and U19-AG024904. S.C.J. and R.L. are supported by NIH grant P30-AG062715. T.J.H. and L.D. are supported by NIH grant P20-AG068082. Funding agencies were not involved in study design, data collection, data analysis, interpretation of data or writing of manuscript.
Competing interests
The following authors declare that they have no competing interests: A.S., J.W., C.P., C.D., G.E., T.J.H., L.D., M.B., S.M.R., L.A.R.-R., R.L., T.B., J.C.M. and J.F. C.L.M. and M.S.A. are advisors to Eli Lilly. S.C.J. has served as an advisor to Roche Diagnostics. S.M.L. is a scientific advisor to Cytox Ltd.
Data availability
The plan is to archive the PAC datafiles at the National Archive of Computerized Data on Aging (NACDA). Investigators interested in accessing the data should contact the PAC Coordinating Center at Johns Hopkins University for details.
References
- 1. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jack CR Jr, Bennett DA, Blennow K, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Attems J, Jellinger KA. The overlap between vascular disease and Alzheimer’s disease—Lessons from pathology. BMC Med. 2014;12:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee S, Viqar F, Zimmerman ME, et al. White matter hyperintensities are a core feature of Alzheimer’s disease: Evidence from the dominantly inherited Alzheimer network. Ann Neurol. 2016;79(6):929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S, Zimmerman ME, Narkhede A, et al. White matter hyperintensities and the mediating role of cerebral amyloid angiopathy in dominantly-inherited Alzheimer’s disease. PLoS One. 2018;13(5):e0195838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moghekar A, Kraut M, Elkins W, et al. Cerebral white matter disease is associated with Alzheimer pathology in a prospective cohort. Alzheimers Dement. 2012;8(5 Suppl):S71–S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phuah CL, Chen Y, Strain JF, et al. Association of data-driven white matter hyperintensity spatial signatures with distinct cerebral small vessel disease etiologies. Neurology. 2022;99(23):e2535–e2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soldan A, Pettigrew C, Lu Y, et al. Relationship of medial temporal lobe atrophy, APOE genotype, and cognitive reserve in preclinical Alzheimer’s disease. Hum Brain Mapp. 2015;36(7):2826–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pettigrew C, Soldan A, Zhu Y, et al. Cortical thickness in relation to clinical symptom onset in preclinical AD. Neuroimage Clin. 2016;12:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soldan A, Pettigrew C, Zhu Y, et al. Cognitive reserve and midlife vascular risk: Cognitive and clinical outcomes. Ann Clin Transl Neurol. 2020;7(8):1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyle PA, Yu L, Fleischman DA, et al. White matter hyperintensities, incident mild cognitive impairment, and cognitive decline in old age. Ann Clin Transl Neurol. 2016;3(10):791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. [DOI] [PubMed] [Google Scholar]
- 13. Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 14. Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. [DOI] [PubMed] [Google Scholar]
- 15. Suri S, Heise V, Trachtenberg AJ, Mackay CE. The forgotten APOE allele: A review of the evidence and suggested mechanisms for the protective effect of APOE ɛ2. Neurosci Biobehav Rev. 2013;37(10 Pt 2):2878–2886. [DOI] [PubMed] [Google Scholar]
- 16. Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desikan RS, Fan CC, Wang Y, et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14(3):e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mormino EC, Sperling RA, Holmes AJ, et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology. 2016;87(5):481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riaz M, Huq A, Ryan J, et al. Effect of APOE and a polygenic risk score on incident dementia and cognitive decline in a healthy older population. Aging Cell. 2021;20(6):e13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan CH, Hyman BT, Tan JJX, et al. Polygenic hazard scores in preclinical Alzheimer disease. Ann Neurol. 2017;82(3):484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chauhan G, Adams HHH, Bis JC, et al. Association of Alzheimer’s disease GWAS loci with MRI markers of brain aging. Neurobiol Aging. 2015;36(4):1765. e7–1765. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. den Heijer T, Oudkerk M, Launer LJ, van Duijn CM, Hofman A, Breteler MM. Hippocampal, amygdalar, and global brain atrophy in different apolipoprotein E genotypes. Neurology. 2002;59(5):746–748. [DOI] [PubMed] [Google Scholar]
