Abstract
Recent advances in statistical methods and genomic technologies have ushered in a new era in mapping clinically important quantitative traits. However, many refinements and novel statistical approaches are required to enable greater successes in this mapping. The possible impact of recent findings pertaining to the structure of the human genome on efforts to map quantitative traits is yet unclear.
Mapping quantitative trait loci
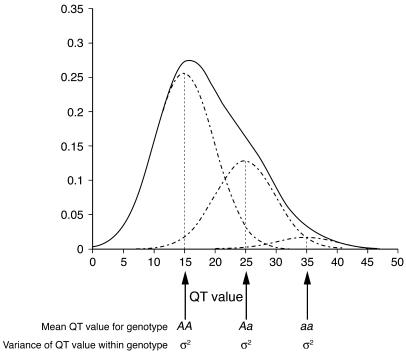
Clinical end points are usually binary — affected or unaffected. Such binary end points almost invariably have quantitative precursor states. Myocardial infarction — a binary end point — for example, has many known quantitative precursors, such as blood pressure and cholesterol levels, which determine the end-point risk. Such quantitative traits (QTs) almost always have strong genetic determinants (that is, are highly heritable). It is, therefore, of considerable interest to map the genes underlying a QT. The traditional viewpoint has been that tens or even hundreds of genes determine a QT, each gene contributing a tiny fraction to the overall variation of the QT. If this were true, then efforts to map a QT locus (QTL) would be futile. However, with the availability of precisely mapped high-density DNA markers for many species, early QTL mapping efforts revealed a much simpler genetic architecture for many QTs (1–4). The emerging paradigm was that even if there were many genes determining the value of a QT, some would have major effects, and hence their chromosomal locations could potentially be determined. Figure 1 illustrates the effects of a single locus with 2 alleles on a QT.
Figure 1.
Genetic effects of a single QTL with 2 alleles. The variances of QT values within genotypes can be unequal. The differences in mean values between AA and Aa genotypes need not be the same as the difference in the mean values between Aa and aa genotypes. If the mean value of the QT for genotype Aa is exactly in the middle of the mean values for genotypes AA and aa, then the 2 alleles A and a have additive effects. If the heterozygote mean is shifted toward the mean value of either of the homozygotes, then there is a dominance effect.
Unfortunately, consistent successes in QTL mapping have been achieved only in species in which inbred strains or lines could be developed. In inbred or experimental populations, the parental origin of each allele is known unambiguously, and all offspring have parents with the same genotypes. These 2 features enable pooling of data across families and testing for equality of mean values of the QT in different genotype classes, using standard ANOVA procedures. Even environmental heterogeneity can be largely controlled experimentally. In humans, unambiguous identification of parental origin of alleles or control for environmental heterogeneity are not possible. Even in inbred strains, efforts at fine mapping of QTLs have revealed unforeseen complexities and have resulted in many failures (5–6), and there are many unresolved issues pertaining to study design and statistical analyses (7–8). In humans, and in other outbred species, QTL mapping has only had limited success (see Taste sensitivity to phenylthiocarbamide for a success story). In this review, we shall focus only on QTL mapping in humans.
Approaches to human QTL mapping
The 2 broad approaches — not mutually exclusive, but complementary — are the candidate gene approach and the marker locus approach. In the candidate gene approach, genes that are physiologically or biochemically relevant to the QT (candidate genes) are screened, and the effects of variant alleles on the QT are investigated. This approach cannot lead to the detection of new QTLs. Further, it is often difficult to choose candidate genes. Although this approach seems attractive, there is not yet sufficient evidence to support its general utility.
The availability of polymorphic markers and refinements of statistical methods (9) have made the marker locus approach very popular. The density of markers and the throughput of marker genotyping have increased over the years, and the cost of marker genotyping has decreased, further facilitating QTL mapping by marker locus approach. Thus, availability of dense markers, high-throughput genotyping, and cost are no longer limiting factors for performing genome-wide scans for positional mapping of QTLs (10–13). The major problems at this time seem to be the difficulty of gathering high-quality phenotype data in a sample of adequate size using an appropriate study design and the analysis of these data using a method with high statistical power.
