Abstract
PURPOSE
Indolent prostate cancer (PCa) is prevalent in the intended use population (adults age 50-79 years) for blood-based multicancer early detection (MCED) tests. We examined the detectability of PCa by a clinically validated, targeted methylation–based MCED test.
METHODS
Detectability by Gleason grade group (GG), clinical stage, association of detection status with tumor methylated fraction (TMeF), and overall survival (OS) were assessed in substudy 3 of Circulating Cell-Free Genome Atlas (CCGA; ClinicalTrials.gov identifier: NCT02889978) and PATHFINDER (ClinicalTrials.gov identifier: NCT04241796) studies.
RESULTS
Test sensitivity for PCa in substudy 3 of CCGA was 11.2% (47/420). The test detected 0 (0%) of 58 low-grade (GG1), 3 (1.9%) of 157 favorable intermediate-grade (GG2), 4 (5.1%) of 78 unfavorable intermediate-grade (GG3), and 36 (31.9%) of 113 high-grade (GG4 and 5) cancers and 3 (3.2%) of 95 stage I, 11 (4.7%) of 235 stage II, 7 (14.9%) of 47 stage III, and 22 (81.5%) of 27 stage IV cases. The median TMeF was higher for detected than nondetected cases (2,106.0 parts per million [PPM]; IQR, 349.8-24,376.3 v 24.4 PPM; IQR, 17.8-38.5; P < .05). Nondetected cases had better OS (P < .05; hazard ratio [HR], 0.263 [95% CI, 0.104 to 0.533]) and detected cases had similar survival (P = .2; HR, 0.672 [95% CI, 0.323 to 1.21]) compared with SEER adjusted for age, GG, and stage. Performance was similar in PATHFINDER, with no detected GG1/2 (0/13) or stage I/II (0/16) cases.
CONCLUSION
This MCED test preferentially detects high-grade, clinically significant PCa. Use in population-based screening programs in addition to standard-of-care screening is unlikely to exacerbate overdiagnosis of indolent PCa.
GRAIL’s Galleri test preferentially detects more deadly prostate cancer over indolent cases
INTRODUCTION
Overdiagnosis of a disease or condition that is unlikely to cause harm during a patient's lifetime is an inherent risk of all screening tests. In oncology, overdiagnosis of prostate cancer (PCa) is a common problem, estimated to account for 23%-42% of all screen-detected cases.1 This problem arises largely because of the high population prevalence of low-grade, indolent cancers coupled with the low specificity of prostate-specific antigen (PSA).1 Overdiagnosis leads to unnecessary harms, including heightened patient anxiety and overtreatment of biologically insignificant cancers that may cause undesirable urinary, bowel, and sexual side effects; unnecessary cost; and overuse of health care resources.
CONTEXT
Key Objective
Does multicancer early detection (MCED) testing affect overdiagnosis of indolent prostate cancers?
Knowledge Generated
The MCED test assessed in two large-scale clinical studies (substudy 3 of Circulating Cell-Free Genome Atlas [ClinicalTrials.gov identifier: NCT02889978] and PATHFINDER [ClinicalTrials.gov identifier: NCT04241796]) preferentially detected high-grade and high-stage prostate cancers (93% GG3-5 and 67% stage III or IV), notably detecting no GG1, 1.9% of GG2, and 4.2% of stage I and II prostate cancers in these studies. Nondetected prostate cancers had better survival than would be expected on the basis of the National Cancer Institute SEER data adjusted for age, grade, and stage.
Relevance
MCED testing is unlikely to exacerbate overdiagnosis of indolent prostate cancers. Furthermore, a cancer signal detected result with prediction of prostate origin strongly suggests the presence of aggressive disease that warrants prompt diagnostic workup.
