Abstract
Dipterocarpus retusus Blume is an endangered species on the IUCN Red List. In this study, we reported the complete chloroplast (cp) genome of D. retusus (GenBank accession number: OP271853). The cp genome was 154,303 bp long, with a large single-copy (LSC) region of 85,586 bp and a small single-copy (SSC) region of 20,273 bp separated by a pair of inverted repeats (IRs) of 24,222 bp. It encodes 128 genes, including 84 protein-coding genes, 36 tRNA genes, and eight ribosomal RNA genes. We also reconstructed the cp genome phylogeny of Dipterocarpus, which indicated D. retusus was closely related with the sympatric species D. gracilis. This study may contribute valuable information to the phylogenetic relationships within the genus Dipterocarpus.
Keywords: Dipterocarpus retusus, Dipterocarpus, chloroplast genome, phylogenomics
Introduction
Species of Dipterocarpaceae are regarded as the symbolic species of South-east Asian tropical rain forests and many seasonally dry forests (Brearley et al. 2017). Many species in Dipterocarpaceae are the most important and valuable source in the timber market for producing large quantity and high-quality wood (Schulte and Schöne 1996). The borneol obtained from some Dipterocarpaceae trees has been widely used in the fields of medicine, pesticide, and chemical industry (Yang et al. 2020; Dong et al. 2021). Although Dipterocarpaceae is important to forest ecology, medicine, and conservation, the classifications of Dipterocarpaceae (e.g. the delineation of genera Parashorea and Shorea) still remain controversial (Cvetković et al. 2019; Zhu and Sun 2019; Yu et al. 2021). Over the past few decades, next-generation sequencing has experienced rapid advancements, providing accurate, comprehensive, and cost-effective approaches for de novo assembly of organelle genomes, and the gene-rich plastid genome has been widely used for inferring phylogenetic relationships of closely related plant species (Jin et al. 2020; Tao et al. 2020). Dipterocarpus retusus Blume 1868 is an endangered species on the IUCN Red List (Bodos et al. 2019) which has a native range from Assam to China (W. & SE. Yunnan), Indo-China to Lesser Sunda Islands. This species has high economic value as an important timber species and ecological value as a key structural component of the rain forest. Therefore, we assembled and characterized the complete chloroplast (cp) genome of D. retusus, with the aim of establishing a solid foundation for future phylogenetic investigations and medical applications of Dipterocarpus.
Materials and methods
Leaf material of D. retusus (Figure 1) was collected from the Linhai campus of Taizhou University, Linhai, Zhejiang province, China (28.8808N, 121.1722E). A voucher specimen was deposited at Herbarium of Taizhou University (https://www.tzc.edu.cn/; collector: Zhong-Shuai Sun, sun2143998@163.com) under the voucher number Sun2108001. Total genomic DNA was extracted as reported in Chen et al. (2022).
Figure 1.
The morphology of D. retusus. The leaves of D. retusus are broadly ovate in shape, and the petioles are devoid of hair, unlike the same genus species of Dipterocarpus gracilis Blume, which exhibits oblong leaves and stellate hairy petioles. Pictures were taken by the author Zhong-Shuai Sun on 2022-06-01 in Linhai campus of Taizhou University that located in Zhejiang province of China.
Sequencing was performed with an Illumina NovaSeq platform (Illumina, San Diego, CA). The cp genome was assembled via NOVOPlasty 2.6.3 (Dierckxsens et al. 2017), using the Dipterocarpus tempehes (NC026839, Undaharta et al. 2020) as reference. The cp genome annotation was performed online using GeSeq v.1.59 (Tillich et al. 2017) by comparing the cp genome sequence of D. tempehes (NC026839, Undaharta et al. 2020). Geneious R11 (Biomatters Ltd., Auckland, New Zealand) was used for inspecting the cp genome structure (Kearse et al. 2012). Circular cp genome map was visualized with CPGView software (Liu et al. 2023).
The phylogenetic relationship with Dipterocarpus was evaluated based on an alignment of concatenated protein-coding regions (CDS) among all predicted protein coding genes by the software MAFFT v7.475 (Katoh and Standley 2013). The CDS sequences were extracted from 18 complete cp genomes of Dipterocarpaceae. Shorea zeylanica (NC040965, Heckenhauer et al. 2019) and Parashorea chinensis (NC046579, Zhu and Sun 2019) were used as outgroups. A maximum-likelihood (ML) analysis was conducted using RAxML-HPC v.8.2.10 on the CIPRES cluster (Miller et al. 2010). GTRCAT model and rapid bootstrap analysis (1000 replicates) were employed for ML inference.
