Abstract
Pseudomonas aeruginosa is a common nosocomial pathogen and a major cause of morbidity and mortality in hospitalized patients. Multiple reports highlight that P. aeruginosa gastrointestinal colonization may precede systemic infections by this pathogen. Gaining a deeper insight into the dynamics of P. aeruginosa gastrointestinal carriage is an essential step in managing gastrointestinal colonization and could contribute to preventing bacterial transmission and progression to systemic infection. Here, we present a clinically relevant mouse model relying on parenteral vancomycin pretreatment and a single orogastric gavage of a controlled dose of P. aeruginosa. Robust carriage was observed with multiple clinical isolates, and carriage persisted for up to 60 days. Histological and microbiological examination of mice indicated that this model indeed represented carriage and not infection. We then used a barcoded P. aeruginosa library along with the sequence tag-based analysis of microbial populations (STAMPR) analytic pipeline to quantify bacterial population dynamics and bottlenecks during the establishment of the gastrointestinal carriage. Analysis indicated that most of the P. aeruginosa population was rapidly eliminated in the stomach, but the few bacteria that moved to the small intestine and the caecum expanded significantly. Hence, the stomach constitutes a significant barrier against gastrointestinal carriage of P. aeruginosa, which may have clinical implications for hospitalized patients.
INTRODUCTION
In 2019, one in eight global deaths were attributable to bacterial infections (1). A handful of bacteria were responsible for half of these deaths, including Pseudomonas aeruginosa, which causes a wide range of healthcare-associated infections such as pneumonia, bloodstream infections, wound or surgical site infections, and urinary tract infections (1, 2). These infections are especially life-threatening for individuals who are hospitalized, immunocompromised, or have chronic lung diseases. In addition to being one of the leading causes of nosocomial infections (3), P. aeruginosa is also highly resistant to antimicrobial agents, making these infections difficult to treat (4). Some P. aeruginosa isolates are resistant to nearly all available antibiotics, including carbapenems, which has led to the CDC classifying multidrug-resistant P. aeruginosa as a serious threat (5).
P. aeruginosa is rarely part of the gastrointestinal (GI) microbiome of healthy individuals (4%) (6); however, it more efficiently colonizes patients in the intensive care unit (ICU) (10–55%) (6–8), with cancer (31%-74% of hospitalized patients) (9, 10), or in long-term care facilities (52%) (11). Importantly, GI carriage of P. aeruginosa is a key risk factor for subsequent development of infection (7, 12–14). As part of a prospective study, Gómez-Zorrilla et al. determined that after a 14-day stay in the ICU the probability of developing a P. aeruginosa infection was 26% for carriers versus 5% for noncarriers (13). In addition to the risk for infection, GI carriage can facilitate transmission of P. aeruginosa to other patients (15, 16). Thus, gaining a deeper understanding of P. aeruginosa GI carriage is crucial to prevent infections, manage rising rates of antibiotic resistance, and improve overall patient safety in healthcare settings.
While several animal models are available for the investigation of P. aeruginosa virulence and dissemination (17–24), fewer models have focused on GI carriage. In patients, antibiotic use has been correlated with an increased risk of P. aeruginosa GI colonization (12, 25–27). Previous studies have exploited this correlation to develop animal models that have provided important information on how P. aeruginosa establishes carriage. However, these models have relied on extended exposure to P. aeruginosa or the use of immunocompromised mice (18, 28, 29). It is estimated that around two-thirds of patients in the ICU are immunocompetent (30, 31). By using antibiotic pretreatment and immunocompetent animals, we aimed to develop an animal model of P. aeruginosa GI carriage that better mimics a typical ICU patient.
Here, we describe a murine model of P. aeruginosa GI carriage that is facilitated by the daily intraperitoneal (IP) injection of vancomycin for seven days and by a single dose of P. aeruginosa delivered via oral gavage. With this model, robust GI carriage was observed in both female and male mice, occurred with multiple clinical P. aeruginosa isolates, and persisted for up to 60 days. Additionally, to investigate the population dynamics of GI carriage, we used barcoded P. aeruginosa bacteria and determined that the stomach constituted a major barrier against GI carriage of P. aeruginosa. Bacteria that passed through the stomach were able to efficiently replicate in the small intestine and caecum, facilitating excretion of high numbers of P. aeruginosa. These barcoding experiments yielded interesting insights into the dynamics of P. aeruginosa GI carriage.
RESULTS
Vancomycin promotes gastrointestinal carriage of P. aeruginosa
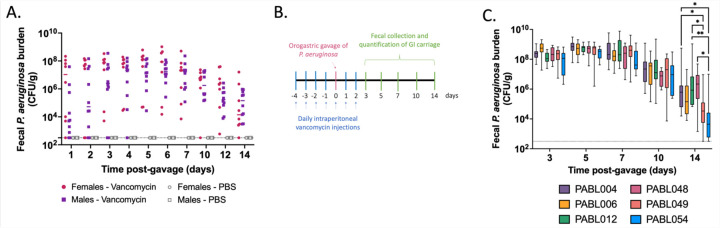
Our goal was to develop a clinically relevant animal model that recapitulates the asymptomatic P. aeruginosa GI carriage observed in hospitalized patients. First, we tested the ability of the P. aeruginosa clinical isolate PABL048, delivered by orogastric gavage, to be carried in the GI tract of mice in the absence of antibiotic pretreatment. The extent of the carriage was assessed by fecal collection and plating on selective medium followed by CFU enumeration. Following pretreatment with IP phosphate-buffered saline (PBS), GI carriage of P. aeruginosa did not occur (Fig. 1A). Because antibiotic exposure correlates with an increased risk of GI colonization in patients (12, 25–27), we investigated the effect of IP vancomycin injection on the carriage of P. aeruginosa in mice. Vancomycin was chosen because it is one of the most commonly used antibiotics in the U.S. (32, 33) and does not have activity against P. aeruginosa (34). Three regimens of IP vancomycin treatment combined with orogastric gavage of P. aeruginosa were tested (all with a daily dose of 370 mg/kg of vancomycin – equivalent to a human dose of 30 mg/kg (35)). All three regimens supported the carriage of P. aeruginosa (Supplemental Fig.1). While vancomycin pretreatment for either 3 or 5 days prior to the orogastric gavage led to similar levels of GI carriage of P. aeruginosa, higher fecal burdens were observed when vancomycin injections were continued for two days after the orogastric gavage (Supplemental Fig.1). For all subsequent experiments, we therefore chose a regimen consisting of vancomycin on days −4 to −1, vancomycin and P. aeruginosa on day 0, and vancomycin on days +1 and +2 (Fig. 1B), which cumulatively corresponds to a typical 7-day course of vancomycin commonly prescribed to patients (36). When mice were treated with this regimen of vancomycin and challenged with 105.6 CFU of the P. aeruginosa isolate PABL048, bacterial shedding averaged 106-108 CFU/g of feces during the first week (Fig. 1A). Although recovered CFU decreased somewhat during the second week post-inoculation, carriage levels remained between 103 and 107 CFU/g of feces. GI carriage of P. aeruginosa was similar in male and female mice (Fig. 1A).
Figure 1: Murine model of P. aeruginosa GI carriage.
(A) PABL048 fecal burden after an orogastric gavage with 105.6 CFU. Male (square) or female (circle) mice received either PBS (gray symbols) or vancomycin (colored symbols) injections, and an orogastric gavage with 105.6 CFU of PABL048. The experiment was performed twice for vancomycin treated mice (combined results are shown; n ≥ 8), and once for PBS treated mice (n = 5). Each symbol represents one mouse. Solid horizontal lines indicate medians. No significant differences in fecal CFU were found at any time point between male and female mice (multiple t-tests). (B) Timeline schematic of the model. Mice were intraperitoneally injected daily with vancomycin for 7 days at a dose of 370 mg/kg. On the fifth day of vancomycin treatment (day 0), mice received a defined dose of P. aeruginosa through orogastric gavage. On selected days, feces were collected to assess the extent of GI carriage, estimated by CFU counts. (C) Fecal burden of six clinical isolates of P. aeruginosa during GI carriage. Vancomycin and bacterial delivery (inoculum sizes: 105.4+/-0.2 CFU PABL004, 105.6+/-0.3 CFU PABL006, 106+/-0.2 CFU PABL012, 105.4+/-0.1 CFU PABL048, 106+/-0.1 PABL049 or 106+/-0.1 CFU PABL054) were performed as in B. Box plots are shown with boxes extending from the 25th to 75th percentiles, whiskers representing minimum and maximum values and lines indicating medians. Experiments were performed at least twice, and combined results are shown (n ≥ 10). *p ≤ 0.05, **p ≤ 0.01 (t-tests with Holm-Sidak correction for multiple comparisons). The dotted line indicates the limit of detection.
A substantial proportion of the P. aeruginosa genome is accessory (i.e., varies from strain to strain) (37). To examine whether these genomic differences allowed some strains of P. aeruginosa to establish higher or lower levels of GI carriage in this model, we individually inoculated mice with six clinical isolates: PABL004, PABL006, PABL012, PABL048, PABL049 and PABL054. These isolates are genetically diverse and exhibit differing levels of virulence in a bloodstream infection model (38) (Supplementary Table 1). Despite these differences, bacterial loads detected in the feces were similar for all strains over the first 10 days of the experiment (Fig. 1C). While more variability was observed on day 14, persistent carriage of all strains was detected. These results, taken together with the establishment of GI carriage in both sexes, demonstrate that vancomycin pretreatment produces a robust and reliable model to investigate P. aeruginosa carriage.