Study designs
The study designs for QTs are, in the main, similar to those for binary complex traits, that is, binary traits with multilocus determination and possibly with environmental influences. Many conceptual and statistical issues are also similar.
Broadly, there are 2 classes of study designs: study designs in which large sets of relatives from extended or nuclear families are sampled and study designs in which pairs of relatives are sampled (e.g., sibling pairs). Often, sampling is not done randomly. For example, when a sibpair design is adopted, often both siblings are chosen from one tail (upper or lower) of the distribution of the QT (concordant siblings) or one sibling is chosen from the upper tail and the other sibling is chosen from the lower tail (discordant siblings). Another sampling design could include a pair of siblings, one chosen from the upper or lower tail of the distribution and the other chosen randomly from among the remaining siblings (single selection; ref. 14). Even when nuclear or extended families are sampled, the ascertainment of a family may be through an individual who belongs to the upper tail or exceeds a predetermined cutoff point of the distribution of the QT. Alternatively, if the study pertains to a QT that is known to be a precursor of a clinical end point (e.g., blood pressure level and myocardial infarction), a family may be ascertained through an individual who has encountered the clinical end point. Any nonrandom sampling scheme obviously entails the screening of a large number of potential sampling units to obtain the requisite number of units that satisfy the inclusion criteria. This is expensive in terms of time, effort, and money. However, the adoption of a nonrandom sampling strategy is statistically more powerful for QTL mapping. Such sampling strategies often require modifications of the standard statistical methods for QTL mapping because the resulting distribution of trait values is no longer the same as in the original source population.
Central issues in QTL mapping
The ability to map a QTL depends on the magnitude of its effect, as measured by the proportion of genetic variance of the QT explained by the putative QTL. Whether or not a QTL can be successfully mapped also depends on the study design, sample size, and statistical method used to analyze the data. In general, even in experimental populations, it has been estimated that under the second filial generation (F2) design, a QTL with an effect of 5–15% can be detected with a reasonable (80–90%) statistical power if the sample size is between 200 and 300 individuals (8, 15). In natural populations, such as in humans, sample-size estimation is difficult and involves a lot of assumptions, some of which are discussed below.
The second major issue is the nature and distribution of markers on chromosomes. The usefulness of a marker increases with its level of polymorphism (as estimated by the proportion of heterozygous individuals at the locus; see Locus heterozygosity and marker choice). As the density of markers is increased, the precision of estimation of the location and effect of the QTL increases. The recent finding that the human genome has a block-like structure with respect to levels of association among loci (16, 17) may potentially reduce the number of markers required in genome-wide scans for QTL mapping to attain the same level of statistical power, although there are many unresolved issues (17, 18).
There are other issues that are also central, such as the extent of gene-gene interaction (epistasis) and genotype-environment interaction in the determination of trait values. However, these issues have received little attention with respect to human QTL mapping because the statistical intricacies of even the 2 central issues listed above are still being worked out.
Statistical methods
Since the positions of the QTLs in the genome are unknown, one can gather genotype data at a large number of marker loci and analyze these data statistically to test whether there is increased allele sharing at the marker loci among individuals who show similar trait values (see Human QTL mapping: key principles). If there is increased allele sharing, then the QTL probably lies in the vicinity of this marker locus; that is, the QT and marker loci are linked (2-point mapping). If there is increased allele sharing at several consecutive marker loci, as revealed by the joint analysis of the QT data with multiple markers (multipoint mapping), then there is a higher probability that the QTL lies in the interval spanned by these marker loci.
There are then 2 major goals: (a) measuring the expected level of allele sharing at marker loci (based on genotype data) among the sampled sets of relatives and (b) testing whether there is an increased level of sharing among individuals with similar trait values (and, therefore, inferring that the marker and the trait loci are linked).
A pair of relatives can share 0, 1, or 2 alleles that are identical by descent (IBD). (For an allele to be IBD in a pair of relatives, the allele in both the relatives must have been the same allele transmitted by the same ancestor.) A general method for calculating the probabilities of sharing 0, 1, or 2 alleles at a locus was given by Li and Sacks (19); this was then extended by Campbell and Elston (20), and a more general method was developed by Donnelly (21). For pedigrees also, methods for estimating IBD probabilities from genotype data have been developed (22).