Blood-based multicancer early detection (MCED) tests represent a new paradigm for cancer screening. Case-control studies have demonstrated the ability of these tests to detect multiple cancer types from a single blood sample with very low false-positive rates (<1%-2%).2-5 In addition, some tests include the ability to predict cancer type on the basis of specific molecular features. One such test that is based on methylation patterns of circulating cell-free DNA (cfDNA), was developed and clinically validated in the Circulating Cell-Free Genome Atlas study (CCGA; ClinicalTrials.gov identifier: NCT02889978).5,6 In substudy 3 of CCGA, this MCED test detected more than 50 individual cancer types (including urological and nonurological cancers) and accurately predicted cancer signal origin (CSO) in 89% of cases, with a false-positive rate of 0.5%.5 The subsequent PATHFINDER study (ClinicalTrials.gov identifier: NCT04241796) prospectively assessed the use of the same MCED test in an intended use population and confirmed its ability to detect multiple cancer types, including early-stage cancers, with high specificity.7
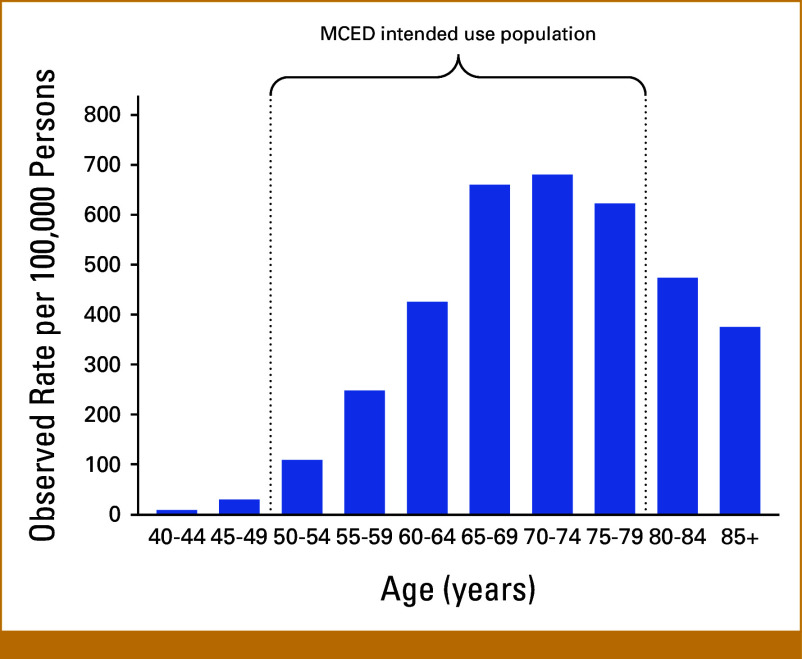
MCED tests are currently under development to be used in addition to existing single-cancer screening protocols in adults 50 years and older.8 Considering the significant age-related incidence of PCa within this demographic (Appendix Fig A1), it is important to evaluate the performance of MCED tests in identifying both indolent and aggressive forms of this disease. This study analyzed the performance of the MCED test used in both substudy 3 of CCGA and PATHFINDER studies in detecting PCa, with a key goal of determining whether its use might contribute to overdiagnosis.
METHODS
MCED Test
The MCED test used in substudy 3 of CCGA and PATHFINDER uses next-generation sequencing to interrogate cfDNA in peripheral blood.9 The test measures the extent and location of genomic DNA methylation patterns that indicate the presence of cancer and are also specific to cancer type. A computational algorithm utilizing locked classifier determines whether a cancer signal is present, and if so, a second algorithm predicts the most likely tumor type, labeled the CSO prediction, which is intended to direct diagnostic evaluations. Technical aspects of the assay and analytical and clinical validation have been previously described.5,9
Tumor Methylated Fraction
Tumor methylated fraction (TMeF) is an estimate of the quantity of tumor-derived cfDNA and is derived from observations of methylation patterns characteristic of tumor DNA. TMeF is a measure of circulating tumor allele fraction and correlates with tumor burden as measured by clinical stage, tumor size, and overall survival (OS) for multiple cancer types.10 TMeF levels were calculated algorithmically by evaluating the presence of differentially methylated regions of cfDNA that are hyper- or hypomethylated in cancers when compared with noncancer individuals and are reported in parts per million (PPM). Technical details of the molecular methods and algorithm have been described previously.10
Study Cohorts
Substudy 3 of CCGA
This cohort was drawn from the larger CCGA study, a multicenter, case-control, observational study with longitudinal follow-up apportioned for discovery (substudy 1 of CCGA),9 training and validation (substudy 2 of CCGA),6 and independent clinical validation (substudy 3 of CCGA)5 of this novel MCED blood test. CCGA enrolled 15,254 participants (8,584 with and 6,670 without cancer) 20 years and older from 142 sites in North America between August 2016 and February 2019. Eligibility criteria for cases required a confirmed cancer diagnosis or high suspicion of cancer with biopsy or surgical resection planned within 6 weeks of study entry. Controls were nontransplant recipients without known current or previous cancer and no acute illness at study entry. Detailed test performance characteristics in substudy 3 of CCGA are listed in Appendix Table A1.
The PCa cohort from substudy 3 of CCGA included in the current study comprises 420 recently diagnosed men.