GI carriage of P. aeruginosa does not cause GI inflammation
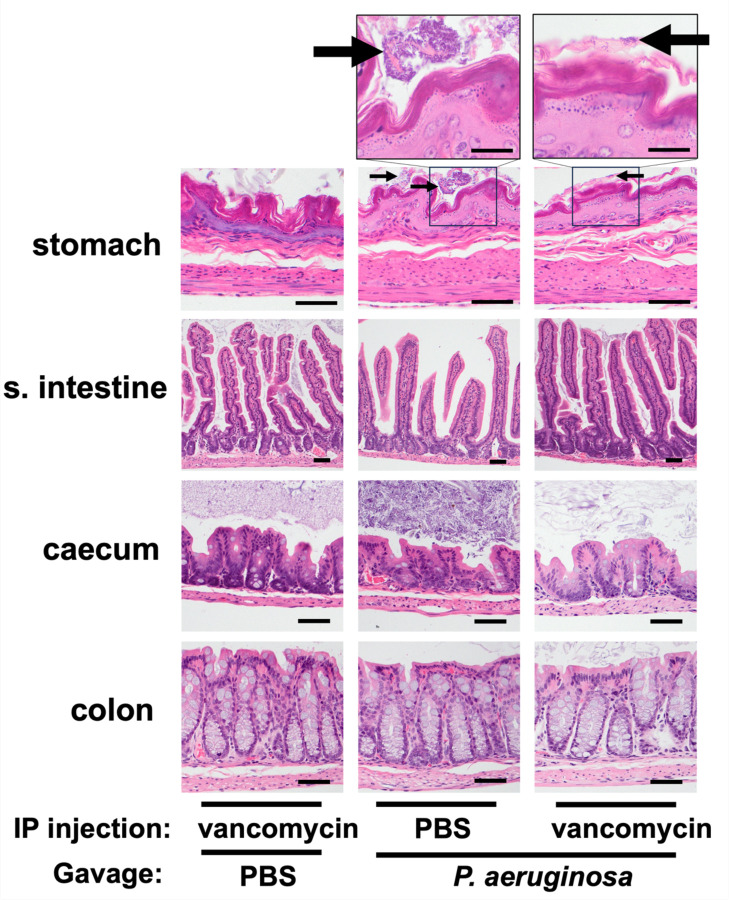
Carriage may be distinguished from infection by the absence of inflammation. To examine the vancomycin-treatment model represented true carriage, we performed histopathological analyses of the GI tract tissues. Mice received daily IP injections of vancomycin or PBS (day −4 to day +2) and were gavaged with either 107.1 CFU of PABL048 or PBS (mock) on day 0. We chose to expose mice to a higher dose of bacteria than used in the previous experiments to maximize the possibility of observing inflammation. On day 3 post-orogastric gavage, mice were sacrificed and organs from the GI tract were harvested for histopathological examination. Organ sections were stained with hematoxylin-eosin (H&E) and screened for inflammation as evidenced by the presence of inflammatory cells or tissue damage. Neither of these were observed in any of the samples, and each section of the GI tract remained histologically normal (Fig. 2). Multifocal clusters of bacteria were observed adjacent to the mucosal surface of the stomach of all 3 mice that received IP PBS treatment and orogastric delivery of P. aeruginosa. These bacteria were primarily rod-shaped, compatible with P. aeruginosa morphology (39), but it was unclear whether they were dead or alive. Despite the presence of these bacteria in the stomach of PBS-treated mice three days after inoculation, P. aeruginosa bacteria were not cultured from their feces (Supplemental Fig. 2). Among mice treated with vancomycin prior to the bacterial inoculation, only one mouse exhibited bacteria adjacent to the surface mucosa of the stomach. While all mice that received vancomycin treatment and P. aeruginosa had feces that grew this bacterium (Supplemental Fig. 2), no bacteria were observed within the bowel walls of the small intestine, the caecum, or the colon of these same animals (Fig. 2), suggesting that P. aeruginosa remained in the lumen of the GI tract and did not invade the intestinal wall. In addition to these histopathological observations, mice exhibited no signs of systemic illness (e.g., decreased activity, ruffled fur) at any point during the experiment. Taken together, these results suggest that this is a model of GI carriage of P. aeruginosa rather than infection.
Figure 2: Tissue histology of the GI tracts of mice with carriage of P. aeruginosa.
Hematoxylin-eosin staining of organ tissues collected at day 3 post-inoculation with either 107.1 CFU of PABL048 or PBS (Mock). Prior to the orogastric gavage, mice received either vancomycin or PBS through intraperitoneal injections. Images in the bottom 4 rows were captured at a 400x magnification (bar = 100 µm). Images on the top row were captured at a 1,000x magnification (bar = 200 µm). (n = 3–4 mice/group). Arrows indicate intraluminal clumps of bacterial bacilli.
P. aeruginosa bacteria remain largely within the GI tract
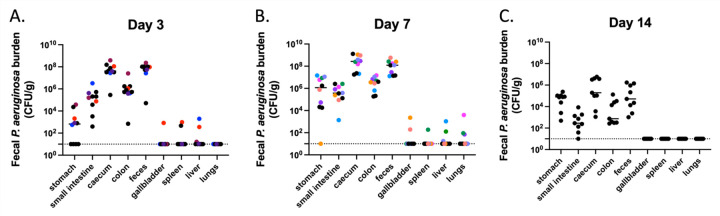
Since the intestinal tract has previously been identified as the main source of P. aeruginosa for the development of infections in immunocompromised patients (12, 40, 41), we assessed whether this model resulted in dissemination of P. aeruginosa to other tissues. Mice were orogastrically inoculated with 107.4 CFU of PABL048, and P. aeruginosa CFU were enumerated from various organs at days 3, 7 and 14 post gavage. Most of the bacteria were detected in the organs of the GI tract, including the stomach, small intestine, caecum, colon, and feces (Fig. 3). Nevertheless, P. aeruginosa was occasionally detected in the gallbladder, spleen, liver, or lungs within the first 7 days post gavage. This suggests that, in this model, escape of bacteria from the gut, while very infrequent, did occasionally occur in the absence of observable signs of systemic illness. However, by two weeks post-inoculation, we did not observe bacteria in any systemic site of the mice. In summary, dissemination of P. aeruginosa from the GI tract is rare in this model of GI carriage.
Figure 3: Dissemination of P. aeruginosa from the GI tract.
P. aeruginosa burden in organs of mice carrying PABL048. Mice were sacrificed at (A) day 3 (n = 10), (B) day 7 (n = 10) or (C) day 14 (n = 9) post-orogastric gavage with 107.4+/-0.2 CFU of PABL048, and bacterial CFU in the organs were enumerated by plating. The experiment was performed twice; combined results are shown. Each symbol represents one mouse. Solid horizontal lines indicate medians. The vertical dashed lines separate GI tract organs (left) from other organs (right). Symbols representing mice with dissemination from the GI tract are colored (one color/mouse). The horizontal dotted line indicates the limit of detection.
P. aeruginosa establishes long-term carriage
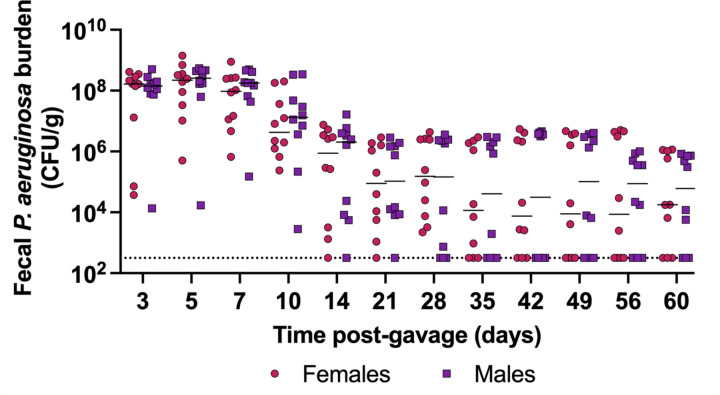
To further characterize this model, we interrogated the duration of carriage following a single orogastric gavage of P. aeruginosa. When inoculated with 105.7 CFU of PABL048, all mice carried P. aeruginosa in their GI tract for at least 10 days (Fig. 4). At day 60, 70% of all mice (7/10 females, 7/10 males) were still shedding P. aeruginosa from their GI tract. These results show that, in this model, P. aeruginosa establishes long-term GI carriage following a single exposure.
Figure 4: Long-term carriage of P. aeruginosa in the GI tract.
PABL048 fecal burdens after an orogastric gavage with 105.7+/-0.3 CFU in male (purple squares) and female (red circles) mice. The experiment was performed twice; combined results are shown (n = 10). Each symbol represents one mouse. Solid horizontal lines indicate medians. No significant differences were found between male and female mice at any time point (multiple t-tests). The dotted line indicates the limit of detection.