- 23. Lupton MK, Strike L, Hansell NK, et al. The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol Aging. 2016;40:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tank R, Ward J, Flegal KE, et al. Association between polygenic risk for Alzheimer’s disease, brain structure and cognitive abilities in UK Biobank. Neuropsychopharmacology. 2022;47(2):564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Veldsman M, Nobis L, Alfaro-Almagro F, Manohar S, Husain M. The human hippocampus and its subfield volumes across age, sex and APOE e4 status. Brain Commun. 2021;3(1):fcaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walhovd KB, Fjell AM, Sorensen O, et al. Genetic risk for Alzheimer disease predicts hippocampal volume through the human lifespan. Neurol Genet. 2020;6(5):e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morrison C, Dadar M, Kamal F, Collins DL; Alzheimer’s Disease Neuroimaging Initiative . Differences in Alzheimer’s disease–related pathology profiles across apolipoprotein groups. J Gerontol A Biol Sci Med Sci 2024;79(2):glad254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Habes M, Janowitz D, Erus G, et al. Advanced brain aging: Relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Transl Psychiatry. 2016;6(4):e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lancaster TM, Hill MJ, Sims R, Williams J. Microglia-mediated immunity partly contributes to the genetic association between Alzheimer’s disease and hippocampal volume. Brain Behav Immun. 2019;79:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crivello F, Lemaitre H, Dufouil C, et al. Effects of ApoE-epsilon4 allele load and age on the rates of grey matter and hippocampal volumes loss in a longitudinal cohort of 1186 healthy elderly persons. Neuroimage. 2010;53(3):1064–1069. [DOI] [PubMed] [Google Scholar]
- 31. Moffat SD, Szekely CA, Zonderman AB, Kabani NJ, Resnick SM. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurology. 2000;55(1):134–136. [DOI] [PubMed] [Google Scholar]
- 32. Regy M, Dugravot A, Sabia S, et al. Association of APOE epsilon4 with cerebral gray matter volumes in non-demented older adults: The MEMENTO cohort study. Neuroimage. 2022;250:118966. [DOI] [PubMed] [Google Scholar]
- 33. Seto M, Mahoney ER, Dumitrescu L, et al. Exploring common genetic contributors to neuroprotection from amyloid pathology. Brain Commun. 2022;4(2):fcac066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan CH, Bonham LW, Fan CC, et al. Polygenic hazard score, amyloid deposition and Alzheimer’s neurodegeneration. Brain. 2019;142(2):460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ge T, Sabuncu MR, Smoller JW, Sperling RA, Mormino EC; Alzheimer’s Disease Neuroimaging Initiative . Dissociable influences of APOE epsilon4 and polygenic risk of AD dementia on amyloid and cognition. Neurology. 2018;90(18):e1605–e1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harrison TM, Mahmood Z, Lau EP, et al. An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro. 2016;3(3):ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brickman AM, Schupf N, Manly JJ, et al. APOE epsilon4 and risk for Alzheimer’s disease: Do regionally distributed white matter hyperintensities play a role? Alzheimers Dement. 2014;10(6):619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lyall DM, Cox SR, Lyall LM, et al. Association between APOE e4 and white matter hyperintensity volume, but not total brain volume or white matter integrity. Brain Imaging Behav. 2020;14(5):1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Leeuw FE, Richard F, de Groot JC, et al. Interaction between hypertension, apoE, and cerebral white matter lesions. Stroke. 2004;35(5):1057–1060. [DOI] [PubMed] [Google Scholar]
- 40. Schilling S, DeStefano AL, Sachdev PS, et al. APOE genotype and MRI markers of cerebrovascular disease: Systematic review and meta-analysis. Neurology. 2013;81(3):292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Habes M, Sotiras A, Erus G, et al. White matter lesions: Spatial heterogeneity, links to risk factors, cognition, genetics, and atrophy. Neurology. 2018;91(10):e964–e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Godin O, Tzourio C, Maillard P, Alperovitch A, Mazoyer B, Dufouil C. Apolipoprotein E genotype is related to progression of white matter lesion load. Stroke. 2009;40(10):3186–3190. [DOI] [PubMed] [Google Scholar]
