Abstract
The wild-type Lactococcus lactis strain LAC460 produces two bacteriocin-like phage lysins, LysL and LysP. This study aimed to produce and secrete LysL in various heterologous hosts and an in vitro cell-free expression system for further functional studies. Initially, the lysL gene from L. lactis LAC460 was cloned into Lactococcus cremoris NZ9000 and L. lactis N8 strains, with and without the usp45 signal sequence (SSusp45), under a nisin-inducible promoter. Active LysL was primarily produced intracellularly in recombinant L. lactis N8, with some secretion into the supernatant. Recombinant L. cremoris NZ9000 lysed upon nisin induction, indicating successful lysL expression. However, fusion with Usp45 signal peptide (SPUsp45–LysL) weakened LysL activity, likely due to incomplete signal peptide cleavage during secretion. Active LysL was also produced in vitro, and analysed in SDS-PAGE, giving a 42-kDa band. However, the yield of LysL protein was still low when produced from recombinant lactococci or by in vitro expression system. Therefore, His-tagged LysL was produced in Escherichia coli BL21(DE3). Western blot confirmed the intracellular production of about 44-kDa His-tagged LysL in E. coli. His-tagged active LysL was then purified by Ni-NTA affinity chromatography yielding sufficient 4.34 mg of protein to be used in future functional studies.
Keywords: Lactococcus lactis, phage lysin, heterologous expression, Usp45 signal peptide, protein purification, inhibitory tests
Expressing the gene and purifying the protein of a novel bacterial enzyme.
Introduction
Lactococcus lactis is a lactic acid bacterium broadly applied as starter culture for fermentation of dairy products. Thus far, more than 40 different bacteriocins, ribosomally synthesized antimicrobial proteins, have been characterized from Lactococcus spp. (Takala et al. 2023). Bacteriocins are generally divided into three classes, although alternative classifications have also been proposed (Soltani et al. 2021). Nearly all lactococcal bacteriocins belong to either class I or II bacteriocins, heat-stable peptides. Class I bacteriocins mostly act by cell membrane disruption and inhibition of peptidoglycan synthesis (Simons et al. 2020). Class II bacteriocins cause cell death through different ways, mainly by disrupting membrane permeability (Lozo et al. 2017). To date, the only class III bacteriocins, heat-labile large antimicrobial proteins, identified from lactococci are prophage-encoded lytic enzymes LysL and LysP from L. lactis LAC460 (Takala et al. 2023).
Lytic enzymes so far characterized from Lactococci are autolysins and phage-related lysins, which either contribute to autolysis or behave like bacteriocins (Visweswaran et al. 2017, Takala et al. 2023). To date, there are four main N-acetylglucosaminidases (AcmA, AcmB, AcmC, and AcmD) and one peptidase (YjgB) identified in Lactococcus spp. genomes. N-acetylglucosaminidases are mostly involved in cell separation and are involved in cell autolysis (Visweswaran et al. 2013). YjgB is an endopeptidase, which degrades the cell wall of lactococci (Redko et al. 2007). More recently, Usp45, the major extracellular protein of Lactococcus spp. was also reported to act as a peptidoglycan hydrolase that mediates cell separation (Hernandez-Valdes et al. 2020). As Usp45 is the most secreted protein in Lactococcus spp., its signal peptide (SPUsp45) has been used for producing many different heterologous proteins in lactococci (Wan et al. 2015, Bıyıklı et al. 2023).
Prophages and prophage-like elements are present in all lactococcal genomes (Aucouturier et al. 2018). For instance, Kelleher et al. (2018) studied 30 strains of Lactococcus spp., and all of them carried 2–10 intact, questionable, or incomplete prophage regions. All intact and some incomplete lactococcal prophages carry genes encoding lytic enzymes called virion associated lysins (VALs) and endolysins (Fernandes and São-José 2018, Abdelrahman et al. 2021). VALs are phage lysins with both enzymatic and structural functions. They help perforate the peptidoglycan layer of target bacteria, allowing the viral genome to enter the target cells. Endolysins, in contrast, act in the end of lytic cycle to degrade the peptidoglycan of the host bacteria (Abdelrahman et al. 2021). Endolysins are usually coexpressed with holins, small proteins facilitating the access of endolysins to peptidoglycan by forming pores in the cell membrane (Saier and Reddy 2015). This process is followed by the lysis of the host cell, ultimately leading to the release of phages. As demonstrated previously (de Ruyter et al. 1997), heterologous expression of lactococcal phage lysins in cheese starter strains accelerates cheese ripening by releasing intracellular enzymes from the lysed starter bacteria. This could be accomplished by producing lysins in a lysin-sensitive cheese starter strain, followed by the lysis of the host, or by secreting the lysins from a lysin-resistant adjunct starter strain.
In our previous work, we found a prophage-encoded bacteriocin-like lysin, LysL, from the culture supernatant of L. lactis strain LAC460 (Takala et al. 2023). LysL is a 385-aa lysozyme + peptidase M23 from a defective prophage. Among 34 Lactococcus strains tested, 11 were sensitive to LysL, while it had no lytic effect on the producer strain. However, there is no experimental evidence about the precise mode of action of LysL or its secretion mechanism. Therefore, in order to study the function of LysL, expression and purification of the enzyme are needed.
In this study, the lysL gene was cloned and the LysL protein was produced in different heterologous hosts, as well as expressed in vitro. In addition, LysL protein was purified, and the secretion of LysL was investigated.
Materials and methods
Plasmids, strains, and culture conditions
Plasmids and bacterial strains used in this study are listed in Table 1. Escherichia coli strains were grown in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) with shaking at 37°C. Lactococcus lactis and L. cremoris strains were incubated in M17 medium (Oxoid Ltd. Basingstoke, UK) supplemented with 0.5% (w/v) glucose (M17G) at 30°C. Solid media were made by adding 1.5% agar to the liquid media. For selecting Lactococcus transformants, erythromycin was used at a concentration of 10 µg/ml (M17GE). For E. coli transformants, 250 µg/ml erythromycin or 150 µg/ml ampicillin was used.