The stomach constitutes the main bottleneck of GI carriage
Investigation across the segments of the GI tract indicated that not all tissues supported the same levels of P. aeruginosa carriage (Fig. 3). Thus, we sought to examine the GI carriage dynamics following orogastric inoculation with P. aeruginosa. In particular, we wanted to identify which segments of the GI tract contributed to population bottlenecks or supported bacterial expansion in this model. The sequence tag-based analysis of microbial populations (STAMP) technique (42), which relies on the generation of a bacterial library with insertions of short, random nucleotide DNA tags into a neutral site of the chromosome, is ideal for this purpose. Animals are inoculated with this library, and barcode frequency and diversity at different locations and times post inoculation are interpreted using the refined framework of STAMP (known as “STAMPR”) (43). This analysis estimates the size of the founding population (Ns), defined as the number of bacterial cells from the inoculum that successfully passed through physical, chemical and immune barriers in the host to establish the population at the site of infection. A low Ns value (a small number of unique barcodes) is indicative of a tight bottleneck, while a high Ns value (a large number of unique barcodes) is reflective of a wide bottleneck. Comparison of CFU obtained from a tissue with the Ns value provides insight into the extent of bacterial expansion; for example, high CFU could be obtained from 1) a wide bottleneck followed by little bacterial replication or 2) a tight bottleneck followed by extensive replication of a small number of founders.
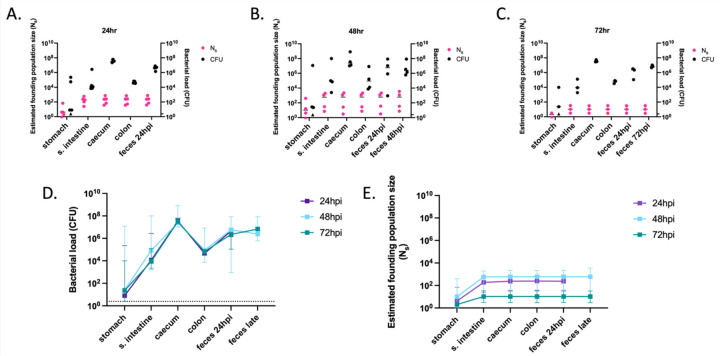
We applied STAMP to the vancomycin-treated mouse model. We used a previously generated barcoded library in the P. aeruginosa clinical isolate PABL012 (~6,000 unique tags, each ~30 bp) that had been validated by Bachta and colleagues (designated “PABL012pool”) (44). Note that fecal shedding of PABL012 was similar to that of PABL048 in the vancomycin-treated mouse model (Fig. 1C). Because we observed stable GI carriage of PABL012 between days 3 and 7, we deduced that major steps of GI carriage establishment were likely to occur within the first 3 days following inoculation. Using the vancomycin-treated mouse model, we delivered 106.1 CFU of the PABL012pool library through orogastric gavage and collected and analyzed segments of the GI tract (stomach, small intestine, caecum, colon, and feces) at 24, 48, or 72 hours post-inoculation (hpi).
As previously observed with PABL048 (Fig. 3), all organs of the GI tract supported the carriage of PABL012pool (Fig. 5A-D). The stomach was the organ with the largest variation in total CFU recovered (Fig. 5 A-D); while P. aeruginosa was no longer recovered from the stomach of some mice, others carried 104–5 CFU. The caecum and the feces had the highest bacterial burdens during the first 3 days of GI carriage. Median CFU loads recovered from all sites were stable over the first 3 days (Fig. 5D), suggesting that GI carriage is established during the first 24 hours following P. aeruginosa delivery and maintained for the next two days.
Figure 5: Founding populations and bacterial loads of P. aeruginosa in the GI tract.
A total of 106.1 CFU of PABL012pool were delivered to single-caged mice by orogastric gavage. P. aeruginosa CFU in 250 µL of resuspensions of collected homogenized tissues (one-fourth of tissue homogenates of stomach, small intestine [“s. intestine”], caecum, colon, and feces) were enumerated by plating, and founding population sizes (Ns) were estimated using the STAMPR approach. (A-C) Bacterial loads (CFU, black circles) and estimated founding population sizes (Ns, pink circles) were quantified at (A) 24 (n = 5), (B) 48 (n = 4) and (C) 72 hpi (n = 3, except for Ns in stomach, which was n = 2 due to a sequencing issue). Each circle represents an organ from one mouse. Solid horizontal lines indicate medians. Minor ticks on the right Y axis represent the limits of detection for the CFU. Triangles represent samples with no recovered CFU. (D) P. aeruginosa burdens and (E) estimated founding population sizes in different tissues of the GI tract at 24 (purple), 48 (blue) and 72 (green) hpi. For comparison, fecal samples were collected at 24 hpi (“feces 24 hpi”) regardless of the ending timepoint. An additional terminal fecal sample was available for animals harvested at 48 or 72 hpi (“feces late”). Squares represent medians, and error bars represent 95% confidence intervals. The dotted line indicates the limit of detection for CFU. Ns values are not significantly different over time (t-test).
In all organs at all time points, Ns values were low, indicating that a tight bottleneck was encountered by P. aeruginosa following inoculation (Fig. 5A-C, E). Ns values were the lowest in the stomach, with median values below 10 for all three time points. Therefore, nearly all the bacteria initially inoculated into the stomach were either killed or expelled to the small intestine within the first 24 hours. The higher Ns values observed in the distal GI tract suggest that certain clones passed through the stomach but successfully established themselves further along the GI tract. By looking at the inoculum passage through the GI tract at early timepoints (1h and 6h post gavage), we confirmed that P. aeruginosa bacteria were mostly killed in the proximal GI tract rather than rapidly passaged to the distal GI tract and expelled in the feces (Supplemental Fig. 3). The nearly identical founding population sizes at each time point in the distal GI tract indicate that these segments are quite permissive for P. aeruginosa carriage (Fig. 5E). Taken together, these data suggest that nearly all P. aeruginosa bacteria are rapidly (in less than 6 hours) eliminated from the stomach and that a small number of bacteria pass through to the small intestine and downstream segments of the GI tract.
The small intestine and caecum support high replication of P. aeruginosa
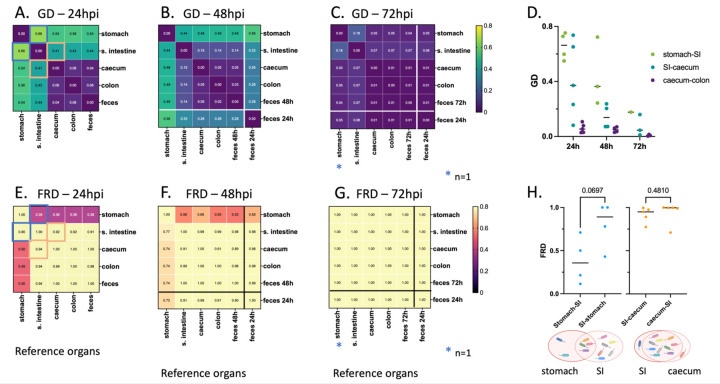
The large CFU counts and corresponding small founding populations in different organs highlighted the ability of P. aeruginosa to replicate in the GI tract (Fig. 5). For each segment of the GI tract, we defined net replication (which includes the combined effects of replication, death, and migration) as the ratio between CFU and Ns. The greatest expansion of P. aeruginosa from a small founding population occurred in the caecum with CFU/Ns ratio greater than 105, but substantial expansion was also observed in the stomach, small intestine and colon (Supplemental Fig. 4). Theoretically, high CFU/Ns could occur solely by local bacterial multiplication, by migration of bacteria en masse from an adjacent portion of the GI tract, or a combination of these two processes. Local multiplication would yield compartmentalized regions of the intestine, where different clones are spatially segregated along the length of the GI tract. In contrast, movement of bacterial populations along the GI tract would yield more similar barcode distributions between regions of the intestine. To distinguish between these possibilities, we quantified the genetic relatedness of P. aeruginosa populations in each segment of the GI tract. Genetic relatedness is determined by comparing the genetic distance (GD) of barcode distributions between two populations (45). GD varies from 0 to 0.9, with low values indicating highly similar barcode distributions between two samples and high values indicating different barcode distributions between samples. For all time points, the caecum, the colon, and the feces contained, on average, highly similar barcode populations of P. aeruginosa (GD ≤ 0.06) (Fig. 6 A-D, dark purple). Additionally, the GD between the small intestine and the caecum, colon and feces decreased over time (Fig. 6D, teal). These findings, along with the high CFU/Ns values observed in the distal GI tract (Supplemental Fig. 4), are consistent with a model in which P. aeruginosa, despite not being viewed as an enteric bacterium, multiplies rapidly and to high numbers in the caecum or the small intestine. These large populations of bacteria then move to the colon and are subsequently expelled in the feces. They also support the conclusion that P. aeruginosa bacteria recovered from fecal samples are most representative of those carried in the distal GI tract and confirm the utility of using fecal sampling to study P. aeruginosa carriage.
Figure 6: Average intra-mouse genetic relatedness of P. aeruginosa populations in the GI tract.