PATHFINDER
The PATHFINDER study was a prospective cohort study of 6,662 adults over age 50 years without clinical suspicion of cancer enrolled from seven US health networks between December 2019 and December 2020. Participants underwent MCED testing using the same test that was used in substudy 3 of CCGA, with results returned to ordering physicians and participants. Test results included the presence or absence of a cancer signal and a CSO prediction for those with a cancer signal detected. The primary objective of the study was to understand diagnostic pathways resulting from a cancer signal–detected result. Participants continued with standard-of-care screening, including PSA, during the trial according to published recommendations and guidance of their medical providers. Participants were followed for 12 months after enrollment, with a determination of cancer status (yes/no) made by results of diagnostic evaluations in those with a cancer signal detected and at study end by review of the electronic health record in all other participants. Detailed test performance characteristics in PATHFINDER are listed in Appendix Table A1.
The PCa cohort from PATHFINDER included in the current study comprises 18 men diagnosed during the study by MCED testing or PSA screening, excluding two with recurrent disease. Individual PSA data were not collected in PATHFINDER.
Statistical Analysis
Assessment of Test Performance
In this post hoc subanalysis of substudy 3 of CCGA and the PATHFINDER study, MCED test performance for detecting PCa was assessed by Gleason grade group (GG) and clinical stage. Assignments of grade and stage were based on case report forms from each study. Differences in TMeF between detected and nondetected cases were analyzed by Wilcoxon rank-sum test (for substudy 3 of CCGA cohort only).
OS Estimates (substudy 3 of CCGA cohort only)
OS estimates for the 420 participants with PCa in substudy 3 of CCGA were stratified by MCED detection status (detected v nondetected). To provide a reference for survival rates, we obtained OS data from a representative population of individuals diagnosed with PCa in 17 regions of the United States from the National Cancer Institute's SEER Program and related SEER*Stat program (version 8.4.1). Using data from patients diagnosed between 2006 and 2015 stratified by age, cancer stage at diagnosis (American Joint Committee on Cancer 6th edition stage I, II, III, IV, or unknown), and cancer grade in SEER, we estimated the expected OS of participants in substudy 3 of CCGA and compared it with the observed OS from SEER. A one-sample proportional hazards model was used to assess the significance of the difference between observed OS and expected OS in cancer-detected and nondetected participants.11 All computations and Kaplan-Meier plots were done using the R software language (version 4.1.2 [2021-11-01]).12
RESULTS
Substudy 3 of CCGA Cohort (N = 420)
Demographic data are presented in Table 1. The median age was 65 years (IQR, 59-70), and 85% self-reported as White, non-Hispanic. The median PSA was higher in detected cases versus nondetected cases (14 ng/mL; IQR, 7-38 v 6 ng/mL; IQR, 5-9; Table 1).
TABLE 1.
Demographic and Clinical Characteristics of Prostate Cancer Cases in Substudy 3 of CCGA and PATHFINDER
| Characteristic | Detecteda (n = 48) | Not Detecteda (n = 390) |
|---|---|---|
| Substudy 3 of CCGA | ||
| Age, median (IQR) | 67 (62-70) | 64 (59-70) |
| Race, No. (%) | ||
| Asian, Native American, or Pacific Islander | 1 (2.1) | 5 (1.3) |
| Black, non-Hispanic | 0 | 37 (9.9) |
| Hispanic | 2 (4.3) | 12 (3.2) |
| Other/unknown | 0 | 8 (2.1) |
| White, non-Hispanic | 44 (94) | 311 (83) |
| PSA, ng/mL, median (IQR) | 14 (7-38) | 6 (5-9) |
| PATHFINDERb | ||
| Age, median (IQR) | 66 (66-66) | 65 (58-71) |
| Race, No. (%) | ||
| Asian, Native American, or Pacific Islander | 0 | 0 |
| Black, non-Hispanic | 0 | 1 (6.2) |
| Hispanic | 0 | 0 |
| Other/unknown | 0 | 0 |
| White, non-Hispanic | 1 (100) | 15 (94) |
Abbreviations: CCGA, Circulating Cell-Free Genome Atlas study; PSA, prostate-specific antigen.
Substudy 3 of CCGA and PATHFINDER combined.
PSA data were not collected in PATHFINDER.