Regression
Haseman-Elston regression was the first statistical method that was developed for human QTL mapping (23). This method is applicable to human sibpair data. This linear regression model (Y = a + bX) includes the squared difference in trait values between members of a sibling pair (Y) as the dependent variable and the number of alleles shared IBD between them (X) at the marker locus (marker IBD score) as the independent variable. If any parent is homozygous at the marker locus or if parental genotypes are missing, then the marker IBD score cannot be determined with certainty. In such a case, the IBD score is estimated by conditioning on either the marker genotypes of parents (if available) and the sibs at the given marker locus (single-point estimate) or an integrated genotype profile based on all available marker loci (multipoint estimate) on a chromosome (24, 25). Under the null hypothesis of no linkage, the regression coefficient is 0, while under linkage it is less than 0. The null hypothesis is easily tested by a standard Student’s t test. This method was extended to other relative pairs and pairs drawn from larger pedigrees (26). However, there are statistical limitations in using the Haseman-Elston regression on pedigree data, and it is not considered to be a method of choice.
The choice of the squared trait value difference between members of a sibling pair wastes valuable information by not using the trait values of the individual siblings. Twenty-five years later, it was shown (27) that the inclusion of the sum of trait values of the siblings, along with the squared trait value difference, in the analysis results in gain of statistical power. It was suggested (28) that these 2 variables (squared trait difference and trait sum) be used as dependents in 2 separate linear regression equations with the estimated IBD score as the independent variable and that the estimated slopes be averaged to draw inferences on linkage. This method relies on several assumptions that have been relaxed to develop statistically more sound methods of combining the 2 slope estimates; use of a mean corrected trait product has been used as a dependent variable in the regression, score tests have been proposed, and various statistical properties of these estimators and methods have been explored (29–36). A summary of these new statistics is provided in ref. 37. In a large sibship, there will be many sibpairs. The squared differences in trait values of these sibpairs will be correlated. To allow the inclusion of multiple sibpairs from a large sibship in the statistical analysis, a generalized linear model that assumes a specific correlation structure of functions of trait values of sibpairs has been proposed (29). To circumvent the problem of assigning weights to different sibpairs, Ghosh and Reich (38) have proposed a linear regression based on a “contrast function” of trait values within a sibship. The maximum-likelihood binomial approach (39), although strictly not a regression method, can also accommodate sibship data without assumption of any specific probability distribution of trait values. The method introduces a latent variable that captures the link between QTs and marker information and tests for linkage via a Bernoulli parameter modeling the transmission of marker alleles from parents to the different sibs within a sibship. These advances in statistical methodologies have resulted in improvements in statistical power to map QTLs, but the regression-based method is applicable only to sibpairs and, under some assumptions, to sibships.
Recently, a novel approach has been proposed (40), in which the IBD scores have been modeled as a function of observed trait values instead of the usual modeling of trait values as a function of IBD scores. This method is applicable to large sibships and also to general pedigrees, but does not necessarily have more statistical power (41) than a competing method called variance components (VC) (discussed below).
In these regression models, the relationship between the dependent and independent variables being linear is an assumption. This assumption is valid when there is no dominance at the trait locus; but in the presence of dominance, the regression can deviate from linearity. This assumption has been relaxed and nonparametric alternatives have been proposed, as discussed later.
Regression methods continue to be widely used because they are computationally easy and efficient, and the standard deviations of parameters can easily be estimated using resampling techniques (42). However, there is no strong statistical reason for using regression methods for QTL mapping, except when the collected data are from pairs of relatives, such as sibling pairs (discussed below).
