Abstract
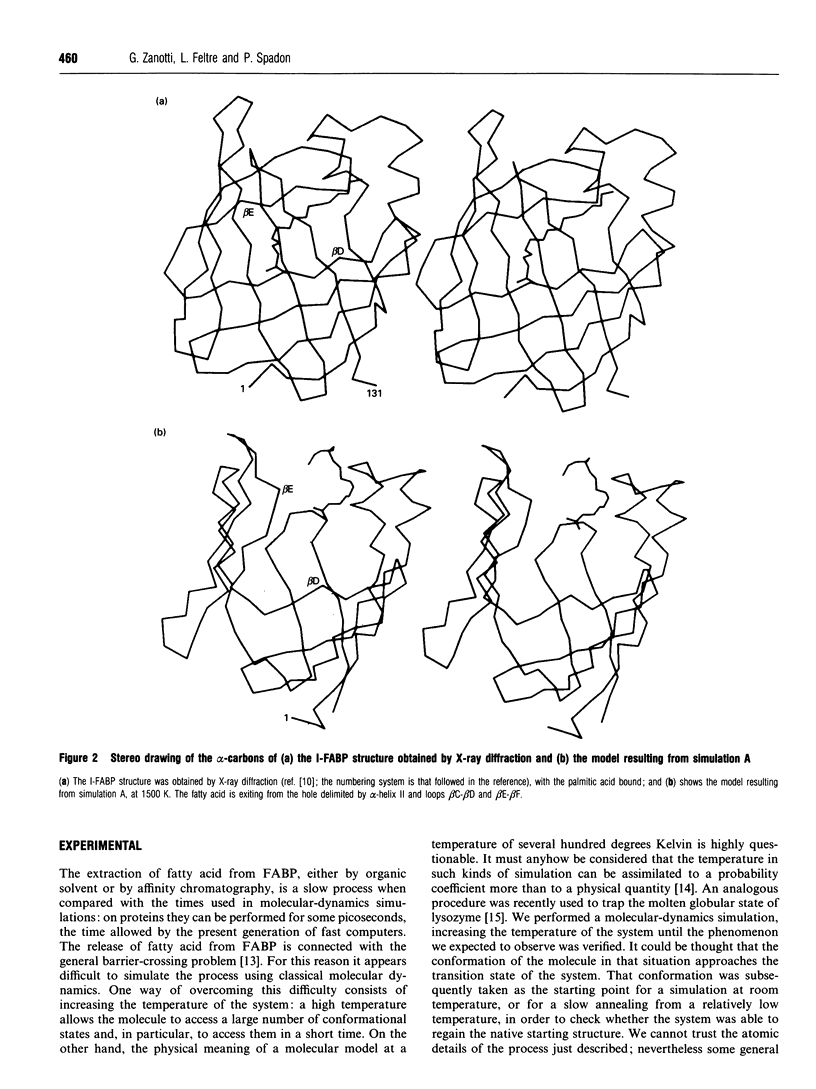
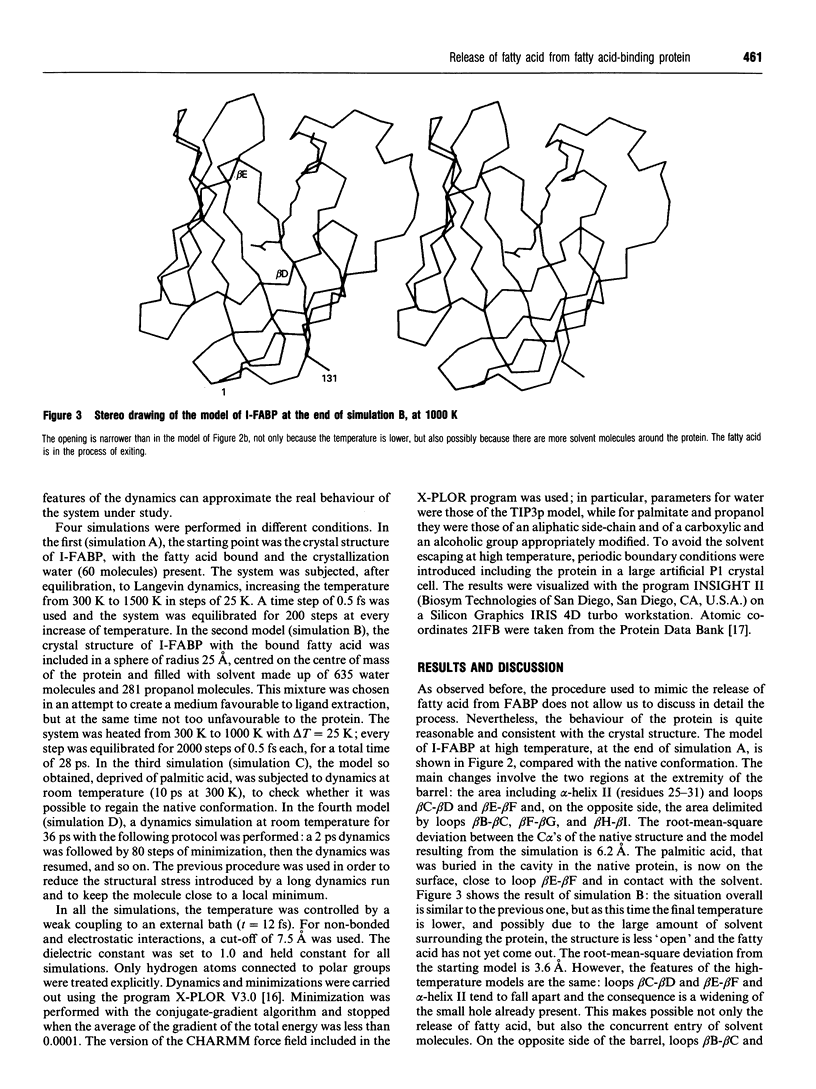
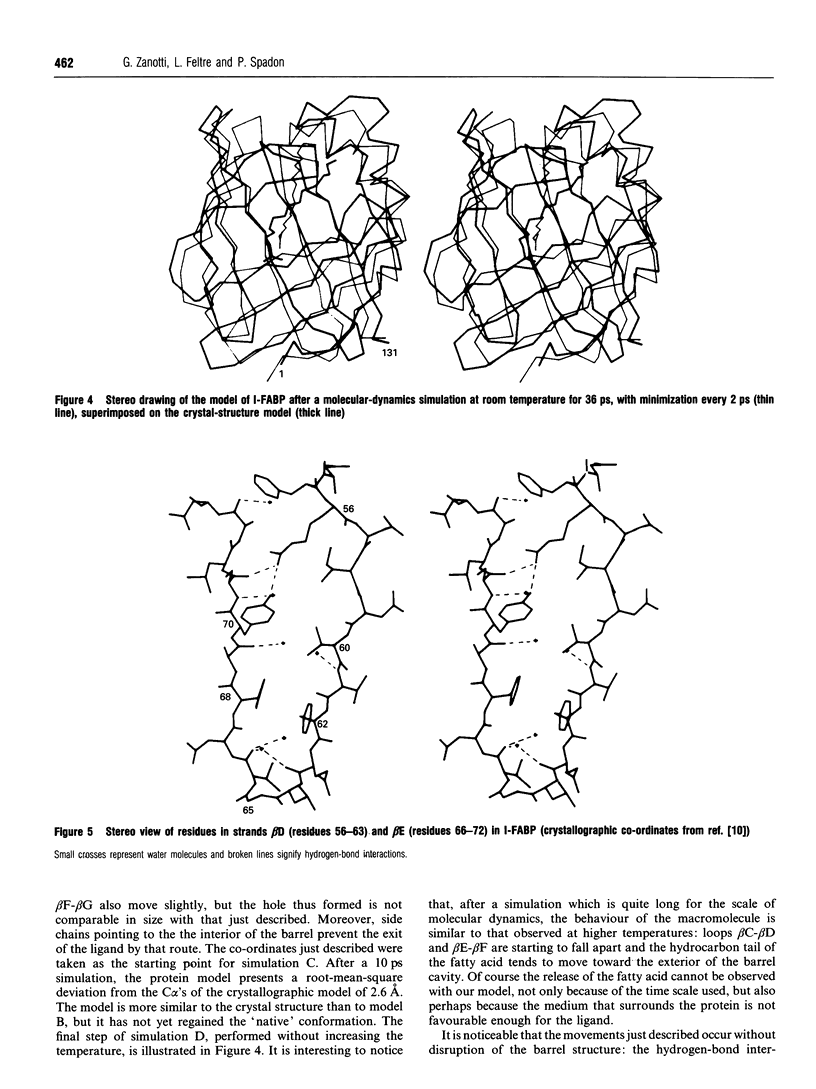
A simulation of the release of fatty acid from intestinal fatty acid-binding protein was attempted, starting with the crystallographic model and using molecular-dynamic processes at different temperatures. The release of the ligand was observed only at high temperature, which perhaps makes the process unreliable in detail. Nevertheless, the overall behaviour of the protein, also confirmed by the simulation performed at room temperature, strongly supports the idea that the fatty acid leaves the protein through an opening formed by alpha-helix II and turns beta C-beta D and beta E-beta F. Additionally, it suggests a role for the lack of hydrogen bonds between the main chains of beta-strands D and E: this feature, observed in all the protein structures of this family which have currently been determined, seems to provide the structure with great flexibility, allowing the barrel to open and close without disruption of the hydrogen-bond network.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benning M. M., Smith A. F., Wells M. A., Holden H. M. Crystallization, structure determination and least-squares refinement to 1.75 A resolution of the fatty-acid-binding protein isolated from Manduca sexta L. J Mol Biol. 1992 Nov 5;228(1):208–219. doi: 10.1016/0022-2836(92)90501-a. