Abstract
Gene defects play a major role in the pathogenesis of degenerative disorders of the nervous system. In fact, it has been the very knowledge gained from genetic studies that has allowed the elucidation of the molecular mechanisms underlying the etiology and pathogenesis of many neurodegenerative disorders. In this review, we discuss the current status of genetic epidemiology of the most common neurodegenerative diseases: Alzheimer disease, Parkinson disease, Lewy body dementia, frontotemporal dementia, amyotrophic lateral sclerosis, Huntington disease, and prion diseases, with a particular focus on similarities and differences among these syndromes.
The complexities of common diseases
Familial aggregation had been recognized as a prominent characteristic of many neurodegenerative disorders decades before the underlying molecular genetic or biochemical properties were known. It was often the identification of specific, disease-segregating mutations in previously unknown genes that directed the attention to certain proteins and pathways that are now considered crucial in the pathogenesis of these diseases. These include mutations in the β-amyloid (Aβ) precursor protein, causing Alzheimer disease (AD); in α-synuclein, causing Parkinson disease (PD); or in microtubule-associated protein tau, causing frontotemporal dementia (FTD) with parkinsonism. Another feature observed in most common neurodegenerative diseases — as well as in other common disorders — is a dichotomy between familial (rare) and seemingly nonfamilial (common) forms. The latter are also frequently described as “sporadic” or “idiopathic,” although there is a growing body of evidence suggesting that a large proportion of these cases are also significantly influenced by genetic factors. These risk genes are likely to be numerous, displaying intricate patterns of interaction with each other as well as with nongenetic variable, and — unlike classical Mendelian (“simplex”) disorders — exhibit no simple or single mode of inheritance. Hence, the genetics of these diseases has been labeled “complex.”
A popular conception regarding the genetic makeup of complex diseases is the “common disease/common variant” (CD/CV) hypothesis (1). According to this theory common disorders are also governed by common DNA variants (such as single nucleotide polymorphisms. These variants significantly increase disease risk but are insufficient to actually cause a specific disorder. Current empirical and theoretical data support this hypothesis, although there remains great uncertainty as to the number of the underlying risk factors and their specific effect sizes. In this context, it is noteworthy that even recent genetic advances in the study of complex diseases such as AD or diabetes likely only represent the most obvious, most extreme cases of the underlying risk spectrum (Figure 1; ref. 2). In AD, for instance, rare, fully penetrant autosomal dominant mutations in 3 genes (i.e. APP, PSEN1, and PSEN2) have been shown to cause the disease, while a common, incompletely penetrant susceptibility variant (i.e., ε4 in APOE; see below) significantly increases the risk for AD. The identification of these genes early in the study of AD genetics was possible due to the combination of several favorable circumstances, such as the presence of multiple independent mutations in the same locus and the availability of extended, multigenerational pedigrees for DNA genotyping and sequencing (in the case of PSEN1) or a large attributable fraction to the overall genetic variance, resulting from relatively high allele frequency and pronounced effect size (in the case of APOE). However, identification of disease genes that make smaller overall contributions to the genetic spectrum (because of only few mutational events; e.g., PSEN2), or risk factors with smaller effect sizes (i.e. odds ratios [ORs] ranging between 2 and 3), will require much larger samples and possibly more sensitive and efficient analytic tools to enable consistent detection across study populations (2).
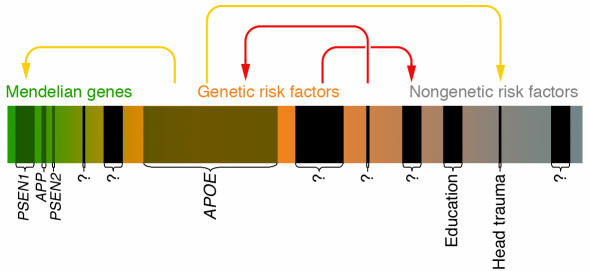
Figure 1.
This scheme depicts the risk spectrum predisposing to common diseases as one continuum, using AD as an example. The continuum extends from the most extreme genetic form (“Mendelian genes”; green) to cases influenced by genetic susceptibility factors (“Genetic risk factors”; orange), until reaching into a less well-defined area of cases that may be caused by genes of lesser penetrance/lower effect size and/or altogether nongenetic factors (“Nongenetic risk factors”; gray). Established Mendelian genes (APP, PSEN1, and PSEN2) or genetic risk factors (APOE-ε4) are represented by shaded boxes and represent the most obvious candidates of AD genetics; the width of these boxes approximately represents the relative contribution to the overall risk for disease. Black boxes indicate still-elusive disease genes/risk factors (“?”). Colored arrows indicate possible gene-gene and gene-environment interaction patterns: yellow arrows represent previously suggested interactions (e.g., between PSEN1 and APOE-ε4). Note that some interactions (red arrows) as well as the number of elusive genes are entirely hypothetical and are depicted for didactic purposes only.
Additional, and commonly cited, problems in finding complex disease genes beyond the most obvious are multiple testing, publication bias, and questionable replication (3–5). Multiple testing can be placed under the larger category of “avoidable false positive” findings, which are also caused by testing insufficiently sized samples, using inappropriate matching of cases versus controls, stratifying populations, and choosing inadequate analysis strategies, etc. Publication bias, on the other hand, which indicates the higher a priori likelihood of a positive finding being published as opposed to a negative one, may have been a possible source of serious bias in the early days. However, there is only relatively little empirical evidence that publication bias actually represents a common or significant source of error in current publications investigating the genetics of a number of disorders (e.g., refs. 6–12). The sheer number of publications in the AD genetics literature, for example, reveals that nearly two-thirds represent “negative” articles, with the rest being “positive” or “suggestive,” so that one can hardly speak of a preponderance of the positive (13). Finally, independent replication of a positive genetic finding is one of the essential requirements to distinguish a genuine from a false-positive gene effect (Figure 2). However, replication — just like the primary detection of disease association — is affected by a number of factors, which include locus heterogeneity, small effect size, high risk allele frequency, population stratification, and poor case-control matching. Thus, the failure to provide independent replication may be meaningless if the association study has not been carefully designed. In the case of multiple conflicting reports, metaanalysis across all published studies and/or the evidence for a biochemical/functional consequence of the putative risk allele can help distinguish real disease genes from their harmless counterparts (Figure 2).