Variance components
Another popular statistical approach is called the VC method, which is applicable to large sibships or pedigrees. In the framework underlying this method, the trait value of an individual is assumed to be determined by a major gene, random polygenic, and environmental effects and covariates. The covariance between trait values of a pair of relatives is an increasing function of the extent of allele sharing, IBD, at the marker locus. The general framework and methodology that is currently popular was developed by Amos (43), although Goldgar (44) first proposed this method in the context of human QTL mapping. Amos (43) derived expressions for the covariances in trait values for a number of common pairs of relatives. The trait values of individuals in a pedigree are assumed to follow a multivariate normal distribution, with the variance-covariance matrix determined by the expressions given in Amos (43) or their straightforward generalizations. The likelihood of the observations on a pedigree or any other set of relatives can then be written down by standard statistical methods. The likelihood is maximized to obtain parameter estimates, and a likelihood ratio test is used to test for linkage.
Various extensions of the basic model and methodology of Amos (43) have been made. These include extensions to permit likelihood calculations to pedigrees of arbitrary sizes and complexity (25), inclusion of gene-gene (45, 46) and gene-environment (47) interactions in the model, and analysis of multiple correlated traits (48). When the model assumptions hold, especially multivariate normality, the VC method is very powerful, considerably more powerful than the Haseman-Elston regression. Further, it is readily applicable to large and complex pedigrees. Thus, for QTL mapping, the method of choice is VC. For sibling pairs, however, it has been shown — both by theory and by simulation — that the computationally simpler regression methods are as powerful as the VC method (32, 33).
We emphasize here that the statistical power and efficiency of the VC method critically depend on whether the assumption that the trait values are normally distributed in the population is satisfied. However, it is often not feasible to verify distributional and other model assumptions. Further, even when the distribution in the population from which sampling units are drawn is normal, if the sampling design is nonrandom, then the distribution of the QT in the data so obtained may be nonnormal, thus violating the assumptions underlying the VC method. When underlying assumptions are violated, parametric methods (that is, methods — such as VC — that rely on models that assume specific forms of the probability distribution of trait values) can result in a high proportion of either false-positive or false-negative inferences. For example, if the trait distribution has a sharp peak (leptokurtic) and if gene-environment interactions are present, then one can get inflated false-positive error rates (49). Some methods based on permutation tests — which do not rely on normality of the test distribution in drawing inferences — have been proposed to obtain P values (50), but these methods entail enormous increase in the computational load. The problems associated with the possible violation of normality continue to be a limiting factor in practical applications of the VC method. Some novel methods have recently been proposed to deal with these problems (51), but the difficulties are far from resolved. VC methods for mapping QTLs have been implemented in several software packages, including Genehunter (52, 53), Merlin (54), Mx (55), and SOLAR (25).
Nonparametric alternatives
When the assumptions underlying the regression (linearity of relationship between the dependent and the independent variables) and the VC (multivariate normality of QT values of family members) methods hold, these methods are statistically quite powerful for QTL mapping. However, it is difficult to ensure that these assumptions are met, especially for pedigree data. Deviations from these assumptions can adversely affect linkage inferences. Therefore, alternative methods that do not rely critically on these model assumptions (model-free approaches) have begun to be developed. In these model-free methods, there is inevitably a loss of statistical power, but these methods provide safeguards against high rates of both false positives and false negatives.
Since the nature of dependence of estimated marker IBD scores on the squared difference in sibpair trait values is a function of the recombination fraction between the marker and the trait loci and other biological parameters, such as interference and dominance at the trait locus, the assumption of a specific form of functional relationship between them may not be a robust strategy. Rank-based statistics (23, 56) have been proposed to deal with this problem. A proposed nonparametric regression procedure based on kernel smoothing (in which the relationship between the dependent and the independent variables is estimated empirically) has been shown to perform well (57, 58) both in simulations and in practical applications. The available nonparametric methods are useful only for sibpair data. Such methods need to be developed for pedigree data also.
A summary of choices
VC method is statistically the method of choice for QTL mapping, provided that the assumption of multivariate normality of trait values within family members is satisfied. This assumption is hard to test, and more importantly, if it is violated, then it is hard to rectify even by using mathematical transformations of QT values, e.g., logarithmic or power transformations. Further, in families that are selected through a member possessing an extreme QT value, there is an even bigger problem of noncompliance with the normality assumption. Fortunately, there are indications (49) that when this assumption is not met, it is the type I error, rather that the type II error, that is inflated to a greater degree. Thus, with the VC method, if linkage is detected, then chances are good that it is not a false inference.