- 43. Sudre CH, Cardoso MJ, Frost C, et al. APOE epsilon4 status is associated with white matter hyperintensities volume accumulation rate independent of AD diagnosis. Neurobiol Aging. 2017;53:67–75. [DOI] [PubMed] [Google Scholar]
- 44. Habes M, Erus G, Toledo JB, et al. White matter hyperintensities and imaging patterns of brain ageing in the general population. Brain. 2016;139(Pt 4):1164–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lyall DM, Munoz Maniega S, Harris SE, et al. APOE/TOMM40 genetic loci, white matter hyperintensities, and cerebral microbleeds. Int J Stroke. 2015;10(8):1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lane CA, Barnes J, Nicholas JM, et al. Associations between vascular risk across adulthood and brain pathology in late life: Evidence from a British birth cohort. JAMA Neurol. 2020;77(2):175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Debette S, Seshadri S, Beiser A, et al. Midlife vascular risk factor exposure accelerates structural brain aging and cognitive decline. Neurology. 2011;77(5):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luo X, Jiaerken Y, Yu X, et al. Affect of APOE on information processing speed in non-demented elderly population: A preliminary structural MRI study. Brain Imaging Behav. 2017;11(4):977–985. [DOI] [PubMed] [Google Scholar]
- 49. Rojas S, Brugulat-Serrat A, Bargallo N, et al. Higher prevalence of cerebral white matter hyperintensities in homozygous APOE-varepsilon4 allele carriers aged 45–75: Results from the ALFA study. J Cereb Blood Flow Metab. 2018;38(2):250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burggren AC, Zeineh MM, Ekstrom AD, et al. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage. 2008;41(4):1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cacciaglia R, Molinuevo JL, Falcon C, et al. Effects of APOE-epsilon4 allele load on brain morphology in a cohort of middle-aged healthy individuals with enriched genetic risk for Alzheimer’s disease. Alzheimers Dement. 2018;14(7):902–912. [DOI] [PubMed] [Google Scholar]
- 52. Fennema-Notestine C, Panizzon MS, Thompson WR, et al. Presence of ApoE epsilon4 allele associated with thinner frontal cortex in middle age. J Alzheimers Dis. 2011;26 Suppl 3(Suppl 3):49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sabuncu MR, Buckner RL, Smoller JW, et al. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22(11):2653–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bunce D, Anstey KJ, Cherbuin N, Gautam P, Sachdev P, Easteal S. APOE genotype and entorhinal cortex volume in non-demented community-dwelling adults in midlife and early old age. J Alzheimers Dis. 2012;30(4):935–942. [DOI] [PubMed] [Google Scholar]
- 55. Cherbuin N, Anstey KJ, Sachdev PS, et al. Total and regional gray matter volume is not related to APOE*E4 status in a community sample of middle-aged individuals. J Gerontol A Biol Sci Med Sci. 2008;63(5):501–504. [DOI] [PubMed] [Google Scholar]
- 56. Habes M, Toledo JB, Resnick SM, et al. Relationship between APOE genotype and structural MRI measures throughout adulthood in the study of health in Pomerania population-based cohort. AJNR Am J Neuroradiol. 2016;37(9):1636–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zheng Y, Habes M, Gonzales M, et al. Mid-life epigenetic age, neuroimaging brain age, and cognitive function: Coronary artery risk development in young adults (CARDIA) study. Aging (Albany NY). 2022;14(4):1691–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Protas HD, Chen K, Langbaum JB, et al. Posterior cingulate glucose metabolism, hippocampal glucose metabolism, and hippocampal volume in cognitively normal, late-middle-aged persons at 3 levels of genetic risk for Alzheimer disease. JAMA Neurol. 2013;70(3):320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Foo H, Thalamuthu A, Jiang J, et al. Associations between Alzheimer’s disease polygenic risk scores and hippocampal subfield volumes in 17,161 UK Biobank participants. Neurobiol Aging. 2021;98:108–115. [DOI] [PubMed] [Google Scholar]
- 60. Fan M, Liu B, Zhou Y, et al. Cortical thickness is associated with different apolipoprotein E genotypes in healthy elderly adults. Neurosci Lett. 2010;479(3):332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y, Paajanen T, Westman E, et al. APOE epsilon2 allele is associated with larger regional cortical thicknesses and volumes. Dement Geriatr Cogn Disord. 2010;30(3):229–237. [DOI] [PubMed] [Google Scholar]
