Abstract
Background:
Advances in targeted therapy development and tumor sequencing technology are reclassifying cancers into smaller biomarker-defined diseases. Randomized controlled trials (RCTs) are often impractical in rare diseases, leading to calls for single-arm studies to be sufficient to inform clinical practice based on a strong biological rationale. However, without RCTs, favorable outcomes are often attributed to therapy but may be due to a more indolent disease course or other biases. When the clinical benefit of targeted therapy in a common cancer is established in RCTs, this benefit may extend to rarer cancers sharing the same biomarker. However, careful consideration of the appropriateness of extending the existing trial evidence beyond specific cancer types is required. A framework for extrapolating evidence for biomarker-targeted therapies to rare cancers is needed to support transparent decision-making.
Objectives:
To construct a framework outlining the breadth of criteria essential for extrapolating evidence for a biomarker-targeted therapy generated from RCTs in common cancers to different rare cancers sharing the same biomarker.
Design:
A series of questions articulating essential criteria for extrapolation.
Methods:
The framework was developed from the core topics for extrapolation identified from a previous scoping review of methodological guidance. Principles for extrapolation outlined in guidance documents from the European Medicines Agency, the US Food and Drug Administration, and Australia’s Medical Services Advisory Committee were incorporated.
Results:
We propose a framework for assessing key assumptions of similarity of the disease and treatment outcomes between the common and rare cancer for five essential components: prognosis of the biomarker-defined cancer, biomarker test analytical validity, biomarker actionability, treatment efficacy, and safety. Knowledge gaps identified can be used to prioritize future studies.
Conclusion:
This framework will allow systematic assessment, standardize regulatory, reimbursement and clinical decision-making, and facilitate transparent discussions between key stakeholders in drug assessment for rare biomarker-defined cancers.
Keywords: biomarker-driven trials, biomarker-guided therapy, biomarkers, clinical trials, companion diagnostics, predictive biomarkers, prognostic biomarkers, response biomarkers, targeted therapy
Introduction
Advances in high-throughput sequencing technology and improved understanding of molecular drivers of carcinogenesis 1 continue to identify potentially targetable molecular alterations and pathways.2–4 As a result, the drug development paradigm is shifting to include approaches that match targeted therapies to specific molecular alterations. Effectiveness of targeted therapy in one histology in the presence of a matching molecular alteration, also referred to herein as a “predictive biomarker,” might then establish a rationale for classifying other cancer types with the same biomarker, as a potentially histology-independent, druggable target (Box). In this way, cancers may be classified into smaller subgroups.
Box.
Glossary of terms.
| Prognostic biomarker—a biomarker variant that describes a population that differs in clinical outcome or natural history from a population without this variant regardless of treatment. Outcomes are typically observed in those on non-targeted control treatment or best supportive care. Predictive biomarker—a biomarker that can indicate (1) a druggable target or oncogenic pathway is present to identify a population that will benefit from a matched therapy with improved health outcomes compared to non-targeted control treatment, or (2) a resistance pathway that identifies a population of patients for which the targeted therapy is ineffective thereby guiding choice of therapy. It is also referred to as “biomarker actionability.” Positive predictive value—proportion of patients who test positive by a test being assessed, who also test positive by a reference test or evidentiary standard test. It can also be thought of as the probability that a test correctly identifies the biomarker. Negative predictive value—proportion of patients who test negative by a test being assessed, who also test negative by a reference test or evidentiary standard test. It can also be thought of as the probability that the test correctly identifies absence of the biomarker. Positive percent agreement—proportion of individuals with the biomarker by the clinical utility standard test who also test positive with the new test. Negative percent agreement—proportion of patients without the biomarker by the clinical utility standard test who also test negative with the new test. |
Small population size presents a critical practical challenge for generating robust evidence within an acceptable timeframe for regulatory approval and reimbursement of novel targeted therapies. Typically, novel therapies are assessed through comparison with standard-of-care treatment on measures of net clinical benefit, such as overall survival (OS) and quality of life, from randomized controlled trials (RCTs).5,6 However, adequately powering RCTs for these measures for each biomarker-defined cancer subgroup may be infeasible. Thus, there are calls for approval of molecularly targeted therapies to be based on non-randomized studies, including basket studies, using intermediate endpoints such as objective response rates (ORRs).7–9 Tempering these calls, reviews of phase III RCTs have shown the clinical benefit of molecularly targeted therapies is often only modest when compared with standard-of-care non-targeted therapies.10–12 Further, post-approval commitments and timely withdrawal of drugs do not always occur when required, potentially exposing patients to therapies that are less effective and/or more harmful than previously assumed.13–15 Uncontrolled histology-agnostic studies provide valuable proof-of-concept evidence for rare cancer populations. However, without randomized comparison, it is difficult to differentiate the clinical benefit of targeted treatment from the natural course of the disease process (“natural history”) for each cancer type, other biases such as selection bias and especially to distinguish any predictive properties from any prognostic properties of the biomarker for each cancer type (Box). RCTs in rare cancers, albeit on intermediate or surrogate outcomes, are ideal and recommended wherever possible. Innovative trial designs are being developed to increase study power to assess the predictive value of a biomarker and include biomarker-adaptive designs using classical or Bayesian methods for randomization according to biomarker status and treatment group.16–19 However, as demonstrated by regulatory decisions based on non-randomized evidence, 20 there is an immediate need for a framework to support transparent decision-making where robust RCT data on clinically relevant outcomes is not available.
In this paper, we address the problem where RCT evidence of the effectiveness of targeted treatment is available for at least one cancer type (referred to herein as the “common cancer”), and the question is whether it can also be recommended in other cancers sharing the same biomarker for which a new RCT is not feasible on clinically relevant outcomes due to small population size (referred to herein as the “rare cancer”). In this context, extrapolation refers to the leveraging or extending process whereby an indication for use of a therapy in a new patient population can be supported by existing clinical data from a related studied patient population.21–23 Extrapolation of evidence from drugs already approved in adults is accepted to support submissions for pediatric use provided that the disease course and treatment response are sufficiently similar in both populations.22,23 Similarly, in rare biomarker-defined cancer populations, it may be possible to extrapolate evidence from similar populations where robust RCT evidence exists for the same targeted therapy to support regulatory approval in the rare cancer. 21 However, careful consideration of the appropriateness of extending the existing trial evidence beyond specific cancer types is required.
In this framework, we outline a series of questions to guide extrapolation of evidence for a molecularly targeted therapy generated from RCTs in common cancers to different rare cancers sharing the same biomarker.
Extrapolation framework
Current approach for evaluation
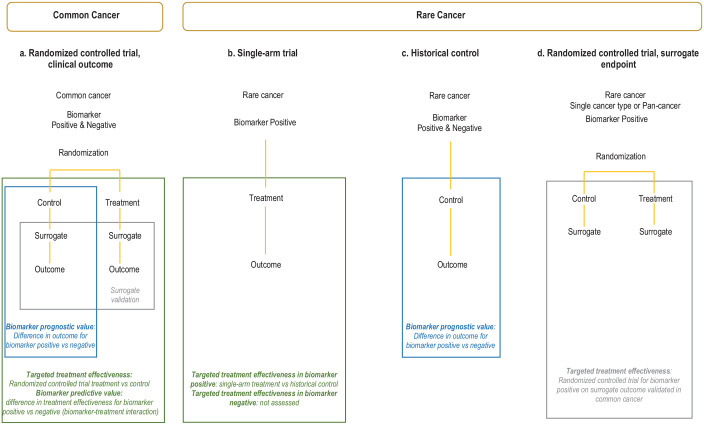
RCTs enrolling patients with biomarker-positive and biomarker-negative disease to compare the targeted therapy against standard non-targeted care can provide definitive evidence to assess treatment effectiveness in both groups and distinguish the prognostic and predictive value of a biomarker (Figure 1(a)). For targeted therapies in the common cancer, RCTs may provide this comprehensive evidence. Where data is restricted to the biomarker-positive population, stronger evidence for treatment efficacy and/or evidence across a range of cancer types is required for extrapolation. For rare biomarker-defined cancers, targeted therapies are typically assessed in single-arm studies in the biomarker-positive population (Figure 1(b)). In this scenario, pragmatic cross-study comparisons with a historical control group are usually relied upon to support claims of treatment effectiveness. Ideally, prognostic studies in patients with the rare cancer, treated with the same standard (non-targeted) treatment or best supportive care, and known biomarker status would be available for these comparisons (Figure 1(c)). These studies allow assessment of both the prognostic value of the biomarker in the rare cancer by comparing outcomes for biomarker-positive and biomarker-negative patients; and provide an untreated historical control group for cross-study comparison with the single-arm study/studies of the targeted therapy to determine treatment effectiveness. Our previous scoping review of methodological guidance showed regulatory agencies, health technology assessment (HTA) bodies, research groups, and others use different approaches for this assessment. 24 Each group incorporated additional topics to guide extrapolation of evidence from common to rare cancers, but we did not identify a framework to promote the explicit assessment of commonly used criteria.
