Abstract
Multigenerational effects of silver nanoparticles (Ag-NPs) on reproduction of the soil nematode Caenorhabditis elegans have been observed previously. However, mechanisms of this reproductive sensitivity are unknown. Here we examine whether epigenetic changes occur as a result of multigenerational exposure to Ag-NPs and whether such modifications can be inherited by unexposed generations. Changes at histone methylation markers, histone 3 lysine 4 dimethylation (H3K4me2) and histone 3 lysine 9 trimethylation (H3K9me3), known to affect reproduction, as well as changes in the expression of the genes encoding demethylases and methyltransferases associated with the selected markers, were investigated. We exposed C. elegans at EC30 to AgNO3, pristine Ag-NPs, and its environmentally transformed product, sulfidized Ag-NPs (sAg-NPs). Histone di-methylation levels at H3K4me2 increase in response to pristine Ag-NP exposure and did not recover after rescue from the exposure, suggesting transgenerational inheritance. Compared to pristine Ag-NPs, exposure to transformed sAg-NPs significantly decreased H3K4me2 and H3K9me3 levels. These changes in the histone methylation were also supported by expression of spr-5 and jmjd-2 (H3K4me2 and H3K9me3 demethylases, respectively) and set-30 (H3K4me2 methyltransferase). Our study demonstrates that multigenerational exposure to Ag-NPs induces epigenetic changes that are inherited by unexposed offspring. However, environmental transformations of Ag-NPs may also reduce toxicity via epigenetic mechanisms, such as changes at histone methylation.
Keywords: Demethylase, Gene expression, Histone methylation, Methyltransferase, Nanomaterials, Nematode
Graphical Abstract
1. Introduction
Silver NPs are valued for their antimicrobial, optical and conductive properties (Mélanie et al., 2009; Rao et al., 2002). This has resulted in the widespread application of Ag-NPs in the industrial and biomedical sectors (Haider and Kang, 2015; Juganson et al., 2015; Salata, 2004). After detailed analysis of consumer products in the market advertised to contain nanomaterials from several countries, Ag-NPs was identified as the most used nanomaterial in the consumer products recorded (Vance et al., 2015). There has been an increasing concern over the release of silver nanoparticles (Ag-NPs) into the environment from the many consumer products containing Ag nanomaterials (Juganson et al., 2015; Vance et al., 2015). Silver nanoparticles can be introduced in the environment during production, as well as during the usage and disposal of consumer products, thereby increasing the environmental concentration of Ag. Most of the Ag-NPs released from consumer products end up in wastewater treatment plants (WWTP), where they mainly partition into sewage sludge (Kaegi et al., 2011; Mueller and Nowack, 2008). The sludge may then be applied on land as fertilizer for agricultural purposes, thus introducing Ag-NPs into soils. A global life cycle release of several engineered nanomaterials showed that most of the engineered nanomaterials eventually end up in landfills (63–91 %) and soils (8–28 %), followed by the aquatic environment (0.4–7 %), and then air (0.1–1.5 %) (Keller et al., 2013).
The release of Ag-NPs into soil environment is ongoing process and it is inevitable that soil organisms will encounter these NPs. Thus, examining effects and mechanisms of exposure to Ag-NPs over multiple generations in a soil invertebrate such as a nematode, Caenorhabditis elegans, represents an environmentally realistic scenario. Nematodes inhabit soil and water environments and are among the most abundant, in terms of numbers, soil-dwelling animal organisms with ability to adapt to a wide range of environmental conditions (Neher and Powers, 2005). Model nematode C. elegans is a free living nematode, about 1 mm in length, with a short life cycle (3 days) and prolific reproduction (up to 300 offspring). For over two decades it has been accepted as a powerful model for ecotoxicity studies including nanotoxicity (Choi et al 2014).
Potential harmful effects of Ag-NPs in the environment have been investigated in different soil organisms (Diez-Ortiz et al., 2015; Jesmer et al.; Shoults-Wilson et al.; Starnes et al., 2015; Velicogna et al., 2016; Waalewijn-Kool et al., 2014; Yang et al., 2014). However, most Ag nanotoxicity studies have focused on acute or sub-chronic exposures. Adequate analysis of the risk posed by Ag-NPs should include more realistic chronic exposures at sub-lethal concentrations, with environmentally transformed forms of Ag-NPs taken into consideration. Prominent environmental transformation of pristine Ag-NPs includes oxidation which results in the release of Ag+ ions and sulfidation to form sulfidized Ag-NPs (sAg-NPs) (Levard et al., 2012; Lowry et al., 2012).
Schultz et al. (Schultz et al., 2016) performed one of the most extensive multigenerational exposure study in which the nematode C. elegans exposed to AgNO3 and Ag-NPs, showed heightened reproductive sensitivity in subsequent generations. Multigenerational reproductive effects of Ag-NPs have also been observed in Drosophila melanogaster (Panacek et al., 2011; Raj et al., 2017). Despite these multigenerational effects, the mode of toxicity transfer from exposed parental generations to subsequent generations has not been thoroughly investigated. The contribution of germline mutations to the previously observed multigenerational toxicity in response to Ag-NPs and Ag ions was investigated in our most recent study (Wamucho et al., 2019). In that study we showed that there was a pattern for increase in the germline mutations observed in all treatments with transformed sAg-NPs inducing significant increase in the number of transversion. This result could not explain the previously observed multigeneration toxicity, where pristine Ag-NPs and AgNO3 induced stronger reproductive toxicity than sAg-NPs. Therefore the results from the genomic mutation study (Wamucho et al., 2019) suggest that other mechanisms of inheritance, such as epigenetics, must be involved in the observed multigenerational toxicity in response to Ag-NPs and AgNO3. Additionally, a previous study with human cell lines showed changes in epigenetic patterns, such as DNA methylation, histone methylation and miRNA expression, in response to Ag-NP exposure (Wong et al., 2017).