Table 1.
Plasmids and bacterial strains used in this study.
| Name | Description | Reference |
|---|---|---|
| pET-22b(+) | E. coli expression vector harboring pBR322 ori, ampR, PT7 | GenScript Biotech (Leiden, Netherlands) |
| pLEB823 | lysL expression plasmid for Lactococcus; repAC, ermC, PnisZ, lysL | This study |
| pLEB824 | SSusp45–lysL expression plasmid for Lactococcus; repAC, ermC, PnisZ, SSusp45–lysL | This study |
| pLEB844 | 6xHislysL in pET-22b(+) under PT7 control | This study |
| pVS2 | Control plasmid; repAC, ermC | von Wright et al. (1987) |
| pWUST25 | Lactococcus expression plasmid; source of repAC, ermC, PnisZ | Wan et al. (2016) |
| E. coli BL21(DE3) | Protein production strain; IPTG inducible gene expression | Studier and Moffatt (1986) |
| E. coli ECO859 | BL21(DE3) carrying lysL expression plasmid pLEB844 | This study |
| E. coli TG1 | Cloning host | Sambrook et al. (2001) |
| L. lactis LAC460 | Wild-type LysL producer; source of lysL gene | Takala et al. (2023) |
| L. cremoris LAC277 | Indicator strain; resistant to nisin and erythromycin, sensitive to LysL | Takala et al. (2004) |
| L. cremoris NZ9000 | Nisin inducible gene expression host; sensitive to LysL | Kuipers et al. (1998) |
| L. cremoris LAC474 | NZ9000 carrying pLEB824; SSusp45–lysL expression, ErmR | This study |
| L. cremoris LAC475 | NZ9000 carrying pLEB823; lysL expression, ErmR | This study |
| L. cremoris LAC476 | NZ9000 carrying pVS2; control strain, ErmR | This study |
| L. lactis N8 | Constitutive gene expression host with nisin promoter; nisin producer, resistant to LysL | Valio Ltd, Helsinki, Finland; (Wan et al. 2021) |
| L. lactis LAC477 | N8 carrying pLEB824; SSusp45–lysL expression, ErmR | This study |
| L. lactis LAC478 | N8 carrying pLEB823; lysL expression, ErmR | This study |
| L. lactis LAC479 | N8 carrying pVS2; control strain, ErmR | This study |
DNA techniques
Gene amplifications were carried out by standard PCR (Eppendorf, Hamburg, Germany) and using Phusion High-Fidelity DNA polymerase (Thermo Scientific, Waltham, MA, USA), according to the instructions of the manufacturers. PCR primers used are listed in Supplementary Table S1. DNA purification of PCR products and from agarose gel were carried out using the GeneJET PCR Purification Kit and GeneJET Gel Extraction Kit (Thermo Scientific), respectively. Plasmid DNA was isolated from the recombinant cultures with the GeneJET Plasmid Miniprep Kit (Thermo Scientific). Transformation of the strains was done by electroporation with Gene Pulser (Bio-Rad Laboratories, Hercules, CA, USA). PCR products were sequenced by an outsourced DNA sequencing service in the Institute of Biotechnology (University of Helsinki, Finland).
Construction of expression vectors
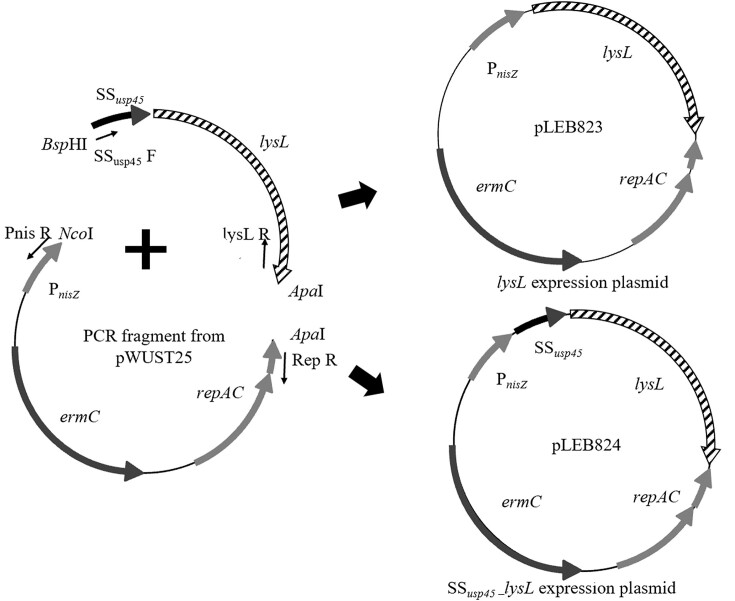
The lysL gene and SSusp45 were amplified by PCR using L. lactis LAC460 culture as template. SSusp45 was fused upstream of lysL (SSusp45–lysL) using gel-extracted amplicons of the previous PCRs as templates in overlap extension PCR (OE-PCR). The vector composed of the replication genes repAC, the erythromycin resistance gene ermC, and the nisin-inducible promoter PnisZ was amplified by PCR from plasmid pWUST25 (Wan et al. 2016). The vector fragment was cut with NcoI/ApaI restriction enzymes, and the lysL and SSusp45–lysL fragments with the compatible enzymes BspHI/ApaI (Thermo Scientific) at 37°C for 30 min, as recommended by the supplier. Digested and purified DNA fragments were ligated using T4 DNA ligase (Thermo Scientific) overnight at room temperature, as instructed by the manufacturer. Figure 1 illustrates the construction of lysL expression plasmid (pLEB823) and SSusp45–lysL expression plasmid (pLEB824). The ligation mixtures were transferred by electroporation into E. coli TG1, essentially according to Zabarovsky and Winberg (1990). Colonies were screened for the right constructs by PCR with PnisZ F and RSF (repAC screening forward) primers (Supplementary Table S1). Plasmids were isolated from the right clones, followed by sequencing to verify correct constructs. The lysL expression plasmids pLEB823 and pLEB824, as well as the control plasmid pVS2 were then electroporated into L. cremoris NZ9000 and L. lactis N8, essentially as described by Holo and Nes (1989). Lactococcus cremoris NZ9000 strains carrying SSusp45–lysL expression plasmid, lysL expression plasmid, or pVS2 were named as LAC474, LAC475, and LAC476, respectively. Lactococcus lactis N8 strains carrying SSusp45–lysL expression plasmid, lysL expression plasmid, or pVS2 were named as LAC477, LAC478, and LAC479, respectively.