Figure 1.
Approach for evaluating biomarker-targeted therapies in common and rare cancers.
In the common cancer, RCTs enrolling patients with biomarker-positive and biomarker-negative diseases to compare the targeted therapy against standard non-targeted care may be feasible. This provides definitive evidence to assess treatment effectiveness in both groups and distinguish the prognostic and predictive value of a biomarker (a). For rare biomarker-defined cancers, targeted therapies are typically assessed in single-arm studies in the biomarker-positive population (b) and rely on cross-study comparisons with a historical control group to support claims of treatment effectiveness (c). RCTs in the rare cancer or across multiple rare cancer types, albeit on surrogate endpoints, may be possible and recommended. Relative treatment benefits from a common cancer could be extrapolated to the rare cancer(s) sharing the same biomarker based on comparable outcomes measured by the same validated surrogate endpoint in both the common and rare cancers (d).
RCT, randomized controlled trial.
Extrapolation framework
We developed the framework from the core topics for extrapolation identified from the scoping review 24 and incorporated the principles outlined in the medicines extrapolation framework of the European Medicines Agency. 21 The framework is presented as a series of questions articulating essential criteria for extrapolation, with illustrative examples. The criteria reflect key assumptions of similarity for the disease definition and treatment outcome between the common and rare cancer (Table 1). To support the assumption of similar treatment outcomes, extrapolation can be more readily considered if the targeted treatment is proposed as last-line therapy in the rare cancer. Extrapolation would be more complex if effective alternative therapies existed. We have further outlined a pragmatic approach for evaluating existing evidence to judge these criteria for regulatory approval, reimbursement, and clinical decisions (Figure 2). The level of uncertainty for each criterion can be judged based on existing approaches for evidence-based decision-making. 25 Each criterion should be considered individually and then the overall assessment should be made based on the totality of the evidence to judge whether adequate to conclude treatment effectiveness and favorable benefit–risk profile 26 in the rare cancer. The evidence available for each criterion may increase or decrease uncertainty for the overall judgment (Table 2). Knowledge gaps where limited evidence exists should inform future research to acquire additional data. The framework is intended to describe the criteria that need to be explicitly addressed for decision-making. It is not intended to be prescriptive since the suitability for data extrapolation will likely vary for different biomarker-targeted therapy-cancer scenarios. The reporting of this study conforms to the RIGHT statement modified for a research framework 27 (Supplemental Material).
Table 1.
Extrapolation criteria.
| Disease definition |
|---|
| (1) Prognosis |
| Criterion (1a) Is the prognosis of the biomarker-positive rare cancer adequately described and estimated with adequate precision? |
| Criterion (1b) Could favorable outcomes in single-arm studies in the biomarker-defined rare cancer be due to better prognosis? |
| (2) Analytical validity |
| Criterion (2a) If the biomarker test proposed in the rare cancer is the same test used in the common cancer pivotal trial, have the performance characteristics of the test been assessed in the rare cancer? |
| Criterion (2b) Can the scoring criteria or molecular grouping strategy to define the biomarker-positive and biomarker-negative subgroups established in the common cancer be directly applied to the rare cancer or does it require modification? |
| Criterion (2c) What is the prevalence of the biomarker in the rare cancer? Does this prevalence change over the course of the disease? What is the performance of the proposed test in low-prevalence biomarker-positive rare cancers? |
| Criterion (2d) Is the test proposed in the rare cancer different to the test used in the common cancer? If so, has the new/alternative test been analytically validated against the evidentiary standard test in the rare cancer? |
| (3) Biomarker actionability |
| Criterion (3a) How strong is the evidence supporting biomarker actionability in the rare cancer? |
| Criterion (3b) Is there evidence that suggests treatment effect in the rare cancer may differ from the common cancer thereby not supporting extrapolation? |
| Treatment outcome |
| (4) Efficacy |
| Criterion (4) Is there a validated surrogate endpoint that can be used to extrapolate the clinical benefit of targeted therapy from the common cancer to the rare cancer? Are estimates of targeted therapy efficacy based on this surrogate endpoint similar between the common and rare cancer? |
| (5) Safety |
| Criterion (5) Are the adverse events experienced in the rare cancer similar to those experienced in the common cancer? Are there any clinically meaningful differences between cancers? |
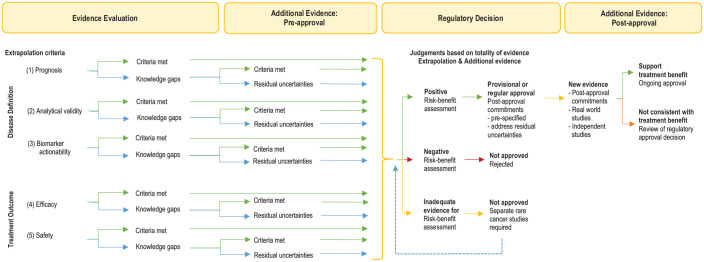
Figure 2.
Extrapolation framework: decision tree.
A pragmatic approach for evaluating existing evidence for molecularly targeted therapies in rare biomarker-defined cancers for regulatory and clinical decision-making. Existing evidence for the same targeted therapy from common cancers sharing the same biomarker is considered and judged according to extrapolation criteria for applicability to the rare cancer histotype.
Table 2.
Assessment of uncertainty when extrapolating evidence for transparent decision-making.
| Judgment a | Evidence assessed for each criterion a | Decision assessed from evidence for all criteria b |
|---|---|---|
| Important uncertainty | No research evidence identified or searched for | Use criteria to identify or plan studies for later reassessment |
| Possibly important uncertainty | Judgment Responses from other extrapolation criteria increase uncertainty |
Identify additional evidence required pre-approval |
| Probably no important uncertainty | Judgment Responses from other extrapolation criteria decrease uncertainty |
Provisional or regular approval, define post-approval commitments |
Source: Adapted from Piggott et al. 28
Judgment for the level of uncertainty for extrapolation should be made individually for each criterion. Judgments from other extrapolation criterion either increase or decrease certainty of each criterion.
The final decision should be made based on the totality of the evidence. If there is probably no important uncertainty for most of the criteria, then there is likely sufficient evidence to support regulatory approval. Any substantial knowledge gaps identified resulting in possibly important uncertainty for one or more criteria should define additional studies required pre-approval. If there is important uncertainty for many or most of the criteria, further studies are required to address knowledge gaps for later reassessment. Judgment for decision-making should be individualized and consider estimated benefits versus risks of targeted therapy compared to alternative therapies if available.
Components
We propose assessing disease similarity under three extrapolation components: (1) “Prognosis” addressing clinical outcomes of biomarker-defined cancer in the absence of targeted treatment to inform control data, (2) “Analytical Validity” addressing the performance characteristics of the test used to identify the biomarker, and (3) “Biomarker Actionability” addressing the evidence that the biomarker represents a dominant targetable molecular pathway and predicts the effect of the therapy being assessed. We propose assessing similarity of treatment outcomes under two components: (4) “Efficacy” addressing predictions of similar clinical benefit between cancers based on signals of efficacy on intermediate/surrogate outcomes in the rare cancer and (5) “Safety” addressing similarity of the safety profile between cancers and methods to augment safety data in the rare cancer. The order of the components is not fixed, and a pragmatic approach may be to start with identifying the best available evidence of treatment outcomes in the rare cancer first and addressing other criteria later.
Disease definition
Prognosis
Criterion (1a) Is the prognosis of the biomarker-positive rare cancer adequately described and estimated with adequate precision for use as a historical control?
The prognosis of biomarker-positive rare cancers describes the natural history in the absence of targeted therapies. Natural history may be available for the histology-defined cancer without biomarker information. However, targeted therapies are developed to reverse or inactivate an aberrant biological pathway and the related biomarker may be associated with unfavorable, favorable, or neutral prognosis. For example, HER2 gene amplification or overexpression is a poor prognostic factor in breast cancer 29 and anti-HER2 therapies, such as trastuzumab, have been shown to reverse the natural history of this poor prognosis disease. 30
Historical control data for the biomarker-positive population could be obtained from retrospective biomarker analyses of RCTs or cohort studies testing non-targeted therapy, and real-world studies annotated with biomarker data (e.g. electronic health record data or registries). Critical requirements for such prognostic studies include unbiased patient selection, large sample size, uniform treatment, high-quality data collection for marker status at baseline, identification of potential confounders, complete and long-term follow-up for clinical outcome assessment, and outcome ascertainment with sufficient precision and replicability.31,32 The Reporting Recommendations for Tumor Marker Prognostic Studies checklist for reporting prognostic marker studies details important issues for study design and conduct.33,34
When natural history data are only available for the rare cancer type without biomarker stratification, extrapolating data on prognosis of the biomarker-positive tumor from the common to rare cancer might provide the best available evidence but it is associated with high level of uncertainty. Statistical modeling techniques such as propensity score matching to generate synthetic control arms, and adjusting for known prognostic factors including differences in histotypes, could be used to better estimate prognosis, but such approaches are still limited as it is not possible to account for all possible confounders.35–43
Criterion (1b) Could favorable outcomes in single-arm studies in the biomarker-defined rare cancer be due to better prognosis?