When the normality assumption is not met, then it may be better to use a nonparametric regression method based on sibpair data, even though there will be loss of statistical power. In this case, the false-positive error rate will be lower. However, no results are yet available on the statistical properties of this method when siblings are selected based on some inclusion criteria, e.g., siblings belonging to opposite extremes of the trait distribution — discordant sibpairs (see, however, Peng and Siegmund; ref. 59).
Unselected samples have low statistical power. Selection of discordant sibpairs yields a high statistical power. This property is also true of families ascertained through a member with an extreme QT value. These selection strategies can be very expensive and difficult to implement in practice. A compromise solution is to select 1 sibling with an extreme value and choose another sibling randomly from among the remaining siblings in the sibship. This selection strategy — less expensive and easier to implement than selecting discordant sibpairs — has comparable, albeit slightly lower, statistical power (14). However, in studies based on sibling pairs in which the focus of interest is on trait-allele relationships at an individual level rather than on allele-sharing in families (association analysis), a crucial criterion for success of QTL mapping is that the frequencies of marker and trait alleles should be in the same ballpark. This means that generating polymorphic markers with high frequencies may not result in greater success of QTL mapping unless the trait alleles have matching frequencies (14). Similar results have also been obtained with respect to association study designs that pertain to unrelated individuals (not siblings) selected from opposite tails of the distribution of QT values (60). This lack of greater success in QTL mapping unless the trait alleles have matching frequencies is not encouraging. While considerable efforts are being made to generate markers that will ease genome-wide association mapping of QTLs, if the allele frequencies at the QTL are very skewed, efforts in mapping the QTL may be unsuccessful. This is in addition to the fact that a QTL that explains less than 10% of the variance of trait values is very hard to map. The recent efforts of the HapMap project (16) to provide markers that may be the most informative for association mapping will not be a panacea for overcoming these limitations of human QTL mapping. As we have discussed, there is also a great need to devise statistical methods for human QTL mapping that do not critically depend on model assumptions. In association mapping, population stratification (61) is a major issue, and therefore, although designs involving unrelated individuals are easier to implement, these are best avoided for human QTL mapping. Further, the statistical power of QTL mapping using association analysis declines very rapidly with the decrease of nonrandom association between the QTL and the marker locus (62). Notwithstanding the caveats listed above, efforts to map human QTLs using a combination of family-based association and linkage analysis methods are continuing and should continue. Successes in practice will crucially depend on refinements of statistical methods and developments of novel approaches to handle interactions among QTLs as well as the effects of environmental factors.
Supplementary Material
Footnotes
Nonstandard abbreviations used: IBD, identical by descent; QT, quantitative trait; QTL, QT locus; VC, variance components.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Paterson AH, et al. Resolution of quantitative traits into Mendelian factors using a complete RFLP linkage map. Nature. 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- 2.Todd JA, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 3.Hilbert P, et al. Chromosomal mapping of two genetic loci associated with blood pressure regulation in hereditary hypertensive rats. Nature. 1991;353:521–529. doi: 10.1038/353521a0. [DOI] [PubMed] [Google Scholar]
- 4.Jacob HJ, et al. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell. 1991;67:213–224. doi: 10.1016/0092-8674(91)90584-l. [DOI] [PubMed] [Google Scholar]
- 5.Podolin PL, et al. Localization of two insulin-dependent diabetes (Idd) genes to the Idd10 region on mouse chromosome 3. Mamm. Genome. 1998;9:283–286. doi: 10.1007/s003359900749. [DOI] [PubMed] [Google Scholar]