Figure 1.
Schematic overview of the construction procedure of the lactococcal lysL expression plasmid pLEB823, and the SSusp45–lysL expression plasmid pLEB824. ermC, erythromycin resistance gene; repAC, plasmid replication genes; and PnisZ, nisin-inducible promoter.
Activity assay
Lactococcus cremoris LAC277 was chosen as the indicator for LysL activity tests because it is LysL sensitive, and nisin and erythromycin resistant without nisin production. Inhibitory activity of the recombinant L. lactis N8 strains carrying the lysL expression plasmids against L. cremoris LAC277 was determined using spot-on-lawn method as described by Wan et al. (2015). Briefly, five times sterile water-diluted overnight culture of L. cremoris LAC277 in M17GE was spread and left to dry onto an M17GE agar plate. Cell-free supernatant (CFS) of the overnight sample cultures were obtained by centrifugation at 5000 × g for 10 min followed by filtering through 0.22 µm sterilized filter. 10 ml of CFSs were concentrated using 30 kDa centrifugal filters (Merck, Darmstadt, Germany) according to manufacturer’s instructions. After centrifuging (5000 × g, 10 min), the upper phase was washed twice with 5 ml of PBS, and finally centrifuged to a final volume of 500 µl.
The centrifuged cell pellets of the overnight sample cultures were used for obtaining the cell lysates. After centrifugation of the cultures, the pellets were resuspended in the same volume of PBS (pH 7.2). 1 ml of each cell suspension was added to a 2-ml tube containing 0.1 mm glass beads (Omni International, Kennesaw, GA, USA) and placed in Omni bead mill homogenizer (Bead Ruptor Elite, Omni International). The cell breakage was performed at the speed of 6 m/s for 40 s at 0°C. The tubes’ contents were then centrifuged at 4000 × g for 10 min, and the cell lysates were obtained by passing each supernatant through 0.22 µm sterilized filter. 10 µl of the concentrated CFS or cell lysate samples were then spotted on the indicator plates and incubated at 30°C overnight.
In the case of L. cremoris NZ9000, the recombinant colonies were cultivated in M17GE and incubated at 30°C until the OD600 value reached to 0.5 (about 5 h). The induction was then applied with nisin at the concentration of 0.5 IU/ml and the incubation was continued for another few hours.
In vitro expression
Cell-free protein biosynthesis of LysL was done using the rapid translational system (RTS) 100 E. coli HY Kit (Biotechrabbit GmbH, Berlin, Germany). Using the forward primer lysL F OE T7 carrying overlap extension to T7 promoter, with the reverse primer lysL R OE RTS (Supplementary Table S1), lysL containing homologous regions of the RTS forward adaptor sequence at the 5′ ends, and RTS reverse adaptor sequence at the 3′ ends was amplified. The purified PCR product was then used as a template for the second PCR with the primers RTS T7 F and RTS R (Supplementary Table S1), which bear RTS forward and RTS reverse adaptor sequences, respectively. Thereby, T7 promoter and RTS forward adaptor sequences were added upstream of lysL gene, and the RTS reverse adaptor sequence was fused downstream of that, forming the RTS linear expression template. Finally, 0.5 µg of the purified expression template was used for in vitro protein synthesis reaction in a volume of 50 µl. The reaction was incubated for 6 h at 30°C according to the manufacturer’s instructions. One neutral reaction was also run in parallel without template, serving as the control.
SDS-PAGE
SDS-PAGE was performed according to the method essentially described by Laemmli with modifications (Laemmli. 1970). The in vitro translation reactions were mixed with SDS-PAGE loading buffer (Bio-Rad Laboratories) and heated at 95°C for 5 min. 10 µl of the samples were then loaded to a gradient SDS-PAGE gel (4%–20%; Bio-Rad Laboratories) and electrophoresed at 4°C under 200 V for 30 min. The gel was then fixed in a fixing solution (100 mM ammonium acetate, 10% acetic acid, and 50% methanol) for 30 min with gentle agitating. It was then stained with Coomassie Brilliant Blue R-250 (Bio-Rad Laboratories) for 1 h, followed by destaining for 2 h in a 10% acetic acid solution.
Cloning of lysL gene into the pET-22b(+) vector
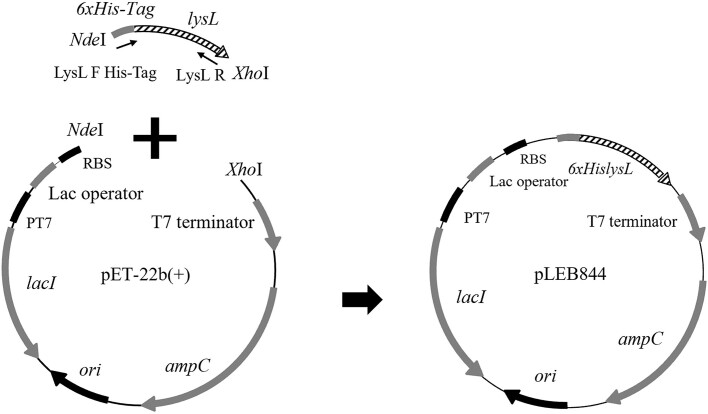
Heterologous expression of lysL in E. coli BL21(DE3) was done using pET-22b(+) as the expression vector. The gene lysL was amplified using the primers LysL F His-Tag Xa NdeI and LysL R XhoI (Supplementary Table S1), which amplify lysL with 6 × His codons plus the Factor Xa protease cleavage site for removal of His-Tag in the N-terminus of LysL. The vector and insert were then cut using NdeI/XhoI restriction enzymes at 37°C for 30 min and ligated, resulting in the plasmid pLEB844 (pET-22b(+)-lysL) (Fig. 2). Then, the ligation mixture was electroporated into E. coli BL21(DE3) similarly as described above for E. coli TG1. Colony PCR screening using T7 F and LysL R XhoI primers (Supplementary Table S1), followed by sequencing of the insert, confirmed correct construct. The recombinant strain was stored as E. coli ECO859.