Evidence that a biomarker has no or worse prognostic impact in rare cancer provides greater confidence that favorable outcomes from a single-arm study may be attributable to the targeted therapy. Even so, other biases, such as selection bias, may lead to better outcomes in single-arm studies. Any given biomarker may be prognostic but not predictive, predictive but not prognostic, both prognostic and predictive, or neither prognostic nor predictive. HER2 overexpression in breast cancer is an example of a biomarker that is both prognostic and predictive.29,30 When biomarker expression is associated with good prognosis, such as in the case of hormone receptor-positive breast cancer, the benefit of targeted treatments will be difficult to establish in the absence of RCTs. Favorable clinical outcomes from single-arm studies are often assumed to be the effect of the targeted therapy but may, in fact, be due to the indolent natural history of cancer.
Analytical validity
Analytical validity refers to the analytical performance characteristics of a test to reliably detect the biomarker in a biological specimen. Measures include concordance, sensitivity, and specificity against a validated test, and reproducibility. Assessment of the analytical validity of the biomarker test (assay/technology) is distinct to, but predicated on, the clinical utility of the biomarker to predict treatment benefit. The pivotal RCTs that established targeted treatment effectiveness in the common cancer, also establish the clinical utility of the biomarker in the common cancer. 44 As such, the biomarker test used in the pivotal RCT is generally regarded as the “clinical utility standard” (or evidentiary standard) test for assessment of analytical performance. However, the analytical performance characteristics of the test established in common cancers may or may not be directly relevant to rare cancers.44–46 Two central issues are: first, without an RCT to validate the biomarker predicts treatment benefit in the rare cancer, assessing the analytical validity of the biomarker test for use in the rare cancer will be more complex. Second, as technology evolves, the biomarker test proposed in the rare cancer may not be the same as that used in the pivotal RCT in the common cancer. These and other issues that should be considered when evaluating analytical validity of the biomarker test are outlined below:
Criterion (2a) If the biomarker test proposed in the rare cancer is the same test used in the common cancer pivotal trial, have the performance characteristics of the test been assessed in the rare cancer?
The test proposed in the rare cancer may be the same test used in the RCT of the common cancer. Pre-analytic factors that affect quality of analytes include specimen type (e.g. core tumor biopsy vs blood), preservation (e.g. fresh vs formalin-fixed paraffin-embedded), tissue fixation methods (e.g. time to fixation, duration and temperature of fixation, fixing agent), and specimen age. These factors are specified for the clinical utility standard test and influence the usefulness of the assay. Even so, biological differences between the cancers may alter the test’s performance characteristics, potentially limiting applicability in rare cancers. For example, excessive melanin pigment in some melanomas can interfere with DNA polymerases used in polymerase chain reaction (PCR) methods and invalidate test results. 47 Testing should be undertaken in accredited laboratories. Sufficient concordance and reproducibility across laboratories should be confirmed.
Criterion (2b) Can the scoring criteria or grouping strategy to define the biomarker-positive and biomarker-negative subgroups established in the common cancer be directly applied to the rare cancer or does it require modification?
Scoring criteria
For some binary biomarkers such as DNA point mutations, the same criteria to define the biomarker-positive and biomarker-negative subpopulations in one cancer could be directly applied to another. For example, in the Kirsten rat sarcoma viral oncogene homolog gene, a single-nucleotide variation, where glycine is substituted by cysteine at codon 12 (KRAS G12C), results in activation of downstream signaling pathways. This mutation is found in some non-small cell lung cancers (NSCLC),48,49 colorectal cancers (CRC), 50 and pancreatic adenocarcinoma 51 and the same criteria could be used to classify patients across the different cancer types.
For other biomarkers, such as some quantitative biomarkers or gene signatures, existing criteria in one cancer type will always need to be modified for use in another cancer type. For example, in breast cancer, HER2 gene amplification induces HER2 protein overexpression on tumor cell membrane and is known to be oncogenic.52,53 Although no “gold” standard exists for detecting HER2 alterations, 54 the scoring algorithm based on HER2 amplification, using HER2 gene copies per nucleus or the HER2 gene signals to chromosome 17 centromere ratio as detected by fluorescent or silver in situ hybridization, and HER2 protein overexpression as detected on immunohistochemistry, have been widely validated in breast cancer as a predictive biomarker for various HER2-targeted therapies.54–56 This scoring algorithm required modification before applying to gastric/gastroesophageal junction cancers due to differences in pattern of HER2 expression.57,58 The HER2 scoring systems for CRC 59 and endometrial serous carcinoma 60 are also modified and differ slightly from each of the other cancers.
Grouping strategy
Different but related molecular alterations involving one or more genes affecting a common pathway can result in the same clinical disease. 61 The grouping of alterations may be accepted if there is strong rationale that the group will respond similarly to therapy based on clinical, preclinical, or in silico (computational) mechanistic evidence. 61 In this way, where various molecular alterations comprise the biomarker-defined disease in the common cancer, the same grouping strategy may be used to define the disease in the rare cancer. To illustrate, numerous “deleterious” mutations of the breast cancer susceptibility genes 1 and 2 affect a common DNA repair pathway resulting in a similar phenotype that predicts treatment benefit with poly(adenosine diphosphate–ribose) polymerase enzyme inhibitors in breast, ovarian, prostate, and pancreatic cancers.62–66 Depending on the strength of scientific rationale, it may be reasonable to either expand or restrict the alterations included in the common cancer trial when applying the data to the rare cancer.
Establishing databases of rare cancers annotated with comprehensive genomic profile data would be very useful for assay development and validation of scoring. The criteria or grouping strategy to define the biomarker-positive and biomarker-negative subgroups in the rare cancer should initially be established a priori by consensus based on available information from common cancers. Rare cancer databases that also capture the natural history and clinical outcomes of targeted therapies can also be used to validate the biomarker criteria established in the rare cancer. Modification of criteria may be necessary depending on findings from validation studies (4).
Criterion (2c) What is the prevalence of the biomarker in the rare cancer? Does this prevalence change over the course of the disease? What is the performance of the proposed test in low-prevalence biomarker-positive rare cancers?
Biomarker prevalence can vary widely across different cancer types, stages, treatments, and disease trajectories. 67 For example, mismatch repair deficiency (dMMR) results from mutations in a family of genes involved in DNA repair. This biomarker is considered to be predictive of immunotherapy benefit, and pembrolizumab, a programmed death 1 (PD-1) inhibitor, has been approved for solid tumors with dMMR following progression on prior treatment.68,69 The prevalence of dMMR varies widely across histotypes, ranging from approximately 28% in endometrial cancer 70 to 0.04% in breast cancer. 71 Furthermore, within the same cancer, such as CRC, prevalence of dMMR can also vary between early-stage disease (10%–20%) and advanced-stage disease (3%–4%). 72 Biomarker status can also change over the course of the disease as part of the disease trajectory and/or result of previous treatment.44,73
For a test with a given analytical sensitivity and specificity, changes in biomarker prevalence can significantly alter its positive predictive value (PPV) and negative predictive value (NPV) 44 (Box). A test with high sensitivity and specificity will have poorer PPV in cancers where biomarker prevalence is low compared to other cancers with higher prevalence of the same biomarker. The same test will have poorer NPV in cancers where biomarker prevalence is high compared to cancers with lower biomarker prevalence.45,73 Incorrect classification of a patient (a false positive or false negative result) can potentially result in incorrect treatment recommendations. 73
The prevalence range of the biomarker in the rare cancer as determined by the proposed test for the disease setting should be assessed. The PPV and NPV of the test can be calculated using estimates of sensitivity and specificity. 44 In rare cancers with low biomarker prevalence, a test with sensitivity and specificity approaching 100% should be used whenever possible to minimize the false negative and false positive rates, respectively. 73 Tolerance of a higher false positive rate would depend on the potential for treatment harm, treatment costs, and delays to more effective alternative therapies if available.
Criterion (2d) Is the test proposed in the rare cancer different to the test used in the common cancer? If so, has the new/alternative test been analytically validated against the evidentiary standard test in the rare cancer?