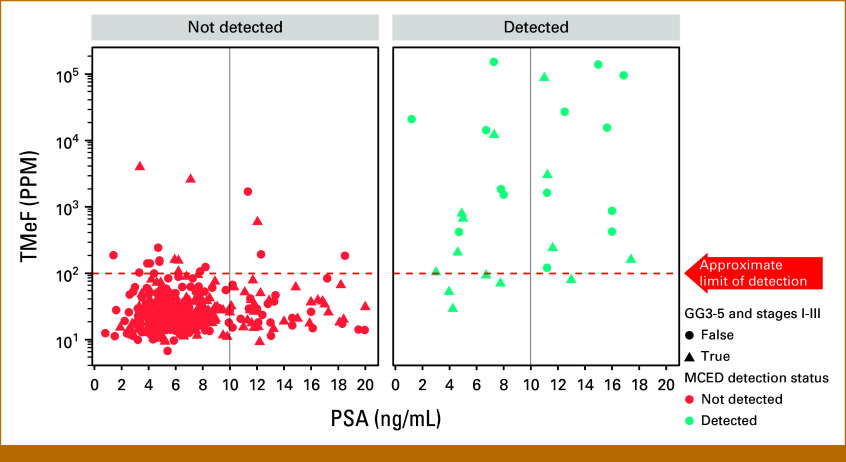
The overall MCED test sensitivity for PCa detection was 11.2% (47/420), and the CSO prediction accuracy was 91.5% (43/47). Of the 420 cases, 14 (3.3%) were missing GG assignment and two (0.47%) were missing staging information. PCa detectability increased with increasing GG (Fig 1A) and clinical stage (Fig 1B). The MCED test detected no (0/58) low-grade (GG1), 1.9% (3/157) favorable intermediate-grade (GG2), 5.1% (4/78) unfavorable intermediate-grade (GG3), and 31.9% (36/113) high-grade (GG4 and 5) cancers. Detection by stage was 3.2% (3/95) for stage I disease, 4.7% (11/235) for stage II disease, 14.9% (7/47) for stage III disease, and 81.5% (22/27) for stage IV disease. Of the detected cases with complete data, 34% (15/44) had a PSA <10 ng/mL (Fig 2) and 31.9% (15/47) had unfavorable intermediate- (GG3) or high-grade (GG ≥3) early-stage (I-III) disease.
FIG 1.
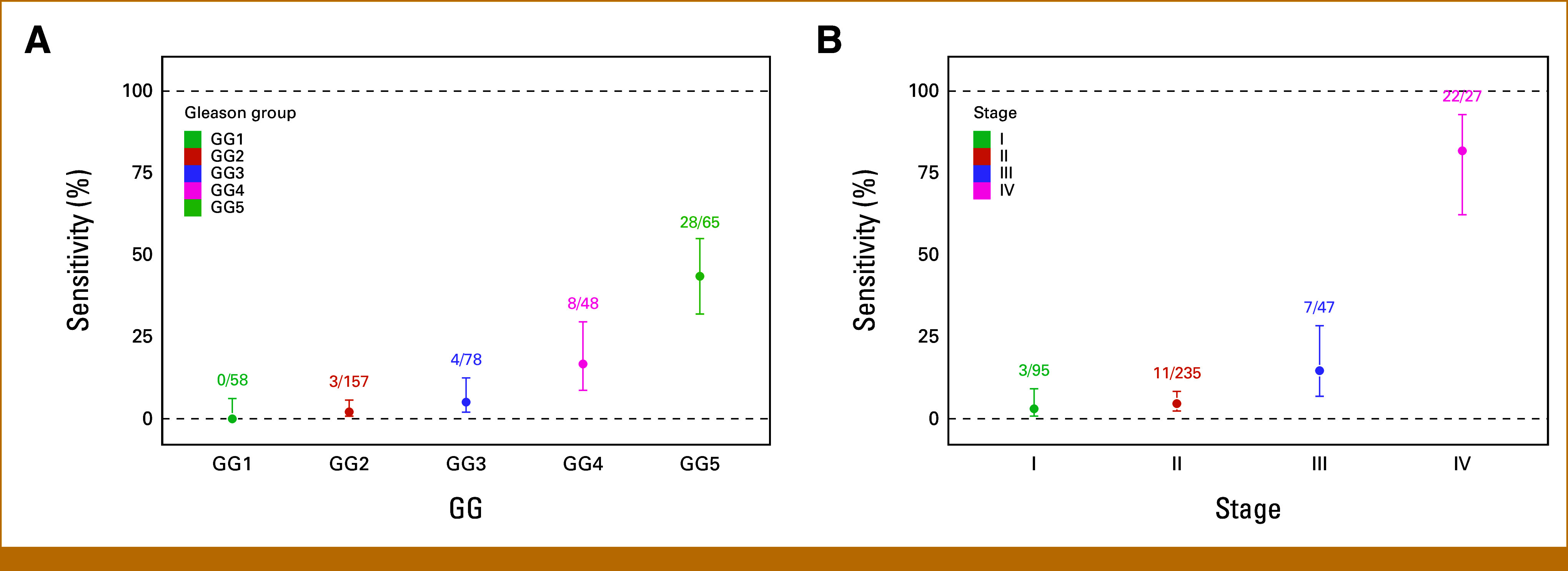
Sensitivity of prostate cancer detection by (A) GG and (B) clinical stage in substudy 3 of CCGA. The number of MCED-detected cases out of total cases in each grade or stage group is reported. Error bars designate 95% CIs. CCGA, Circulating Cell-Free Genome Atlas study; GG, Gleason grade group; MCED, multicancer early detection.