Figure 2.
Construction of the E. coli lysL expression plasmid pLEB844. The lysL gene was amplified, digested, and ligated into the pET-22b(+) backbone via NdeI/XhoI restriction sites. The vector contains T7 promoter and ampicillin resistance gene, ampC.
Western blot
200 ml LB Amp150 was inoculated with an overnight incubated seed of E. coli ECO859, then incubated at 37°C with shaking (200 rpm). The induction of protein expression was performed by adding 1 mM isopropylthio-β-galactoside (IPTG) after OD600 reached about 0.5. The incubation was then continued for protein production for another 20 h under the same conditions. The cells were then collected by centrifugation (5000 × g, 10 min). CFS was prepared by filtering the supernatant through 0.22 µm sterilized filter. The cell pellet was dissolved in 50 ml of PBS (pH 7.2) and lysed four times with an Emulsiflex C3 high-pressure homogenizer (Avestin Inc., Ottawa, Canada). Then, cell debris was separated by centrifugation (30 000 × g, 4°C, 20 min).
Western blotting was done according to Towbin et al. (1979) and Burnette (1981) by the Protein Service core facility of the Tampere University. SDS-PAGE electrophoresis was performed for 30 µl of the CFS and the pellet lysate, excluding the staining and destaining steps. Briefly, the samples were separately mixed with Tris–glycine SDS-PAGE loading buffer (Bio-Rad Laboratories) with the ratio of 5:1 and heated at 95°C for 5 min. 5 µl of the samples were then loaded into a gradient SDS-PAGE gel (AnykD; Bio-Rad Laboratories) and electrophoresed at room temperature under 200 V for 30 min. The protein bands were electrotransferred from SDS gel to a cellulose nitrate membrane under 120 V at 4°C for 40 min. The membrane was then incubated in blocking solution (10% (w/v) bovine serum albumin) for 1 h with mild agitation. Next, the membrane was probed with 1:10 000 diluted in 5% Milk-TBST primary mouse anti-His.H8 (Thermo Scientific) at 4°C overnight with mild agitation. It was then incubated with horse antimouse IgG(H + L) Peroxidase (Vector Laboratories, Oxfordshire, UK) at a 1:20 000 dilution in TBST buffer (0.1% Tween 20, 25 mM Tris, 150 mM NaCl, pH 7.5) at room temperature for 1 h. After the last three steps, the membrane underwent three washes, each lasting 5 min, with TBST buffer. Protein bands were lastly detected using WesternBright ECL HRP substrate (Advansta, San Jose, CA, USA) with the ChemiDoc MP Imaging System (Bio-Rad Laboratories) according to the producer’s instructions.
Protein purification
LysL originating from 900 ml of IPTG-induced culture of E. coli ECO859 was purified with affinity chromatography using 6 × His-Tag as the purification tag by outsourced protein purification service (Protein Service core facility of the Tampere University). Prior to the chromatography step, 10 mM imidazole was separately added to the prepared CFS and pellet lysate samples (previous section) to prevent unspecific binding. The samples were then bound to HisPur™ Ni-NTA agarose (Thermo Scientific) in a batch mode for 1 h at 4°C. The bound protein was washed with 16 column volumes of wash buffer (PBS, 250 mM NaCl, 50 mM imidazole, pH 7.2) before stepwise elution with Elution Buffer (PBS, 250 mM NaCl, pH 7.2), which had an increasing concentration of imidazole (100, 150, 200, and 250 mM). The concentration (mg/ml) of the protein was obtained using the equation C = c(molar) × MW, in which, c(molar) = Abs280 nm/extinction coefficient.
Results
Heterologous expression of lysL in Lactococcus spp
As LysL originates from L. lactis, and the nisin-inducible heterologous gene expression method for lactococci is known to be strong, we aimed to overproduce LysL in Lactococcus strains. The lysL gene was cloned in L. cremoris NZ9000 and L. lactis N8, which carry nisin regulatory genes needed for gene expression with the nisin promoter. Also, in order to secrete LysL to the supernatant of the recombinant cultures, SSusp45 was fused with the lysL gene (SSusp45–lysL), and parallel to lysL, was cloned into the bacteria under the control of a nisin-inducible promoter.
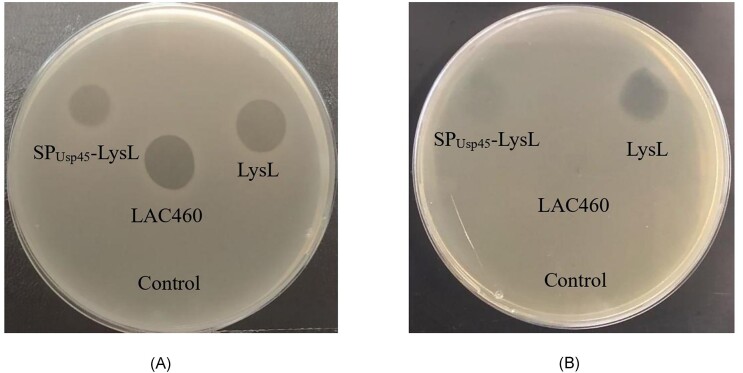
As shown in Fig. 3(A), both concentrated CFSs from the recombinant L. lactis N8 cultures gave halos on the indicator plates, demonstrating that the expression of lysL, both with and without SSusp45, was successful, and active LysL was secreted to supernatant. The halo formed by SPUsp45–LysL was weaker than that of native LysL.
Figure 3.