With advancements in diagnostic technology following the pivotal treatment trial in the common cancer, a new test may be considered a more valid measure of the biological target. The new test proposed to identify the biomarker in the rare cancer may use similar (e.g. two different commercially developed PCR tests) or different (e.g. panel point mutations vs whole exome sequencing) technology. When the test proposed for the rare cancer is not the same test used in the common cancer, it may result in discordance between the biomarker-defined populations using each test. 74 Retrospective testing of patient samples from the pivotal trial to assess concordance with an accepted clinical utility standard test and linked with clinical outcome data is ideal and should be done if possible75,76 but may not be feasible. 77 Concordance measures include positive percent agreement, negative percent agreement, and overall percent agreement (Box). However, different organizations have adopted different criterion and the extent of sufficient agreement is an unresolved issue.78–80 Discrepancies are resolved using another orthogonal method. 77 Intra-observer, inter-observer, and inter-laboratory reproducibility is assessed where appropriate.45,81 Where discordance exists between the two tests, there would be insufficient evidence of clinical utility of the biomarker as defined by the new test in rare cancer.
Biomarker actionability
A biomarker is potentially “actionable” if it represents (i) a molecular pathway driving oncogenesis and tumor progression that can be mitigated or reversed by targeted therapy to improve clinical outcomes and is not also affected by (ii) a resistance pathway so that targeted therapy rapidly becomes ineffective. Critical to demonstrating actionability is evidence of the ability of the biomarker to predict clinical outcomes. Methods to validate a predictive biomarker within a specific cancer type utilizing trial designs to assess for the biomarker-treatment interaction are established. 82 When assessing the predictive value of a biomarker in rare cancers utilizing Bayesian adaptive trial designs, increased biological understanding may reasonably shift Bayesian priors. In this paper, we assume the predictive value of the biomarker has been validated in the common cancer. Scenarios where this does not hold are beyond the scope of this work.
A principal assumption for extrapolation is that the biomarker is equally actionable for both the common and rare cancers, but this might not be the case. Assessment of biomarker actionability in the rare cancer may be informed by considering the two questions outlined below:
Criterion (3a) How strong is the evidence supporting biomarker actionability in rare cancer?
Frameworks ranking biomarkers according to strength of evidence supporting actionability have been published and can inform this assessment in rare cancers.83–89 Top-tier evidence of biomarker actionability for matching targeted therapy is established in prospective, adequately powered RCTs on measures of net clinical benefit—often established for common cancers. In rare cancers where RCTs utilizing these outcome measures are not possible, these frameworks make recommendations for ranking the strength of evidence supporting actionability and include (i) retrospective studies showing clinical benefit from targeted therapy in the biomarker-positive versus biomarker-negative group, (ii) prospective studies showing increased tumor responsiveness without data on survival endpoints, (iii) evidence for a top-tier association but in a different cancer histotype, (iv) preclinical models predicting sensitivity to matched therapy without clinical data, and (v) in silico evidence predicting functional impact similar to that seen for a biomarker-therapy match in different histotypes. Evidence supporting the biological rationale in the rare cancer should be ranked according to strength of clinical validity using these frameworks. Extending these frameworks, we propose considerations for downgrading the strength of the evidence for actionability in the rare cancer below.
Criterion (3b) Is there evidence that suggests the treatment effect in the rare cancer may differ from that in the common cancer thereby not supporting extrapolation?
Clinical, preclinical, and mechanistic evidence for different actionability across cancers should be assessed.
Cellular context and tumor microenvironment
Complex interactions between the biomarker and the cellular context or tumor microenvironment unique to a specific tumor type may exist and alter the actionability of the biomarker. Pembrolizumab received histology-agnostic FDA approval for the treatment of advanced solid tumors with a high tumor mutational burden (TMB-H) for patients who have no other alternative therapeutic options based on the non-randomized, open-label KEYNOTE-158 study. 90 However, clinical benefit was shown to differ across TMB-H tumors where ORR in endometrial cancer was 47% while in anal cancer only 7%, suggesting that tumor microenvironments may influence treatment response and that the predictive ability of TMB may not be uniform across different cancer types.91–94
Compensatory resistance pathways
Even if a molecular alteration is a driver across multiple cancers and these are treated with the same targeted agent(s), emergence of compensatory resistant pathways may differ across cancer types. 95 For example, v-raf murine sarcoma viral oncogene homolog B1 (BRAF) inhibitor vemurafenib has been evaluated in a range of BRAF V600-mutant histotypes including melanoma, NSCLC,96,97 and CRC. 96 In melanoma, RCTs have demonstrated vemurafenib improved OS compared to dacarbazine chemotherapy.98,99 In a non-randomized basket trial, response rates in NSCLC (42%) 96 were comparable to melanoma 99 but no responses were seen in CRC. 96 Subsequent preclinical studies have shown that vemurafenib monotherapy results in rapid acquired resistance to BRAF inhibition in CRC but not in the other cancer types.95,100 This finding has been subsequently confirmed in a prospective RCT with dual inhibition of BRAF and epidermal growth factor receptor pathways. 101
Significant differences in the cellular or tumor microenvironment and/or compensatory resistance pathways between common and rare cancers downgrade the strength of the evidence for actionability and raise uncertainty about extrapolation.
Treatment outcomes
Efficacy
A principal assumption of extrapolation is that the common and rare cancers sharing the same biomarker are similar in prognosis and response to targeted treatment such that the same treatment effect could be expected. 102 When efficacy of targeted therapy is only evaluated in single-arm trials, relative treatment benefit could be extrapolated from the common cancer to the rare cancer provided that: (i) RCT data confirms net clinical benefit in the common cancer, and (ii) signals of efficacy from single-arm or randomized studies in the rare cancer are comparable between the common and rare cancer based on the same validated surrogate endpoint measure(s). 102 Similarly, where clinical benefit of targeted therapy has been demonstrated in a range of heterogeneous cancer types grouped together by the same actionable biomarker profile in a “pan-cancer” study, clinical benefit may reasonably be extrapolated to each rare cancer type provided signals of efficacy on the surrogate measures are similar. For example, fam-trastuzumab deruxtecan is a HER2 directed antibody–drug conjugate which has been shown to improve ORR, duration of response (DOR), and OS compared to physician’s choice chemotherapy in HER2 overexpressed/amplified, previously treated, metastatic breast cancer (ORR 70% vs 29%, median DOR 19.6 months vs 8.3 months, HR for OS 0.66, p = 0.0021). 103 HER2 overexpression is found across diverse cancer types but prevalence rates can be low. In endometrial and cervical cancers, the prevalence of HER2 overexpression in these tumors is approximately 4%. 104 In April 2024, the FDA granted accelerated tumor-agnostic approval to fam-trastuzumab deruxtecan for patients with previously treated, advanced HER2-positive (Immunohistochemistry (IHC) 3+) cancers who have no satisfactory alternative treatment options. 105 This approval was based on a pan-cancer single-arm basket trial showing a comparable ORR of 61.3% and median DOR of 22.1 months. 106 Magnitude of benefit was particularly high in IHC 3+ endometrial (ORR 84.6%, DOR not reached) and cervical cohorts (ORR 75%, DOR 14.2 months). 106 These results compare favorably to historical controls where survival outcomes are poor and chemotherapy response rates are low.107,108 However, there was no observed benefit in the pancreatic cohort (ORR 4%, DOR 5.7 months). 106 Concurrent control comparison may not be feasible in all rare cancer cohorts. Hence uncertainties will remain when the overall treatment effect is applied in each of the different rare cancer cohorts.
Surrogate measures may include progression-free survival (PFS), ORR, pharmacokinetic/pharmacodynamic (PK/PD) properties, circulating tumor DNA levels, and functional imaging responses. Heterogeneity of treatment effect on surrogate measures may be tested. This approach has been supported by regulatory and HTA bodies including the The Food and Drug Administration (FDA), The European Medicines Agency (EMA), United Kingdom’s National Institute for Health and Care Excellence and Australia’s Medical Services Advisory Committee.22,44,61,109
RCTs in rare cancers are ideal wherever possible, including the use of novel trial designs such as randomized basket trials using intermediate outcomes, to strengthen the evidence of efficacy as compared with relying solely on non-randomized trials. Beyond clinical trials in rare cancers, the organized collection of clinical outcome data from post-marketing studies, 110 registries, and real-world studies should be prioritized to continuously build the body of evidence. When considering extrapolation of relative treatment effect, the following questions should be considered:
Criterion (4) Is there a validated surrogate endpoint that can be used to extrapolate the clinical benefit of targeted therapy from the common cancer to the rare cancer? Are estimates of targeted therapy efficacy based on this surrogate endpoint similar between the common and rare cancer?
Treatment outcomes based on surrogate endpoints, such as PFS or ORR, used for extrapolation should be adequately assessed to determine whether they reliably predict treatment benefits for OS.111–114 It is generally not feasible to adequately validate surrogate endpoints in rare cancer studies, particularly in the absence of trials of randomized design. However, these surrogate endpoints should, at a minimum, be validated in the common cancer trials.