FIG 2.
Relationship between cancer detectability, TMeF, and PSA. Red = cases not detected; blue = cases detected. Triangles indicate GG3-5, stage I-III cases. GG, Gleason grade group; MCED, multicancer early detection; PPM, parts per million; PSA, prostate-specific antigen; TMeF, tumor methylated fraction.
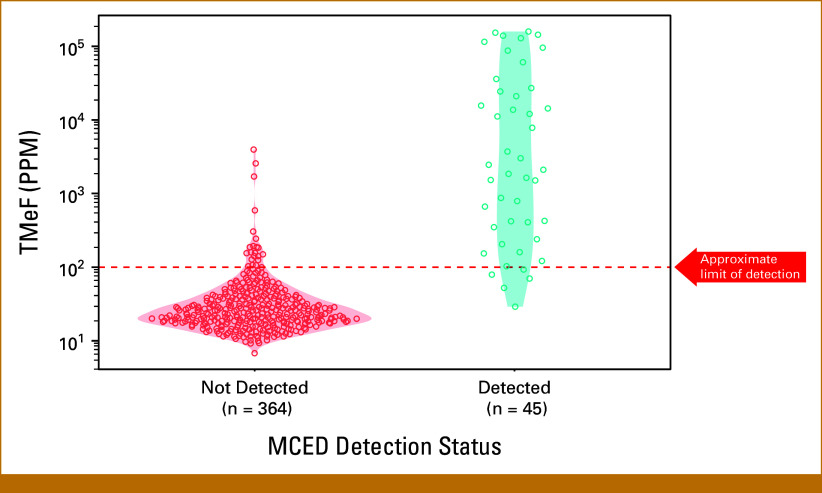
The median TMeF was higher for detected cases than nondetected cases (2,106.0 PPM; IQR, 349.8-24,376.3 v 24.4 PPM; IQR, 17.8-38.5; P < .05; Fig 3) and was correlated with serum PSA levels for detected cases (Spearman's rank correlation coefficient r = 0.42; Fig 2).
FIG 3.
TMeF of prostate cancer cases in substudy 3 of CCGA by MCED test detection status. CCGA, Circulating Cell-Free Genome Atlas study; MCED, multicancer early detection; PPM, parts per million; TMeF, tumor methylated fraction.
Compared with expected OS estimated from SEER, nondetected cases had roughly three times better OS (P < .05; hazard ratio [HR], 0.263 [95% CI, 0.104 to 0.533]), and detected cases had similar survival (P = .2; HR, 0.672 [95% CI, 0.323 to 1.21]) when adjusted for age, GG, and clinical stage (Fig 4).
FIG 4.
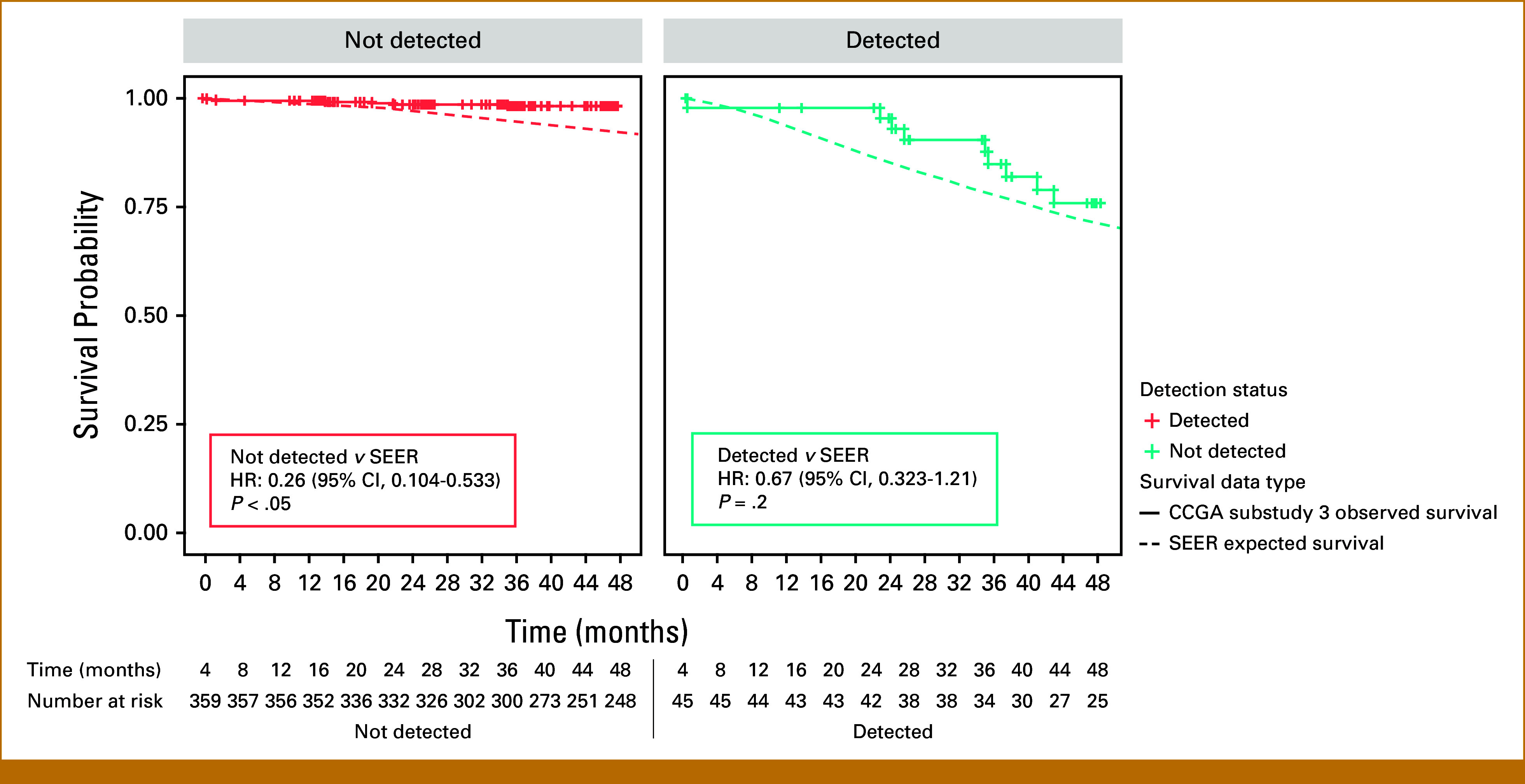
Observed versus expected OS of prostate cancer cases in substudy 3 of CCGA compared with SEER adjusted for age, stage, and grade. Nondetected cases had better OS (P < .05; HR, 0.263 [95% CI, 0.104 to 0.533]), and detected cases had similar survival (P = .2; HR, 0.672 [95% CI, 0.323 to 1.21]) compared with SEER estimates adjusted for age, GG, and clinical stage. CCGA, Circulating Cell-Free Genome Atlas study; GG, Gleason grade group; HR, hazard ratio; OS, overall survival.
PATHFINDER Cohort (N = 18)
Demographic data are presented in Table 1. The median age was 65 years (IQR, 58-70), and 89% self-reported as White, non-Hispanic and 5.6% as Black, non-Hispanic. The overall episode sensitivity for PCa detection was 5.6% (1/18), including no (0/13) GG1-2 and 33% (1/3) GG3-5 cases detected. No stage I or II (0/16) and 50% (1/2) stage III and IV cases were detected.
Because only a single case of PCa was detected in PATHFINDER, CSO prediction accuracy, TMeF, and survival estimates were not calculated.
DISCUSSION
MCED tests are being developed as population-level screening tools in asymptomatic individuals to be used in addition to single-cancer standard-of-care screening, including PSA for PCa. They are intended for use in the screen-eligible population, which in the United States is generally considered to be adults age 50-79 years.13 As PCa is the most common visceral cancer in men in this age range, it is important to understand how specific MCED tests perform in detecting this cancer.