Inhibition of L. cremoris LAC277 by LysL from recombinant L. lactis N8 and wild-type L. lactis LAC460 concentrated CFSs (A) and cell lysates (B). 10 µl of the lysates/20-fold concentrated CFSs were spotted onto L. cremoris LAC277 indicator lawn on M17GE and incubated overnight at 30°C. LysL, L. lactis N8 transformed with the lysL expression plasmid; SPUsp45–LysL, L. lactis N8 transformed with the SSusp45–lysL expression plasmid; LAC460, L. lactis LAC460; and control, L. lactis N8 transformed with pVS2. A clearer and larger halo was created by the CFS of LysL producing L. lactis N8 than that of SPUsp45–LysL producer. The cell lysate containing LysL caused a clear inhibition zone whereas the SPUsp45–LysL lysate created a very diffuse halo. In case of LAC460, only CFS gave inhibition halo. The figure was digitally processed to enhance their quality.
Next, the cells were lysed to investigate the activity of LysL and SPUsp45–LysL inside the cell. As seen in Fig. 3(B), the cell lysate containing LysL gave a clear halo on the indicator plate, while SPUsp45–LysL resulted in much weaker inhibition. Hence, either most of the SPUsp45–LysL protein was secreted, or the activity of the SPUsp45–LysL fusion is lower than native LysL. The strong halo from concentrated LAC460 CFS but not from its lysate shows that in the wild-type strain LAC460, LysL is completely secreted. However, when expressed in heterologous L. lactis N8, most of the produced LysL is found inside the cells, as the clear halo observed in Fig. 3(A) results from 20-fold concentrated CFS, representing about 200 µl supernatant, while the halo in Fig. 3(B) represents cells from about 10 µl culture.
The attempt to produce LysL in the LysL-sensitive strain L. cremoris NZ9000 caused lysis of the producer after nisin induction. Figure 4 shows the lysis extent of NZ9000 producing LysL and SPUsp45–LysL after induction with nisin. Compared with the control NZ9000(pVS2), the culture of LysL producing NZ9000 turned completely clear within 80 min, whereas SPUsp45–LysL producing strain lysed only partially. Further incubation of the latter culture also resulted in complete lysis (result not shown). The slower lysis could be explained by incomplete cleavage of SPUsp45 during secretion, decreasing the activity of LysL. The result is comparable with that of L. lactis N8, where native LysL presented higher activity in CFS than when fused with SPUsp45. It can be concluded that in heterologous host, LysL is partly secreted without additional signal peptide, and that it is active both inside and outside the cell. Therefore, LysL could be produced in sensitive strain for the host autolysis, but not for purification purposes, as the secreted enzyme lyses the host too fast.
Figure 4.
Autolysis of the LysL-producing recombinant L. cremoris NZ9000 cultures before and 80 min after induction by nisin. LysL, L. cremoris NZ9000 producing native LysL; SPUsp45–LysL, L. cremoris NZ9000 producing SPUsp45–LysL; and control, L. cremoris NZ9000 carrying plasmid vector pVS2. After 5 h (OD600 about 0.5) incubation at 30°C, the cultures were induced by nisin (0.5 IU/ml) followed by 80 min incubation. Native LysL-producing culture turned completely transparent, whereas the SPUsp45–LysL-producing culture remained partly clear. The control strain culture without any LysL production became turbid. The figure was digitally processed to enhance their quality.
In vitro expression and identification of LysL
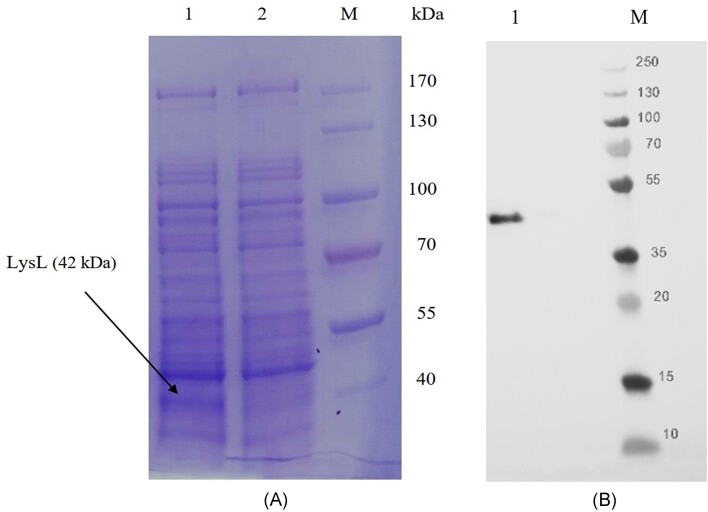
Even though LysL produced from heterologous L. lactis N8 was highly active and functional, the protein production level was still low for studying the mode of action of the enzyme. Therefore, we tested whether the cell-free in vitro gene expression system would give higher yield of LysL. The lysL template with T7 promoter and the adaptor sequences for the in vitro system was produced by OE-PCR, and the reaction was performed in 50 µl volume. Supplementary Figure S2 shows the comparison of the activity of LysL produced from different sources. The in vitro system could produce more LysL than the heterologous L. lactis N8. Since the expression in the cell-free translational system was stronger, the lysin produced in vitro was chosen for visualization on SDS-PAGE gel. The reaction solution of in vitro expression with lysL template, as well as the reaction solution without template as a control were electrophoresed in a gradient gel. Since the RTS 100 E. coli HY Kit is based on E. coli lysate, other intracellular E. coli proteins are also present in addition to the translated ones. As shown in Fig. 5(A), LysL (translated from lysL) presents the only additional band in the gel, when compared with the control. As marked in lane 1 the band is in the expected size of about 42 kDa. The result identifies the LysL protein, and further indicates that the antimicrobial activity of the cell-free translational reaction solution comes from LysL.
Figure 5.
(A) Coomassie blue-stained SDS-PAGE visualizing proteins produced in the cell-free translational system using an RTS 100 E. coli HY Kit. Lane 1, reaction solution using lysL as a template; Lane 2, reaction solution without template as a control; and M, PageRuler Protein Ladder (Thermo Scientific). The figure was digitally processed to enhance their quality. (B) Western blot analysis of the cell lysate of the culture of E. coli BL21(DE3) carrying pET-22b(+)-lysL. The His-Tag antibody used recognized LysL containing a 6 × His-Tag in the N-terminus (lane 1), M, PageRuler Broad Range Protein Ladder (Thermo Scientific).