Results
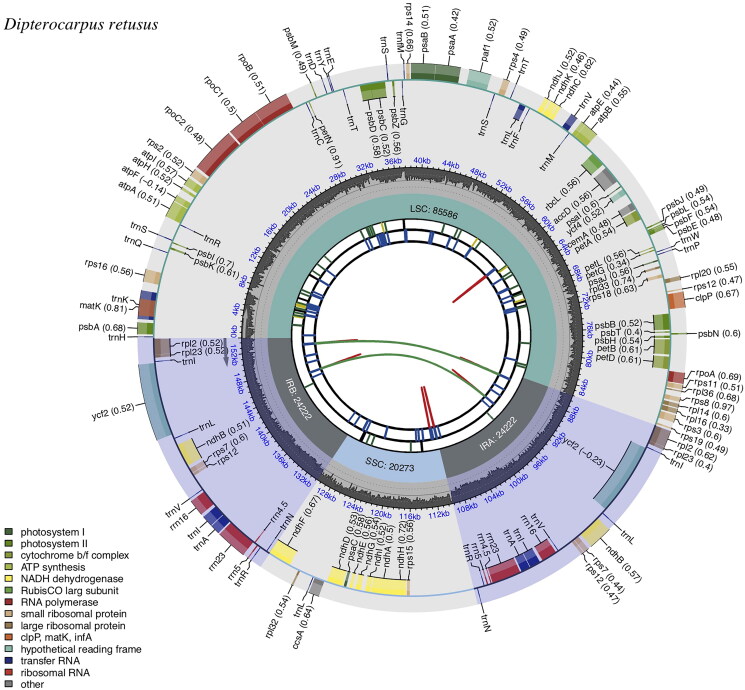
The complete cp genome of D. retusus spanned a length of 154,303 bp. The read coverage depth was sufficient (with an average of 1314×), indicating the robustness of genome assembly (Supplementary Figure 1). It exhibited a typical quadripartite structure (Figure 2) which was common in angiosperms: a large single-copy (LSC) region (85,586 bp), a small single-copy (SSC) region (20,273 bp), and a pair of inverted repeats (IRs) region (24,222 bp). The overall GC contents of the total length, LSC, SSC, and IR regions were 37.0%, 34.8%, 31.5%, and 43.1%, respectively.
Figure 2.
Schematic map of D. retusus chloroplast genome constructed by CPGview (http://www.1kmpg.cn/cpgview/). The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats connected with arcs. The second track shows the long tandem repeats as short bars. The third track shows the short tandem repeats or microsatellite sequences as short bars. The small single-copy (SSC), inverted repeat (IRs), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification is shown at the bottom left.
In total, 128 genes were annotated in the genome, including 84 protein-coding genes (PCGs), 36 tRNA genes, and eight rRNA genes. Among these genes, 13 genes (clpP, ndhA, ndhB, petB, petD, rpl2, rpoC1, rps16, trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) had one intron and two genes (rps12 and pafI) had two introns. Most genes were single-copy genes, while six PCGs (ndhB, rpl2, rpl23, rps7, rps12, and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) in IR regions were duplicated. The genome contained 11 cis-splicing genes and one trans-splicing gene (Supplementary Figures 2 and 3).
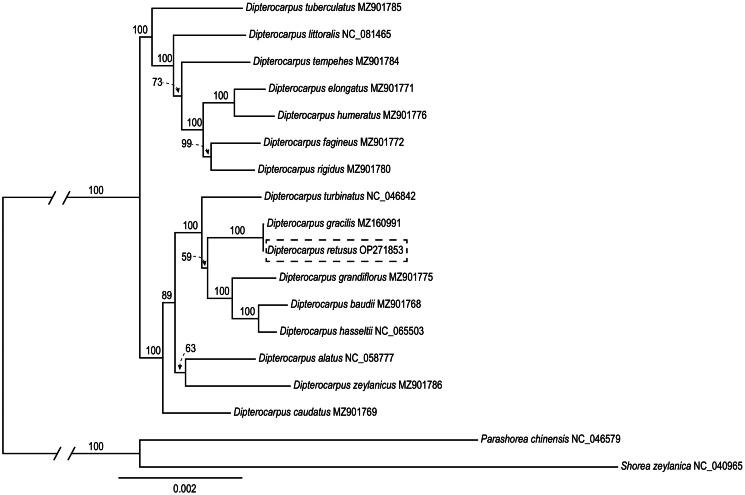
A robust phylogenetic tree of Dipterocarpus was obtained based on the CDS data; most nodes in the phylogenetic tree were highly supported (Figure 3). According to the phylogenetic tree, Dipterocarpus was resolved as a monophyletic clade consisting two well supported clades. Among the Dipterocarpus species analyzed, D. retusus was observed to be closely related with D. gracilis according to the phylogenetic tree, and the relationship was fully supported.