(A-C) Heatmaps representing the average intra-mouse genetic distances (GDs) of P. aeruginosa from organs of the mice described in figure 5, at (A) 24, (B) 48, and (C) 72 hpi. Lower values of GD (purple) indicate a higher frequency of barcode sharing between the samples, with 0 reflecting identical populations. (D) GD values over time between: stomach and small intestine (“SI”) (green), small intestine and caecum (teal), and caecum and colon (purple). Each symbol represents one mouse. Lines indicate medians. (E-G) Heatmaps representing the average intra-mouse Fractional Resilient Genetic Distances (FRD) of P. aeruginosa from organs of the mice described in figure 5, at (E) 24, (F) 48 and (G) 72 hpi. The FRD is calculated using the following formula: where RDA-B is the number of shared barcodes that contribute to genetic similarity between samples A and B. The column names in the FRD heatmaps correspond to the organ of reference (B in the above formula). High FRD values (yellow) indicate that most bacterial barcodes are shared between samples. Thick lines in panels B-C, F-G separate the samples collected at the time of dissection (top/left) from samples of feces collected from the same animals at an earlier time point (“feces 24h”) (bottom/right). Samples outlined by blue and orange squares in panels A and E indicate pairs that are detailed in panel H. (H) FRD values for bacteria from the stomach/small intestine (SI) (blue) and small intestine/caecum (orange) pairs at 24 hpi. Each symbol represents one mouse. Lines indicate medians. p-values are indicated (two-tailed paired t test). The Venn diagrams under the graph are visual representations of the averaged proportion of barcodes shared between two adjacent organs (circles). Diagrams created using Biorender.com. As observed in figure 5, no bacteria could be detected in the stomach of some mice, leading to variation in the number of samples used for this analysis: A, E, H: n = 5 (except for the stomach; n = 4), B, F: n = 4 (except for the stomach; n = 3), C, G: n = 3 (except for the stomach; n = 1), D: see panels A-C.
Genetic similarity between two sites can be achieved through different population patterns. For example, two GI segments can be highly similar owing to a single dominant clone shared between both sites or due to underlying sharing of hundreds of clones, each with low abundance. To define the number of clones that contribute to genetic similarity, we calculated a metric known as “resilient” genetic distance (RD), which quantifies the number of shared clones that contribute to genetic similarity between two samples (0.8 threshold, see Methods) (43). Genetically similar samples with many shared clones have high RD values, whereas genetically similar samples which share only a few clones have low RD values.
The interpretation of whether RD is “low” or “high” is relative to the number of barcodes in a sample. To normalize RD values, the natural logarithm (ln) of RD is divided by the ln of the number of distinct barcodes, creating a fractional RD (FRD). The FRD represents the number of shared barcodes in a pair of samples (samples A and B) relative to the number of distinct barcodes in a reference sample (sample B) (43). For example, FRDA-B is calculated as measuring the ratio of shared barcodes between samples A and B relative to the number of distinct barcodes in sample B. Therefore, an FRDA-B of ≈ 1 indicates that nearly all barcodes shared between sample A and B are found in sample B, while low FRDA-B indicates that shared clones represent a low fraction of barcodes in sample B. When contextualized with GD, FRD provides a normalized metric to interpret the number of shared clones that contribute to similarity and to suggest the possible directionality of clone transfer.
We next compared GD and FRD to decipher how clonal sharing of P. aeruginosa along the intestine changes over time. At 24 hpi, the P. aeruginosa populations in the stomach and the small intestine had only moderate similarity to those of the caecum, large intestine, and feces (average GDs = 0.54 and 0.41, respectively) (Fig. 6A). Using FRD values, we identified distinct patterns driving the genetic distances between the segments of the GI tract. At 24 hpi, the stomach and the small intestine had moderate genetic distance (average GD = 0.66) (Fig. 6A, D), indicating that the populations between these environments were relatively different. The median FRDstomach-small intestine (0.36) was lower than FRDsmall intestine-stomach (0.89) (Fig. 6H), illustrating that the shared barcodes constitute a smaller proportion of the total barcodes in the small intestine than in the stomach; the small intestine possesses a greater number of unique clones. The greater number of unique clones in the small intestine suggests either (1) reflux of a subpopulation of bacteria from the small intestine to the stomach or (2) initial seeding of the stomach and small intestine with more similar populations followed by rapid elimination of a portion of the population in the stomach. The decreasing GD between the stomach and the small intestine over time supports the idea of bacterial reflux from the small intestine. Very little similarity was present between P. aeruginosa populations in the feces and the stomach, indicating that coprophagia did not significantly contribute to reseeding of the stomach (Supplemental Fig. 5). Both the average FRDsmall intestine-caecum and FRDcaecum-small intestine were greater than 0.9, indicating that relatedness between the small intestine and caecum is driven by a large portion of shared barcodes (Fig. 6E, H). The differential expansion of a small number of clones likely accounted for the genetic distance (average GD = 0.41) between these two sites (Fig. 6A, Supplemental Fig. 5). The high FRD and low GD values between the small intestine and the caecum suggest that P. aeruginosa clones efficiently trafficked between these two compartments, either through natural peristalsis or retrograde movement, before continuing expansion at both sites. On the other hand, the extremely low GD values between the caecum, colon, and feces (Fig. 6A-C) with corresponding FRD values ≥ 0.89 (Fig. 6E-G) suggest that bacterial populations move freely between these sites. These observations, together with the fact that CFU are consistently higher than Ns values, indicate that vancomycin pretreatment robustly enables P. aeruginosa replication and movement along the GI tract, rather than simply facilitating the transient transfer of an initial inoculum through the GI tract.
Overall, our findings suggest that (i) the vast majority of orogastrically administered P. aeruginosa bacteria are rapidly (within 6 hours) killed in the stomach, (ii) less than 0.01% of P. aeruginosa from the inoculum persists in the intestine over the first 72 hours, (iii) robust P. aeruginosa replication occurs in the small intestine and the caecum, and (iv) bacterial populations subsequently migrate along the distal GI tract and are expelled in the feces.
DISCUSSION
In this study, we established a murine model of P. aeruginosa GI carriage that mimics patients receiving antibiotics in the hospital setting. This model is clinically relevant, as it utilizes vancomycin, and has been validated with both sexes and multiple clinical isolates. The absence of GI tract inflammation confirmed that this model represents carriage, not infection. Nevertheless, occasional low-level escape of P. aeruginosa to other organs suggests a possible route by which GI carriage may lead to subsequent infection at remote sites (7, 12–14). Long-term GI carriage was established after a single dose of P. aeruginosa, with 70% of mice still carried the bacterium after 60 days. Using barcoded bacteria, we found that most of the bacterial inoculum was eliminated within the first 6 hours, primarily in the stomach. However, once P. aeruginosa reached the small intestine and the caecum, bacteria replicated robustly, leading to significant fecal excretion as winnowed bacterial populations migrated unimpeded through the caecum and colon.
Confirming previously reported results (28), we found that untreated mice did not support GI carriage of P. aeruginosa. In contrast, seven days of vancomycin delivered through IP injection promoted high-level and prolonged GI carriage of P. aeruginosa. Several studies have investigated the impact of orally administered vancomycin on the gut microbiota and have reported a decrease in Bacteroidetes and a subsequent increase in Proteobacteria and Fusobacteria phyla (46–50). Firmicutes levels were also altered by oral vancomycin treatments, with the directionality of the impact varying across bacterial species. We speculate that IP delivered vancomycin achieves relatively high concentrations in the lumen of the GI tract, that it has a similar effect on the mouse microbiome, and that depletion of some microbiome constituents from the GI tract creates a niche for the establishment of P. aeruginosa. While we did not observe histopathological changes following vancomycin treatment, it is also possible that this antibiotic facilitates P. aeruginosa carriage through a direct effect on the host, such as immunomodulation, independent from a modification of the microbiome (49). As a first step in understanding the mechanism by which vancomycin facilitates P. aeruginosa carriage, we are currently characterizing the changes in the microbiome over time following administration of vancomycin in this model.
All six clinical isolates of P. aeruginosa tested in our study established carriage in the GI tract to a similar extent. These isolates are genetically diverse and include both exoU+ and exoS+ strains, as well as high-risk and non-high-risk clones (38). Robust colonization by different isolates suggests that the ability for P. aeruginosa to establish GI carriage depends on a set of features encoded by the core genome of the bacterium. Genes encoding metabolic factors, transcriptional regulators, adhesins, secretion systems, membrane homeostasis proteins and bile resistance factors have been identified as important for the GI carriage of other bacterial species (51–55). We suspect that P. aeruginosa carriage requires a similar set of features. The reliance on additional strain-specific strategies is nonetheless not excluded. Treatment with vancomycin for seven days likely caused a severe dysbiosis, masking the need for strain-specific factors that contribute to carriage in the presence of a less perturbed microbiome. Additionally, we observed that GI carriage can last for several weeks following a single exposure to the bacterium and cessation of vancomycin. Several studies have reported that the long-term carriage of P. aeruginosa in the lungs of individuals with cystic fibrosis or chronic obstructive pulmonary disease is accompanied by genetic adaptations of the bacterium (56–58). It is possible that the set of genes or alleles contributing to carriage of P. aeruginosa evolves as bacteria transition from the early stages of carriage to long-term colonization.