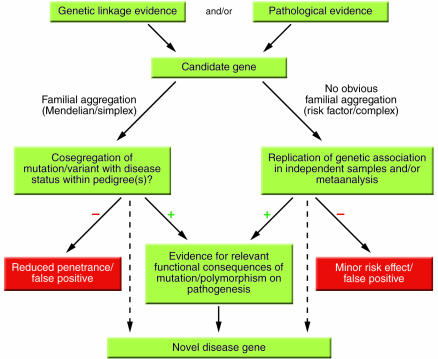
Figure 2.
Flow chart of current strategies used to identify novel disease genes. This scheme outlines strategies for identifying mutations and/or polymorphisms causing or predisposing to disease. Candidate genes are chosen based on genetic linkage data and/or known or hypothesized pathobiological relevance to disease mechanisms. This procedure is referred to as the “candidate gene approach.” An alternative and inherently similar strategy is based on the detection of formerly unknown genes/proteins according to genetic linkage data and is referred to as “positional cloning.” Dashed lines indicate “shortcuts” allowing the definition of a novel disease gene based on the genetic evidence alone, e.g., APOE-ε4 in AD, of which the precise functional consequences remain unknown despite an established genetic role. Note that there are examples of genes/mutations with reduced penetrance or minor risk effects (red boxes) within bona fide disease genes (e.g., certain mutations in PSEN1 in AD).
Notwithstanding these difficulties, genetic analyses have laid the foundation for understanding a variety of disease mechanisms leading to neurodegeneration and associated symptoms. Likewise, a detailed understanding of their genetic basis will be essential for the development of effective strategies aimed at the early prediction and early prevention/treatment of these devastating diseases. In the following sections, we briefly outline the status of genetic research across a number of common neurodegenerative conditions, with a particular focus on the similarities and differences among disorders.
Alzheimer disease
AD is one of the most serious health problems in the industrialized world. It is an insidious and progressive neurodegenerative disorder that accounts for the vast majority of age-related dementia and is characterized by global cognitive decline and the accumulation of Aβ deposits and neurofibrillary tangles in the brain (Figure 3). Family history is the second-greatest risk factor for the disease after age, and the growing understanding of AD genetics has been central to the knowledge of the pathogenic mechanisms leading to the disease. Genetically, AD is complex and heterogenous and appears to follow an age-related dichotomy: rare and highly penetrant early-onset familial AD (EOFAD) mutations in different genes are transmitted in an autosomal dominant fashion, while late-onset AD (LOAD) without obvious familial segregation is thought to be explained by the CD/CV hypothesis (14).
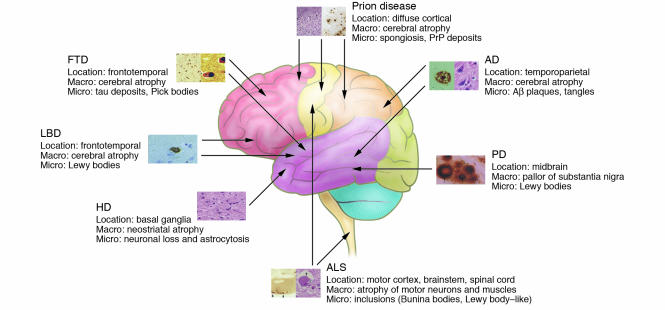
Figure 3.
Overview of the anatomical location of and macroscopic and microscopic changes characteristic of the neurodegenerative disorders discussed in this review. Note that the full neuropathological spectrum of these disorders is much more complex than depicted here. When there is more than one characteristic histopathological feature, these are depicted from left to right, as indicated in the labels listing microscopic changes (e.g., the 2 panels for AD depict an Aβ plaque [left] and neurofibrillary tangles [right]). All histopathological images are reprinted with permission from ISN Neuropath Press (ref. 99).
EOFAD represents only a small fraction of all AD cases (≤5%) and typically presents with onset ages younger than 65 years, showing autosomal dominant transmission within affected families. To date, more than 160 mutations in 3 genes have been reported to cause EOFAD. These include the Aβ precursor protein (APP) on chromosome 21 (15), presenilin 1 (PSEN1) on chromosome 14 (16), and presenilin 2 (PSEN2) on chromosome 1 (17, 18). The most frequently mutated gene, PSEN1, accounts for the majority of AD cases with onset prior to age 50. While these AD-causing mutations occur in 3 different genes located on 3 different chromosomes, they all share a common biochemical pathway, i.e., the altered production of Aβ leading to a relative overabundance of the Aβ42 species, which eventually results in neuronal cell death and dementia. An up-to-date overview of disease-causing mutations in these genes can be found at the Alzheimer Disease & Frontotemporal Dementia Mutation Database (19).
LOAD, on the other hand, is classically defined as AD with onset at age 65 years or older and represents the vast majority of all AD cases. While segregation and twin studies conclusively suggest a major role of genetic factors in this form of AD (20), to date, only 1 such factor has been established, the ε4 allele of the apolipoprotein E gene on chromosome 19q13 (APOE; Table 1; refs. 21, 22). In contrast to all other association-based findings in AD, the risk effect of APOE-ε4 has been consistently replicated in a large number of studies across many ethnic groups, yielding ORs between approximately 3 and approximately 15 for heterozygous and homozygous carriers, respectively, of the ε4 allele in white individuals (for metaanalysis, see ref. 23). In addition to the increased risk exerted by the ε4-allele, a weak, albeit significant, protective effect for the minor allele, ε2, has also been reported in several studies. Unlike the mutations in the known EOFAD genes, APOE-ε4 is neither necessary nor sufficient to cause AD but instead operates as a genetic risk modifier by decreasing the age of onset in a dose-dependent manner. Despite its long-known and well-established genetic association, the biochemical consequences of APOE-ε4 in AD pathogenesis are not yet fully understood but likely encompass Aβ-aggregation/clearance and/or cholesterol homeostasis (Table 1).
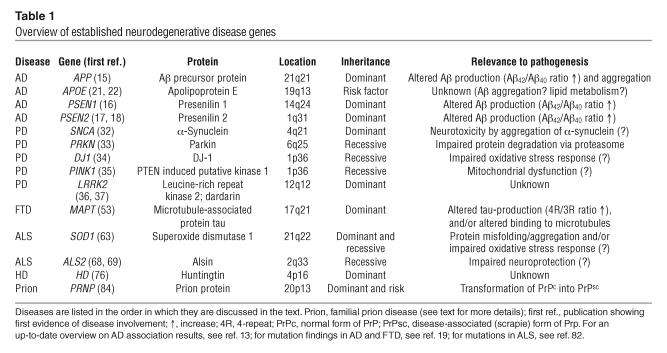
Table 1.