Figure 3.
Maximum-likelihood (ML) tree reconstruction of 18 taxa from Dipterocarpaceae based on 84 shared CDS in the chloroplast genomes. Relative branch lengths are indicated. Support values above the branches are ML bootstrap support. The following sequences were used: NC_046579 (Zhu and Sun 2019), NC_040965 (Heckenhauer et al. 2019), MZ160991 (Yu et al. 2021), NC_046842 (Ci et al. 2019), NC_058777 (direct submission to NCBI), NC_065503 (direct submission to NCBI), NC_081465 (direct submission to NCBI), MZ901768 (Cvetković et al. 2022), MZ901769 (Cvetković et al. 2022), MZ901771 (Cvetković et al. 2022), MZ901772 (Cvetković et al. 2022), MZ901775 (Cvetković et al. 2022), MZ901776 (Cvetković et al. 2022), MZ901780 (Cvetković et al. 2022), MZ901784 (Cvetković et al. 2022), MZ901785 (Cvetković et al. 2022), and MZ901786 (Cvetković et al. 2022).
Discussion and conclusions
The cp genome of D. retusus was assembled using short-read data in this study. It showed that the genome size, GC content, and gene composition of the cp genome sequence of D. retusus were similar to those of other species of the genus Dipterocarpus (Ci et al. 2019; Yu et al. 2021; Cvetković et al. 2022). Our phylogenetic result coincides with previous molecular phylogeny studies (Jacqueline et al. 2017; Yu et al. 2021), but provides additional insights of the phylogeny of Dipterocarpus for involved more Dipterocarpus species. Notably, D. retusus is closely with a sympatric species D. gracilis among the Dipterocarpus species analyzed with robust bootstrap support (Figure 3). We expect that the cp genome of D. retusus will be a valuable resource for future studies on molecular identification and the better understanding of phylogeny in Dipterocarpus and Dipterocarpaceae.
Supplementary Material
Funding Statement
This work was supported by the Natural Science Foundation of China [No. 31400321].
Author contributions
YTT designed and conceived this article; MY and QQL assembled and annotated the cp genome; ZSS collected and identified the plant material, and revised the manuscript. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Ethical approval
Dipterocarpus retusus is listed as an endangered plant in the IUCN Red List of Threatened Species and the wild populations are strictly protected in China. However, the plant material we used in this study was from a cultivated seedling that planted in the Linhai Campus of Taizhou University (the seeds were obtained from Xishuangbanna Tropical Botanical Garden). Thus, no specific permissions were needed to perform this research since no damage was caused to its natural population. This study was permitted by Zhejiang Provincial Key Laboratory of Plant Evolutionary Ecology and Conservation, Taizhou University, China. All collection and sequencing work were strictly executed under local legislation and related laboratory regulations to protect wild resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP271853. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1054675, SRR27290476, and SAMN38930875, respectively.