Production of LysL in E. coli and purification of the recombinant protein
Although in vitro production of LysL was higher than that by the Lactococcus recombinant strains, the quantity versus costs was not adequate for further investigation of the mode of action. In order to obtain LysL in adequate quantity for further studies, LysL expression was carried out in E. coli and later purified by affinity chromatography using a His-Tag. The sequence encoding 6 × His was added to the 5′-end of lysL, ligated with the vector pET-22b(+), and cloned into E. coli BL21(DE3). Prior to purification, Western blot was carried out to confirm the intracellular location of LysL, as well as to confirm the presence of the N-terminal 6 × His-Tag in the produced LysL. The cell lysate obtained from overnight culture of E. coli ECO859 was used for anti-His antibody detection. The strong distinct band at about 44 kDa represents LysL carrying the 6 × His-Tag recognized by anti-His antibody (Fig. 5A).
LysL was purified using 6 × His-Tag and HisPur™ Ni-NTA chromatography. The optimum concentration of imidazole for elution of the bound protein in the last step was found to be 100 and 150 mM. From the 900 ml of the initial culture of E. coli ECO859, the total purified protein was 4.34 mg at a concentration of 0.7 mg/ml. The activity test of the purified enzyme showed that it was active (Supplementary Figure S3).
Discussion
In this study, we expressed L. lactis prophage lysin gene lysL in different heterologous hosts and in in vitro cell-free gene expression system. Production of active LysL enzyme was successful both in Lactococcus spp. and cell-free in vitro system. However, the protein yields obtained were insufficient for conducting further functional studies. Consequently, LysL was then produced and purified from recombinant E. coli cells.
Phage lysins have previously been expressed in many heterologous bacteria, including E. coli and Lactococcus spp. for various purposes. For example, phage lysins can be produced in cheese starter strains to lyse the starter cells for speeding up the cheese maturation step (de Ruyter et al. 1997). Still, the interest in phage lysins has more commonly been focused on producing lysins from phages of pathogenic or spoilage bacteria, and using the lysins as specific antimicrobials. For instance, prophage-encoded endolysin LysP108 from Staphylococcus aureus XN108, and several phage structural genes from Klebsiella bacteriophage ΦK64-1 have been expressed in E. coli, showing specific antimicrobial activity against the target bacteria (Lu et al. 2021 and Pan et al. 2017). As examples of using Lactococcus spp. as hosts for heterologous lysins, L. cremoris NZ9000 and L. lactis INIA 415 have been used to produce endolysins Endo88 from S. aureus bacteriophage 88, and CTP1 L from Clostridium tyrobutyricum, respectively (Garde et al. 2020, Chandran et al. 2022). Similar to our study here, these Lactococcus studies used nisin promoter to drive the lysin gene expression; constitutive expression in the nisin producing strain INIA 415, and nisin inducible production in the strain NZ9000. The latter strain is undoubtedly the most commonly used Lactococcus for nisin controlled gene expression, also known as the NICE system (Mierau and Kleerebezem 2005). Nisin induction is generally regarded as strong and tightly regulated, and therefore overproducing active LysL in another Lactococcus strain felt a reasonable and safe strategy, in terms of e.g. avoiding protein misfolding. Tight regulation of the gene expression was indeed demonstrated, as the apparent suicide gene lysL could easily be cloned under nisin promoter in LysL sensitive NZ9000. However, the induced production of LysL enzyme was highly detrimental for the host, and hence large-scale production of the protein would be impossible. Nevertheless, successful production and secretion of LysL in different Lactococcus strains demonstrated its potential usefulness in cheese making. Production of LysL in a LysL-sensitive starter strain or a LysL-resistant adjunct starter would be expected to cause the release of starter cells’ cytosolic enzymes to the cheese matrix for accelerated cheese ripening.
To overproduce LysL in heterologous Lactococcus, we aimed to try both intracellular and extracellular production. We hypothesized that addition of the signal peptide of Usp45 to the N-terminus of LysL would improve its secretion, which could also be required for the activity of LysL. SPUsp45 has been widely used for secreting homologous and heterologous proteins in lactococci and other bacteria. For instance, β-1,3–1,4-glucanase from Bacillus sp. SJ-10 (Tak et al. 2019) and glucansucrase from Leuconostoc mesenteroides (Skory and Côté 2015) have been secreted from the recombinant Lactococcus strains by SPUsp45. Replacement of the native signal peptide with SPUsp45 has also been reported to enhance secretion of recombinant proteins. For example, after deletion of the signal peptide's inherent sequence in cbh2, bgl1, and egl3, also using SPUsp45 instead, the secretion in L. lactis was enhanced (Liu et al. 2016). Nevertheless, in some cases, manipulating the signal peptide of the recombinant proteins has impaired secretion. For example, Borrero et al. (2011) reported that, when produced by E. hirae DCH5, antimicrobial activity of the recombinant hiracin JM79, in which the inherent signal peptide was replaced by SPUsp45, was decreased. Therefore, although SPUsp45 has been proven to facilitate extracellular secretion of recombinant proteins in many studies, there is no guarantee that it is better signal peptide than the native one in all cases. Our study also provided an example for SPUsp45 not improving the yield of active enzyme in supernatant, since the fusion of SSusp45 with lysL in recombinant lactococci caused lower inhibitory activity compared with the corresponding native LysL producers. As shown in Fig. 3(B), the lysate of the nisin producing L. lactis host N8 producing LysL created a strong halo on the indicator plate, while the halo from SPUsp45–LysL was significantly fainter. This illustrates that SPUsp45 attached to LysL disturbs the activity of LysL, as all LysL in the lysate is expected to be fused with SPUsp45. Hence, it is possible that SPUsp45 improved the secretion of LysL but decreased its activity due to incomplete cleavage of the leader. The weak halo from SPUsp45–LysL in Fig. 3(A) probably reflects the portion of LysL in the CFS, which has been secreted through the Sec-dependent pathway, and from which the SPUsp45 had been cleaved.