Epigenetic changes such as histone modifications (methylation, acetylation, and phosphorylation), DNA methylation, and non-coding RNAs, which do not involve changes in the underlying DNA sequence, can be inherited. Epigenetic markers regulate vital processes such as development and aging in C. elegans (Gonzalez-Aguilera et al., 2014). Transgenerational fertility defects in C. elegans are known to correlate with changes in certain histone methylation profiles as well as DNA methylation (Greer et al., 2014; Greer et al., 2015). Environmental stressors also induce epigenetic changes that are inherited by subsequent unstressed generations (Kishimoto et al., 2017; Klosin et al., 2017; Yu and Liao, 2016). As such, the heightened reproductive sensitivity observed in subsequent generations after exposures to AgNO3 and Ag-NPs may be explained by changes in heritable epigenetic markers. Previous sub-chronic exposure revealed differences in bio-distribution of Ag and transcriptomic profiles in response to AgNO3, Ag-NPs, and sAg-NPs (Starnes et al., 2016; Starnes et al., 2015). Thus, different epigenetic responses are expected after C. elegans exposure to the different Ag forms.
Changes in epigenetic markers such as histone 3 lysine 4 dimethylation (H3K4me2) and histone 3 lysine 9 trimethylation (H3K9me3), have been shown to correlate with transgenerational fertility in C. elegans. An increase in H3K4me2 and a decrease in H3K9me3 were shown to increase sterility in C. elegans in a transgenerational manner (Greer et al., 2014). Methyltransferases and demethylases associated with these epigenetic markers, which catalyze the addition or removal of methyl groups respectively, have been predicted or identified. Among them are H3K4 mono/dimethyltransferases, set-17 and set-30, and H3K4me2 demethylase, spr-5 (Greer et al., 2014).The demethylase for H3K9me3 is jmjd-2 and the methyltransferase is set-25 (Gonzalez-Aguilera et al., 2014; Greer et al., 2014). Though set-26 was initially shown to trimethylate H3K9 in vitro (Greer et al., 2014) but not in vivo, it is involved in such vital biological processes as transgenerational fertility, life span, development and aging, and also seems to repress H3K4me3 levels (Gonzalez-Aguilera et al., 2014; Ni et al., 2012; Wang et al., 2018). In this study, we investigated the epigenetic changes in histone methylation markers, H3K4me2 and H3K9me3, after multigenerational exposure of C. elegans to pristine and transformed (sulfidized) Ag-NPs (sAg-NPs) as well as AgNO3. The AgNO3 treatment was included as a positive control for complete dissolution of Ag-NPs. Analyses were carried out on nematode populations prior to exposure (F0), after three generations of exposure (F3), and three generations of rescue from exposure (F6) to determine if these epigenetic changes after exposures, if any, are inherited by subsequent unexposed generations. The expression levels of the genes encoding the specific demethylases and methyltransferases were also investigated to determine if they correlate with the histone methylation levels.
2. Materials and Methods
2.1. Silver nanoparticle synthesis and characterization
Polyvinylpyrrolidone (PVP) coated Ag-NPs were synthesized as previously described (Cheng et al., 2011). The same batch of Ag-NPs used by Starnes et al. (Starnes et al., 2015) and Schultz et al. (Schultz et al., 2016) was used in this study. Sulfidation was carried out by combining Ag-NPs with Na2S at a 2:1 molar ratio of S to Ag. The mixture was incubated at room temperature for 4 h open to the atmosphere. Tube was capped and sealed and incubated at room temperature for an additional week. The sAg-NPs were separated from the reaction solution and triple washed with 18MΩ deionized water. Complete sulfidation was confirmed by powder X-ray diffraction (X’Pert Pro, Malvern PANalytical, Malvern, UK).
Characterization of Ag-NPs was described in our previous studies (Schultz et al., 2016; Starnes et al., 2015). The transmission electron microscopy (TEM) primary particle sizes were reported to be 58.3 ± 12.9 nm for PVP coated Ag-NPs and 64.5 ± 19.4 nm for sAg-NPs (Starnes et al., 2015). Modified simulated soil pore water (SSPW: Na 4 mM, Mg 0.5 mM, Al 1 μM, K 1.0 mM, Ca 1.25 mM, NO3 3.5 mM, SO4 0.5 mM, PO4 1.0 μM, and I=10.3 mM) was used for all exposures to mimic natural soil environment for C. elegans (Schultz et al., 2016; Tyne et al., 2013).
Upon addition of Ag-NPs and sAg-NPs into the SSPW(Tyne et al., 2013), the volume weighted sizes were determined by dynamic light scattering as 66.26 ± 34.34 and 60.73 ± 20.67 nm, respectively. The zeta potential of Ag-NPs and sAg-NPs in the SSPW were −5.3 mV and −15.7 mV, respectively. The stability of these Ag nanoparticles over period of exposure of 72 h have been investigated and described in our most recent study (Wamucho et al. 2019). According to that study, after the 72 h period, the volume weighted sizes were 68.87 ± 30.38 nm for Ag-NPs and 68.64 ± 20.78 nm for sAg-NPs. After 72 h zeta potential of Ag-NPs and sAg-NPs in the SSPW were −5.3 and −15.7 mV, respectively. From our previous multigenerational study, which also used the same Ag-NPs in the same media (SSPW), the dissolution determined via ultrafiltration was 1.5 ± 0.1% for Ag-NPs and 0.023 ± 0.002% for sAg-NPs (Schultz et al., 2016).
2.2. Nematode exposures
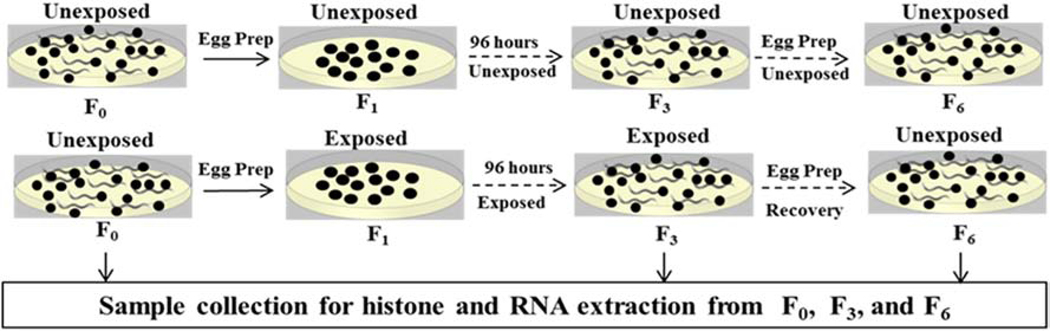
Caenorhabditis elegans (N2) was acquired from Caenorhabditis Genetics Center (University of Minnesota, USA). The exposures were carried out on a population of nematodes which were propagated for multiple generations (Fig. 1). Epigenetic markers (H3K4me2 and H3K9me3) and gene expression levels of the demethylases and methyltransferases associated with these epigenetic markers were analyzed in different generations prior to exposure (F0), after three generations of exposure (F3), and after three generations of rescue from exposure (F6).