In patients, the GI carriage of P. aeruginosa is a predictor for the subsequent development of P. aeruginosa infections at various sites (7, 12, 13). Shortly after bacterial inoculation, we show that low-level dissemination from the gut to the gallbladder, spleen, liver, or lungs occasionally occurs, perhaps explaining these clinical observations. Gut dysbiosis can lead to the development of a leaky intestinal barrier, where pathogen molecules translocate from the gut into the bloodstream (59). The present study utilized healthy mice. However, dissemination to other tissues may be accentuated in immunocompromised animals, leading to more severe infections, as shown by Koh et al. (18). Interestingly, 12 days after cessation of vancomycin administration (14 days after orogastric gavage with P. aeruginosa), P. aeruginosa CFU in the feces remained high but dissemination from the gut was no longer observed. Calderon-Gonzalez et al. showed that Klebsiella pneumoniae dissemination from the GI tract was promoted by antibiotic treatments (51). It is therefore possible that vancomycin may support not only GI carriage but also dissemination of P. aeruginosa, and that dissemination diminishes over time as vancomycin is cleared.
STAMPR has been used by several groups to measure the dynamics of bacterial spread in colonization and systemic dissemination (44, 60, 61). We used STAMPR to show that most P. aeruginosa bacteria are eradicated prior to carriage. Two recent studies indicate that stomach acidity significantly restricts the GI carriage of Citrobacter rodentium and K. pneumoniae (51, 62). While stomach acidity constricts C. rodentium numbers by 10- to 100-fold (62), our results show an even more severe constriction for P. aeruginosa in the stomach. The stomach pH of healthy mice fluctuates between 3–4, while the rest of the GI tract tends to have a more neutral pH of 6–8 (63). P. aeruginosa can grow at a wide range of pH, but its optimal growth pH ranges between 6 and 8 (64). Thus, acidity may be the mechanism underlying the loss of P. aeruginosa barcode diversity in the stomach. This possibility could be tested by pharmacologically neutralizing stomach acid and subsequently measuring the size of downstream founding populations of P. aeruginosa. Interestingly, the rest of the GI tract was quite permissive to carriage. In the absence of vancomycin treatment, no carriage could be established, indicating the presence of a second barrier to P. aeruginosa, perhaps downstream of the stomach in untreated mice. Campbell et al. recently monitored the dynamics of C. rodentium enteric carriage and identified the microbiota as the major factor limiting colonization (62). This supports the idea that vancomycin facilitates the carriage of P. aeruginosa by eliminating a microbiome-mediated barrier, a hypothesis that could be tested with germ-free mice. In this sense, P. aeruginosa is both an “opportunistic pathogen” and an “opportunistic colonizer.”
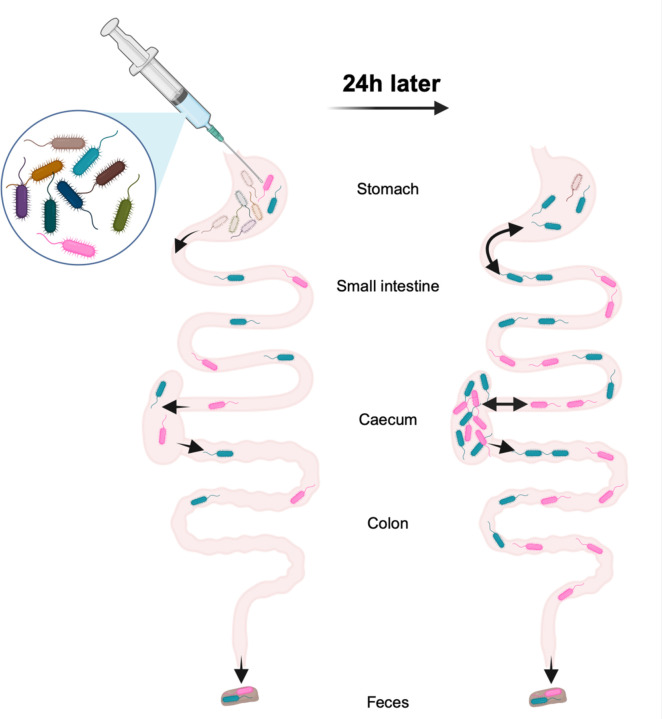
Together, our findings suggest the following model of P. aeruginosa carriage: within a few hours (less than 6 hours), a drastic constriction (less than 0.01% survival on average) of the bacterial inoculum occurs in the stomach (Fig. 7). A very small proportion of the P. aeruginosa inoculum passes through the stomach to reach the small intestine and the caecum. The small number of remaining founders rapidly replicate in both the small intestine and the caecum, and the resulting bacterial populations migrate from the caecum to the colon and are expelled in the feces. In addition to the expected trafficking route from the stomach to the small intestine, a small portion of P. aeruginosa may also reflux from the small intestine back to the stomach. Using STAMPR, we have identified the stomach as the main barrier to P. aeruginosa carriage in this animal model and the small intestine and the caecum as the main sites of bacterial expansion. Overall, this model advances the understanding of P. aeruginosa dynamics during GI carriage and may be useful in studying adaptation of P. aeruginosa during prolonged colonization, persistence following administration of antibiotics other than vancomycin, and identification of P. aeruginosa and host factors that facilitate carriage. In addition, our findings have clinical implications, such as the potential importance of more judicious use of acid suppressing drugs and vancomycin in preventing P. aeruginosa GI carriage.
Figure 7: Model of the population dynamics of P. aeruginosa following orogastric gavage.
Left: soon after the orogastric delivery of P. aeruginosa, most bacteria are eliminated from the stomach, severely constricting the size of the remaining population (less than 0.01% survival). Part of the population passes through the stomach to reach other compartments of the GI tract: small intestine, caecum, colon, and feces. P. aeruginosa does not encounter additional barriers downstream from the stomach. Right: over the first 24 hours, population expansion and/or reflux from the small intestine occurs in the stomach. The small intestine and the caecum support massive expansion of the remaining P. aeruginosa clones, and bacteria freely migrate from the caecum to the colon and feces. This figure was created using Biorender.com.
MATERIALS AND METHODS
Bacterial strains and culture conditions
PABL004, PABL006, PABL012, PABL048, PABL049 and PABL054 are archived P. aeruginosa clinical isolates cultured between 1999 and 2003 from the bloodstream of patients at Northwestern Memorial Hospital in Chicago (65). Relevant characteristics of the strains are listed in Supplementary Table 1. Unless otherwise stated, bacteria were streaked from frozen stocks onto either Lysogeny broth (LB) or Vogel-Bonner minimal (VBM) (66) agar plates and subsequently grown at 37°C in LB medium with shaking. When antibiotic selection for P. aeruginosa was necessary, supplementation with irgasan (irg) at 5 µg/mL was used.
Murine model of gastrointestinal carriage
Six- to eight-week-old C57BL/6 mice (Jackson Laboratory) received either 200 µL (female) or 250 µL (male) of vancomycin (370 mg/kg, Hospira, Lake Forest, IL) daily IP for seven days. The vancomycin dosage was allometrically scaled based on a total human daily dose of 30 mg/kg (35). On the fifth day of antibiotic treatment, mice were gavaged (20 G x 30 mm straight animal feeding needle, Pet Surgical, Phoenix, AZ) with 50 µL of P. aeruginosa prepared as follows: after overnight culture in LB, bacteria were diluted, regrown to exponential phase in LB and resuspended to the desired dose in PBS. When specified, mock IP injections or mock orogastric gavage was performed using PBS (same volumes as the treatment groups). Starting the day after the orogastric gavage, cages were changed daily to limit the impact of coprophagy.
To determine bacterial GI carriage, mice were individually placed in boxes to induce defecation, and feces were collected, weighed, homogenized in 1 mL of PBS using a bead blaster (Benchmark Scientific, Sayreville, NJ), and centrifuged for 30 sec at 1,100 x g. The supernatant was serially diluted and plated on either LB agar supplemented with irgasan or VBM agar for CFU enumeration.
Mice were housed in the containment ward of the Center for Comparative Medicine at Northwestern University. All experiments were approved by the Northwestern University Institutional Animal Care and Use Committee in compliance with all relevant ethical regulations for animal testing and research. Experiments used female mice unless otherwise stated.
Murine GI carriage of PABL012pool
Six- to eight-week-old female mice received IP injection of vancomycin (200 µL, 370 mg/kg) for 5 to 7 days, with each experimental group receiving their last injection 24 h prior to dissection. An aliquot of 50 µL of the PABL012pool library was grown overnight in 5 mL of LB (37°C, 250 rpm) and subcultured (1:40) in 30 mL of LB for 3 h. The bacterial inoculum was prepared as described above, and orogastric gavage was performed on the 5th day of vancomycin treatment, using 106.1 CFU of PABL012pool. Following bacterial inoculation, mice were housed individually. At 24, 48, and 72 hours post-gavage, mice were euthanized, and the stomach, small intestine, caecum, colon and feces were collected. The stomach, small intestine and caecum were processed along with their luminal contents. The colon was emptied, and the colonic contents were added to excreted feces (when available) to constitute the “feces” samples. All samples were weighed, homogenized in 1 mL of PBS using a bead blaster, centrifuged for 30 sec at 1,100 x g, and the supernatant was serially diluted and plated on VBM agar for CFU enumeration. For the estimated founding population sizes (Ns), 250 µL of the organ samples, as well as 250 µL of the inoculum (26 technical replicates) were spread on 150-mm-diameter VBM plates. Plates used for CFU and Ns determination were grown overnight at 37°C and the colonies were counted. CFU counts and Ns values in figure 5 represent those determined in 250 µL (1/4 of the homogenized tissue volume).