- 62. Reiter K, Nielson KA, Durgerian S, et al. Five-year longitudinal brain volume change in healthy elders at genetic risk for Alzheimer’s disease. J Alzheimers Dis. 2017;55(4):1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen S, Zhou W, Chen X, Zhang J; for Alzheimer’s Disease Neuroimaging Initiative . Sex differences in the association of APOE epsilon4 genotype with longitudinal hippocampal atrophy in cognitively normal older people. Eur J Neurol. 2019;26(11):1362–1369. [DOI] [PubMed] [Google Scholar]
- 64. Armstrong NM, An Y, Beason-Held L, et al. Sex differences in brain aging and predictors of neurodegeneration in cognitively healthy older adults. Neurobiol Aging. 2019;81:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Du AT, Schuff N, Chao LL, et al. Age effects on atrophy rates of entorhinal cortex and hippocampus. Neurobiol Aging. 2006;27(5):733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chiang GC, Insel PS, Tosun D, et al. Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology. 2010;75(22):1976–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Coats M, Morris JC. Antecedent biomarkers of Alzheimer’s disease: The adult children study. J Geriatr Psychiatry Neurol. 2005;18(4):242–244. [DOI] [PubMed] [Google Scholar]
- 68. Fowler C, Rainey-Smith SR, Bird S, et al. Fifteen years of the Australian Imaging, Biomarkers and Lifestyle (AIBL) study: Progress and observations from 2,359 older adults spanning the spectrum from cognitive normality to Alzheimer’s disease. J Alzheimers Dis Rep. 2021;5(1):443–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Albert M, Soldan A, Gottesman R, et al. Cognitive changes preceding clinical symptom onset of mild cognitive impairment and relationship to ApoE genotype. Curr Alzheimer Res. 2014;11(8):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Resnick SM, Goldszal AF, Davatzikos C, et al. One-year age changes in MRI brain volumes in older adults. Cereb Cortex. 2000;10(5):464–472. [DOI] [PubMed] [Google Scholar]
- 71. Johnson SC, Koscik RL, Jonaitis EM, et al. The Wisconsin Registry for Alzheimer’s Prevention: A review of findings and current directions. Alzheimers Dement (Amst). 2018;10:130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ellis KA, Bush AI, Darby D, et al. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int Psychogeriatr. 2009;21(4):672–687. [DOI] [PubMed] [Google Scholar]
- 73. Gross AL, Hassenstab JJ, Johnson SC, et al. A classification algorithm for predicting progression from normal cognition to mild cognitive impairment across five cohorts: The preclinical AD consortium. Alzheimers Dement (Amst). 2017;8:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pettigrew C, Nazarovs J, Soldan A, et al. Alzheimer’s disease genetic risk and cognitive reserve in relationship to long-term cognitive trajectories among cognitively normal individuals. Alzheimers Res Ther. 2023;15(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Albert M, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gottesman RF, Schneider AL, Zhou Y, et al. Association between midlife vascular risk factors and estimated brain amyloid deposition. JAMA. 2017;317(14):1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lin Z, Lim C, Jiang D, et al. Longitudinal changes in brain oxygen extraction fraction (OEF) in older adults: Relationship to markers of vascular and Alzheimer’s pathology. Alzheimers Dement. 2023;19(2):569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pettigrew C, Soldan A, Wang J, et al. Association of midlife vascular risk and AD biomarkers with subsequent cognitive decline. Neurology. 2020;95(23):e3093–e3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Soldan A, Pettigrew C, Zhu Y, et al. White matter hyperintensities and CSF Alzheimer disease biomarkers in preclinical Alzheimer disease. Neurology. 2020;94(9):e950–e960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goldberg TE, Huey ED, Devanand DP. Association of APOE e2 genotype with Alzheimer’s and non-Alzheimer’s neurodegenerative pathologies. Nat Commun. 2020;11(1):4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-atlas skull-stripping. Acad Radiol. 