Fig. 1.
Exposure of Caenorhabditis elegans to AgNO3, pristine Ag nanoparticles (Ag-NPs) and sulfidized Ag-NPs (sAg-NPs). Top panel illustrates multigenerational control group and bottom panel illustrate Ag exposures. Total histone and RNA were extracted from generations F0, F3 and F6.
Age synchronization using NaClO/ NaOH solution was performed and eggs were placed in 10 cm SSPW agar plates to start the unexposed F0 populations of four replicates (about 4000 nematodes per replicate) per treatment group (control, AgNO3, pristine Ag-NPs, and sAg-NPs). Uracil deficient E. coli strain, OP50, suspended in 6 ml of SSPW at an optical density (540 nm) of 0.35 was added (10 μl/ml) to each plate as food source and incubated at 20 °C. The amount of E.coli used provided optimal feeding conditions for the nematodes. After 96 h, starting from the egg stage, gravid adults were washed off the plate into 15 ml centrifuge tubes using SSPW and split in half. Half of the nematodes were saved for histone and RNA extraction and half used for age synchronization to obtain eggs for the next generation (F1). Exposures were started at the F1 generation with the SSPW/OP50 food source dosed with equitoxic concentrations of AgNO3 (0.07 mg/L), Ag-NPs (1.5 mg/L), and sAg-NPs (6 mg/L). These concentrations correspond to EC30s for reproduction, as determined from concentration-response experiments previously (Schultz et al., 2016) and also reconfirmed in this experiment (Fig. S1). The EC30 concentrations were selected to examine epigenetic and gene expression changes at sub-lethal equitoxic concentration in all treatments. The SSPW/OP50 solution without NPs or AgNO3 was used for controls throughout the experiment. Exposures were carried out for three generations (F1, F2, and F3). Each generation was exposed for 96 h starting at the egg stage after which age synchronization was performed to obtain eggs for each subsequent generation. The 6 ml of SSPW (for control) or exposure solutions in SSPW were deposited on the agar plates. When exposures are performed in liquid media starting from eggs, there is a delay in C. elegans development. Instead of 64 h required for C. elegans to reach stage of gravid adults, when maintained on a solid K-agar media, it took 96 h to reach this stage in liquid (SSPW) media. After the last exposure (F3), nematodes were washed off the plate with SSPW and half of the nematodes saved for histone and RNA extraction. The other half was used for age synchronization to obtain eggs to start the F4 generation at which point rescue from exposure started by feeding nematodes with SSPW/OP50 solution without NPs or AgNO3. Rescue was carried out for three generations (F4, F5, and F6) at which point the experiment was terminated. The F6 population were washed off the plate with SSPW and used for histone and RNA extraction.
2.3. Histone and RNA extractions
Nematodes were washed three times using DI water with gentle centrifugation at 800 rpm for 1 min to remove residual bacteria. After the final wash, the nematodes were split in two groups for histone and RNA extractions. Nematodes for histone extraction were homogenized by sonication on ice after which the EpiQuik total histone extraction kit (EpiGentek, Farmingdale, NY) was used following manufacturers recommendations. Extracted total histone was quantified by use of the Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, Rockford, IL). RNA extraction was performed using RNeasy Mini Kit with on-column DNase 1 digestion for 15 min at room temperature (QIAGEN, Hilden, Germany) and quantified using a UV-visible spectrophotometer.
2.4. Total H3, global H3K4me2 and global H3K9me3 quantification
Total H3 levels were measured with enzyme-linked immunosorbent assay (ELISA)-based colorimetric EpiQuik Total Histone H3 Quantification Kit (EpiGentek, Farmingdale, NY) using 250 ng of total histone proteins. Global H3K4me2 and global H3K9me3 levels were measured with EpiQuik Global Di-Methyl Histone H3K4 Quantification Kit EpiQuik and Global Tri-Methyl Histone H3K9 Quantification Kit (EpiGentek, Farmingdale, NY), respectively, using 1000 ng of total histone. The tri- and dimethylated histones were captured by anti-dimethyl H3K4 or anti-trimethyl H3K9 antibody and detected with labeled detection antibody. The absorbance were measured at 450 nm using SpectraMax i3. Measurements were performed using three biological replicates per treatment with two technical replicates per sample. Global levels of H3K4me2 and H3K9me3 were normalized to the total H3 levels before analysis. The calculations of the concentrations were performed using a Four Parameter Logistic curve fit, implemented into MyAssays analysis open source software available at myassays.com.
2.5. Quantitative Real-Time Polymerase Chain Reaction Analysis
Expression levels of H3K4me2 demethylase (spr-5) and H3K4 mono/dimethyltransferases (set-17 and set-30) were investigated. H3K9me3 demethylase (jmjd-2) and H3K9 methyltransferase (set-25) expression levels were also analyzed as well as expression levels of set-26. The TaqMan primer/probe assay IDs and their amplification efficiencies are shown in Table S1. Y45F10D.4 gene, a putative iron sulfur cluster assembly enzyme, was used as the reference gene due to its highly stable expression levels (Zhang et al., 2012). Its expression levels were also stable among all Ag treatments used in this study with 0.5–1Ct difference (Fig. S2).
cDNA synthesis was carried out with 500 ng of total RNA by using RevertAid First Strand Synthesis Kit (ThermoFisher Scientific, Vilnius, Lithuania). qRT-PCR reactions were carried out in 10 μL volumes with TaqMan fast advanced master mix, TaqMan gene expression assays for each gene, and cDNA diluted 1:100 (spr-5,set-17,set-30) or 1:19 (jmjd-2, set-25, set-26). The optimal cDNA dilution factor was determined based on the dilution amplification curves from the efficiency tests. StepOne Plus system (Applied Biosystems) was used for all amplifications with a program of 10 min at 95 °C, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C. All treatments for each gene were run in biological and technical triplicates. Negative controls and minus reverse transcriptase (-RT) negative controls were run for every gene/sample to check for DNA contamination. The data were exported into GenEx software (MultiD), and after normalization to the reference gene, the expression levels of the target genes relative to controls were calculated following the Pfaffl method (Pfaffl, 2001). The patterns of the gene expression levels were compared to their respective histone methylation levels observed.