- 6.Legare ME, Bartlett FS, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- 7.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 8.Doerge RW. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES, Botstein D. Strategies for studying heterogeneous genetic traits in humans by using a linkage map of restriction fragment length polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 1986;83:7353–7357. doi: 10.1073/pnas.83.19.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perusse L, et al. A genome-wide scan for abdominal fat assessed by computed tomography in the Québec Family Study. Diabetes. 2001;50:614–621. doi: 10.2337/diabetes.50.3.614. [DOI] [PubMed] [Google Scholar]
- 11.Deng HW, et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am. J. Hum. Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschhorn JN, et al. Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am. J. Hum. Genet. 2001;69:106–116. doi: 10.1086/321287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosse Y, et al. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Québec Family Study. J. Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Abecasis GR, Cookson WO, Cardon LR. The power to detect linkage disequilibrium with quantitative traits in selected samples. Am. J. Hum. Genet. 2001;68:1463–1474. doi: 10.1086/320590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson DL, Fenster CB, Stenoien HK, Price D. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 2004;13:2505–2522. doi: 10.1111/j.1365-294X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- 16.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 18.Schulze TG, et al. Defining haplotype blocks and tag single-nucleotide polymorphisms in the human genome. Hum. Mol. Genet. 2004;13:335–342. doi: 10.1093/hmg/ddh035. [DOI] [PubMed] [Google Scholar]
- 19.Li CC, Sacks J. The derivation of joint distribution and correlation between relatives by the use of stochastic matrices. Biometrics. 1954;10:347–360. [Google Scholar]
- 20.Campbell MA, Elston RC. Relatives of probands: models for preliminary genetic analysis. Ann. Hum. Genet. 1971;35:225–236. doi: 10.1111/j.1469-1809.1956.tb01395.x. [DOI] [PubMed] [Google Scholar]
- 21.Donnelly KP. The probability that related individuals share some section of the genome identical by descent. Theor. Popul. Biol. 1983;23:34–63. doi: 10.1016/0040-5809(83)90004-7. [DOI] [PubMed] [Google Scholar]
- 22.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 23.Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and marker loci. Behav. Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- 24.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson JM, Wijsman EM. Linkage between quantitative trait and marker loci: methods using all relative pairs. Genet. Epidemiol. 1993;10:87–102. doi: 10.1002/gepi.1370100202. [DOI] [PubMed] [Google Scholar]
- 27.Wright FA. The phenotypic difference discards sib-pair QTL information. Am. J. Hum. Genet. 1997;60:740–742. [PMC free article] [PubMed] [Google Scholar]
- 28.Drigalenko E. How sib-pairs reveal linkage. Am. J. Hum. Genet. 1998;63:1242–1245. [PMC free article] [PubMed] [Google Scholar]
- 29.Elston RC, Buxbaum S, Jacobs KB, Olson JM. Haseman and Elston revisited. Genet. Epidemiol. 2000;19:1–17. doi: 10.1002/1098-2272(200007)19:1<1::AID-GEPI1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Weiss S, Xu X, Wei IJ. A unified Haseman-Elston method for testing linkage with quantitative traits. Am. J. Hum. Genet. 2000;67:1025–1028. doi: 10.1086/303081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrest W. Weighting improves the “new Haseman Elston” method. Hum. Hered. 2001;52:47–54. doi: 10.1159/000053353. [DOI] [PubMed] [Google Scholar]
- 32.Sham PC, Purcell S. Equivalence between Haseman-Elston and variance components QTL linkage analysis of selected and non-normal samples: conditioning on trait values. Am. J. Hum. Genet. 2001;68:1527–1532. doi: 10.1086/320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visscher PM, Hopper JL. Power of regression and maximum likelihood methods to map QTL from sib-pair and DZ twin data. Ann. Hum. Genet. 2001;65:583–601. doi: 10.1017/S0003480001008909. [DOI] [PubMed] [Google Scholar]
- 34.Tang H-K, Siegmund D. Mapping quantitative trait loci in oligogenic models. Biostatistics. 2001;2:147–162. doi: 10.1093/biostatistics/2.2.147. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Huang J. A score-statistic approach for the mapping of quantitative-trait loci with sibships of arbitrary size. Am. J. Hum. Genet. 2002;70:412–424. doi: 10.1086/338659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putter H, Sandkuijl LA, van Houwelingen JC. Score test for detecting linkage to quantitative traits. Genet. Epidemiol. 2002;22:345–355. doi: 10.1002/gepi.01104. [DOI] [PubMed] [Google Scholar]