Interestingly, in the wild-type producer strain LAC460, LysL was completely excreted to the environment, as no active LysL was found in the cell lysate (Fig. 3B). The protein seems not to contain a typical signal peptide for secretion, and hence, the mechanism of the secretion is unknown. It is still clear that LysL finds its way out of the LAC460 cells, whereas in the heterologous L. lactis host N8, LysL mostly remains inside the cell, suggesting that some factors needed for efficient secretion are missing in the strain N8. As the production of LysL in L. lactis N8 turned out to be somewhat ineffective,therefore, we then aimed to produce LysL in a cell-free in vitro system.
Cell-free protein synthesis may yield a higher production rate of recombinant proteins than cellular expression systems. The system is also useful for production of toxic proteins, or those that are difficult to express (Jin and Hong 2018), For instance, cell-based production of colicin, a bacteriocin from E. coli, requires coproduction of immunity proteins, if produced in E. coli, and therefore the overexpression of colicin is more convenient in vitro (Jin et al. 2018). In our study, the in vitro production of LysL provided sufficiently protein for antimicrobial tests and SDS-PAGE. However, the total protein yield fell short of expectation, and particularly the quantity versus costs was not reasonable for producing large amounts of LysL by in vitro system. Therefore, the decision was made to overproduce LysL in E. coli as the host. After all, the E. coli approach proved to be both straightforward and successful, resulting in significant quantities of active LysL protein suitable for subsequent functional studies with minimal challenges such as protein misfolding or the formation of inclusion bodies. Escherichia coli has frequently been used as the host for overexpression of lysins (Wang et al. 2020, Chu et al. 2022). For instance, Dreher-Lesnick et al. (2015) demonstrated the use of recombinant Lactobacillus phage lysin LysA2 for purity assays of probiotics, and Chu et al. (2022) highlighted the antimicrobial potential of Abp013 against multidrug-resistant Acinetobacter baumannii, with both studies relying on purified lysin.
There are several possible explanations about the secretion mechanism of LysL in LAC460. To date, many protein secretion pathways have been discovered in Gram-positive bacteria, for example the classical Sec and Tat pathways, and at least seven nonclassical secretion mechanisms (Dai et al. 2022). Nonclassically secreted proteins are proteins without a predictable signal sequence or known secretory pathways, but which are still found in the extracellular space. An apparent secretion mechanism of LysL in its native host LAC460 could be through a holin pore, as prophage holins have been reported to mediate nonlytic secretion of proteins, particularly phage endolysins (Mukherjee et al. 2002, Palmer et al. 2021, Brüser and Mehner-Breitfeld 2022). The type of holin determines whether it contributes to transferring proteins into the cell exterior through secretion, leakage, or membrane lysis (Desvaux 2012).
In any case, dozens of nonclassically secreted proteins have been reported from Lactococcus, Listeria, Streptococcus, and other Gram-positive bacteria (Wang et al. 2016). Perhaps LysL is another nonclassical secretory protein, and its mechanism of secretion remains to be investigated.
Supplementary Material
Acknowledgements
The authors acknowledge the Biocenter Finland and Tampere facility of Protein Services (PS) for their service.
Contributor Information
Samira Mokhtari, Department of Microbiology, University of Helsinki, PO Box 56, FI-00014 Helsinki, Finland.
Per E J Saris, Department of Microbiology, University of Helsinki, PO Box 56, FI-00014 Helsinki, Finland.
Timo M Takala, Department of Microbiology, University of Helsinki, PO Box 56, FI-00014 Helsinki, Finland.
Conflict of interest
None declared.
Funding
Samira Mokhtari was funded by personal working grants from the Niemi Foundation (20200071) and the Finnish Cultural Foundation (27.02.2021).
References
- Abdelrahman F, Easwaran M, Daramola OIet al. . Phage-encoded endolysins. Antibiotics. 2021;10:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucouturier A, Chain F, Langella Pet al. . Characterization of a prophage-free derivative strain of Lactococcus lactis ssp. lactis IL1403 reveals the importance of prophages for phenotypic plasticity of the host. Front Microbiol. 2018;9:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bıyıklı A, Niçin R, Dertli Eet al. . Extracellular recombinant production of 4,6 and 4,3 α-glucanotransferases in Lactococcus lactis. Enzyme Microb Technol. 2023;164:110175. [DOI] [PubMed] [Google Scholar]
- Borrero J, Jiménez JJ, Gútiez Let al. . Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis. Appl Microbiol Biotechnol. 2011;89:131–43. [DOI] [PubMed] [Google Scholar]
- Brüser T, Mehner-Breitfeld D. Occurrence and potential mechanism of holin-mediated non-lytic protein translocation in bacteria. Microb Cell. 2022;9:159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. [DOI] [PubMed] [Google Scholar]
- Chandran C, Tham HY, Abdul Rahim Ret al. . Lactococcus lactis secreting phage lysins as a potential antimicrobial against multi-drug resistant Staphylococcus aureus. PeerJ. 2022;10:e12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJK, Poh WH, Hasnuddin NTBet al. . Novel phage lysin abp013 against Acinetobacter baumannii. Antibiotics. 2022;11:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Li J, Li Qet al. . PncsHub: a platform for annotating and analyzing non-classically secreted proteins in Gram-positive bacteria. Nucleic Acids Res. 2022;50:D848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruyter PG, Kuipers OP, Meijer WCet al. . Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat Biotechnol. 1997;15:976–9. [DOI] [PubMed] [Google Scholar]
- Desvaux M. Contribution of holins to protein trafficking: secretion, leakage or lysis?. Trends Microbiol. 2012;20:259–61. [DOI] [PubMed] [Google Scholar]