Surrogate endpoints are validated for a specified context of use for a specific biomarker, type of therapy, cancer type, and disease setting. Therefore, a validated surrogate endpoint for one cancer type is not necessarily a valid surrogate for a different cancer type. 115 For example, in a study of multiple first-line chemotherapy and hormone therapy trials of advanced cancers, PFS was shown to be an acceptable surrogate for OS in colorectal and ovarian cancers but not in breast and prostate cancers. 116 The minimum size of the surrogate difference or threshold needed to predict a clinical benefit gain (e.g. OS gain) can also differ across cancer types. 117
ORR and disease stabilization measures including DOR and disease control rate (DCR, a combined measure of ORR and stable disease (SD) at a specific time-point) are commonly used as a surrogate for OS in oncology and as a primary endpoint in pivotal trials supporting regulatory approvals in rare cancers.118–120 Tumor shrinkage is regarded to be exceedingly rare in the absence of effective therapy and is widely perceived to precede other clinical improvements, including survival prolongation. However, the validity of ORR as a surrogate for OS has not been established for most settings.121,122 Non-randomized trials have been shown to exaggerate DOR for targeted therapies when compared with RCTs of the same drug for the same setting. 123 DCR as an endpoint also does not completely capture treatment activity as many tumors with indolent natural history will satisfy the criterion of short-term SD. 124
In view of these limitations, alternative endpoints assessing PD response utilizing minimally invasive functional technologies 125 and/or composite endpoints may need to be considered. If validated for the specific context of use, these endpoints may prove useful for extrapolation (Figure 1(d)). Composite endpoints may be particularly useful in rare cancers as they may be more sensitive in detecting the spectrum of treatment effects and reduce sample size requirements.126,127 Composites can also assess more than one aspect of the patient’s health status and incorporate clinically meaningful outcomes. 126 If composite endpoints are used, they should be prespecified, clearly defined, weighted according to clinical relevance, used and reported according to published guidance,128,129 and validated prior to use in different cancer types and clinical settings. Where evidence based on surrogate endpoints does not support similar efficacy between the common and rare cancers, extrapolation may not be appropriate.
Safety
Criterion (5) Are the adverse events experienced in the rare cancer similar to those experienced in the common cancer? Are there any clinically meaningful differences between cancers?
Another important assumption made for extrapolation is that the safety profile of the targeted therapy in rare cancer is similar to that of common cancer. In common cancers, safety data of targeted therapy from RCTs provide an unbiased comparison of adverse events (AEs) related to treatment and differentiate these from disease-related or other non-treatment-related AEs. Safety data is also augmented by post-marketing and real-world studies that capture use in populations outside those highly selected for RCTs.
AEs are likely to be common across multiple cancer types. However, differences can exist across cancer types because of differences in co-existing environmental exposures, comorbidities, organ-specific tumor burden, and prior systemic and local therapies resulting in differing tolerance to treatment-related toxicity.130,131 For example, a meta-analysis of 20 PD-1 inhibitor trials showed significantly higher incidence of pneumonitis in NSCLC and renal cell carcinoma compared to melanoma. 130 Safety data across cancer types should be assessed to judge whether differences are clinically meaningful and are of significant magnitude to represent important high uncertainty, limiting extrapolation. Additional sources that may augment safety data in the rare cancer include natural history studies, auxiliary safety cohorts, expanded access programs, and real-world studies capturing off-label use132,133 and PK/PD data. 125 In controlled “pan-cancer” trials, safety data of the combined control arm may be heterogeneous due to the varying control treatments.
Decision tree
We recommend addressing all criteria for the five components necessary for extrapolation to inform decision-making for the targeted therapy in the rare cancer. Explicit judgments about the level of uncertainty for each component based on an assessment of the supportive evidence will result in a more transparent approach to regulatory decisions. We propose that certainty for all or most of the criteria is required to extrapolate the treatment benefit of targeted therapy from the common to the rare cancer. During the process of evidence evaluation, knowledge gaps may be identified in one or more component(s). Depending on the clinical impact of these gap(s), further research may be needed before extrapolation can be used (Figure 2).
When there is sufficient evidence for provisional or regular regulatory approval, uncertainties may remain regarding the longer-term clinical benefits, safety in broader rare cancer populations, and spectrum of uncommon AEs. Detailed plans for post-approval commitments addressing specific residual uncertainties identified during pre-approval evaluation should be outlined (Figure 2).
Strengths
The framework is an important first step to outline the breadth of criteria essential for evidence assessment for rare biomarker-defined cancers. It is an initial conceptual construct for stimulating multidisciplinary discourse toward developing a validated and reproducible tool that can be incorporated into the HTA process, clinical practice guidelines, and clinical decision-making. Five essential components of evidence assessment from multidisciplinary fields have been incorporated into a single framework. These components should be, but are not commonly, considered as a whole. However, consideration of only one or few components, such as efficacy without addressing prognosis or the analytic validity of the biomarker test, would be incomplete.
Limitations
There are several limitations. The utility of the framework and validity of the approach for judging extrapolation criteria and uncertainty has not yet been assessed. The applicability of the framework across a wide range of targeted therapy-cancer histotype scenarios, as well as reproducibility and consistency of uncertainty judgments require testing.
Future work
As evidence for histology-agnostic-targeted therapies accumulates, the extrapolation criteria may be refined, and anchors developed to guide uncertainty judgments. A transparent process should be developed to assess consistency and reproducibility of uncertainty judgments by independent assessors. This could be undertaken by seeking expert consensus on trialed examples and used to develop a guidance document. Future studies evaluating the utility of this framework for regulatory and reimbursement decisions should be conducted. Outcome measures for these studies may include completeness of evidence assessment and transparency of decisions, time taken from initial targeted therapy approval in a common cancer to additional approvals in other rare cancers sharing the same biomarker, clinical benefit of drugs approved using this framework, and proportion of subsequent withdrawals.
Conclusion
We have proposed a framework for extrapolating evidence of treatment effects for molecularly targeted therapies from common to rare cancers sharing the same predictive biomarker. This framework supports systematic assessment, standardized decision-making, and transparent discussions between key stakeholders. Where there is still insufficient evidence for extrapolation, our approach will also help better target future research to address critical gaps. This will ultimately inform clinical practice and will benefit patients with rare biomarker-defined cancers to access safe and effective targeted therapies.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359241273062 for Criteria for assessing evidence for biomarker-targeted therapies in rare cancers—an extrapolation framework by Doah Cho, Sarah J. Lord, Robyn Ward, Maarten IJzerman, Andrew Mitchell, David M. Thomas, Saskia Cheyne, Andrew Martin, Rachael L. Morton, John Simes and Chee Khoon Lee in Therapeutic Advances in Medical Oncology
Acknowledgments
None.
Footnotes
ORCID iDs: Doah Cho  https://orcid.org/0000-0002-8045-0751
https://orcid.org/0000-0002-8045-0751
Sarah J. Lord  https://orcid.org/0000-0003-2763-5949
https://orcid.org/0000-0003-2763-5949
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Doah Cho, National Health and Medical Research Council Clinical Trials Centre, Faculty of Medicine and Health, University of Sydney, Australia; Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Locked Bag 77, Camperdown, NSW 1450, Australia.
Sarah J. Lord, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia
Robyn Ward, Faculty of Medicine and Health, University of Sydney, Camperdown, NSW, Australia.
Maarten IJzerman, Faculty of Medicine, Dentistry and Health Sciences, Centre for Health Policy, University of Melbourne Centre for Cancer Research, Parkville, VIC, Australia; Erasmus School of Health Policy and Management, Erasmus University Rotterdam, Rotterdam, The Netherlands.
Andrew Mitchell, Department of Health Economics Wellbeing and Society, The Australian National University, Canberra, ACT, Australia.
David M. Thomas, Centre for Molecular Oncology, University of New South Wales, Sydney, NSW, Australia
Saskia Cheyne, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia.
Andrew Martin, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia; Centre for Clinical Research, University of Queensland, St Lucia, QLD, Australia.
Rachael L. Morton, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia
John Simes, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia.
Chee Khoon Lee, Faculty of Medicine and Health, National Health and Medical Research Council Clinical Trials Centre, University of Sydney, Camperdown, NSW, Australia.
Declarations
Ethics approval and consent to participate: Not applicable as this study does not involve human participants, human data, or human tissue.
Consent for publication: Not applicable as this study does not involve human participants.
Author contributions: Doah Cho: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Visualization; Project administration; Writing – original draft; Writing – review & editing.
Sarah J. Lord: Conceptualization; Formal analysis; Investigation; Methodology; Supervision; Visualization; Writing – original draft; Writing – review & editing.
Robyn Ward: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
Maarten IJzerman: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
Andrew Mitchell: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
David M. Thomas: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
Saskia Cheyne: Conceptualization; Formal analysis; Investigation; Methodology; Writing – review & editing.
Andrew Martin: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
Rachael L. Morton: Conceptualization; Formal analysis; Methodology; Supervision; Writing – review & editing.