2.6. Data analysis
Statistical analyses were conducted in SAS. The data are presented as mean and standard errors of the mean. The statistical significance among treatments was determined after applying one-way ANOVA with Tukey’s post hoc test. The differences were accepted as statistically significant at p < 0.05.
3. Results and Discussion
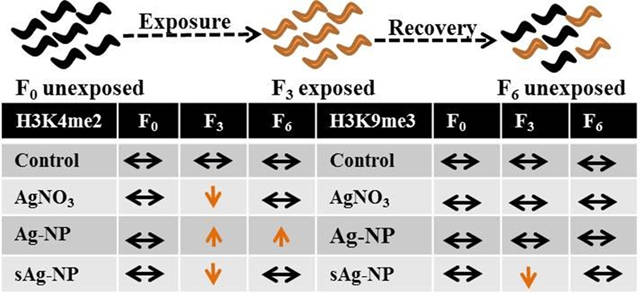
This study was designed to investigate the potential for changes in epigenetic modifications and their transgenerational inheritance in C. elegans exposed to AgNO3, Ag-NPs, and sAg-NPs. We anticipate that our results provide insight into the mechanisms of previously observed transgenerational toxicity to Ag-NPs and AgNO3 (Schultz et al., 2016). Overall, our results indicate that all three different Ag treatments induce differential epigenetic changes in global levels of H3K4me2 and H3K9me3 in C. elegans and this may have different implications in regards to toxicity. Differential gene expression patterns identified previously after sub-chronic exposures of C. elegans within single generation to these Ag treatments, suggested different mechanisms of toxicity (Starnes et al., 2016). Epigenetic modifications, such as histone methylation, are known to regulate gene expression (Dong and Weng, 2013; Liu et al., 2011; Pokholok et al., 2005), and it is possible that the differential histone methylation patterns observed after exposure to different forms of Ag play a role in such differential gene expression. In addition, the differences in epigenetic changes observed in response to pristine and transformed Ag-NPs (sAg-NPs) in the current study correlate with the different effects on reproduction identified in our previous multigenerational study (Schultz et al., 2016). This suggests that the epigenetic mechanism, such as histone methylation, is likely involved in the multigenerational reproductive toxicity. We have also observed that changes in histone methylation persist even in the nematode populations after rescue from pristine Ag-NP treatment (Fig. 2) suggesting that these modifications have been inherited by the unexposed generations. Our histone methylation results correlate with the changes in the expression of the genes, shown or predicted to play a role in these epigenetic modifications, thus providing additional support for the results of this study.
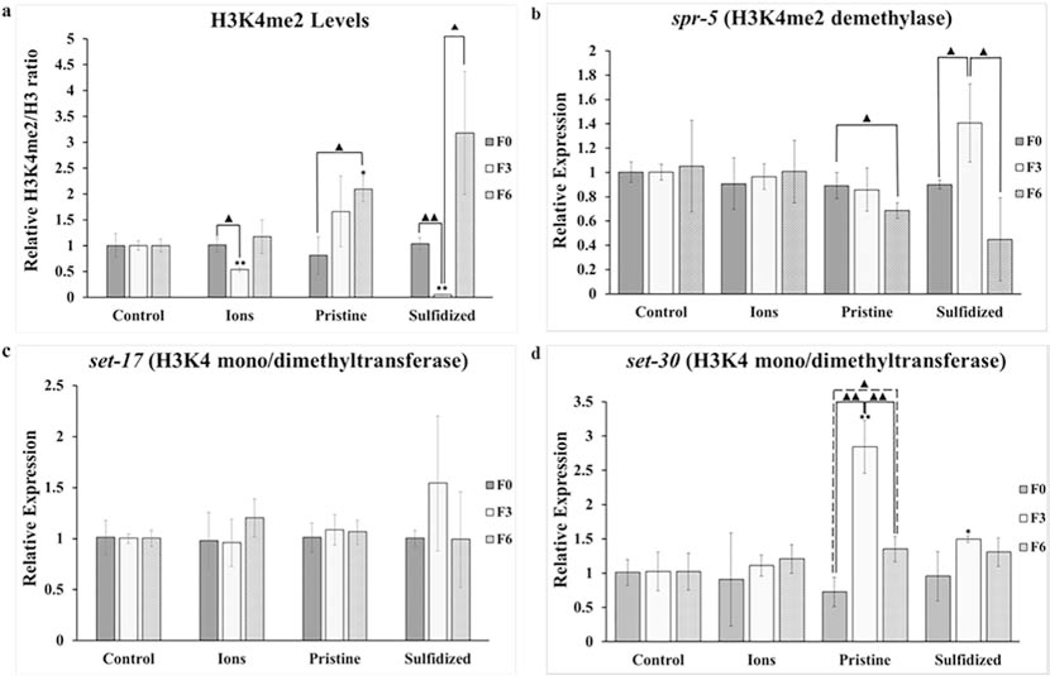
Fig. 2.
Changes in global levels of histone di-methylation at H3K4 marker (a) and levels of gene expression at spr-5 (b), set-17 (c), and set-30 (d) after exposure of Caenorhabditis elegans to AgNO3 (Ions), pristine Ag nanoparticles (Ag-NPs) and sulfidized Ag-NPs (sAg-NPs). Bars represent standard error of the means. *, ** signifies difference with control at p ≤ 0.05 and p ≤ 0.01 respectively.▲, ▲▲signifies difference between generations of the same treatment at p ≤ 0.05 and p ≤ 0.01, respectively.
3.1. Global H3K4me2 levels and expression of H3K4me2 demethylase and methyltransferase
Global H3K4me2 levels prior to exposure (F0), after three generations of exposure (F3), and after three generations of rescue from exposure (F6) are shown in Fig. 2a. Exposure to AgNO3 and sAg-NPs resulted in a significant decrease (p = 0.030 and p = 0.001 respectively) in H3K4me2 levels compared to the unexposed F0 with recovery after rescue from the exposure at F6 (p = 0.0674 and p = 0.146 respectively). For Ag-NP treatment, an increase in H3K4me2 was observed after three generations of exposure compared to the unexposed F0, though not significant (p = 0.334). Interestingly, H3K4me2 level was significantly higher in F6 (p = 0.043) after three generations of rescue from Ag-NP exposure compared to the unexposed F0 and not significantly different (p = 0.585) from the F3 generation suggesting transgenerational inheritance of the H3K4me2 levels.
