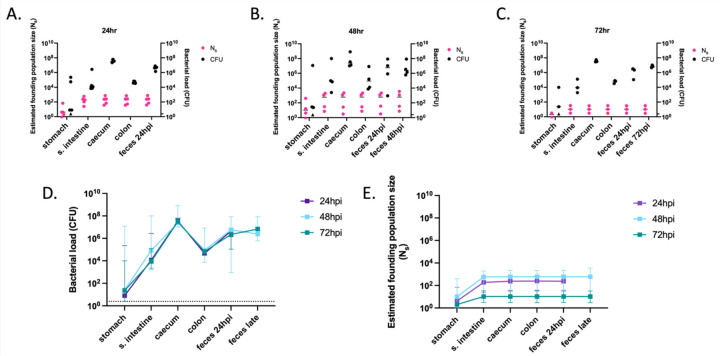
Figure 5: Founding populations and bacterial loads of P. aeruginosa in the GI tract.
A total of 106.1 CFU of PABL012pool were delivered to single-caged mice by orogastric gavage. P. aeruginosa CFU in 250 µL of resuspensions of collected homogenized tissues (one-fourth of tissue homogenates of stomach, small intestine [“s. intestine”], caecum, colon, and feces) were enumerated by plating, and founding population sizes (Ns) were estimated using the STAMPR approach. (A-C) Bacterial loads (CFU, black circles) and estimated founding population sizes (Ns, pink circles) were quantified at (A) 24 (n = 5), (B) 48 (n = 4) and (C) 72 hpi (n = 3, except for Ns in stomach, which was n = 2 due to a sequencing issue). Each circle represents an organ from one mouse. Solid horizontal lines indicate medians. Minor ticks on the right Y axis represent the limits of detection for the CFU. Triangles represent samples with no recovered CFU. (D) P. aeruginosa burdens and (E) estimated founding population sizes in different tissues of the GI tract at 24 (purple), 48 (blue) and 72 (green) hpi. For comparison, fecal samples were collected at 24 hpi (“feces 24 hpi”) regardless of the ending timepoint. An additional terminal fecal sample was available for animals harvested at 48 or 72 hpi (“feces late”). Squares represent medians, and error bars represent 95% confidence intervals. The dotted line indicates the limit of detection for CFU. Ns values are not significantly different over time (t-test).