John Simes: Conceptualization; Formal analysis; Funding acquisition; Methodology; Supervision; Writing – review & editing.
Chee Khoon Lee: Conceptualization; Formal analysis; Investigation; Methodology; Supervision; Visualization; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by an NHMRC Postgraduate Research Scholarship (Medical—Dental) (SC0798), Postgraduate Research Supplementary Scholarship in Oncology, Postgraduate Research Supplementary Scholarship in Decision Making Frameworks in Rare Cancers, and NHMRC Program Grant.
All authors declare no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years, and no other relationships or activities that could appear to have influenced the submitted work. D.C. declares receiving an NHMRC Postgraduate Research Scholarship (Medical—Dental) (SC0798), Postgraduate Research Supplementary Scholarship in Oncology and NHMRC Program Grant. She has also received support from Novartis for attending scientific meetings. R.W. declares that she is the Chair of Australia’s Medical Service Advisory Committee and in that capacity an employee of the Commonwealth of Australia. M.I. declares unrestricted research funding from Illumina. He was a member of ESC/MSAC until June 2022. A.M. declares advisory role for national advisory committees for health technologies as an employee of the Department of Health and Aged Care of Australia. S.J.L., S.C., R.L.M., and A.M. declare no conflict of interests. D.M.T. declares he is the chief executive officer of Omico (non-profit). He/Omico declares grants, consultancies, or research support (Roche, AstraZeneca, Pfizer, Eisai, Illumina, Beigene, Elevation Oncology, RedX Pharmaceuticals, SunPharma, Bayer, Abbvie, George Clinical, Janssen, Merck, Kinnate, Microba, BioTessellate, Australian Unity, Foundation Medicine, Guardant Health). J.S. declares research grants to the University of Sydney (Bayer, Roche, BMS, AstraZeneca, Abbvie, MSD, Pfizer). C.K.L. declares honoraria (AstraZeneca, Pfizer, Amgen, Takeda, Yuhan, Boehringer Ingelheim, Roche, MSD Oncology), consulting or advisory role (Novartis, Boehringer Ingelheim, Takeda, AstraZeneca, Yuhan, Amgen), and research funding (AstraZeneca, Roche, Merck KGaA).
Contributor selection: All authors are members of the NHMRC Clinical Trials Centre and/or Clinical Trials: Impact & Quality (CT:IQ) group. This work was undertaken as part of Doah Cho’s PhD studies.
Availability of data and materials: Not applicable.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 2. Dancey JE, Bedard PL, Onetto N, et al. The genetic basis for cancer treatment decisions. Cell 2012; 148: 409–420. [DOI] [PubMed] [Google Scholar]
- 3. Barrett JC, Frigault MM, Hollingsworth S, et al. Are companion diagnostics useful? Clin Chem 2013; 59: 198–201. [DOI] [PubMed] [Google Scholar]
- 4. Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics of cancer: genome sequencing and beyond. Annu Rev Genomics Hum Genet 2011; 12: 407–430. [DOI] [PubMed] [Google Scholar]
- 5. U.S. Food and Drug Administration. Guidance for industry providing clinical evidence of effectiveness for human drug and biological products, https://www.fda.gov/media/71655/download (1998, accessed 14 June 2023).
- 6. European Medicines Agency. General considerations for clinical trials, https://www.ema.europa.eu/en/ich-e8-general-considerations-clinical-studies-scientific-guideline (1998, accessed 14 June 2023). [Google Scholar]
- 7. Sharma MR, Schilsky RL. Role of randomized phase III trials in an era of effective targeted therapies. Nat Rev Clin Oncol 2011; 9: 208–214. [DOI] [PubMed] [Google Scholar]
- 8. Selaru P, Tang Y, Huang B, et al. Sufficiency of single-arm studies to support registration of targeted agents in molecularly selected patients with cancer: lessons from the clinical development of crizotinib. Clin Transl Sci 2016; 9: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simon R, Blumenthal GM, Rothenberg ML, et al. The role of nonrandomized trials in the evaluation of oncology drugs. Clin Pharmacol Ther 2015; 97: 502–507. [DOI] [PubMed] [Google Scholar]
- 10. Cho D, Roncolato FT, Man J, et al. Clinical equipoise for trials of novel biologic therapies, therapeutic success rates, and predictors of success: a meta-analysis. JCO Precis Oncol 2017; 1: 1–12. [DOI] [PubMed] [Google Scholar]
- 11. Seruga B, Ocana A, Amir E, et al. Failures in phase III: causes and consequences. Clin Cancer Res 2015; 21: 4552–4560. [DOI] [PubMed] [Google Scholar]
- 12. Del Paggio JC, Azariah B, Sullivan R, et al. Do contemporary randomized controlled trials meet ESMO thresholds for meaningful clinical benefit? Ann Oncol 2017; 28: 157–162. [DOI] [PubMed] [Google Scholar]
- 13. Yao JC, Meric-Bernstam F, Lee JJ, et al. Accelerated approval and breakthrough therapy designation: oncology drug development on speed? Clin Cancer Res 2013; 19: 4305–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beaver JA, Howie LJ, Pelosof L, et al. A 25-year experience of US Food and Drug Administration accelerated approval of malignant hematology and oncology drugs and biologics: a review. JAMA Oncol 2018; 4: 849–856. [DOI] [PubMed] [Google Scholar]
- 15. Gill J, Prasad V. A reality check of the accelerated approval of immune-checkpoint inhibitors. Nat Rev Clin Oncol 2019; 16: 656–658. [DOI] [PubMed] [Google Scholar]
- 16. Renfro LA, Mallick H, An MW, et al. Clinical trial designs incorporating predictive biomarkers. Cancer Treat Rev 2016; 43: 74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S-J, Hung HMJ. Adaptive enrichment with subpopulation selection at interim: methodologies, applications and design considerations. Contemp Clin Trials 2013; 36: 673–681. [DOI] [PubMed] [Google Scholar]
- 18. Jenkins M, Stone A, Jennison C. An adaptive seamless phase II/III design for oncology trials with subpopulation selection using correlated survival endpoints. Pharm Stat 2011; 10: 347–356. [DOI] [PubMed] [Google Scholar]
- 19. Freidlin B, McShane LM, Polley MY, et al. Randomized phase II trial designs with biomarkers. J Clin Oncol 2012; 30: 3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Odogwu L, Mathieu L, Blumenthal G, et al. FDA approval summary: dabrafenib and trametinib for the treatment of metastatic non-small cell lung cancers harboring BRAF V600E mutations. Oncologist 2018; 23: 740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. European Medicines Agency. Concept paper on extrapolation of efficacy and safety in medicine development, https://www.ema.europa.eu/en/extrapolation-efficacy-safety-medicine-development (2013, accessed 10 January 2023).
- 22. European Medicines Agency. Reflection paper on the use of extrapolation in the development of medicines for paediatrics, https://www.ema.europa.eu/en/extrapolation-efficacy-safety-paediatric-medicine-development (2018, accessed 10 January 2023).
- 23. U.S. Food and Drug Administration. Leveraging existing clinical data for extrapolation to pediatric uses of medical devices, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/leveraging-existing-clinical-data-extrapolation-pediatric-uses-medical-devices (2016, accessed 10 January 2023).