An increase in histone methylation levels may be explained by a down-regulation in the demethylase and/or up-regulation in the methyltransferase associated with the histone methylation mark. However a decrease in histone methylation may be explained by an up-regulation in the demethylase and/or a down-regulation in the methyltransferase. The expression level of spr-5, an H3K4me2 demethylase, is shown in Fig. 2b. No significant changes in spr-5 expression levels was observed for AgNO3 for all three generations analyzed despite the significant decrease in H3K4me2 levels observed after exposures at F3 when compared to F0. Though no significant down-regulation (p = 0.792) in spr-5 was observed after exposures to Ag-NP at F3, a significant down-regulation (p = 0.045) in spr-5 levels was observed after rescue from exposure (F6) when compared to the unexposed F0, which corresponds to the higher levels of H3K4me2 observed at F6. This also suggests transgenerational gene regulation, which can be achieved through the transfer of small non-coding RNAs to subsequent generations through the germline that may affect gene expression in subsequently unexposed populations (Rechavi and Lev, 2017) but this was not investigated in this study. There was a borderline significant up-regulation (p = 0.053) in spr-5 levels after three generations of exposure (F3) to sAg-NPs when compared to F0. The spr-5 expression levels recovered with a significant decrease (p = 0.024) from the F3 levels at F6 to unexposed F0 levels after rescue from the exposure. This correlates with the H3K4me2 levels which was significantly decreased after exposure and returned to the F0 levels after rescue.
When comparing treatments to control group, there were significant differences in H3K4me2 levels. However, since each treatment started with a different population of nematodes with potentially different histone methylation backgrounds, unexposed F0 within each treatment served as its respective “control’. In addition, there were no significant differences among F0, F3, and F6 generations in the levels of H3K4me2 in our main control group.
Expression levels of set-17 and set-30, H3K4 mono/dimethyltransferases, were also tested as their expression levels may explain the observed H3K4me2 levels in addition to the spr-5 expression levels. No significant differences in expression levels were observed for set-17, previously identified as mono/dimethyltransferase for H3K4 (Greer et al., 2014), after exposure to any of the three Ag forms or after recovery from the exposure (Fig. 2c). For set-30, no significant changes were observed among generations for AgNO3 and sAg-NP treatments after exposure and recovery. However, for Ag-NP treatment, a significant up-regulation (p = 0.001) in set-30 expression levels was observed after exposures (F3) as seen in Fig. 2d. Though recovery from the increased levels was observed at F6 with a significant decrease (p = 0.03) when compared to F3, the expression levels remain significantly higher (p = 0.019) than F0, again suggesting transgenerational gene regulation. These expression levels of set-30 correspond with the H3K4me2 levels observed for the Ag-NP treatment.
For the Ag-NP and sAg-NP treatments, correlation observed between the gene expression levels of the demethylases and methyltransferases with the global H3K4me2 levels provides another level of evidence to support the H3K4me2 levels observed. However, despite the significant decrease in H3K4me2 levels after exposures to AgNO3 treatment, none of the gene expression levels corresponded to the H3K4me2 trend. Given the good correlation between the expression levels of demethylases and methyltransferases for Ag-NP and sAg-NP treatments, it is possible that genes encoding other enzymes, which have not been identified, may be at play in the response of C. elegans to AgNO3. Our previous findings suggested that some of the mutigenerational reproductive toxicity was due to release of Ag+ ions from pristine Ag-NPs while mainly particle specific toxicity was indicated for sAg-NPs (Schultz et al., 2016). The epigenetic changes observed in this study seem to indicate that some of these modifications in pristine Ag-NP treatment are also particle-specific, which is possible, given their low dissolution of 1.5% in the SSPW exposure media.
Exposure of a single generation of C. elegans to another environmental toxin arsenic in arsenite form was shown to cause an increase in H3K4me2 levels and down-regulation of spr-5 in up to three subsequent unexposed generations (Yu and Liao, 2016), which also suggests inheritance of H3K4me2 levels or transgenerational regulation of spr-5 expression levels. In our study, this transgenerational effect was observed only for Ag-NP exposures. The trend in H3K4me2 was different for Ag-NPs compared to sAg-NPs. This suggests that an environmental transformation, such as sulfidation of pristine Ag-NPs, is likely to reduce their toxic effects by affecting histone methylation differently than pristine Ag-NPs. Genomic regions with high H3K4me2 levels have been linked with active transcriptional activity (Bernstein et al., 2002; Pokholok et al., 2005) and as such, the changes observed may affect gene expression in these nematodes.
3.2. Global H3K9me3 levels and expression of H3K9me3 demethylase and methyltransferase
Global H3K9me3 levels for the F0, F3, and F6 populations are shown in Fig. 3a. Exposure to AgNO3 did not cause any significant changes among the generations. In the case of Ag-NP exposure, an increase was observed at F3 when compared to F0 but was not significant (p = 0.114) and the levels recovered to almost F0 levels after recovery from exposure (F6). sAg-NP exposure caused a significant decrease (p = 0.002) in H3K9me3 levels after exposures (F3) when compared to the unexposed F0. Recovery was observed at F6 with a borderline significant increase (p = 0.052) in H3K9me3 levels when compared to F3 and not significantly different (p = 0.316) from F0 levels.