- 37.Cuenco KT, Szatkiewicz JP, Feingold E. Recent advances in human quantitative-trait-locus mapping: comparison of methods for discordant sibling pairs. Am. J. Hum. Genet. 2003;73:874–885. doi: 10.1086/378590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, Reich T. Integrating sibship data for mapping quantitative trait loci. Ann. Hum. Genet. 2002;66:169–182. doi: 10.1017/S0003480002001070. [DOI] [PubMed] [Google Scholar]
- 39.Alcais A, Abel L. Maximum-likelihood-binomial method for genetic model-free linkage analysis of quantitative traits in sibships. Genet. Epidemiol. 1999;17:102–117. doi: 10.1002/(SICI)1098-2272(1999)17:2<102::AID-GEPI2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am. J. Hum. Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Knott SA, Visscher PM. Theoretical and empirical power of regression and maximum-likelihood methods to map quantitative trait loci in general pedigrees. Am. J. Hum. Genet. 2004;75:17–26. doi: 10.1086/421845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am. J. Hum. Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 44.Goldgar DE. Multipoint analysis of human quantitative genetic variation. Am. J. Hum. Genet. 1990;47:957–967. [PMC free article] [PubMed] [Google Scholar]
- 45.Stern MP, et al. Evidence for linkage of regions on chromosomes 6 and 11 to plasma glucose concentrations in Mexican Americans. Genome Res. 1996;6:724–734. doi: 10.1101/gr.6.8.724. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell BD, Ghosh S, Schneider JL, Birznicks G, Blangero J. Power of variance component linkage analysis to detect epistasis. Genet. Epidemiol. 1997;14:1017–1022. doi: 10.1002/(SICI)1098-2272(1997)14:6<1017::AID-GEPI76>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.Towne B, Siervogel RM, Blangero J. Effects of genotype-by-sex interaction on quantitative trait linkage analysis. Genet. Epidemiol. 1997;14:1053–1058. doi: 10.1002/(SICI)1098-2272(1997)14:6<1053::AID-GEPI82>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 48.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet. Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Allison DB, Fernandez JR, Heo M, Beasley TM. Testing the robustness of the new Haseman-Elston quantitative-trait loci-mapping procedure. Am. J. Hum. Genet. 2000;67:249–252. doi: 10.1086/302966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itturia SJ, Williams JT, Almasy L, Dyer TD, Blangero J. An empirical test of the significance of an observed quantitative trait locus effect that preserves additive genetic variation. Genet. Epidemiol. 1999;17(Suppl.):S169–S173. doi: 10.1002/gepi.1370170729. [DOI] [PubMed] [Google Scholar]
- 51.Shete S, et al. Effect of winsorization on power and type 1 error of variance components and related methods of QTL detection. Behav. Genet. 2004;34:153–159. doi: 10.1023/B:BEGE.0000013729.26354.da. [DOI] [PubMed] [Google Scholar]
- 52.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 53.Pratt SC, Daly MJ, Kruglyak L. Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am. J. Hum. Genet. 2000;66:1153–1157. doi: 10.1086/302830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abecasis G, Cherny S, Conkson W, Cardon L. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 55.Neale, M.C., Boker, S.M., Xie, G., and Mael, H.H. 2002. Mx: statistical modeling. 6th edition. Department of Psychiatry, Virginia Commonwealth University. Richmond, Virginia, USA. 217 pp.
- 56.Kruglyak L, Lander ES. A nonparametric approach for mapping quantitative trait loci. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh S, Majumder PP. A two-stage variable-stringency semiparametric method for mapping quantitative trait loci with the use of genomewide scan data on sib-pairs. Am. J. Hum. Genet. 2000;66:1046–1061. doi: 10.1086/302815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh S, et al. Linkage mapping of beta 2 EEG waves via non-parametric regression. Am. J. Med. Genet. 2003;118:66–71. doi: 10.1002/ajmg.b.10057. [DOI] [PubMed] [Google Scholar]
- 59.Peng J, Siegmund D. Mapping quantitative traits with random and with ascertained sibships. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7845–7850. doi: 10.1073/pnas.0401713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am. J. Hum. Genet. 2000;67:1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 62.Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am. J. Hum. Genet. 2000;66:1616–1630. doi: 10.1086/302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim U, et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.