The current study provides insight into the PCa performance of the MCED test that was developed and used in two large clinical studies and is now available commercially (Galleri, GRAIL, Inc). Detection of PCa was low (test sensitivity of 11.2% in substudy 3 of CCGA and episode sensitivity of 5.6% in PATHFINDER). This MCED test preferentially detected high-grade and high-stage PCas, notably detecting no GG1 cancers, 1.9% of GG2 cancers, and only 4.2% of stage I and II cancers across both studies combined. These results are unsurprising given the preponderance of early-stage disease (79% stage I or II) in these two studies and previous observations that cfDNA concentrations in men with localized PCa are low and similar to concentrations in healthy individuals without cancer.14,15 The results are also consistent with a prospective study of men with localized disease undergoing radical prostatectomy, where circulating tumor DNA (ctDNA) was assessed using the variant-based integration of variant reads approach.16 In that study, 16% of preoperative blood samples had detectable ctDNA, and detectability was associated with higher grade and stage disease and the risk of both biochemical recurrence and metastasis-free survival.17
Together with the observation that nondetected PCas had better survival than age, stage, and grade-comparable SEER controls, these data strongly suggest that use of this test in population-based screening programs in addition to standard-of-care screening tests is unlikely to exacerbate overdiagnosis of indolent PCa. Conversely, the observation that the test preferentially detects clinically significant PCas (93% were GG3-5 and 67% were stage III or IV) with a CSO prediction accuracy for PCa >90% suggests that individuals with a cancer signal detected and a prostate CSO prediction should undergo a prompt diagnostic evaluation to establish or rule out the presence of high-grade or high-stage disease for which treatment is generally indicated.18 Notably, about one third of the detected cases in substudy 3 of CCGA had GG3-5, nonmetastatic (stage I-III) disease that is potentially amenable to cure, supported by the observation that the OS for detected cases was not worse than expected.