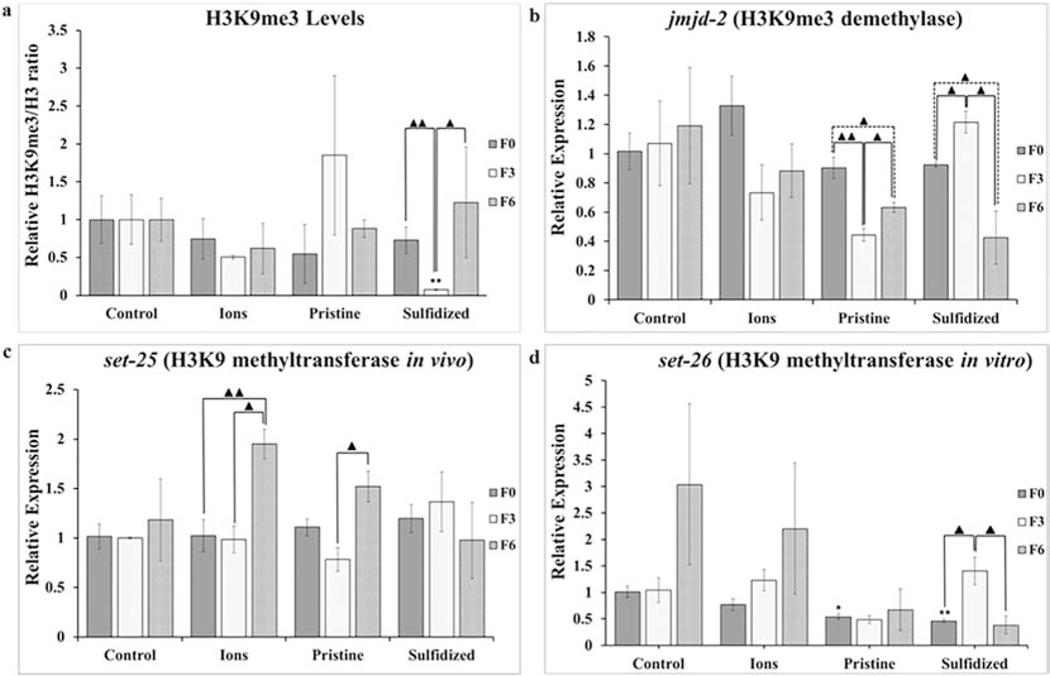
Fig. 3.
Changes in global levels of histone tri-methylation at H3K9 marker (a) and levels of gene expression at jmjd-2 (b), set-25 (c), and set-26 (d) after exposure of Caenorhabditis elegans to AgNO3 (Ions), pristine Ag-NPs (Ag-NP) and sulfidized Ag-NPs (sAg-NP). Bars represent standard error of the means. *, ** signifies difference with control at p ≤ 0.05 and p ≤ 0.01 respectively.▲, ▲▲signifies difference between generations of the same treatment at p ≤ 0.05 and p ≤ 0.01 respectively.
A significant down-regulation (p = 0.006) in H3K9me3 demethylase, jmjd-2, expression levels was observed after exposure to Ag-NPs at F3 (Fig. 3b). Even though a significant recovery (p = 0.027) was observed at F6, the levels still remained significantly lower (p = 0.029) compared to F0, again suggesting transgenerational gene regulation for Ag-NP exposure. Though no significant differences in H3K9me3 levels were observed for Ag-NP exposure, the trend observed correlates with the jmjd-2 expression levels. For transformed sAg-NPs, a significant up-regulation (p = 0.019) in jmjd-2 levels was observed after exposure in F3 when compared to the unexposed F0, which also supports the decrease in H3K9me3 levels. The jmjd-2 levels recovered in F6 with a significant increase (p = 0.016) when compared to F3. The recovery overcompensates such that the jmjd-2 levels in F6 were significantly lower than the F0 levels. This again correlates with the H3K9me3 levels observed in response to the sAg-NP exposure. In addition to serving as demethylase for H3K9me3, jmjd-2 is also demethylase for H3K36me3 (Camacho et al., 2018). The H3K36me3 levels were not measured in this study but the changes in the jmjd-2 levels may be indicative of the impact of Ag-NPs on H3K36me3 levels. The trends observed for the different treatments suggest that an environmental transformation of Ag-NPs alters their toxicity and toxicity mechanisms. For AgNO3 treatment, an insignificant down-regulation (p = 0.099) was observed after exposures in F3 compared to the F0 generations. No significant differences were detected among any of the AgNO3 generations for jmjd-2 expression levels as expected, based on the H3K9me3 levels, suggesting that exposure to AgNO3 does not affect H3K9me3 levels.
The expression levels of H3K9 methyltransferase set-25 are shown in Fig. 3c. No significant differences were observed in the expression levels after exposures at F3 compared to the unexposed F0 levels for all treatments. This suggests that the changes in H3K9me3 levels observed after exposures to Ag-NPs and sAg-NPs are primarily due to the changes observed for jmjd-2. Despite the significant down-regulation of jmjd-2 in F3 for Ag-NPs, the insignificant increase in H3K9me3 levels might be explained by the slight but insignificant down-regulation of set-25 (p = 0.091) observed after exposures. Surprisingly, after rescue from exposure at F6, there was a significant up-regulation of set-25 for AgNO3 when compared to F0 (p = 0.013) and F3 (p = 0.008) but this does not correlate with H3K9me3 levels. The recovery population of the Ag-NP exposed worms also showed a significant up-regulation of set-25 when compared to F3 (p = 0.019) but not F0 (p = 0.077). Some redundancy in the activity of the methyltransferases might be expected and may also play a role in histone methylation at other lysine sites, as has been shown for human histone methyltransferases (Volkel and Angrand, 2007). It has been shown previously that set-25 can mediate all three modifications at H3K9 including me1, me2 and me3(Padeken et al., 2019). Therefore, it is likely that set-25 expression levels in F6 in AgNO3 and Ag-NP treatments is due to the effect of the exposures on the methylation levels at the other two marks. This suggests that the changes observed in the H3K9me3 levels are primarily due to jmjd-2 expression levels as the expression pattern clearly matches the H3K9me3 changes observed for Ag-NP and sAg-NP exposures.
Though set-26 was shown to have H3K9 methyltransferase activity in vitro (Greer et al., 2014), it does not have the same activity in vivo. However, it plays a role in the regulation of lifespan, transgenerational fertility, development, and ageing (Ni et al., 2012; Wang et al., 2018). No significant differences in terms of the gene expression levels were observed among generations for the AgNO3 and Ag-NP treatments (Fig. 3d). Despite the increase observed at F6 for controls and AgNO3, there was a large amount of variation among replicates and the increases were not significant when compared to the corresponding F0 (p = 0.253 and p = 0.311 respectively) or F3 generations (p = 0.263 and p = 0.479 respectively). Exposure to sAg-NPs caused a significant up-regulation of set-26 (p = 0.022) after exposures at F3 compared to F0 (Fig. 3d). After recovery from exposure (F6), there was a significant down-regulation (p = 0.029) when compared to F3 but no significant difference (p =0.661) compared to the unexposed F0. This gene expression pattern of set-26 does not correspond with H3K9me3 levels observed for sAg-NPs suggesting that set-26 indeed may not have H3K9 methyltransferase activity in vivo. Interestingly, in our previous multigenerational study by Schultz et al. (Schultz et al., 2016), sAg-NPs showed a significant decrease in lifespan and set-26 is known to correlate negatively with lifespan (Ni et al., 2012).