Overview of established neurodegenerative disease genes
Several lines of evidence suggest that numerous additional LOAD loci (24) — and probably also EOFAD loci (25, 26) — remain to be identified, since the 4 known genes together account for probably less than 50% of the genetic variance of AD. As outlined above, it is currently unclear how many of these loci will prove to be risk factors as opposed to causative variants. As candidates for the former, more than 3 dozen genes have been significantly associated with AD in the past (27, 28). Despite the more than 500 independent association studies, however, no single gene has been shown to be a risk factor with even nearly the same degree of replication or consistency as has APOE-ε4. An up-to-date overview of the status of these and other potential AD candidate genes, including metaanalyses across published association studies, can be found at the Alzheimer Research Forum genetic database (13). One of the conclusions to be drawn from currently available data, as well as from the few independently performed metaanalyses on putative AD risk factors, is that even if some of the published associations were genuine, their overall effect size is likely to be only minor, i.e. with ORs not exceeding 2.
Parkinson disease
PD is the second most common neurodegenerative disease of adult onset. Histopathologically, it is characterized by a severe loss of dopaminergic neurons in the substantia nigra and cytoplasmic inclusions consisting of insoluble protein aggregates (Lewy bodies; Figure 3), which lead to a progressive movement disorder including the classic triad of tremor, bradykinesia, and rigidity, with an average onset age between 50 and 60 years. Although the heritability – and thus the contribution of genetic factors to the overall prevalence – of PD is likely smaller than that of AD, genetics has played a major role in elucidating the causes of nigrostriatal neuronal loss across a wide spectrum of clinically and histopathologically heterogenous PD cases. As in AD, there appears to be an age-dependent dichotomy: the majority of individuals with an early or even juvenile onset show typical Mendelian inheritance. However, unlike in AD, these cases show a predominantly autosomal-recessive mode of inheritance, and there is an ongoing debate as to whether genetic factors play any substantial role in contributing to disease risk in cases with onset beyond approximately 50 years (29–31).
Notwithstanding these uncertainties, there is a plethora of genetic studies on both forms of the disease, and mutations in at least 5 genes have now been shown to cause familial early-onset parkinsonism (α-synuclein [SNCA or PARK1; ref. 32]; parkin [PRKN or PARK2; ref. 33]; DJ-1 [DJ1 or PARK7; ref. 34]; PTEN-induced putative kinase I [PINK1 or PARK6; ref. 35]; and leucine-rich repeat kinase 2 or dardarin [LRRK2 or PARK8; refs. 36, 37]), with several other linkage regions pending characterization and/or replication. As was the case in the study of AD, the first locus to be characterized – PARK1, on chromosome 4q21 – involves the protein that is the major constituent of one of the classic neuropathological hallmarks of the disease, i.e., α-synuclein (32), which can be found at the core of Lewy bodies. While the exact mechanisms underlying α-synuclein toxicity currently remain only incompletely understood, recent evidence suggests that some SNCA mutations may change normal protein function quantitatively rather than qualitatively, via duplication or triplication of the α-synuclein gene (38, 39). Very recently, mutations in a second gene with dominant inheritance have been identified by several different laboratories (LRRK2; refs. 36, 37). While the functional consequences of LRRK2 mutations are still unknown, it was suggested that at least some mutations could interfere with the protein’s kinase activity (40).
While changes in SNCA and LRRK2 are the leading causes of autosomal-dominant forms of PD, the majority of affected pedigrees actually show a recessive mode of inheritance (Table 1). The most frequently involved gene in recessive parkinsonism is parkin (PRKN) on chromosome 6q25 (33, 41), which causes nearly half of all early-onset PD cases. Parkin is a ubiquitin ligase that is involved in the ubiquitination of proteins targeted for degradation by the proteasomal system. The spectrum of parkin mutations ranges from amino acid–changing single base mutations to complex genomic rearrangements and exon deletions, which probably result in a loss of protein function. It has been speculated that this may trigger cell death by rendering neurons more vulnerable to cytotoxic insults, e.g., the accumulation of glycosylated α-synuclein (42). In addition to parkin mutations, genetic analyses of 2 non-parkin early-onset, autosomal-recessive PD pedigrees revealed 2 independent, homozygous mutations in DJ1 (34) on chromosome 1p36 (43). Both mutations result in a loss of function of DJ-1, a protein that is suggested to be involved in oxidative stress response. While several studies have independently confirmed the presence of DJ-1 mutations in other PD cases, the frequency of disease-causing variants in this gene is estimated to be low (∼1%; ref. 44). Less than 13 Mb toward the long arm of the same chromosome, additional PD-causing mutations were subsequently discovered in PINK1 (35) following positive linkage evidence to this region (45). This enzyme is expressed with particularly high levels in brain, and the first 2 identified mutations (G309D and W437ter) were predicted to lead to a loss of function that may render neurons more vulnerable to cellular stress, similar to the effects of parkin mutations. While Lewy bodies are typically not found in brains of patients bearing parkin mutations, it is currently unclear whether these are present in PD cases with mutations in DJ1 and PINK1.
At least 6 additional candidate PD loci have been described, including putative disease-causing mutations in the ubiquitin carboxy-terminal hydrolase L1 (UCHL1) on chromosome 4p14 (46), and in a nuclear receptor of subfamily 4 (NR4A2, or NURR1; ref. 47) located on 2q22. However, and unlike the previously outlined PD genes, neither of these maps to known PD linkage regions, nor were they independently confirmed beyond the initial reports. However, polymorphisms in both genes have been – albeit inconsistently – associated with PD in some case-control studies. A recent metaanalysis of the S18Y polymorphism in UCHL1 showed a modest but significant protective effect of the Y allele (11), which suggests that this gene may actually be a susceptibility factor rather than a causal PD gene.