- Dreher-Lesnick SM, Schreier JE, Stibitz S. Development of phage lysin lysa2 for use in improved purity assays for live biotherapeutic products. Viruses. 2015;7:6675–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, São-José C. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses. 2018;10:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garde S, Calzada J, Sánchez Cet al. . Effect of Lactococcus lactis expressing phage endolysin on the late blowing defect of cheese caused by Clostridium tyrobutyricum. Int J Food Microbiol. 2020;329:108686. [DOI] [PubMed] [Google Scholar]
- Hernandez-Valdes JA, Huang C, Kok Jet al. . Another breaker of the wall: the biological function of the Usp45 protein of Lactococcus lactis. Appl Environ Microb. 2020;86:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microb. 1989;55:3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Hong SH. Cell-free protein synthesis for producing ‘difficult-to-express’ proteins. Biochem Eng J. 2018;138:156–64. [Google Scholar]
- Jin X, Kightlinger W, Kwon YCet al. . Rapid production and characterization of antimicrobial colicins using Escherichia coli-based cell-free protein synthesis. SYNBIO. 2018;3:ysy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher P, Mahony J, Schweinlin Ket al. . Assessing the functionality and genetic diversity of lactococcal prophages. Int J Food Microbiol. 2018;272:29–40. [DOI] [PubMed] [Google Scholar]
- Kuipers OP, de Ruyter PG, Kleerebezem Met al. . Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- Liu SN, Takala TM, Reunanen Jet al. . Investigation of Listeria phage endolysin cell-wall binding domain (CBD) surface display in Escherichia coli. In: Proceedings of the International Conference on Biological Sciences and Technology, Dordrecht: Atlantis Press, 2016, 23–28. [Google Scholar]
- Lozo J, Mirkovic N, O'Connor PMet al. . Lactolisterin BU, a novel class II broad-spectrum bacteriocin from Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4. Appl Environ Microb. 2017;83:e01519–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang Y, Wang Jet al. . Phage endolysin lysp108 showed promising antibacterial potential against methicillin-resistant Staphylococcus aureus. Front Cell Infect Microbiol. 2021;11:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68:705–17. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Karlsson S, Burman LGet al. . Proteins released during high toxin production in Clostridium difficile. Microbiology. 2002;148:2245–53. [DOI] [PubMed] [Google Scholar]
- Palmer T, Finney AJ, Saha CKet al. . A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol Microbiol. 2021;115:345–55. [DOI] [PubMed] [Google Scholar]
- Pan YJ, Lin TL, Chen CCet al. . Klebsiella phage φk64-1 encodes multiple depolymerases for multiple host capsular types. J Virol. 2017;91:e02457–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y, Courtin P, Mézange Cet al. . Lactococcus lactis gene yjgB encodes a gamma-D-glutaminyl-L-lysyl-endopeptidase which hydrolyzes peptidoglycan. Appl Environ Microb. 2007;73:5825–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Reddy BL. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol. 2015;197:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW, Irwin Net al. . Molecular Cloning: A Laboratory Manual. 3th edn.Vol 2. New York: Cold Spring Harbor Laboratory Press, 2001. [Google Scholar]
- Simons A, Alhanout K, Duval RE. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. 2020;8:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skory CD, Côté GL. Secreted expression of Leuconostoc mesenteroides glucansucrase in Lactococcus lactis for the production of insoluble glucans. Appl Microbiol Biotechnol. 2015;99:10001–10. [DOI] [PubMed] [Google Scholar]
- Soltani S, Hammami R, Cotter PDet al. . Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. FEMS Microbiol Rev. 2021;45:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–30. [DOI] [PubMed] [Google Scholar]
- Tak JY, Jang WJ, Lee JMet al. . Expression in Lactococcus lactis of a β-1,3-1,4-glucanase gene from Bacillus sp. SJ-10 isolated from fermented fish. Protein Expr Purif. 2019;162:18–23. [DOI] [PubMed] [Google Scholar]
- Takala T, Koponen O, Qiao Met al. . Lipid-free NisI: interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol Lett. 2004;237:171–7. [Google Scholar]
- Takala TM, Mokhtari S, Ahonen SLet al. . Wild-type Lactococcus lactis producing bacteriocin-like prophage lysins. Front Microbiol. 2023;14:1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GRR, Kurek D, Szeliga Met al. . Expression of prophage-encoded endolysins contributes to autolysis of Lactococcus lactis. Appl Microbiol Biotechnol. 2017;101:1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GRR, Steen A, Leenhouts Ket al. . AcmD, a homolog of the major autolysin AcmA of Lactococcus lactis, binds to the cell wall and contributes to cell separation and autolysis. PLoS One. 2013;8:e72167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wright A, Tynkkynen S, Suominen M. Cloning of a Streptococcus lactis subsp. lactis chromosomal fragment associated with the ability to grow in milk. Appl Environ Microb. 1987;53:1584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Saris PEJ, Takala TM. Genetic characterization and expression of leucocin B, a class IId bacteriocin from Leuconostoc carnosum 4010. Res J Microbiol. 2015;166:494–503. [DOI] [PubMed] [Google Scholar]
- Wan X, Takala TM, Qiao Met al. . Complete genome sequence of nisin producing Lactococcus lactis subsp. lactis N8. Microbiol Resour Announc. 2021;10:e01147–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Usvalampi AM, Saris PEet al. . A counterselection method for Lactococcus lactis genome editing based on class IIa bacteriocin sensitivity. Appl Microbiol Biotechnol. 2016;100:9661–9. [DOI] [PubMed] [Google Scholar]
- Wang F, Zhang G, Peng Jet al. . High cell-density fermentation, expression and purification of bacteriophage lysin TSPphg, a thermostable antimicrobial protein from extremophilic Thermus bacteriophage TSP4. Protein Expr Purif. 2020;174:105676. [DOI] [PubMed] [Google Scholar]
- Wang G, Xia Y, Song Xet al. . Common non-classically secreted bacterial proteins with experimental evidence. Curr Microbiol. 2016;72:102–11. [DOI] [PubMed] [Google Scholar]
- Zabarovsky RE, Winberg G. High efficiency electroporation of ligated DNA into bacteria. Nucl Acids Res. 1990;18:5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.