References
- Bodos V, Kusumadewi Y, Robiansyah I, Randi A, Tsen S, Pereira J, Maycock CR.. 2019. Dipterocarpus tempehes. The IUCN red list of threatened species. T33378A125628354. doi: 10.2305/IUCN.UK.2019-3.RLTS.T33378A125628354.en. [DOI]
- Brearley FQ, Banin LF, Saner P.. 2017. The ecology of the Asian Dipterocarps. Plant Ecol Divers. 9(5–6):429–436. doi: 10.1080/17550874.2017.1285363. [DOI] [Google Scholar]
- Chen C, Qu D-H, Shan F-Q, Jin Z-X, Sun Z-S.. 2022. Complete chloroplast genome of Lonicera crassifolia Batalin (Caprifoliaceae) and its phylogenetic implications. Mitochondrial DNA B Resour. 7(5):732–734. doi: 10.1080/23802359.2022.2068979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci X, Peng J, Shi C, Zhu Z, Cai N, Duan A, Wang D.. 2019. The complete chloroplast genome of Dipterocarpus turbinatus Gaertn. F. Mitochondrial DNA B Resour. 4(2):3636–3637. doi: 10.1080/23802359.2019.1677192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetković T, Hinsinger DD, Strijk JS.. 2019. Exploring evolution and diversity of Chinese Dipterocarpaceae using next-generation sequencing. Sci Rep. 9(1):11639. doi: 10.1038/s41598-019-48240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetković T, Hinsinger DD, Thomas DC, Wieringa JJ, Velautham E, Strijk JS.. 2022. Phylogenomics and a revised tribal classification of subfamily Dipterocarpoideae (Dipterocarpaceae). Taxon. 71(1):85–102. doi: 10.1002/tax.12648. [DOI] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Zhao Y, Chen J, Chang M, Wang X, Jin Q, Wang X.. 2021. Enzymatic lipophilization of d-borneol extracted from Cinnamomum camphora chvar. Borneol seed. LWT. 148:111801. doi: 10.1016/j.lwt.2021.111801. [DOI] [Google Scholar]
- Heckenhauer J, Paun O, Chase MW, Ashton PS, Kamariah AS, Samuel R.. 2019. Molecular phylogenomics of the tribe Shoreeae (Dipterocarpaceae) using whole plastid genomes. Ann Bot. 123(5):857–865. doi: 10.1093/aob/mcy220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacqueline H, Rosabelle S, Peter SA, Barbara T, Michael HJB, Tae-Soo J, Eva MT, Jamie M, Kamariah AS, Achala SA, et al. . 2017. Phylogenetic analyses of plastid DNA suggest a different interpretation of morphological evolution than those used as the basis for previous classifications of Dipterocarpaceae (Malvales). Bot J Linn Soc. 185(1):1–26. doi: 10.1093/botlinnean/box044. [DOI] [Google Scholar]
- Jin J, Tao Y-T, Ding X-B, Guo W-P, Ruan L, Yang Q-L, Chen P-C, Yao H, Zhang H-B, Chen X.. 2020. Predicted yeast interactome and network-based interpretation of transcriptionally changed genes. Yeast. 37(11):573–583. doi: 10.1002/yea.3516. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C.. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop 14; p. 1–8. doi: 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- Schulte A, Schöne D.. 1996. Dipterocarp forest ecosystems: towards sustainable management. London: World Scientific. doi: 10.1142/3160. [DOI] [Google Scholar]
- Tao Y-T, Ding X-B, Jin J, Zhang H-B, Guo W-P, Ruan L, Yang Q-L, Chen P-C, Yao H, Chen X.. 2020. Predicted rat interactome database and gene set linkage analysis. Database. 2020:baaa086. doi: 10.1093/database/baaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, , Lehwark P, , Pellizzer T, , Ulbricht-Jones ES, , Fischer A, , Bock R, , Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undaharta NKE, Cvetković T, Wee AKS.. 2020. Complete plastome sequence of endangered timber species Dipterocarpus tempehes Slooten. Mitochondrial DNA B. 5(3):3136–3138. doi: 10.1080/23802359.2020.1800428. [DOI] [Google Scholar]
- Yang L, Zhan C, Huang X, Hong L, Fang L, Wang W, Su J.. 2020. Durable antibacterial cotton fabrics based on natural borneol-derived anti‐MRSA agents. Adv Healthc Mater. 9(11):e2000186. doi: 10.1002/adhm.202000186. [DOI] [PubMed] [Google Scholar]
- Yu Y, Han Y, Peng Y, Tian Z, Zeng P, Zong H, Zhou T, Cai J.. 2021. Comparative and phylogenetic analyses of eleven complete chloroplast genomes of Dipterocarpoideae. Chin Med. 16(1):125. doi: 10.1186/s13020-021-00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-F, Sun Y.. 2019. The complete chloroplast genome of the endangered tree Parashorea chinensis (Dipterocarpaceae). Mitochondrial DNA B. 4(1):1163–1164. doi: 10.1080/23802359.2019.1591236. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bodos V, Kusumadewi Y, Robiansyah I, Randi A, Tsen S, Pereira J, Maycock CR.. 2019. Dipterocarpus tempehes. The IUCN red list of threatened species. T33378A125628354. doi: 10.2305/IUCN.UK.2019-3.RLTS.T33378A125628354.en. [DOI]
Supplementary Materials
Data Availability Statement
The genome sequence data of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OP271853. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1054675, SRR27290476, and SAMN38930875, respectively.