Unlike early-onset PD, the heritability of late-onset PD is probably low (29). Despite this caveat, while a number of whole-genome screens across several late-onset PD family samples have been performed, only a few overlapping genomic intervals have been identified. One of the more extensively studied regions is 17q21, near the gene encoding the microtubule-associated protein tau (MAPT; ref. 48). Previously, it had been shown that rare missense mutations in MAPT lead to a syndrome of frontotemporal dementia with parkinsonism (FTD with parkinsonism linked to chromosome 17 [FTDP-17]; see below), but to date no mutations have been identified as causing parkinsonism without frontotemporal degeneration. However, haplotype analyses of the tau gene have revealed some evidence of genetic association of the H1 haplotype with both PD (ref. 49; for metaanalysis see ref. 50) and a related syndrome, progressive supranuclear palsy (PSP; ref. 51). Despite the lack of evidence for genetic linkage to chromosome 19q13, variants in APOE have also been tested for a role in PD and related syndromes. Across the nearly 3 dozen different studies available to date, some authors report a significant risk effect of APOE-ε4 for PD, while others only see association with certain PD phenotypes or even a risk effect of the ε2 allele, which is protective in AD (see above). A recent metaanalysis on the effects of APOE in PD concluded that only the ε2-related increase in PD risk remains significant when all published studies are considered jointly (12). Finally, and in addition to the findings in autosomal-dominant familial PD, there is also some support for a potential role of SNCA variants on the risk for late-onset PD (52).
Lewy body dementia
According to some investigators, Lewy body dementia (LBD) is the second most common type of degenerative dementia in the elderly, possibly accounting for up to 15% of all dementia cases in autopsy samples (53). Clinically, LBD is characterized by progressive cognitive impairment with fluctuating course, recurrent visual hallucinations, and parkinsonism. Although formal clinical criteria have been proposed (53), there is a pronounced clinical and neuropathological overlap with AD as well as PD with dementia (PDD). The predominant histological feature of LBD is the presence of cortical and subcortical Lewy bodies (Figure 3), but many patients with LBD also have AD pathology, i.e., cortical amyloid plaques and neurofibrillary tangles. Conversely, Lewy bodies are also frequently observed in cases of classic AD, including in patients with mutations in APP, PSEN1, and PSEN2 (54).
While a familial aggregation of LBD has been described (55), the identification of specific LBD genetic factors is complicated by its still-uncertain phenotypic classification, in particular its distinction from AD and PDD. The little genetic evidence that has accrued to date shows — not unexpectedly — substantial overlap with that for AD and PD. For instance, follow-up analyses to the original AD full-genome screen that led to the description of linkage to chromosome 12 (56) found evidence for considerable genetic heterogeneity in the original study population (57). In particular, the authors found that the better part of the AD linkage signal on chromosome 12q13 near 50 Mb was actually caused by a subset of families fulfilling neuropathological criteria for LBD (i.e., 8 of 54 families). However, these families were linked to a more proximal region, i.e., 12p11 with a maximum linkage signal at approximately 27 Mb. Interestingly, this same region was also implicated in a large Japanese pedigree with autosomal dominant PD (PARK8 [ref. 58], which has now been identified as being caused by mutations in LRRK2 [refs. 36, 37]) and lies only slightly distal to an AD linkage region on 12p12, which was found by 2 different research groups in a sample different from the original chromosome 12 linkage report (59, 60). While these observations could indicate the presence of common genetic risk factors across these 3 syndromes, they could also be purely accidental or even artificial. As for several of the neurodegenerative disorders, a potential association has also been observed with APOE-ε4, albeit inconsistently. However, a moderate risk effect of this allele was recently supported by metaanalysis on LBD case-control studies from 2000 to 2004 (61). Finally and not surprisingly, recent reports also indicate a potential role of α-synuclein in LBD pathogenesis, based on the observation that the occurrence of cortical Lewy bodies and dementia in PD may be dependent on α-synuclein gene dose (see above; 38, 39).
Frontotemporal dementia
FTD is a heterogenous group of syndromes defined clinically by a gradual and progressive change in behavior and personal conduct and/or by a gradual and progressive language dysfunction (62). The initial symptoms typically occur without affecting other cognitive domains, such as memory, and rarely present with an onset age beyond 75 years. In some instances, deficits in behavior and language are also accompanied by parkinsonism or progressive motor neuron disease. Neuropathologically, FTD is caused by neurodegeneration in the frontal and/or temporal lobes (Figure 3). Affected neurons frequently display intracellular, tau-positive inclusions that are distinct from the neurofibrillary tangles observed in AD (63). While 25–40% of all FTD cases are believed to be familial (64), the clinical and neuropathological variability of the syndrome suggests the existence of several distinct genetic factors underlying or modifying pathogenesis. On the other hand, recent advances in the genetics of FTDP-17 (see below) have shown that different mutations in the same gene (or even exon) can lead to a diverse spectrum of FTD-type syndromes, which provides genetic support for merging the apparently diverse clinical entities into 1 overarching category.
The first FTD mutations were identified in cases accompanied by parkinsonism and showing genetic linkage to chromosome 17q21, near the tau gene (FTDP-17). Subsequently, disease-causing mutations were identified in tau (gene: MAPT; Table 1) (ref. 65), currently more than 30 in over 100 families worldwide (for an up-to-date overview, see the Alzheimer Disease & Frontotemporal Dementia Mutation Database; ref. 19). The phenotype observed with mutations in MAPT is variable and ranges from classic FTDP-17 to corticobasal degeneration (CBD), PSP, and frontotemporal lobar degeneration (DLDH; ref. 64). Although, to date, no mutations in MAPT have been reported in pathologically proven AD cases, there have been sporadic observations of early progressive memory loss reminiscent of AD, e.g., segregating with the R406W mutation in exon 13 (66, 67). However, the same mutation was also found in families segregating more typical FTD (65). Molecular genetic studies have shown that the biochemical consequences of the various mutations at the protein level are quite diverse, including reduction or increase in the binding of tau to microtubules, enhancement of tau aggregation, and alterations in the ratio of specific tau isoforms (i.e., an increased ratio of 4-repeat to 3-repeat tau owing to changes in alternative splicing; reviewed in ref. 68; Table 1). Interestingly, it appears that MAPT mutations almost exclusively lead to FTD, with immunohistochemical evidence of both 3- and 4-repeat tau, while classic Pick disease (PiD), in which the 4-repeat isoform is lacking, has not yet been conclusively linked to MAPT or any other genetic defect (69). The correlation between 4-repeat tau and genetic variants in MAPT is further supported by genetic association studies showing almost unanimous support for an MAPT risk haplotype (H1) in samples from patients with PSP or CBD, both characterized by the abundance of 4-repeat tau. This is the same haplotype that has also frequently been associated with PD (for a recent metaanalysis, see ref. 50), which possibly suggests common and as-yet-uncharacterized tau-related pathogenic mechanisms shared by FTD and late-onset PD.