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Bergfors T., Sedzik J., Unge T. The three-dimensional structure of P2 myelin protein. EMBO J. 1988 Jun;7(6):1597–1604. doi: 10.1002/j.1460-2075.1988.tb02985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A. E., van Gunsteren W. F. Simulation of the thermal denaturation of hen egg white lysozyme: trapping the molten globule state. Biochemistry. 1992 Sep 1;31(34):7745–7748. doi: 10.1021/bi00149a001. [DOI] [PubMed] [Google Scholar]
- Müller-Fahrnow A., Egner U., Jones T. A., Rüdel H., Spener F., Saenger W. Three-dimensional structure of fatty-acid-binding protein from bovine heart. Eur J Biochem. 1991 Jul 15;199(2):271–276. doi: 10.1111/j.1432-1033.1991.tb16120.x. [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Gordon J. I., Banaszak L. J. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J Mol Biol. 1989 Jul 20;208(2):327–339. doi: 10.1016/0022-2836(89)90392-6. [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Gordon J. I., Banaszak L. J. The structure of crystalline Escherichia coli-derived rat intestinal fatty acid-binding protein at 2.5-A resolution. J Biol Chem. 1988 Apr 25;263(12):5815–5819. [PubMed] [Google Scholar]
- Scapin G., Gordon J. I., Sacchettini J. C. Refinement of the structure of recombinant rat intestinal fatty acid-binding apoprotein at 1.2-A resolution. J Biol Chem. 1992 Feb 25;267(6):4253–4269. doi: 10.2210/pdb1ifc/pdb. [DOI] [PubMed] [Google Scholar]
- Scapin G., Spadon P., Mammi M., Zanotti G., Monaco H. L. Crystal structure of chicken liver basic fatty acid-binding protein at 2.7 A resolution. 1990 Oct 15-Nov 8Mol Cell Biochem. 98(1-2):95–99. doi: 10.1007/BF00231372. [DOI] [PubMed] [Google Scholar]
- Wilkinson T. C., Wilton D. C. Studies on fatty acid-binding proteins. The binding properties of rat liver fatty acid-binding protein. Biochem J. 1987 Oct 15;247(2):485–488. doi: 10.1042/bj2470485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter N. S., Bratt J. M., Banaszak L. J. Crystal structures of holo and apo-cellular retinol-binding protein II. J Mol Biol. 1993 Apr 20;230(4):1247–1259. doi: 10.1006/jmbi.1993.1239. [DOI] [PubMed] [Google Scholar]
- Xu Z., Bernlohr D. A., Banaszak L. J. Crystal structure of recombinant murine adipocyte lipid-binding protein. Biochemistry. 1992 Apr 7;31(13):3484–3492. doi: 10.1021/bi00128a024. [DOI] [PubMed] [Google Scholar]
- Zanotti G., Scapin G., Spadon P., Veerkamp J. H., Sacchettini J. C. Three-dimensional structure of recombinant human muscle fatty acid-binding protein. J Biol Chem. 1992 Sep 15;267(26):18541–18550. doi: 10.2210/pdb2hmb/pdb. [DOI] [PubMed] [Google Scholar]