Supplementary Material
IMPORTANCE.
While P. aeruginosa is rarely part of the normal human microbiome, carriage of the bacterium is quite frequent in hospitalized patients and residents of long-term care facilities. P. aeruginosa carriage is a precursor to infection. Options for treating infections caused by difficult-to-treat P. aeruginosa strains are dwindling, underscoring the urgency to better understand and impede pre-infection stages, such as colonization. Here, we use vancomycin-treated mice to model antibiotic-treated patients who become colonized with P. aeruginosa in their gastrointestinal tracts. We identify the stomach as a major barrier to the establishment of gastrointestinal carriage. These findings suggest that efforts to prevent gastrointestinal colonization should focus not only on judicious use of antibiotics but also on investigation into how the stomach eliminates orally ingested P. aeruginosa.
ACKNOWLEDMENTS
Support for this work was provided by the National Institutes of Health awards RO1 AI118257, K24 AI04831, R21 AI129167 and R21 AI153953 (all to ARH) and U19 AI135964 (RGW and ARH). Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory which is supported by NCI P30-CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center.
REFERENCES
- 1.Ikuta KS, Swetschinski LR, Aguilar GR, Sharara F, Mestrovic T, Gray AP, Weaver ND, Wool EE, Han C, Hayoon AG, Aali A, Abate SM, Abbasi-Kangevari M, Abbasi-Kangevari Z, Abd-Elsalam S, Abebe G, Abedi A, Abhari AP, Abidi H, Aboagye RG, Absalan A, Ali HA, Acuna JM, Adane TD, Addo IY, Adegboye OA, Adnan M, Adnani QES, Afzal MS, Afzal S, Aghdam ZB, Ahinkorah BO, Ahmad A, Ahmad AR, Ahmad R, Ahmad S, Ahmad S, Ahmadi S, Ahmed A, Ahmed H, Ahmed JQ, Rashid TA, Ajami M, Aji B, Akbarzadeh-Khiavi M, Akunna CJ, Hamad HA, Alahdab F, Al-Aly Z, Aldeyab MA, Aleman AV, Alhalaiqa FAN, Alhassan RK, Ali BA, Ali L, Ali SS, Alimohamadi Y, Alipour V, Alizadeh A, Aljunid SM, Allel K, Almustanyir S, Ameyaw EK, Amit AML, Anandavelane N, Ancuceanu R, Andrei CL, Andrei T, Anggraini D, Ansar A, Anyasodor AE, Arabloo J, Aravkin AY, Areda D, Aripov T, Artamonov AA, Arulappan J, Aruleba RT, Asaduzzaman M, Ashraf T, Athari SS, Atlaw D, Attia S, Ausloos M, Awoke T, Quintanilla BPA, Ayana TM, Azadnajafabad S, Jafari AA, B DB, Badar M, Badiye AD, Baghcheghi N, Bagherieh S, Baig AA, Banerjee I, Barac A, Bardhan M, Barone-Adesi F, Barqawi HJ, Barrow A, Baskaran P, Basu S, Batiha A- MM, Bedi N, Belete MA, Belgaumi UI, Bender RG, Bhandari B, Bhandari D, Bhardwaj P, Bhaskar S, Bhattacharyya K, Bhattarai S, Bitaraf S, Buonsenso D, Butt ZA, Santos FLC dos, Cai J, Calina D, Camargos P, Cámera LA, Cárdenas R, Cevik M, Chadwick J, Charan J, Chaurasia A, Ching PR, Choudhari SG, Chowdhury EK, Chowdhury FR, Chu D-T, Chukwu IS, Dadras O, Dagnaw FT, Dai X, Das S, Dastiridou A, Debela SA, Demisse FW, Demissie S, Dereje D, Derese M, Desai HD, Dessalegn FN, Dessalegni SAA, Desye B, Dhaduk K, Dhimal M, Dhingra S, Diao N, Diaz D, Djalalinia S, Dodangeh M, Dongarwar D, Dora BT, Dorostkar F, Dsouza HL, Dubljanin E, Dunachie SJ, Durojaiye OC, Edinur HA, Ejigu HB, Ekholuenetale M, Ekundayo TC, El-Abid H, Elhadi M, Elmonem MA, Emami A, Bain LE, Enyew DB, Erkhembayar R, Eshrati B, Etaee F, Fagbamigbe AF, Falahi S, Fallahzadeh A, Faraon EJA, Fatehizadeh A, Fekadu G, Fernandes JC, Ferrari A, Fetensa G, Filip I, Fischer F, Foroutan M, Gaal PA, Gadanya MA, Gaidhane AM, Ganesan B, Gebrehiwot M, Ghanbari R, Nour MG, Ghashghaee A, Gholamrezanezhad A, Gholizadeh A, Golechha M, Goleij P, Golinelli D, Goodridge A, Gunawardane DA, Guo Y, Gupta RD, Gupta S, Gupta VB, Gupta VK, Guta A, Habibzadeh P, Avval AH, Halwani R, Hanif A, Hannan MA, Harapan H, Hassan S, Hassankhani H, Hayat K, Heibati B, Heidari G, Heidari M, Heidari-Soureshjani R, Herteliu C, Heyi DZ, Hezam K, Hoogar P, Horita N, Hossain MM, Hosseinzadeh M, Hostiuc M, Hostiuc S, Hoveidamanesh S, Huang J, Hussain S, Hussein NR, Ibitoye SE, Ilesanmi OS, Ilic IM, Ilic MD, Imam MT, Immurana M, Inbaraj LR, Iradukunda A, Ismail NE, Iwu CCD, Iwu CJ, J LM, Jakovljevic M, Jamshidi E, Javaheri T, Javanmardi F, Javidnia J, Jayapal SK, Jayarajah U, Jebai R, Jha RP, Joo T, Joseph N, Joukar F, Jozwiak JJ, Kacimi SEO, Kadashetti V, Kalankesh LR, Kalhor R, Kamal VK, Kandel H, Kapoor N, Karkhah S, Kassa BG, Kassebaum NJ, Katoto PD, Keykhaei M, Khajuria H, Khan A, Khan IA, Khan M, Khan MN, Khan MA, Khatatbeh MM, Khater MM, Kashani HRK, Khubchandani J, Kim H, Kim MS, Kimokoti RW, Kissoon N, Kochhar S, Kompani F, Kosen S, Koul PA, Laxminarayana SLK, Lopez FK, Krishan K, Krishnamoorthy V, Kulkarni V, Kumar N, Kurmi OP, Kuttikkattu A, Kyu HH, Lal DK, Lám J, Landires I, Lasrado S, Lee S, Lenzi J, Lewycka S, Li S, Lim SS, Liu W, Lodha R, Loftus MJ, Lohiya A, Lorenzovici L, Lotfi M, Mahmoodpoor A, Mahmoud MA, Mahmoudi R, Majeed A, Majidpoor J, Makki A, Mamo GA, Manla Y, Martorell M, Matei CN, McManigal B, Nasab EM, Mehrotra R, Melese A, Mendoza-Cano O, Menezes RG, Mentis A- FA, Micha G, Michalek IM, Sá ACMGN de, Kostova NM, Mir SA, Mirghafourvand M, Mirmoeeni S, Mirrakhimov EM, Mirza-Aghazadeh-Attari M, Misganaw AS, Misganaw A, Misra S, Mohammadi E, Mohammadi M, Mohammadian-Hafshejani A, Mohammed S, Mohan S, Mohseni M, Mokdad AH, Momtazmanesh S, Monasta L, Moore CE, Moradi M, Sarabi MM, Morrison SD, Motaghinejad M, Isfahani HM, Khaneghah AM, Mousavi-Aghdas SA, Mubarik S, Mulita F, Mulu GBB, Munro SB, Muthupandian S, Nair TS, Naqvi AA, Narang H, Natto ZS, Naveed M, Nayak BP, Naz S, Negoi I, Nejadghaderi SA, Kandel SN, Ngwa CH, Niazi RK, Sá ATN de, Noroozi N, Nouraei H, Nowroozi A, Nuñez-Samudio V, Nutor JJ, Nzoputam CI, Nzoputam OJ, Oancea B, Obaidur RM, Ojha VA, Okekunle AP, Okonji OC, Olagunju AT, Olusanya BO, Bali AO, Omer E, Otstavnov N, Oumer B, A MP, Padubidri JR, Pakshir K, Palicz T, Pana A, Pardhan S, Paredes JL, Parekh U, Park E-C, Park S, Pathak A, Paudel R, Paudel U, Pawar S, Toroudi HP, Peng M, Pensato U, Pepito VCF, Pereira M, Peres MFP, Perico N, Petcu I-R, Piracha ZZ, Podder I, Pokhrel N, Poluru R, Postma MJ, Pourtaheri N, Prashant A, Qattea I, Rabiee M, Rabiee N, Radfar A, Raeghi S, Rafiei S, Raghav PR, Rahbarnia L, Rahimi-Movaghar V, Rahman M, Rahman MA, Rahmani AM, Rahmanian V, Ram P, Ranjha MMAN, Rao SJ, Rashidi M-M, Rasul A, Ratan ZA, Rawaf S, Rawassizadeh R, Razeghinia MS, Redwan EMM, Regasa MT, Remuzzi G, Reta MA, Rezaei N, Rezapour A, Riad A, Ripon RK, Rudd KE, Saddik B, Sadeghian S, Saeed U, Safaei M, Safary A, Safi SZ, Sahebazzamani M, Sahebkar A, Sahoo H, Salahi S, Salahi S, Salari H, Salehi S, Kafil HS, Samy AM, Sanadgol N, Sankararaman S, Sanmarchi F, Sathian B, Sawhney M, Saya GK, Senthilkumaran S, Seylani A, Shah PA, Shaikh MA, Shaker E, Shakhmardanov MZ, Sharew MM, Sharifi-Razavi A, Sharma P, Sheikhi RA, Sheikhy A, Shetty PH, Shigematsu M, Shin JI, Shirzad-Aski H, Shivakumar KM, Shobeiri P, Shorofi SA, Shrestha S, Sibhat MM, Sidemo NB, Sikder MK, Silva LMLR, Singh JA, Singh P, Singh S, Siraj MS, Siwal SS, Skryabin VY, Skryabina AA, Socea B, Solomon DD, Song Y, Sreeramareddy CT, Suleman M, Abdulkader RS, Sultana S, Szócska M, Tabatabaeizadeh S-A, Tabish M, Taheri M, Taki E, Tan K-K, Tandukar S, Tat NY, Tat VY, Tefera BN, Tefera YM, Temesgen G, Temsah M-H, Tharwat S, Thiyagarajan A, Tleyjeh II, Troeger CE, Umapathi KK, Upadhyay E, Tahbaz SV, Valdez PR, Eynde JV den, Doorn HR van, Vaziri S, Verras G-I, Viswanathan H, Vo B, Waris A, Wassie GT, Wickramasinghe ND, Yaghoubi S, Yahya GATY, Jabbari SHY, Yigit A, Yiğit V, Yon DK, Yonemoto N, Zahir M, Zaman BA, Zaman SB, Zangiabadian M, Zare I, Zastrozhin MS, Zhang Z-J, Zheng P, Zhong C, Zoladl M, Zumla A, Hay SI, Dolecek C, Sartorius B, Murray CJL, Naghavi M. 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet 400:2221–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for the Disease Control and Prevention. 2023. Pseudomonas aeruginosa Infection | HAI | CDC. https://www.cdc.gov/hai/organisms/pseudomonas.html. Retrieved 8 March 2024.