Similar to the examples in the neurodegenerative diseases, tau mutations likely represent the first and most obvious candidates in the puzzle of FTD genetics. They probably account for less than half of the genetic variance in familial FTD (64). In addition to linkage to chromosome 9q21 in a syndrome of FTD coupled with familial amyotrophic lateral sclerosis (ALS; see below), association has been observed between FTD and APOE, albeit with highly variable results. Interestingly, and similar to equivalent studies done in PD, a recent metaanalysis on all data published for APOE in FTD detected a significant risk effect associated with the ε2-allele but no significant results with ε4 (70). While this observation may be purely incidental, it is similar to findings on the H1-tau haplotype, which has also been associated in some FTD syndromes as well as PD. Collectively, these findings are still too preliminary to allow speculation on any functional consequences of the underlying genetic variants in the pathogenesis of FTD. Finally, recent reports have suggested that some cases of FTD may also be caused by mutations in PSEN1 (71). However, a more rigorous proof of familial segregation and pathogenetic mechanism of these variants is needed before they can be considered established.
Amyotrophic lateral sclerosis
ALS (also known as motor neuron disease or Lou Gehrig’s disease) is characterized by a rapidly progressive degeneration of motor neurons in the brain and spinal chord, which ultimately leads to paralysis and premature death. Overall, the prevalence of ALS is low (approximately 5 in 100,000 individuals), but incidence increases with age, showing a peak between 55 and 75 years. Neuropathological features of ALS include intracellular accumulations and perikaryal inclusions of neurofilament (NF) and intracellular inclusions such as Bunina bodies and Lewy body–like cytoplasmic inclusions (Figure 3). Cognitive impairment and dementia coexist with ALS in at least 5% of the cases, and it was actually in a family displaying FTD with parkinsonism and amyotrophy where evidence of linkage to chromosome 17 was first described (see above).
Familial ALS (FALS) is observed in approximately 10% of all cases, but substantially more ALS cases are suspected to be influenced by genetic factors (72). In addition to the variants in MAPT (see above), mutations in 2 genes (SOD1 and ALS2; Table 1) have been shown to cause FALS. Two years after genetic linkage to chromosome 21q was described in 1991 (73), ALS-causing mutations were identified in the gene encoding superoxide dismutase 1 (SOD1; ref. 74), which catalyzes the conversion of superoxide radicals into hydrogen peroxide. Meanwhile, more than 100 mutations in SOD1 have been described in over 200 pedigrees with FALS worldwide, and all but 1 of the known SOD1 mutations are inherited in an autosomal dominant fashion (75). Collectively, these mutations account for approximately 20% of FALS cases and for up to 10% of the sporadic cases of ALS, i.e., those not showing an obvious familial segregation (76). Mutations in SOD1 have been hypothesized as leading to neurodegeneration through protein misfolding, impaired oxidative stress response, cytoskeletal dysfunctions, and glutamatergic excitotoxicity (Table 1; for review, see refs. 77, 78). Recently, mutations in a second gene (ALS2; encoding alsin) were identified in independent families with a rare, juvenile-onset autosomal recessive form of ALS and primary lateral sclerosis, a syndrome restricted to upper motor neuron degeneration (79, 80). Additional mutations in ALS2 have also been described in families suffering from infantile-onset ascending hereditary spastic paralysis, which suggests considerable phenotypic variability of the ALS2 mutations. Functionally, there is evidence that physiologic expression of alsin is neuroprotective in the presence of SOD1 mutations (81); thus, it is conceivable that ALS2 mutations abrogate the protective role of this protein. An up-to-date overview of the status of these and other potential ALS genes can be found at the ALS Online Database (82).
Several other putative FALS loci have been detected by means of linkage analysis in individual families or larger FALS samples, but none of the underlying gene defects have been conclusively proven to be causal. A recent full-genome screen in FALS has pinpointed significant linkage to chromosome 9q21 in families with ALS and FTD (83). This overlaps with a location shown to be linked to AD (84), which possibly indicates a common pathophysiological basis for neurodegeneration or dementia in these 2 disorders. Furthermore, various genetic associations with mostly nonfamilial ALS have been claimed but have been met with only inconsistent replication to date. Specifically — unlike for most of the neurodegenerative disorders discussed in this review— there is virtually no evidence for an association between APOE-ε4 and risk or disease progression of ALS. One potential ALS-specific candidate is the gene encoding the NF heavy chain gene (NFH) on chromosome 22q12, a component of the neuronal inclusions observed histopathologically. However, the genetic data supporting the postulated association between ALS and variants in NFH remain scarce, despite the fact that the association initially was described more than a decade ago (85).
Huntington disease
Huntington disease (HD) is caused by degeneration of neurons in the basal ganglia and then in cortical regions (Figure 3), leading clinically to involuntary movements (chorea), psychiatric symptoms, and dementia. Its prevalence is similar to that of ALS but much less than that of most of the other dementing illnesses discussed above. Approximately 90% of HD cases are hereditary and transmitted in an autosomal dominant fashion. As a matter of fact, the HD gene was the first autosomal disease locus to be mapped by genetic linkage analysis (to chromosome 4q16), in 1983 (86). It took 10 more years to identify the underlying gene defect, which proved to be a poly-CAG (encoding glutamine [Q]) repeat in exon 1 of a 350-kDa protein (huntingtin; gene: HD; Table 1; ref. 87). The mean repeat length in HD patients is 40–45, although variability is quite wide, ranging from 35 to 120 repeats (88), displaying an inverse correlation with onset age. Interestingly, approximately 10% of all HD cases are considered “de novo,” i.e., these cases originate from asymptomatic parents with normal repeat lengths that have expanded to symptomatic range (see below). The precise function of huntingtin remains elusive, but cloning experiments show that it is highly conserved throughout evolution, which suggests an essential functional role of this protein in neuronal development and homoeostasis.