4. Conclusions
The results of this study demonstrate that changes in epigenetic modifications, such as global levels of H3K4me2 and H3K9me3, are likely to play an important role in toxicity when C. elegans is exposed to Ag nanomaterials. These epigenetic changes can be inherited and may explain, at least in part, some of the transgenerational toxicity that has been observed (Schultz et al., 2016). The results also show that environmental transformation of Ag-NPs to sAg-NPs has an effect on the biological responses of C. elegans. While pristine Ag-NPs seem to cause an increase in levels of the histone methylation markers assessed, sAg-NPs have an opposite effect by decreasing the levels of the histone methylation markers. When compared to pristine Ag-NPs, AgNO3 may have a different mode of toxicity despite both treatments inducing transgenerational reproductive toxicity. This study examined changes in the global levels of histone methylations and future studies of the locus-specific histone methylation patterns as well as other histone methylation marks, implicated in transgenerational inheritance are warranted to gain a deeper understanding of epigenetic memory of the reproductive stress induced by the exposure to Ag nanomaterials.
Supplementary Material
Highlights.
Multigenerational exposure to Ag-NPs induces epigenetic changes
Changes in histone methylation differ between pristine and sulfidized Ag-NPs
Pristine Ag-NPs increase histone methylation at H3K4me2 marker
The histone methylation at H3K4me2 is inherited by unexposed offspring
Acknowledgements:
The authors gratefully acknowledge assistance of Jason Unrine, Shristi Shrestha, Stuart Lichtenberg, Tyler Bair, Jieran Li, and Zeinah El Baddar. Caenorhabditis elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the U.S. National Science Foundation (NSF) and U.S. Environmental Protection Agency (EPA) under EF-0830093, DBI-1266252, and CBET-1530594. This work was also supported by the USDA National Institute of Food and Agriculture Multistate Project NC 1194. Any opinions, findings, conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NSF or the EPA. This work has not been subjected to EPA review and no official endorsement should be inferred. AH was supported by Summer Undergraduate Research in Environmental Sciences (SURES) funded by the National Institute of Environmental Health Sciences (NIEHS) R25ES027684.
Footnotes
Credit Author Statement
Anye Wamucho: Methodology, Investigation, Writing - Original draft, Formal Analysis. Allison Heffley: Investigation. Olga Tsyusko: Supervision, Conceptualization, Methodology, Writing - Reviewing and Editing.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflicts of Interest
The authors declare that they have no conflicts of interests with respect to this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, and Schreiber SL (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proceedings of the National Academy of Sciences 99, 8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho J, Truong L, Kurt Z, Chen YW, Morselli M, Gutierrez G, Pellegrini M, Yang X, and Allard P. (2018). The Memory of Environmental Chemical Exposure in C. elegans Is Dependent on the Jumonji Demethylases jmjd-2 and jmjd-3/utx-1. Cell reports 23, 2392–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Yin L, Lin S, Wiesner M, Bernhardt E, and Liu J. (2011). Toxicity reduction of polymer-stabilized silver nanoparticles by sunlight. The Journal of Physical Chemistry C 115, 4425–4432. [Google Scholar]
- Diez-Ortiz M, Lahive E, George S, Ter Schure A, Van Gestel CA, Jurkschat K, Svendsen C, and Spurgeon DJ (2015). Short-term soil bioassays may not reveal the full toxicity potential for nanomaterials; bioavailability and toxicity of silver ions (AgNO(3)) and silver nanoparticles to earthworm Eisenia fetida in long-term aged soils. Environmental pollution (Barking, Essex : 1987) 203, 191–198. [DOI] [PubMed] [Google Scholar]
- Dong X, and Weng Z. (2013). The correlation between histone modifications and gene expression. Epigenomics 5, 113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Aguilera C, Palladino F, and Askjaer P. (2014). C. elegans epigenetic regulation in development and aging. Briefings in functional genomics 13, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, Aristizabal-Corrales D, Chen S, Badeaux AI, Jin Q, et al. (2014). A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell reports 7, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, Hsu CH, Aravind L, He C, and Shi Y. (2015). DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider A, and Kang I-K (2015). Preparation of Silver Nanoparticles and Their Industrial and Biomedical Applications: A Comprehensive Review. Advances in Materials Science and Engineering 2015, 1–16. [Google Scholar]
- Jesmer AH, Velicogna JR, Schwertfeger DM, Scroggins RP, and Princz JA-O The toxicity of silver to soil organisms exposed to silver nanoparticles and silver nitrate in biosolids-amended field soil. [DOI] [PubMed] [Google Scholar]
- Juganson K, Ivask A, Blinova I, Mortimer M, and Kahru A. (2015). NanoE-Tox: New and in-depth database concerning ecotoxicity of nanomaterials. Beilstein J Nanotechnol 6, 1788–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaegi R, Voegelin A, Sinnet B, Zuleeg S, Hagendorfer H, Burkhardt M, and Siegrist H. (2011). Behavior of Metallic Silver Nanoparticles in a Pilot Wastewater Treatment Plant. Environmental science & technology 45, 3902–3908. [DOI] [PubMed] [Google Scholar]
- Keller A, McFerran S, Lazareva A, and Suh S. (2013). Global life cycle releases of engineered nanomaterials. Journal of Nanoparticle Research 15, 1–17. [Google Scholar]
- Kishimoto S, Uno M, Okabe E, Nono M, and Nishida E. (2017). Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nature communications 8, 14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, and Lehner B. (2017). Transgenerational transmission of environmental information in C. elegans. Science 356, 320–323. [DOI] [PubMed] [Google Scholar]