Previous work from CCGA has shown that higher TMeF is associated with biologic aggressiveness, including mitotic and metabolic activity and invasiveness.19 The preferential detection of aggressive PCas in substudy 3 of CCGA is consistent with observations across more than 20 cancer types,6,20 as is the high OS (relative to expected) of nondetected cancers.
Strengths of this study include use of observed detectability of PCa from two independent clinical studies using a locked machine learning classifier and isolation of cfDNA detectability as a prognostic factor using synthetic controls matched on age, grade, and stage from SEER, which avoids confounding detectability with clinical factors. Limitations include the small number of cases in PATHFINDER; that PSA, TMeF, and OS data were not available for that cohort; and that GG and stage were assigned on the basis of determinations by multiple study sites and not subject to central review or confirmation. In addition, given that overdiagnosis of PCa due to PSA levels has been shown to disproportionately affect Black men, the generalizability of these findings is limited by the fact that the vast majority (>85%) of the study cohorts evaluated here were self-reported as White, non-Hispanic.21,22
In conclusion, a clinically validated targeted methylation–based MCED test preferentially detects high-grade and high-stage clinically significant PCa. A cancer signal detected test result with a prostate CSO prediction strongly suggests the presence of aggressive disease and warrants a prompt diagnostic workup. Because the test rarely detects low-grade disease, its use in population-based screening programs in addition to standard-of-care screening is unlikely to exacerbate overdiagnosis of indolent PCa.
ACKNOWLEDGMENT
Editorial assistance provided by Jennifer Hepker, PhD, and Alexandra Thomas, PhD, Prescott Medical Communications Group, a Citrus Health Group, Inc. Company (Chicago, IL).
APPENDIX
TABLE A1.
Performance Characteristics for All Cancer Types of a Targeted Methylation MCED Test in Substudy 3 of CCGA and PATHFINDER Studies
| Study | Substudy 3 of CCGA5 | PATHFINDER7 |
|---|---|---|
| Study type | Case-control | Prospective interventional |
| Number of participants, N | 4,077a | 6,621b |
| Specificity, % | 99.5 | 99.1 |
| Sensitivity, % | 51.5c | Not calculated |
| CSO prediction accuracy, % | 88.7 | 88.0 |
| PPV, % | 44.4 | 43.1 |
Abbreviations: CCGA, Circulating Cell-Free Genome Atlas study; CSO, cancer signal origin; MCED, multicancer early detection; PPV, positive predictive value.
2823 cancer cases and 1,254 noncancer controls.
121 cancer cases.
Ranging from 16.8% in stage I to 90.1% in stage IV.
FIG A1.
Age-adjusted PCa incidence in the United States overlaps with the MCED intended use population. Observed SEER PCa incidence rates by age at diagnosis, years 2016-2020. Figure generated with SEER.23 MCED, multicancer early detection; PCa, prostate cancer.
PRIOR PRESENTATION
Presented as an oral presentation as part of the “Multi-Cancer Early Detection Testing: Where Are We?” minisymposium at the 2024 AACR Annual Meeting, San Diego, CA, April 7, 2024.
SUPPORT
Supported by GRAIL, Inc, Menlo Park, CA.
DATA SHARING STATEMENT
The datasets used in this analysis are available upon reasonable request by email to the corresponding author.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Collection and assembly of data: Brandon A. Mahal, Matthew Margolis, Earl Hubbell, Eric A. Klein
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Brandon A. Mahal
Honoraria: Foundation Medicine
Other Relationship: Prostate Cancer Foundation, Department of Defense-Prostate Cancer Research Program, American Society for Radiation Oncology
Matthew Margolis
Employment: GRAIL
Stock and Other Ownership Interests: Roche, Illumina
Honoraria: Sanofi/Regeneron
Earl Hubbell
Employment: GRAIL
Stock and Other Ownership Interests: Illumina
Research Funding: GRAIL
Patents, Royalties, Other Intellectual Property: GRAIL: filed patent applications
Travel, Accommodations, Expenses: GRAIL
Cheng Chen
Employment: GRAIL
Stock and Other Ownership Interests: GRAIL
Travel, Accommodations, Expenses: GRAIL
Jeffrey M. Venstrom
Employment: Foundation Medicine, Genentech/Roche, GRAIL
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Consulting or Advisory Role: Synkrino Biotherapeutics
Travel, Accommodations, Expenses: Foundation Medicine, Genentech/Roche
Other Relationship: GRAIL
John Abran
Employment: Exact Sciences, GRAIL
Stock and Other Ownership Interests: Exact Sciences, GRAIL
Jordan J. Karlitz
Employment: GRAIL
Stock and Other Ownership Interests: GI OnDEMAND
Travel, Accommodations, Expenses: GRAIL
Alexander W. Wyatt
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: Janssen, AstraZeneca, Merck, Pfizer, EMD Serono, Genentech
Research Funding: ESSA (Inst), Tyra Biosciences (Inst)
Eric A. Klein
Employment: GRAIL
No other potential conflicts of interest were reported.