In contrast to all other diseases reviewed here, HD is virtually always attributable to a defect in a single gene, i.e., poly-Q expansion in huntingtin, although such defects only account for 50% of the interindividual onset age variation. Thus, recent genetic analyses of HD have mainly focused on the search for factors affecting the onset of the disease. A recent full-genome screen aimed at identifying these genes has revealed several suggestive linkage regions (89). The most promising of these is located on chromosome 6q25, close to the glutamate receptor, ionotropic, kainate 2 (GRIK2), which has been associated with a younger HD onset age in some studies; this potentially supports the notion of glutamate-induced excitotoxicity in the pathogenesis of HD (90, 91). However, this finding awaits further replication and functional characterization. Only a small number of studies have investigated a potential onset-age effect of the APOE polymorphisms in HD, and as is the case with ALS, the results have been largely negative.
Creutzfeld-Jacob disease and other prion diseases
Prion diseases include a rare and heterogenous spectrum of clinical and histopathological phenotypes, which are unique in the group of neurodegenerative diseases, as they can be familial (e.g., familial Creutzfeld-Jakob disease [fCJD], fatal familial insomnia [FFI], Gerstmann-Sträussler-Scheinker syndrome [GSS]), sporadic (e.g., Creutzfeld-Jakob disease [CJD], sporadic fatal insomnia [sFI]), or acquired (e.g., kuru, iatrogenic CJD, variant CJD). Most forms are characterized by a rapidly progressing neurodegeneration with spongiosis and amyloid plaques consisting of prion protein (PrP) aggregates, probably created via self-propagation of aberrant or misfolded PrP (92, 93; Figure 3). While only a relatively small subset of cases with prion disease exhibits familial aggregation, genetics has played a crucial role in elucidating the molecular mechanisms underlying these disorders and has facilitated the clinical classification of their various subtypes (94).
As in AD, both causative mutations and risk-conferring gene variants have been identified for the different prion diseases. However, both mutations and risk variants are located within the same locus, i.e., the gene encoding PrP (PRNP), on chromosome 20p13 at approximately 5 Mb (Table 1; ref. 95). First, more than 2 dozen different amino acid–changing mutations in the coding region of PRNP have been identified as causing familial prion diseases, transmitted in an autosomal dominant fashion with nearly 100% penetrance. There is remarkable heterogeneity in the sense that different mutations throughout the gene can give rise to a variety of different phenotypes associated with all 3 familial forms of prion diseases, i.e. fCJD, FFI, and GSS (reviewed in ref. 94). In addition to these point mutations, there are also rare cases of fCJD and GSS caused by variable numbers of octapeptide (i.e., 24 base-pair) repeats within the coding sequence of PRNP. Second, both clinical presentation and disease progression of these familial forms are further modified by a common polymorphism at codon 129, which leads to a nonsynonymous amino acid substitution (from methionine [M] to valine [V]). Most mutated PRNP missense alleles are on the same haplotype as the 129M allele, which occurs in virtually all forms of fCJD. In the rare cases where they co-occur with the 129V allele, they lead to a distinct clinical phenotype, and at D178N, even to a different disease entity within the complex of prion diseases: while the D178N-129V haplotype leads to typical fCJD, the 178N-129M haplotype represents the only currently known genetic cause of FFI, which presents with a quite distinct clinical picture.
In addition to its effects on familial forms of prion diseases, the M129V polymorphism also increases the risk for sporadic forms of CJD (sCJD; refs. 96, 97). Specifically, it was found that the homozygous state for either allele (i.e. M/M or V/V) is disoportionally more frequent in sCJD than the M/V genotype (96). Furthermore, homozygosity at this polymorphism leads to a faster disease progression than heterozygosity in nearly all genetic as well as sporadic (including iatrogenic) forms of prion diseases, and in most instances, the M/M genotype is associated with the most aggressive course of disease (98). Interestingly, almost all individuals thus far known to be affected by the newly described “variant” form of CJD (vCJD), which is characterized by a prion protein isotype resembling that found in BSE, also carry only the M/M genotype. Furthermore, there is some evidence suggesting an overrepresentation of the M allele in AD cases versus controls as well (13). Only a few other genetic risk factors for the nonfamilial forms of CJD have been investigated, and none of them has shown any noteworthy results to date. This includes APOE-ε4, which was found to increase risk and/or accelerate disease progression in some studies, although the majority of samples failed to replicate either of these effects.
Conclusions
While displaying a diverse array of clinical and histopathological characteristics, the neurodegenerative disorders discussed in this review share a variety of epidemiologic and genetic aspects. First, with the exception of HD, they all feature an etiologic dichotomy, with relatively rare familial forms on the one hand and more frequent multifactorial — and usually later-onset — forms on the other. It is possible (and likely) that a substantial number of cases that were hitherto considered nonfamilial and sporadic will eventually prove to originate from specific disease-causing mutations or genetic risk factors (like APOE-ε4 in AD). Second, in some cases, the same mutations and polymorphisms have been linked and associated across clinically and neuropathologically diverse disease entities. For instance, according to recent metaanalyses, the APOE polymorphism may contribute to risk not only for AD, but also for PD and FTD (albeit with different alleles). If confirmed, these observations could point to 1 or several common genetic and mechanistic denominators for neuronal cell death. Finally, genetics has been essential for elucidating the molecular and biochemical pathways leading to neurodegeneration for almost all of the discussed syndromes and disease entities. Likewise, a detailed understanding of the genetic basis of neurodegeneration will be essential for the design and development of effective early prediction and early prevention/treatment strategies, with the prospect of largely decreasing the incidence of these devastating disorders.
Acknowledgments
This work was sponsored by grants from the National Institute of Mental Health, the National Institute on Aging (Alzheimer’s Disease Research Center), and the Alzheimer’s Association. L. Bertram was funded by Deutsche Forschungsgemeinschaft (DFG), Harvard Center for Neurodegeneration and Repair (HCNR), and the National Alliance for Research on Schizophrenia and Depression (NARSAD).