- 24. Cho D, Cheyne S, Lord SJ, et al. Extrapolating evidence for molecularly targeted therapies from common to rare cancers: a scoping review of methodological guidance. BMJ Open 2022; 12(7): E058350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alonso-Coello P, Oxman AD, Moberg J, et al. GRADE Evidence to Decision (EtD) frameworks: a systematic and transparent approach to making well informed healthcare choices. 2: Clinical practice guidelines. BMJ 2016; 353: i2089. [DOI] [PubMed] [Google Scholar]
- 26. U.S. Food and Drug Administration. Benefit-risk assessment in drug regulatory decision-making. Draft PDUFA VI implementation plan (FY 2018–2022), US Department of Health and Human Services, 2018. [Google Scholar]
- 27. Chen Y, Yang K, Marušic A, et al. A reporting tool for practice guidelines in health care: the RIGHT statement. Ann Intern Med 2017; 166: 128–132. [DOI] [PubMed] [Google Scholar]
- 28. Piggott T, Baldeh T, Dietl B, et al. Standardized wording to improve efficiency and clarity of GRADE EtD frameworks in health guidelines. J Clin Epidemiol 2022; 146: 106–122. [DOI] [PubMed] [Google Scholar]
- 29. Ross JS, Slodkowska EA, Symmans WF, et al. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 2009; 14: 320–368. [DOI] [PubMed] [Google Scholar]
- 30. Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344: 783–792. [DOI] [PubMed] [Google Scholar]
- 31. Sargent DJ, Mandrekar SJ. Statistical issues in the validation of prognostic, predictive, and surrogate biomarkers. Clin Trials 2013; 10: 647–652. [DOI] [PubMed] [Google Scholar]
- 32. Amur S, LaVange L, Zineh I, et al. Biomarker qualification: toward a multiple stakeholder framework for biomarker development, regulatory acceptance, and utilization. Clin Pharmacol Ther 2015; 98: 34–46. [DOI] [PubMed] [Google Scholar]
- 33. McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005; 97: 1180–1184. [DOI] [PubMed] [Google Scholar]
- 34. Sauerbrei W, Taube SE, McShane LM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst 2018; 110: 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davi R, Chandler M, Elashoff B, et al. Non-small cell lung cancer (NSCLC) case study examining whether results in a randomized control arm are replicated by a synthetic control arm (SCA). J Clin Oncol 2019; 37: 9108. [Google Scholar]
- 36. Davies J, Martinec M, Delmar P, et al. Comparative effectiveness from a single-arm trial and real-world data: alectinib versus ceritinib. J Comp Eff Res 2018; 7: 855–865. [DOI] [PubMed] [Google Scholar]
- 37. Austin PC. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res 2011; 46: 399–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Austin PC, Stuart EA. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies. Stat Med 2015; 34: 3661–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berry DA, Elashoff M, Blotner S, et al. Creating a synthetic control arm from previous clinical trials: application to establishing early end points as indicators of overall survival in acute myeloid leukemia (AML). J Clin Oncol 2017; 35: 7021. [Google Scholar]
- 40. Menefee ME, Gong Y, Mishra-Kalyani PS, et al. Project Switch: docetaxel as a potential synthetic control in metastatic non-small cell lung cancer (mNSCLC) trials. J Clin Oncol 2019; 37: 9105. [Google Scholar]
- 41. Kanapuru B, Gong Y, Mishra-Kalyani PS, et al. Project Switch: lenalidomide and dexamethasone (Len-Dex) as a potential synthetic control arm (SCA) in relapsed or refractory multiple myeloma (rrMM). J Clin Oncol 2019; 37: 8047. [Google Scholar]
- 42. Lee CK, Lord SJ, Stockler MR, et al. Historical cross-trial comparisons for competing treatments in advanced breast cancer—an empirical analysis of bias. Eur J Cancer 2010; 46: 541–548. [DOI] [PubMed] [Google Scholar]
- 43. Bouttell J, Craig P, Lewsey J, et al. Synthetic control methodology as a tool for evaluating population-level health interventions. J Epidemiol Community Health 2018; 72: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morona JK, Wyndham A, Scott P, et al. Discussion paper on pan-tumour biomarker testing to determine eligibility for targeted treatment. https://database.inahta.org/article/19075, 2020.
- 45. Jennings L, Van Deerlin VM, Gulley ML, et al. Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med 2009; 133: 743–755. [DOI] [PubMed] [Google Scholar]
- 46. Wiktor AE, Van Dyke DL, Stupca PJ, et al. Preclinical validation of fluorescence in situ hybridization assays for clinical practice. Genet Med 2006; 8: 16–23. [DOI] [PubMed] [Google Scholar]
- 47. Anderson S, Bloom KJ, Vallera DU, et al. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med 2012; 136: 1385–1391. [DOI] [PubMed] [Google Scholar]
- 48. Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48: 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jordan EJ, Kim HR, Arcila ME, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 2017; 7: 596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giannakis M, Jasmine Mu X, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 2016; 15: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531: 47–52. [DOI] [PubMed] [Google Scholar]
- 52. Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182. [DOI] [PubMed] [Google Scholar]
- 53. Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244: 707–712. [DOI] [PubMed] [Google Scholar]
- 54. Wolff AC. Guideline summary: American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor HER2 testing in breast cancer. J Oncol Pract 2007; 3: 48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013; 31: 3997–4013. [DOI] [PubMed] [Google Scholar]
- 56. Wolff AC, Hammond MEH, Allison KH, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. J Clin Oncol 2018; 36: 2105–2122. [DOI] [PubMed] [Google Scholar]
- 57. Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008; 52: 797–805. [DOI] [PubMed] [Google Scholar]
- 58. Bartley AN, Washington MK, Colasacco C, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and the American Society of Clinical Oncology. J Clin Oncol 2017; 35: 446–464. [DOI] [PubMed] [Google Scholar]
- 59. Valtorta E, Martino C, Sartore-Bianchi A, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol 2015; 28: 1481–1491. [DOI] [PubMed] [Google Scholar]
- 60. Buza N. HER2 testing in endometrial serous carcinoma: time for standardized pathology practice to meet the clinical demand. Arch Pathol Lab Med 2021; 145: 687–691. [DOI] [PubMed] [Google Scholar]
- 61. U.S. Food and Drug Administration. Developing targeted therapies in low-frequency molecular subsets of a disease, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/developing-targeted-therapies-low-frequency-molecular-subsets-disease (2018, accessed 10 January 2023). [DOI] [PMC free article] [PubMed]
- 62. Robson M, Im S-A, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017; 377: 523–533. [DOI] [PubMed] [Google Scholar]
- 63. Tutt ANJ, Garber JE, Kaufman B, et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med 2021; 384: 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018; 379: 2495–2505. [DOI] [PubMed] [Google Scholar]
- 65. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020; 382: 2091–2102. [DOI] [PubMed] [Google Scholar]
- 66. Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019; 381: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tan DSW, Mok TSK, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol 2016; 34: 91–101. [DOI] [PubMed] [Google Scholar]
- 68. Marcus L, Lemery SJ, Keegan P, et al. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res 2019; 25: 3753–3758. [DOI] [PubMed] [Google Scholar]
- 69. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gordhandas S, Kahn RM, Gamble C, et al. Clinicopathologic features of endometrial cancer with mismatch repair deficiency. Ecancermedicalscience 2020; 14: 1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mills AM, Dill EA, Moskaluk CA, et al. The relationship between mismatch repair deficiency and PD-L1 expression in breast carcinoma. Am J Surg Pathol 2018; 42: 183–191. [DOI] [PubMed] [Google Scholar]
- 72. Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009; 100: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ramsey SD, Shankaran V, Sullivan SD. Basket cases: how real-world testing for drugs approved based on basket trials might lead to false diagnoses, patient risks, and squandered resources. J Clin Oncol 2019; 37(36): JCO.18.02320. [DOI] [PubMed] [Google Scholar]
- 74. Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol 2017; 35: 1453–1486. [DOI] [PubMed] [Google Scholar]
- 75. Harbison CT, Horak CE, Ledeine JM, et al. Validation of companion diagnostic for detection of mutations in codons 12 and 13 of the KRAS gene in patients with metastatic colorectal cancer: analysis of the NCIC CTG CO.17 trial. Arch Pathol Lab Med 2013; 137: 820–827. [DOI] [PubMed] [Google Scholar]
- 76. Sharma A, Zhang G, Aslam S, et al. Novel approach for clinical validation of the cobas KRAS mutation test in advanced colorectal cancer. Mol Diagn Ther 2016; 20: 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. U.S. Food and Drug Administration. Statistical guidance on reporting results from studies evaluating diagnostic tests—guidance for industry and FDA staff, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/statistical-guidance-reporting-results-studies-evaluating-diagnostic-tests-guidance-industry-and-fda (2007, accessed 15 February 2023).
- 78. Centers for Disease Control and Prevention. Clinical laboratory improvement amendments, https://www.cdc.gov/clia/index.html (1988, accessed 2 November 2022).
- 79. Australian Government Department of Health and Aged Care. National pathology accreditation advisory council, https://www1.health.gov.au/internet/main/publishing.nsf/Content/health-npaac-index.htm (2021, accessed 2 November 2022).
- 80. National Association of Testing Authorities. National association of testing authorities, https://www.anzpaa.org.au/forensic-science/forensic-sciences/forensic-groups/nata (2018, accessed 2 November 2022).
- 81. Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 2017; 19: 341–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sargent DJ, Conley BA, Allegra C, et al. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol 2005; 23: 2020–2027. [DOI] [PubMed] [Google Scholar]
- 83. Mateo J, Chakravarty D, Dienstmann R, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann Oncol 2018; 29: 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy [Review]. J Natl Cancer Inst 2015; 107(7): djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Andre F, Mardis E, Salm M, et al. Prioritizing targets for precision cancer medicine. Ann Oncol 2014; 25: 2295–2303. [DOI] [PubMed] [Google Scholar]
- 86. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017; 2017: PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vidwans SJ, Turski ML, Janku F, et al. A framework for genomic biomarker actionability and its use in clinical decision making. Oncoscience 2014; 1: 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Van Allen EM, Wagle N, Stojanov P, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 2014; 20: 682–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017; 19: 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. U.S. Food and Drug Administration. FDA approves pembrolizumab for adults and children with TMB-H solid tumors, https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors (2020, accessed 15 February 2022).