- 3.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (U.S.). 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.). [Google Scholar]
- 6.Hu Y, Wang S, Zhang Y, Wu Y, Liu C, Ju X, Zhou H, Cai C, Zhang R. 2023. A comparative study of intestinal Pseudomonas aeruginosa in healthy individuals and ICU inpatients. One Health Adv 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen R, Babushkin F, Cohen S, Afraimov M, Shapiro M, Uda M, Khabra E, Adler A, Ben Ami R, Paikin S. 2017. A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob Resist Infect Control 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Zorrilla S, Camoez M, Tubau F, Periche E, Cañizares R, Dominguez MA, Ariza J, Peña C. 2014. Antibiotic Pressure Is a Major Risk Factor for Rectal Colonization by Multidrug-Resistant Pseudomonas aeruginosa in Critically Ill Patients. Antimicrob Agents Chemother 58:5863–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andremont A, Marang B, Tancrède C, Baume D, Hill C. 1989. Antibiotic treatment and intestinal colonization by Pseudomonas aeruginosa in cancer patients. Antimicrob Agents Chemother 33:1400–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willmann M, Klimek AM, Vogel W, Liese J, Marschal M, Autenrieth IB, Peter S, Buhl M. 2014. Clinical and treatment-related risk factors for nosocomial colonisation with extensively drug-resistant Pseudomonas aeruginosa in a haematological patient population: a matched case control study. BMC Infect Dis 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martak D, Gbaguidi-Haore H, Meunier A, Valot B, Conzelmann N, Eib M, Autenrieth IB, Slekovec C, Tacconelli E, Bertrand X, Peter S, Hocquet D, Guther J. 2022. High prevalence of Pseudomonas aeruginosa carriage in residents of French and German long-term care facilities. Clin Microbiol Infect 28:1353–1358. [DOI] [PubMed] [Google Scholar]
- 12.Ohara T, Itoh K. 2003. Significance of Pseudomonas aeruginosa Colonization of the Gastrointestinal Tract. Intern Med 42:1072–1076. [DOI] [PubMed] [Google Scholar]
- 13.Gómez-Zorrilla S, Camoez M, Tubau F, Cañizares R, Periche E, Dominguez MA, Ariza J, Peña C. 2015. Prospective Observational Study of Prior Rectal Colonization Status as a Predictor for Subsequent Development of Pseudomonas aeruginosa Clinical Infections. Antimicrob Agents Chemother 59:5213–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheatley RM, Caballero JD, van der Schalk TE, De Winter FHR, Shaw LP, Kapel N, Recanatini C, Timbermont L, Kluytmans J, Esser M, Lacoma A, Prat-Aymerich C, Oliver A, Kumar-Singh S, Malhotra-Kumar S, Craig MacLean R. 2022. Gut to lung translocation and antibiotic mediated selection shape the dynamics of Pseudomonas aeruginosa in an ICU patient. 1. Nat Commun 13:6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denton M, Kerr K, Mooney L, Keer V, Rajgopal A, Brownlee K, Arundel P, Conway S. 2002. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr Pulmonol 34:257–261. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand X, Thouverez M, Talon D, Boillot A, Capellier G, Floriot C, Hélias J. 2001. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med 27:1263–1268. [DOI] [PubMed] [Google Scholar]
- 17.Comolli JC, Hauser AR, Waite L, Whitchurch CB, Mattick JS, Engel JN. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun 67:3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh AY, Priebe GP, Pier GB. 2005. Virulence of Pseudomonas aeruginosa in a Murine Model of Gastrointestinal Colonization and Dissemination in Neutropenia. Infect Immun 73:2262–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cash HA, Woods DE, McCullough B, Johanson WG, Bass JA. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am Rev Respir Dis 119:453–459. [DOI] [PubMed] [Google Scholar]
- 20.van Heeckeren AM, Schluchter MD. 2002. Murine models of chronic Pseudomonas aeruginosa lung infection. Lab Anim 36:291–312. [DOI] [PubMed] [Google Scholar]
- 21.Cole N, Bao S, Stapleton F, Thakur A, Husband AJ, Beagley KW, Willcox MDP. 2003. Pseudomonas aeruginosa keratitis in IL-6-deficient mice. Int Arch Allergy Immunol 130:165–172. [DOI] [PubMed] [Google Scholar]
- 22.Wood SJ, Kuzel TM, Shafikhani SH. 2023. Pseudomonas aeruginosa: Infections, Animal Modeling, and Therapeutics. 1. Cells 12:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldufsky J, Wood SJ, Jayaraman V, Majdobeh O, Chen L, Qin S, Zhang C, DiPietro LA, Shafikhani SH. 2015. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair Regen 23:557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pennington JE, Ehrie MG. 1978. Pathogenesis of Pseudomonas aeruginosa pneumonia during immunosuppression. J Infect Dis 137:764–774. [DOI] [PubMed] [Google Scholar]
- 25.Pettigrew MM, Gent JF, Kong Y, Halpin AL, Pineles L, Harris AD, Johnson JK. 2019. Gastrointestinal Microbiota Disruption and Risk of Colonization With Carbapenem-resistant Pseudomonas aeruginosa in Intensive Care Unit Patients. Clin Infect Dis 69:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepelletier D, Caroff N, Riochet D, Bizouarn P, Bourdeau A, Le Gallou F, Espaze E, Reynaud A, Richet H. 2006. Role of hospital stay and antibiotic use on Pseudomonas aeruginosa gastrointestinal colonization in hospitalized patients. Eur J Clin Microbiol Infect Dis 25:600–603. [DOI] [PubMed] [Google Scholar]
- 27.Hoang S, Georget A, Asselineau J, Venier A-G, Leroyer C, Rogues AM, Thiébaut R. 2018. Risk factors for colonization and infection by Pseudomonas aeruginosa in patients hospitalized in intensive care units in France. PLoS ONE 13:e0193300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pier GB, Meluleni G, Neuger E. 1992. A murine model of chronic mucosal colonization by Pseudomonas aeruginosa. Infect Immun 60:4768–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janapatla RP, Dudek A, Chen C-L, Chuang C-H, Chien K-Y, Feng Y, Yeh Y-M, Wang Y-H, Chang H-J, Lee Y-C, Chiu C-H. 2023. Marine prebiotics mediate decolonization of Pseudomonas aeruginosa from gut by inhibiting secreted virulence factor interactions with mucins and enriching Bacteroides population. J Biomed Sci 30:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zebian G, Kreitmann L, Houard M, Piantoni A, Piga G, Ruffier des Aimes S, Holik B, Wallet F, Labreuche J, Nseir S. 2024. Immunosuppression at ICU admission is not associated with a higher incidence of ICU-acquired bacterial bloodstream infections: the COCONUT study. Ann Intensive Care 14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreitmann L, Helms J, Martin-Loeches I, Salluh J, Poulakou G, Pène F, Nseir S. 2024. ICU-acquired infections in immunocompromised patients. Intensive Care Med 50:332–349. [DOI] [PubMed] [Google Scholar]