REFERENCES
- 1.Loeb S, Bjurlin M, Nicholson J, et al. : Overdiagnosis and overtreatment of prostate cancer. Eur Urol 65:1046-1055, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen JD, Li L, Wang Y, et al. : Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359:926-930, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristiano S, Leal A, Phallen J, et al. : Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570:385-389, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, et al. : Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein EA, Richards D, Cohn A, et al. : Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol 32:1167-1177, 2021 [DOI] [PubMed] [Google Scholar]
- 6.Liu MC, Oxnard GR, Klein EA, et al. : Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 31:745-759, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrag D, Beer TM, McDonnell CH, et al. : Blood-based tests for multicancer early detection (PATHFINDER): A prospective cohort study. Lancet 402:1251-1260, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein EA, Madhavan S, Beer TM, et al. : Dying to find out: The cost of time at the dawn of the multicancer early detection era. Cancer Epidemiol Biomarkers Prev 32:1003-1010, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamshidi A, Liu MC, Klein EA, et al. : Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 40:1537-1549.e12, 2022 [DOI] [PubMed] [Google Scholar]
- 10.Melton CA, Freese P, Zhou Y, et al. : A novel tissue-free method to estimate tumor-derived cell-free DNA quantity using tumor methylation patterns. Cancers 16:82, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein DM, Muzikansky A, Schoenfeld DA: Comparing survival of a sample to that of a standard population. J Natl Cancer Inst 95:1434-1439, 2003 [DOI] [PubMed] [Google Scholar]
- 12.R Core Team : R: A language and environment for statistical computing. https://www.R-project.org
- 13.Hackshaw A, Cohen SS, Reichert H, et al. : Estimating the population health impact of a multi-cancer early detection genomic blood test to complement existing screening in the US and UK. Br J Cancer 125:1432-1442, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennigan ST, Trostel SY, Terrigino NT, et al. : Low abundance of circulating tumor DNA in localized prostate cancer. JCO Precis Oncol 10.1200/PO.19.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen E, Cario CL, Leong L, et al. : Cell-free DNA concentration and fragment size as a biomarker for prostate cancer. Sci Rep 11:5040, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan JCM, Heider K, Gale D, et al. : ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med 12:eaaz8084, 2020 [DOI] [PubMed] [Google Scholar]
- 17.Pope B, Park G, Lau E, et al. : Ultrasensitive detection of circulating tumour DNA enriches for patients with a greater risk of recurrence of clinically localised prostate cancer. Eur Urol 85:407-410, 2024 [DOI] [PubMed] [Google Scholar]
- 18.American Cancer Society : Initial treatment of prostate cancer, by stage and risk group. American Cancer Society. https://www.cancer.org/cancer/types/prostate-cancer/treating/by-stage.html
- 19.Bredno J, Lipson J, Venn O, et al. : Clinical correlates of circulating cell-free DNA tumor fraction. PLoS One 16:e0256436, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredno J, Venn O, Chen X, et al. : Circulating tumor DNA allele fraction: A candidate biological signal for multicancer early detection tests to assess the clinical significance of cancers. Am J Pathol 192:1368-1378, 2022 [DOI] [PubMed] [Google Scholar]
- 21.Down L, Barlow M, Bailey SER, et al. : Association between patient ethnicity and prostate cancer diagnosis following a prostate-specific antigen test: A cohort study of 730,000 men in primary care in the UK. BMC Med 22:82, 2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brawley OW, Ramalingam R: The enigma of race and prostate cancer. Cancer 130:179-181, 2024 [DOI] [PubMed] [Google Scholar]
- 23.SEER*Explorer: An interactive website for SEER cancer statistics. Surveillance Research Program, National Cancer Institute. https://seer.cancer.gov/statistics-network/explorer/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in this analysis are available upon reasonable request by email to the corresponding author.