- Levard C, Hotze EM, Lowry GV, and Brown GE Jr. (2012). Environmental transformations of silver nanoparticles: impact on stability and toxicity. Environmental science & technology 46, 6900–6914. [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung M-S, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. (2011). Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry GV, Gregory KB, Apte SC, and Lead JR (2012). Transformations of nanomaterials in the environment. Environmental science & technology 46, 6893–6899. [DOI] [PubMed] [Google Scholar]
- Mélanie A, Jérôme R, Jean-Yves B, Gregory VL, Jean-Pierre J, and Mark RW (2009). Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nature Nanotechnology 4, 634. [DOI] [PubMed] [Google Scholar]
- Mueller N, and Nowack B. (2008). Exposure Modeling of Engineered Nanoparticles in the Environment. Environmental science & technology 42, 4447–4453. [DOI] [PubMed] [Google Scholar]
- Neher DA, and Powers TO (2005). NEMATODES. In Encyclopedia of Soils in the Environment, Hillel D, ed. (Oxford: Elsevier; ), pp. 1–6. [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, and Lee SS (2012). Two SET-domain containing genes link epigenetic changes and aging in C. elegans. Aging Cell 11, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Zeller P, Towbin B, Katic I, Kalck V, Methot SP, and Gasser SM (2019). Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev 33, 436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panacek A, Prucek R, Safarova D, Dittrich M, Richtrova J, Benickova K, Zboril R, and Kvitek L. (2011). Acute and chronic toxicity effects of silver nanoparticles (NPs) on Drosophila melanogaster. Environmental science & technology 45, 4974–4979. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45-e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527. [DOI] [PubMed] [Google Scholar]
- Raj A, Shah P, and Agrawal N. (2017). Dose-dependent effect of silver nanoparticles (AgNPs) on fertility and survival of Drosophila: An in-vivo study. PloS one 12, e0178051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CNR, Kulkarni GU, Thomas PJ, and Edwards PP (2002). Size- Dependent Chemistry: Properties of Nanocrystals. Chemistry – A European Journal 8, 28–35. [DOI] [PubMed] [Google Scholar]
- Rechavi O, and Lev I. (2017). Principles of transgenerational small RNA nnheritance in Caenorhabditis elegans. In Curr. Biol, pp. R720–R730. [DOI] [PubMed] [Google Scholar]
- Salata OV (2004). Applications of nanoparticles in biology and medicine. Journal of nanobiotechnology 2, 3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CL, Wamucho A, Tsyusko OV, Unrine JM, Crossley A, Svendsen C, and Spurgeon DJ (2016). Multigenerational exposure to silver ions and silver nanoparticles reveals heightened sensitivity and epigenetic memory in Caenorhabditis elegans. Proceedings Biological sciences 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoults-Wilson WA, Reinsch Bc Fau - Tsyusko OV, Tsyusko Ov Fau - Bertsch PM, Bertsch Pm Fau - Lowry GV, Lowry Gv Fau - Unrine JM, and Unrine JM. Effect of silver nanoparticle surface coating on bioaccumulation and reproductive toxicity in earthworms (Eisenia fetida). [DOI] [PubMed] [Google Scholar]
- Starnes DL, Lichtenberg SS, Unrine JM, Starnes CP, Oostveen EK, Lowry GV, Bertsch PM, and Tsyusko OV (2016). Distinct transcriptomic responses of Caenorhabditis elegans to pristine and sulfidized silver nanoparticles. Environ Pollut 213, 314–321. [DOI] [PubMed] [Google Scholar]
- Starnes DL, Unrine JM, Starnes CP, Collin BE, Oostveen EK, Ma R, Lowry GV, Bertsch PM, and Tsyusko OV (2015). Impact of sulfidation on the bioavailability and toxicity of silver nanoparticles to Caenorhabditis elegans. Environ Pollut 196, 239–246. [DOI] [PubMed] [Google Scholar]
- Tyne W, Lofts S, Spurgeon DJ, Jurkschat K, and Svendsen C. (2013). A new medium for Caenorhabditis elegans toxicology and nanotoxicology studies designed to better reflect natural soil solution conditions. Environmental toxicology and chemistry 32, 1711–1717. [DOI] [PubMed] [Google Scholar]
- Vance ME, Kuiken T, Vejerano EP, McGinnis SP, Hochella MF, Rejeski D, and Hull MS (2015). Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory. Beilstein J Nanotechnol 6, 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicogna JR, Ritchie EE, Scroggins RP, and Princz JI (2016). A comparison of the effects of silver nanoparticles and silver nitrate on a suite of soil dwelling organisms in two field soils. Nanotoxicology 10, 1144–1151. [DOI] [PubMed] [Google Scholar]
- Volkel P, and Angrand PO (2007). The control of histone lysine methylation in epigenetic regulation. Biochimie 89, 1–20. [DOI] [PubMed] [Google Scholar]
- Waalewijn-Kool P, Klein K, Forniés R, and Gestel C. (2014). Bioaccumulation and toxicity of silver nanoparticles and silver nitrate to the soil arthropod Folsomia candida. Ecotoxicology 23, 1629–1637. [DOI] [PubMed] [Google Scholar]
- Wamucho A, Unrine JM, Kieran TJ, Glenn TC, Schultz CL, Farman M, Svendsen C, Spurgeon DJ, and Tsyusko OV (2019). Genomic mutations after multigenerational exposure of Caenorhabditis elegans to pristine and sulfidized silver nanoparticles. Environmental Pollution 254, 113078. [DOI] [PubMed] [Google Scholar]
- Wang W, Chaturbedi A, Wang M, An S, Santhi Velayudhan S, and Lee SS (2018). SET-9 and SET-26 are H3K4me3 readers and play critical roles in germline development and longevity. eLife 7, e34970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong BSE, Hu Q, and Baeg GH (2017). Epigenetic modulations in nanoparticle-mediated toxicity. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 109, 746–752. [DOI] [PubMed] [Google Scholar]
- Yang X, Jiang C, Hsu-Kim H, Badireddy AR, Dykstra M, Wiesner M, Hinton DE, and Meyer JN (2014). Silver nanoparticle behavior, uptake, and toxicity in Caenorhabditis elegans: effects of natural organic matter. Environ Sci Technol 48, 3486–3495. [DOI] [PubMed] [Google Scholar]
- Yu CW, and Liao VH (2016). Transgenerational reproductive effects of arsenite are associated with H3K4 dimethylation and SPR-5 downregulation in Caenorhabditis elegans. Environmental science & technology 50, 10673–10681. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen D, Smith MA, Zhang B, and Pan X. (2012). Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PloS one 7, e31849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.