Footnotes
Nonstandard abbreviations used: AD, Alzheimer disease; ALS, amyotrophic lateral sclerosis; CJD, Creutzfeld-Jakob disease; EOFAD, early-onset familial AD; FALS, familial ALS; FFI, fatal familial insomnia; FTD, frontotemporal dementia; FTDP-17, FTD with parkinsonism linked to chromosome 17; HD, Huntington disease; LBD, Lewy body dementia; OR, odds ratio; PD, Parkinson disease; PrP, prion protein.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 2.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Myers A, Wavrant-De Vriese F. Problems and solutions in the genetic analysis of late-onset Alzheimer’s disease. Neurodegenerative Diseases. 2004;1:213–217. doi: 10.1159/000080988. [DOI] [PubMed] [Google Scholar]
- 4.Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res. 2004;129:39–44. doi: 10.1016/j.psychres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuznetsova T, et al. Antihypertensive treatment modulates the association between the D/I ACE gene polymorphism and left ventricular hypertrophy: a meta-analysis. J. Hum. Hypertens. 2000;14:447–454. doi: 10.1038/sj.jhh.1001055. [DOI] [PubMed] [Google Scholar]
- 7.Faraone SV, Doyle AE, Mick E, Biederman J. Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. Am. J. Psychiatry. 2001;158:1052–1057. doi: 10.1176/appi.ajp.158.7.1052. [DOI] [PubMed] [Google Scholar]
- 8.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 9.Lasky-Su JA, Faraone SV, Glatt SJ, Tsuang MT. Meta-analysis of the association between two polymorphisms in the serotonin transporter gene and affective disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;133:110–115. doi: 10.1002/ajmg.b.30104. [DOI] [PubMed] [Google Scholar]
- 10.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat. Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 11.Maraganore DM, et al. UCHL1 is a Parkinson’s disease susceptibility gene. Ann. Neurol. 2004;55:512–521. doi: 10.1002/ana.20017. [DOI] [PubMed] [Google Scholar]
- 12.Huang X, Chen PC, Poole C. APOE-epsilon2 allele associated with higher prevalence of sporadic Parkinson disease. Neurology. 2004;62:2198–2202. doi: 10.1212/01.wnl.0000130159.28215.6a. [DOI] [PubMed] [Google Scholar]
- 13.Alzheimer Research Forum. 2005. AlzGene Database for published Alzheimer disease candidate genes. http://www.alzgene.org.
- 14.Tanzi RE. A genetic dichotomy model for the inheritance of Alzheimer’s disease and common age-related disorders. J. Clin. Invest. 1999;104:1175–1179. doi: 10.1172/JCI8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 16.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 17.Rogaev EI, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 18.Levy-Lahad E, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 19.Alzheimer Disease & Frontotemporal Dementia Mutation Database. http://www.molgen.ua.ac.be/admutations/.
- 20.Mayeux R, Sano M, Chen J, Tatemichi T, Stern Y. Risk of dementia in first-degree relatives of patients with Alzheimer’s disease and related disorders. Arch. Neurol. 1991;48:269–273. doi: 10.1001/archneur.1991.00530150037014. [DOI] [PubMed] [Google Scholar]
- 21.Strittmatter WJ, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmechel DE, et al. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 24.Daw EW, et al. The number of trait loci in late-onset Alzheimer disease. Am. J. Hum. Genet. 2000;66:196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruts M, et al. Estimation of the genetic contribution of presenilin-1 and -2 mutations in a population-based study of presenile Alzheimer disease. Hum. Mol. Genet. 1998;7:43–51. doi: 10.1093/hmg/7.1.43. [DOI] [PubMed] [Google Scholar]
- 26.Campion D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am. J. Hum. Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finckh U, et al. Association of late-onset Alzheimer disease with a genotype of PLAU, the gene encoding urokinase-type plasminogen activator on chromosome 10q22.2. Neurogenetics. 2003;4:213–217. doi: 10.1007/s10048-003-0157-9. [DOI] [PubMed] [Google Scholar]
- 28.Tanzi RE, Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- 29.Tanner CM, et al. Parkinson disease in twins: an etiologic study. JAMA. 1999;281:341–346. doi: 10.1001/jama.281.4.341. [DOI] [PubMed] [Google Scholar]
- 30.Maher NE, et al. Epidemiologic study of 203 sibling pairs with Parkinson’s disease: the GenePD study. Neurology. 2002;58:79–84. doi: 10.1212/wnl.58.1.79. [DOI] [PubMed] [Google Scholar]
- 31.de la Fuente-Fernandez R. A note of caution on correlation between sibling pairs [author reply] Neurology. 2003;60:1561. doi: 10.1212/wnl.60.9.1561. [DOI] [PubMed] [Google Scholar]
- 32.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 33.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 34.Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 35.Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 36.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Paisan-Ruiz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 39.Chartier-Harlin MC, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 40.Albrecht M. LRRK2 mutations and Parkinsonism. Lancet. 2005;365:1230. doi: 10.1016/S0140-6736(05)74810-8. [DOI] [PubMed] [Google Scholar]
- 41.Matsumine H, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am. J. Hum. Genet. 1997;60:588–596. [PMC free article] [PubMed] [Google Scholar]
- 42.Petrucelli L, et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 43.van Duijn CM, et al. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am. J. Hum. Genet. 2001;69:629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson’s disease. Ann. Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 45.Valente EM, et al. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am. J. Hum. Genet. 2001;68:895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leroy E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 47.Le WD, et al. Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 2003;33:85–89. doi: 10.1038/ng1066. [DOI] [PubMed] [Google Scholar]
- 48.Scott WK, et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- 49.Martin ER, et al. Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA. 2001;286:2245–2250. doi: 10.1001/jama.286.18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Healy DG, et al. Tau gene and Parkinson’s disease: a case-control study and meta-analysis. J. Neurol. Neurosurg. Psychiatr. 2004;75:962–965. doi: 10.1136/jnnp.2003.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad C, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann. Neurol. 1997;41:277–281. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 52.Farrer M, et al. alpha-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum. Mol. Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 53.McKeith IG, et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): report of the consortium on DLB international workshop. Neurology. 1996;47:1113–1124. doi: 10.1212/wnl.47.5.1113. [DOI] [PubMed] [Google Scholar]