- 91. Rousseau B, Foote MB, Maron SB, et al. The spectrum of benefit from checkpoint blockade in hypermutated tumors. N Engl J Med 2021; 384: 1168–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. McGrail DJ, Pilié PG, Rashid NU, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021; 32: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Strickler JH, Hanks BA, Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res 2021; 27: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chan KK-W, Tannock IF. Should basket trials be pathways to drug registration for biomarker-defined subgroups of advanced cancers? J Clin Oncol 2021; 39(22): JCO.21.00552. [DOI] [PubMed] [Google Scholar]
- 95. Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012; 483: 100–103. [DOI] [PubMed] [Google Scholar]
- 96. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 2015; 373: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mu Y, Yang K, Hao X, et al. Clinical characteristics and treatment outcomes of 65 patients with BRAF-mutated non-small cell lung cancer. Front Oncol 2020; 10: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chapman PB, Robert C, Larkin J, et al. Vemurafenib in patients with BRAFV600 mutation-positive metastatic melanoma: final overall survival results of the randomized BRIM-3 study. Ann Oncol 2017; 28: 2581–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012; 2: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019; 381: 1632–1643. [DOI] [PubMed] [Google Scholar]
- 102. Cheyne S, Cho D, Lee CK, et al. A scoping review of guidance for assessment of biomarker directed therapies for personalised medicine. Paper presented at the Australian Clinical Trials Alliance 2021 Annual Scientific Meeting, Virtual Meeting, 10–12 November 2021. [Google Scholar]
- 103. André F, Hee Park Y, Kim S-B, et al. Trastuzumab deruxtecan versus treatment of physician’s choice in patients with HER2-positive metastatic breast cancer (DESTINY-Breast02): a randomised, open-label, multicentre, phase 3 trial. Lancet 2023; 401: 1773–1785. [DOI] [PubMed] [Google Scholar]
- 104. Yan M, Schwaederle M, Arguello D, et al. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev 2015; 34: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. U.S. Food and Drug Administration. FDA grants accelerated approval to fam-trastuzumab deruxtecan-nxki for unresectable or metastatic HER2-positive solid tumors, https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-fam-trastuzumab-deruxtecan-nxki-unresectable-or-metastatic-her2 (2024, accessed 29 May 2024).
- 106. Meric-Bernstam F, Makker V, Oaknin A, et al. Efficacy and safety of trastuzumab deruxtecan in patients with HER2-expressing solid tumors: primary results from the DESTINY-PanTumor02 phase II trial. J Clin Oncol 2024; 42: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gadducci A, Cosio S. Pharmacological treatment of patients with metastatic, recurrent or persistent cervical cancer not amenable by surgery or radiotherapy: state of art and perspectives of clinical research. Cancers 2020; 12: 2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Diver EJ, Foster R, Rueda BR, et al. The therapeutic challenge of targeting HER2 in endometrial cancer. Oncologist 2015; 20: 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ciani O, Buyse M, Drummond M, et al. Time to review the role of surrogate end points in health policy: state of the art and the way forward. Value Health 2017; 20: 487–495. [DOI] [PubMed] [Google Scholar]
- 110. Cipriani A, Ioannidis JPA, Rothwell PM, et al. Generating comparative evidence on new drugs and devices after approval. Lancet 2020; 395: 998–1010. [DOI] [PubMed] [Google Scholar]
- 111. Prentice RL. Surrogate endpoints in clinical trials: definition and operational criteria. Stat Med 1989; 8: 431–440. [DOI] [PubMed] [Google Scholar]
- 112. Fleming TR, DeMets DL. Surrogate end points in clinical trials: are we being misled? Ann Intern Med 1996; 125: 605–613. [DOI] [PubMed] [Google Scholar]
- 113. Buyse M, Molenberghs G, Burzykowski T, et al. The validation of surrogate endpoints in meta-analyses of randomized experiments. Biostatistics 2000; 1: 49–67. [DOI] [PubMed] [Google Scholar]
- 114. U.S. Food and Drug Administration. Surrogate endpoint resources for drug and biologic development, https://www.fda.gov/drugs/development-resources/surrogate-endpoint-resources-drug-and-biologic-development (accessed 15 July 2021).
- 115. Baker SG. Five criteria for using a surrogate endpoint to predict treatment effect based on data from multiple previous trials. Stat Med 2018; 37: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Buyse M. Use of meta-analysis for the validation of surrogate endpoints and biomarkers in cancer trials. Cancer J 2009; 15: 421–425. [DOI] [PubMed] [Google Scholar]
- 117. Johnson KR, Ringland C, Stokes BJ, et al. Response rate or time to progression as predictors of survival in trials of metastatic colorectal cancer or non-small-cell lung cancer: a meta-analysis. Lancet Oncol 2006; 7: 741–746. [DOI] [PubMed] [Google Scholar]
- 118. Johnson JR, Ning YM, Farrell A, et al. Accelerated approval of oncology products: the food and drug administration experience. J Natl Cancer Inst 2011; 103: 636–644. [DOI] [PubMed] [Google Scholar]
- 119. Oxnard GR, Wilcox KH, Gonen M, et al. Response rate as a regulatory end point in single-arm studies of advanced solid tumors. JAMA Oncol 2016; 2: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chen EY, Raghunathan V, Prasad V. An overview of cancer drugs approved by the US Food and Drug Administration based on the surrogate end point of response rate. JAMA Intern Med 2019; 179: 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Haslam A, Hey SP, Gill J, et al. A systematic review of trial-level meta-analyses measuring the strength of association between surrogate end-points and overall survival in oncology. Eur J Cancer 2019; 106: 196–211. [DOI] [PubMed] [Google Scholar]
- 122. Mushti SL, Mulkey F, Sridhara R. Evaluation of overall response rate and progression-free survival as potential surrogate endpoints for overall survival in immunotherapy trials. Clin Cancer Res 2018; 24: 2268–2275. [DOI] [PubMed] [Google Scholar]
- 123. Gyawali B, D’Andrea E, Franklin JM, et al. Response rates and durations of response for biomarker-based cancer drugs in nonrandomized versus randomized trials. J Natl Compr Canc Netw 2020; 18: 36–43. [DOI] [PubMed] [Google Scholar]
- 124. Le Tourneau C, Paoletti X, Coquan E, et al. Critical evaluation of disease stabilization as a measure of activity of systemic therapy: lessons from trials with arms in which patients do not receive active treatment. J Clin Oncol 2014; 32: 260–263. [DOI] [PubMed] [Google Scholar]
- 125. Workman P, Aboagye EO, Chung Y-L, et al. Minimally invasive pharmacokinetic and pharmacodynamic technologies in hypothesis-testing clinical trials of innovative therapies. J Natl Cancer Inst 2006; 98: 580–598. [DOI] [PubMed] [Google Scholar]
- 126. Cordoba G, Schwartz L, Woloshin S, et al. Definition, reporting, and interpretation of composite outcomes in clinical trials: systematic review. BMJ 2010; 341: c3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lesko LJ, Atkinson AJ., Jr. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu Rev Pharmacol Toxicol 2001; 41: 347–366. [DOI] [PubMed] [Google Scholar]
- 128. EUnetHTA. Endpoints used for relative effectiveness assessment of pharmaceuticals composite endpoints, https://www.eunethta.eu/wp-content/uploads/2018/01/Endpoints-used-for-Relative-Effectiveness-Assessment-Composite-endpoints_Amended-JA1-Guideline_Final-Nov-2015_0.pdf (2013, accessed 15 July 2021).
- 129. U.S. Food and Drug Administration. Multiple endpoints in clinical trials guidance for industry draft guidance, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/multiple-endpoints-clinical-trials (2017, accessed 15 July 2021).
- 130. Nishino M, Giobbie-Hurder A, Hatabu H, et al. Incidence of programmed cell death 1 inhibitor-related pneumonitis in patients with advanced cancer: a systematic review and meta-analysis. JAMA Oncol 2016; 2: 1607–1616. [DOI] [PubMed] [Google Scholar]
- 131. Sukari A, Nagasaka M, Alhasan R, et al. Cancer site and adverse events induced by immune checkpoint inhibitors: a retrospective analysis of real-life experience at a single institution. Anticancer Res 2019; 39: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. European Medicines Agency. Guideline on clinical trials in small populations, https://www.tga.gov.au/sites/default/files/2024-06/guideline-clinical-trials-small-populations-ema.pdf (2006, accessed 15 July 2021).
- 133. U.S. Food and Drug Administration. Rare diseases common issues in drug development guidance for industry, https://www.fda.gov/media/119757/download (2019, accessed 15 July 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359241273062 for Criteria for assessing evidence for biomarker-targeted therapies in rare cancers—an extrapolation framework by Doah Cho, Sarah J. Lord, Robyn Ward, Maarten IJzerman, Andrew Mitchell, David M. Thomas, Saskia Cheyne, Andrew Martin, Rachael L. Morton, John Simes and Chee Khoon Lee in Therapeutic Advances in Medical Oncology