2013;20(12):1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Doshi J, Erus G, Ou Y, et al. MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage. 2016;127:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Erus G, Doshi J, An Y, Verganelakis D, Resnick SM, Davatzikos C. Longitudinally and inter-site consistent multi-atlas based parcellation of brain anatomy using harmonized atlases. Neuroimage. 2018;166:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Habes M, Pomponio R, Shou H, et al. The Brain Chart of Aging: Machine-learning analytics reveals links between brain aging, white matter disease, amyloid burden, and cognition in the iSTAGING consortium of 10,216 harmonized MR scans. Alzheimers Dement. 2021;17(1):89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moonen JEF, Nasrallah IM, Detre JA, et al. Race, sex, and mid-life changes in brain health: Cardia MRI substudy. Alzheimers Dement. 2022;18(12):2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Srinivasan D, Erus G, Doshi J, et al. A comparison of Freesurfer and multi-atlas MUSE for brain anatomy segmentation: Findings about size and age bias, and inter-scanner stability in multi-site aging studies. Neuroimage. 2020;223:117248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pomponio R, Erus G, Habes M, et al. Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. Neuroimage. 2020;208:116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Doshi J, Erus G, Habes M, Davatzikos C. DeepMRSeg: A convolutional deep neural network for anatomy and abnormality segmentation on MR images. ArXiv. abs/1907.02110. 10.48550/arXiv.1907.02110, 2019, preprint: not peer reviewed. [DOI]
- 91. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. Springer International Publishing; 2015:234–241. [Google Scholar]
- 92. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-ResNet and the impact of residual connections on learning. In: Proceedings of the thirty-first AAAI conference on artificial intelligence. AAAI Press; 2017.
- 93. Da X, Toledo JB, Zee J, et al. Integration and relative value of biomarkers for prediction of MCI to AD progression: Spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin. 2014;4:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Eavani H, Habes M, Satterthwaite TD, et al. Heterogeneity of structural and functional imaging patterns of advanced brain aging revealed via machine learning methods. Neurobiol Aging. 2018;71:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cole JH, Franke K. Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci. 2017;40(12):681–690. [DOI] [PubMed] [Google Scholar]
- 96. Vidal-Pineiro D, Wang Y, Krogsrud SK, et al. Individual variations in ‘brain age’ relate to early-life factors more than to longitudinal brain change. Elife. 2021;10:e69995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Armstrong NM, An Y, Beason-Held L, et al. Predictors of neurodegeneration differ between cognitively normal and subsequently impaired older adults. Neurobiol Aging. 2019;75:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jansen WJ, Janssen O, Tijms BM, et al. Prevalence estimates of amyloid abnormality across the Alzheimer disease clinical spectrum. JAMA Neurol. 2022;79(3):228–243. [DOI] [PubMed] [Google Scholar]
- 99. Pletnikova O, Kageyama Y, Rudow G, et al. The spectrum of preclinical Alzheimer’s disease pathology and its modulation by ApoE genotype. Neurobiol Aging. 2018;71:72–80. [DOI] [PubMed] [Google Scholar]
- 100. Betthauser TJ, Bilgel M, Koscik RL, et al. Multi-method investigation of factors influencing amyloid onset and impairment in three cohorts. Brain. 2022;145(11):4065–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kulason S, Xu E, Tward DJ, et al. Entorhinal and transentorhinal atrophy in preclinical Alzheimer’s disease. Front Neurosci. 2020;14:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Shaw P, Lerch JP, Pruessner JC, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: An observational study. Lancet Neurol. 2007;6(6):494–500. [DOI] [PubMed] [Google Scholar]
- 103. Montembeault M, Migliaccio R. Atypical forms of Alzheimer’s disease: Patients not to forget. Curr Opin Neurol. 2023;36(4):245–252. [DOI] [PubMed] [Google Scholar]