- 54.Hamilton RL. Lewy bodies in Alzheimer’s disease: a neuropathological review of 145 cases using alpha-synuclein immunohistochemistry. Brain Pathol. 2000;10:378–384. doi: 10.1111/j.1750-3639.2000.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuang DW, DiGiacomo L, Bird TD. Familial occurrence of dementia with Lewy bodies. Am. J. Geriatr. Psychiatry. 2004;12:179–188. [PMC free article] [PubMed] [Google Scholar]
- 56.Pericak-Vance MA, et al. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- 57.Scott WK, et al. Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am. J. Hum. Genet. 2000;66:922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Funayama M, et al. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 59.Kehoe P, et al. A full genome scan for late onset Alzheimer’s disease. Hum. Mol. Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 60.Curtis D, North BV, Sham PC. A novel method of two-locus linkage analysis applied to a genome scan for late onset Alzheimer’s disease. Ann. Hum. Genet. 2001;65:473–481. doi: 10.1017/S0003480001008776. [DOI] [PubMed] [Google Scholar]
- 61.Bang OY, Kwak YT, Joo IS, Huh K. Important link between dementia subtype and apolipoprotein E: a meta-analysis. Yonsei. Med. J. 2003;44:401–413. doi: 10.3349/ymj.2003.44.3.401. [DOI] [PubMed] [Google Scholar]
- 62.McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch. Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 63.Brun A. Frontal lobe degeneration of non-Alzheimer type. I. Neuropathology. Arch. Gerontol. Geriatr. 1987;6:193–208. doi: 10.1016/0167-4943(87)90021-5. [DOI] [PubMed] [Google Scholar]
- 64.Bird T, et al. Epidemiology and genetics of frontotemporal dementia/Pick’s disease. Ann. Neurol. 2003;54(Suppl 5):S29–S31. doi: 10.1002/ana.10572. [DOI] [PubMed] [Google Scholar]
- 65.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 66.Reed LA, Wszolek ZK, Hutton M. Phenotypic correlations in FTDP-17. Neurobiol. Aging. 2001;22:89–107. doi: 10.1016/s0197-4580(00)00202-5. [DOI] [PubMed] [Google Scholar]
- 67.Rademakers R, et al. Tau (MAPT) mutation Arg406Trp presenting clinically with Alzheimer disease does not share a common founder in Western Europe. Hum. Mutat. 2003;22:409–411. doi: 10.1002/humu.10269. [DOI] [PubMed] [Google Scholar]
- 68.Ingelsson M, Hyman BT. Disordered proteins in dementia. Ann. Med. 2002;34:259–271. doi: 10.1080/078538902320322529. [DOI] [PubMed] [Google Scholar]
- 69.Morris HR, et al. Analysis of tau haplotypes in Pick’s disease. Neurology. 2002;59:443–445. doi: 10.1212/wnl.59.3.443. [DOI] [PubMed] [Google Scholar]
- 70.Verpillat P, et al. Apolipoprotein E gene in frontotemporal dementia: an association study and meta-analysis. Eur. J. Hum. Genet. 2002;10:399–405. doi: 10.1038/sj.ejhg.5200820. [DOI] [PubMed] [Google Scholar]
- 71.Dermaut B, et al. A novel presenilin 1 mutation associated with Pick’s disease but not beta-amyloid plaques. Ann. Neurol. 2004;55:617–626. doi: 10.1002/ana.20083. [DOI] [PubMed] [Google Scholar]
- 72.Majoor-Krakauer D, Willems PJ, Hofman A. Genetic epidemiology of amyotrophic lateral sclerosis. Clin. Genet. 2003;63:83–101. doi: 10.1046/j.0009-9163.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 73.Siddique T, et al. Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N. Engl. J. Med. 1991;324:1381–1384. doi: 10.1056/NEJM199105163242001. [DOI] [PubMed] [Google Scholar]
- 74.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 75.Andersen PM, et al. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat. Genet. 1995;10:61–66. doi: 10.1038/ng0595-61. [DOI] [PubMed] [Google Scholar]
- 76.Kato, S., Shaw, P., Wood-Allum, C., Leigh, P.N., and Shaw, C. 2003. Amyotrophic Lateral Sclerosis. In Neurodegeneration — the molecular pathology of dementia and movement disorders. D. Dickson, editor. ISN Neuropath Press. Basel, Switzerland. 350–368.
- 77.Stathopulos PB, et al. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wood JD, Beaujeux TP, Shaw PJ. Protein aggregation in motor neurone disorders. Neuropathol. Appl. Neurobiol. 2003;29:529–545. doi: 10.1046/j.0305-1846.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 79.Yang Y, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 80.Hadano S, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 81.Kanekura K, et al. Alsin, the product of ALS2 gene, suppresses SOD1 mutant neurotoxicity through RhoGEF domain by interacting with SOD1 mutants. J. Biol. Chem. 2004;279:19247–19256. doi: 10.1074/jbc.M313236200. [DOI] [PubMed] [Google Scholar]
- 82.Institute of Psychiatry. 2004. The ALS online database for mutations in ALS related genes. http://www.alsod.org.
- 83.Hosler BA, et al. Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA. 2000;284:1664–1669. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- 84.Blacker D, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum. Mol. Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 85.Figlewicz DA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3:1757–1761. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 86.Gusella JF, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 87.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 88.Gusella JF, MacDonald ME. Huntington’s disease: CAG genetics expands neurobiology. Curr. Opin. Neurobiol. 1995;5:656–662. doi: 10.1016/0959-4388(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 89.Li JL, et al. A genome scan for modifiers of age at onset in Huntington disease: the HD MAPS study. Am. J. Hum. Genet. 2003;73:682–687. doi: 10.1086/378133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rubinsztein DC, et al. Genotypes at the GluR6 kainate receptor locus are associated with variation in the age of onset of Huntington disease. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3872–3876. doi: 10.1073/pnas.94.8.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MacDonald ME, et al. Evidence for the GluR6 gene associated with younger onset age of Huntington’s disease. Neurology. 1999;53:1330–1332. doi: 10.1212/wnl.53.6.1330. [DOI] [PubMed] [Google Scholar]
- 92.Prusiner SB. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bieschke J, et al. Autocatalytic self-propagation of misfolded prion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12207–12211. doi: 10.1073/pnas.0404650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br. Med. Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 95.Hsiao K, et al. Linkage of a prion protein missense variant to Gerstmann-Straussler syndrome. Nature. 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 96.Palmer MS, Dryden AJ, Hughes JT, Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature. 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 97.Collinge J, Palmer MS, Dryden AJ. Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 98.Pocchiari M, et al. Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004;127:2348–2359. doi: 10.1093/brain/awh249. [DOI] [PubMed] [Google Scholar]
- 99.Dickson, D., editor. 2003. Neurodegeneration: the molecular pathology of dementia and movement disorders. ISN Neuropath Press. Basel, Switzerland. 414 pp.