- 32.Baggs J, Fridkin SK, Pollack LA, Srinivasan A, Jernigan JA. 2016. Estimating National Trends in Inpatient Antibiotic Use Among US Hospitals From 2006 to 2012. JAMA Intern Med 176:1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magill SS, O’Leary E, Ray SM, Kainer MA, Evans C, Bamberg WM, Johnston H, Janelle SJ, Oyewumi T, Lynfield R, Rainbow J, Warnke L, Nadle J, Thompson DL, Sharmin S, Pierce R, Zhang AY, Ocampo V, Maloney M, Greissman S, Wilson LE, Dumyati G, Edwards JR, Emerging Infections Program Hospital Prevalence Survey Team. 2021. Antimicrobial Use in US Hospitals: Comparison of Results From Emerging Infections Program Prevalence Surveys, 2015 and 2011. Clin Infect Dis 72:1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauser AR. 2018. Antibiotic Basics for Clinicians: The ABCs of Choosing the Right Antibacterial Agent, 3rd ed. Lippincott Williams & Wilkins (LWW). [Google Scholar]
- 35.U.S. Department of Health and Human Services. 2005. Guidance for Industry - Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers. Food and Drug Administration. [Google Scholar]
- 36.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, J. Rybak M, Talan DA, Chambers HF. 2011. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children: Executive Summary. Clin Infect Dis 52:285–292. [DOI] [PubMed] [Google Scholar]
- 37.Klockgether J, Cramer N, Wiehlmann L, Davenport CF, Tümmler B. 2011. Pseudomonas aeruginosa Genomic Structure and Diversity. Front Microbiol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen JP, Ozer EA, Minasov G, Shuvalova L, Kiryukhina O, Anderson WF, Satchell KJF, Hauser AR. 2020. A comparative genomics approach identifies contact-dependent growth inhibition as a virulence determinant. Proc Natl Acad Sci 117:6811–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglewski BH. 1996. Pseudomonas, p. In Baron S (ed.), Medical Microbiology, 4th ed. University of Texas Medical Branch at Galveston, Galveston (TX). [Google Scholar]
- 40.Okuda J, Hayashi N, Okamoto M, Sawada S, Minagawa S, Yano Y, Gotoh N. 2010. Translocation of Pseudomonas aeruginosa from the intestinal tract is mediated by the binding of ExoS to an Na,K-ATPase regulator, FXYD3. Infect Immun 78:4511–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall JC, Christou NV, Meakins JL. 1993. The gastrointestinal tract. The “undrained abscess” of multiple organ failure. Ann Surg 218:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abel S, Abel zur Wiesch P, Chang H-H, Davis BM, Lipsitch M, Waldor MK. 2015. Sequence tag-based analysis of microbial population dynamics. Nat Methods 12:223–226, 3 p following 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hullahalli K, Pritchard JR, Waldor MK. Refined Quantification of Infection Bottlenecks and Pathogen Dissemination with STAMPR. mSystems 6:e00887–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachta KER, Allen JP, Cheung BH, Chiu C-H, Hauser AR. 2020. Systemic infection facilitates transmission of Pseudomonas aeruginosa in mice. 1. Nat Commun 11:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavalli-Sforza LL, Edwards AWF. 1967. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 46.Rosa CP, Pereira JA, de Melo Cristina Santos N, Brancaglion GA, Silva EN, Tagliati CA, Novaes RD, Corsetti PP, de Almeida LA. 2020. Vancomycin-induced gut dysbiosis during Pseudomonas aeruginosa pulmonary infection in a mice model. J Leukoc Biol 107:95–104. [DOI] [PubMed] [Google Scholar]
- 47.Ray P, Pandey U, Aich P. 2021. Comparative analysis of beneficial effects of vancomycin treatment on Th1- and Th2-biased mice and the role of gut microbiota. J Appl Microbiol 130:1337–1356. [DOI] [PubMed] [Google Scholar]
- 48.Nazzal L, Soiefer L, Chang M, Tamizuddin F, Schatoff D, Cofer L, Aguero-Rosenfeld ME, Matalon A, Meijers B, Holzman R, Lowenstein J. 2021. Effect of Vancomycin on the Gut Microbiome and Plasma Concentrations of Gut-Derived Uremic Solutes. Kidney Int Rep 6:2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, Soeters MR, Blaak EE, Dallinga-Thie GM, Reijnders D, Ackermans MT, Serlie MJ, Knop FK, Holst JJ, van der Ley C, Kema IP, Zoetendal EG, de Vos WM, Hoekstra JBL, Stroes ES, Groen AK, Nieuwdorp M. 2014. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 60:824–831. [DOI] [PubMed] [Google Scholar]
- 50.Kim E, Kim AH, Lee Y, Ji SC, Cho J-Y, Yu K-S, Chung J-Y. 2021. Effects of vancomycin-induced gut microbiome alteration on the pharmacodynamics of metformin in healthy male subjects. Clin Transl Sci 14:1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calderon-Gonzalez R, Lee A, Lopez-Campos G, Hancock SJ, Sa-Pessoa J, Dumigan A, McMullan R, Campbell EL, Bengoechea JA. 2023. Modelling the Gastrointestinal Carriage of Klebsiella pneumoniae Infections. mBio 14:e03121–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung BH, Alisoltani A, Kochan TJ, Lebrun-Corbin M, Nozick SH, Axline CMR, Bachta KER, Ozer EA, Hauser AR. 2023. Genome-wide screens reveal shared and strain-specific genes that facilitate enteric colonization by Klebsiella pneumoniae. mBio 14:e0212823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westerman TL, McClelland M, Elfenbein JR. 2021. YeiE Regulates Motility and Gut Colonization in Salmonella enterica Serotype Typhimurium. mBio 12: 10.1128/mbio.03680-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krypotou E, Townsend GE, Gao X, Tachiyama S, Liu J, Pokorzynski ND, Goodman AL, Groisman EA. 2023. Bacteria require phase separation for fitness in the mammalian gut. Science 379:1149–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43:1471–1491. [DOI] [PubMed] [Google Scholar]
- 56.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci 103:8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic Adaptation of Pseudomonas aeruginosa to the Airways of Cystic Fibrosis Patients Is Catalyzed by Hypermutation. J Bacteriol 190:7910–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eklöf J, Misiakou MA, Sivapalan P, Armbruster K, Browatzki A, Nielsen TL, Lapperre TS, Andreassen HF, Janner J, Ulrik CS, Gabrielaite M, Johansen HK, Jensen A, Nielsen TV, Hertz FB, Ghathian K, Calum H, Wilcke T, Seersholm N, Jensen J- US, Marvig RL. 2022. Persistence and genetic adaptation of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Clin Microbiol Infect 28:990–995. [DOI] [PubMed] [Google Scholar]
- 59.Chancharoenthana W, Kamolratanakul S, Schultz MJ, Leelahavanichkul A. 2023. The leaky gut and the gut microbiome in sepsis – targets in research and treatment. Clin Sci Lond Engl 1979 137:645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hullahalli K, Waldor MK. 2021. Pathogen clonal expansion underlies multiorgan dissemination and organ-specific outcomes during murine systemic infection. eLife 10:e70910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Louie A, Zhang T, Becattini S, Waldor MK, Portnoy DA. 2019. A Multiorgan Trafficking Circuit Provides Purifying Selection of Listeria monocytogenes Virulence Genes. mBio 10: 10.1128/mbio.02948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell IW, Hullahalli K, Turner JR, Waldor MK. 2023. Quantitative dose-response analysis untangles host bottlenecks to enteric infection. Nat Commun 14:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Y, Koyama Y, Shimada S. 2022. Measurement of intraluminal pH changes in the gastrointestinal tract of mice with gastrointestinal diseases. Biochem Biophys Res Commun 620:129–134. [DOI] [PubMed] [Google Scholar]
- 64.Tsuji A, Kaneko Y, Takahashi K, Ogawa M, Goto S. 1982. The Effects of Temperature and pH on the Growth of Eight Enteric and Nine Glucose Non-Fermenting Species of Gram-Negative Rods. Microbiol Immunol 26:15–24. [DOI] [PubMed] [Google Scholar]
- 65.Scheetz MH, Hoffman M, Bolon MK, Schulert G, Estrellado W, Baraboutis IG, Sriram P, Dinh M, Owens LK, Hauser AR. 2009. Morbidity Associated with Pseudomonas aeruginosa Bloodstream Infections. Diagn Microbiol Infect Dis 64:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.