- 104. Cox SR, Lyall DM, Ritchie SJ, et al. Associations between vascular risk factors and brain MRI indices in UK Biobank. Eur Heart J. 2019;40(28):2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lamar M, Boots EA, Arfanakis K, Barnes LL, Schneider JA. Common brain structural alterations associated with cardiovascular disease risk factors and Alzheimer’s dementia: Future directions and implications. Neuropsychol Rev. 2020;30(4):546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rabin JS, Pruzin J, Scott M, et al. Association of beta-amyloid and vascular risk on longitudinal patterns of brain atrophy. Neurology. 26 2022;99(3):e270–e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Launer LJ, Lewis CE, Schreiner PJ, et al. Vascular factors and multiple measures of early brain health: CARDIA brain MRI study. PLoS One. 2015;10(3):e0122138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Arenaza-Urquijo EM, Landeau B, La Joie R, et al. Relationships between years of education and gray matter volume, metabolism and functional connectivity in healthy elders. Neuroimage. 2013;83:450–457. [DOI] [PubMed] [Google Scholar]
- 109. Liu Y, Julkunen V, Paajanen T, et al. Education increases reserve against Alzheimer’s disease—Evidence from structural MRI analysis. Neuroradiology. 2012;54(9):929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vemuri P, Lesnick TG, Przybelski SA, et al. Effect of lifestyle activities on Alzheimer disease biomarkers and cognition. Ann Neurol. 2012;72(5):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lo RY, Jagust WJ. Alzheimer’s disease neuroimaging I. Effect of cognitive reserve markers on Alzheimer pathologic progression. Alzheimer Dis Assoc Disord. 2013;27(4):343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pettigrew C, Soldan A, Zhu Y, et al. Cognitive reserve and rate of change in Alzheimer’s and cerebrovascular disease biomarkers among cognitively normal individuals. Neurobiol Aging. 2020;88:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Walters MJ, Sterling J, Quinn C, et al. Associations of lifestyle and vascular risk factors with Alzheimer’s brain biomarker changes during middle age: A 3-year longitudinal study in the broader New York City area. BMJ Open. 2018;8(11):e023664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Salvado G, Ferreira D, Operto G, et al. The protective gene dose effect of the APOE epsilon2 allele on gray matter volume in cognitively unimpaired individuals. Alzheimers Dement. 2022;18(7):1383–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Low A, Su L, Stefaniak JD, et al. Inherited risk of dementia and the progression of cerebral small vessel disease and inflammatory markers in cognitively healthy midlife adults: The PREVENT-Dementia study. Neurobiol Aging. 2021;98:124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Newton P, Tchounguen J, Pettigrew C, et al. Regional white matter hyperintensities and Alzheimer’s disease biomarkers among older adults with normal cognition and mild cognitive impairment. J Alzheimers Dis. 2023;92(1):323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. van Westen D, Lindqvist D, Blennow K, et al. Cerebral white matter lesions—Associations with Abeta isoforms and amyloid PET. Sci Rep. 2016;6:20709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shams S, Granberg T, Martola J, et al. Cerebrospinal fluid profiles with increasing number of cerebral microbleeds in a continuum of cognitive impairment. J Cereb Blood Flow Metab. 2016;36(3):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Groot C, Sudre CH, Barkhof F, et al. Clinical phenotype, atrophy, and small vessel disease in APOEepsilon2 carriers with Alzheimer disease. Neurology. 2018;91(20):e1851–e1859. [DOI] [PubMed] [Google Scholar]
- 120. Salvado G, Brugulat-Serrat A, Sudre CH, et al. Spatial patterns of white matter hyperintensities associated with Alzheimer’s disease risk factors in a cognitively healthy middle-aged cohort. Alzheimers Res Ther. 2019;11(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kim H, Devanand DP, Carlson S, Goldberg TE. Apolipoprotein E genotype e2: Neuroprotection and its limits. Front Aging Neurosci. 2022;14:919712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chandler H, Wise R, Linden D, et al. Alzheimer’s genetic risk effects on cerebral blood flow across the lifespan are proximal to gene expression. Neurobiol Aging. 2022;120:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plan is to archive the PAC datafiles at the National Archive of Computerized Data on Aging (NACDA). Investigators interested in accessing the data should contact the PAC Coordinating Center at Johns Hopkins University for details.