Abstract
PURPOSE
Given the high heterogeneity in survival for patients with multiple myeloma, it would be clinically useful to quantitatively predict the individual survival instead of attributing patients to two to four risk groups as in current models, for example, revised International Staging System (R-ISS), R2-ISS, or Mayo-2022-score.
PATIENTS AND METHODS
Our aim was to develop a quantitative prediction tool for individual patient's 3-/5-year overall survival (OS) probability. We integrated established clinical and molecular risk factors into a comprehensive prognostic model and evaluated and validated its risk discrimination capabilities versus R-ISS, R2-ISS, and Mayo-2022-score.
RESULTS
A nomogram for estimating OS probabilities was built on the basis of a Cox regression model. It allows one to translate the individual risk profile of a patient into 3-/5-year OS probabilities by attributing points to each prognostic factor and summing up all points. The nomogram was externally validated regarding discrimination and calibration. There was no obvious bias or overfitting of the prognostic index on the validation cohort. Resampling-based and external evaluation showed good calibration. The c-index of the model was similar on the training (0.76) and validation cohort (0.75) and significantly higher than for the R-ISS (P < .001) or R2-ISS (P < .01).
CONCLUSION
In summary, we developed and validated individual quantitative nomogram-based OS prediction. Continuous risk assessment integrating molecular prognostic factors is superior to R-ISS, R2-ISS, or Mayo-2022-score alone.
Better survival prediction in multiple myeloma—multimodal and continuous nomogram (instead of few arbitrary groups)
INTRODUCTION
Multiple myeloma is a malignant hematological disease characterized by accumulation of clonal plasma cells in the bone marrow and associated clinical signs and symptoms, especially those related to displacement of normal hematopoiesis and osteolytic bone disease.1
CONTEXT
Key Objective
Given the high heterogeneity in survival for patients with myeloma per se and within the two to four risk groups used in current models, eg, revised International Staging System (R-ISS), R2-ISS, or MAYO-2022-score, we aimed to develop a quantitative prediction tool for individual patient's 3-/5-year overall survival (OS) probability.
Knowledge Generated
We integrated established clinical and molecular risk factors into a comprehensive prognostic model and evaluated and validated its risk discrimination capabilities. The nomogram allows to translate the patient's individual risk profile into continuous 3-/5-year OS probabilities by attributing points to each prognostic factor and summing up all points.
Relevance
We developed and validated individual quantitative nomogram-based prediction of survival in myeloma which can be used in clinical routine. Continuous risk assessment overcomes heterogeneous grouping of patients with different individual risk to discrete groups. Integration of molecular prognostic factors gives significantly superior prediction versus published scores.
Prognosis of individual patients is highly heterogeneous ranging from few months to 15 years or more.2-6 Risk stratification in clinical routine is performed by combining the presence of high-risk chromosomal aberrations as detected by interphase fluorescence in-situ hybridization (iFISH) and the International Staging System (ISS).7-9 Widely accepted standard is the revised ISS-score (R-ISS) including serum β2-microglobulin, albumin, lactate dehydrogenase (LDH), and adverse prognostic chromosomal aberrations, ie, deletion 17p13 (del17p13) and/or translocation t(4;14) and/or t(14;16)7 delineating three risk groups. Its recent suggested modification, R2-ISS, includes chromosome 1q21-gain, delineating four R2-ISS risk groups.10 Prognostic power is increased by assessing gene expression, eg, high-risk scores11-15 or proliferation16 by DNA microarrays (gene expression profiling [GEP]) or next-generation sequencing (NGS) techniques like RNA sequencing.6 Current risk prediction models attribute patients to two to four arbitrary groups, ie, high versus intermediate (-high) versus (intermediate-) low risk. Group size and survival rates largely vary between different systems. This implies that patient's risk within a specific group is considered similar for those attributed to the lower or higher end of the respective group, ie, for a patient scored medium risk, either being almost low or almost high risk. It would, therefore, be clinically very useful to quantitatively predict survival on a continuous scale.
The aims of our study were to (1) develop quantitative prediction of individual myeloma patient's 3-/5-year overall survival (OS) probability, (2) integrate prognostic factors into a comprehensive model, and (3) evaluate its risk discrimination capabilities in relation to R-ISS as current gold standard, and two recently suggested modifications, that is, R2-ISS10 and the Mayo-2022-score.17
PATIENTS AND METHODS
Study Cohort
Six hundred fifty-seven patients presenting with previously untreated, therapy-requiring multiple myeloma were included in the study approved by the ethics committee of the University of Heidelberg (229/2003 and S-152/2010) between June 2005 and June 2015. We obtained written informed consent from all patients for treatment and sample procurement. Patients were treated with bortezomib-based induction regimen (bortezomib, adriamycin, dexamethasone, or bortezomib, cyclophosphamide, dexamethasone, respectively) and intended to undergo high-dose chemotherapy followed by autologous stem cell transplantation (ASCT) either as part of the HOVON65/GMMG-HD43,18 (EudraCT no. 2004-000944-26) or GMMG-MM5 trial19,20 (EudraCT no. 2010-019173-16) or outside clinical trials.
Samples
CD138+ plasma cells were isolated from bone marrow aspirates using anti-CD138 immunobeads and an autoMACS Separator (Miltenyi Biotec, Bergisch Gladbach, Germany).16,21-27 Purity was assessed by flow cytometry (Becton Dickinson, Heidelberg, Germany) using antibodies against CD38 (clone HB-7; Becton Dickinson) and CD138 (clone B-B4; Miltenyi Biotec). Aliquots of CD138+ plasma cells were subjected to cytospin preparation for iFISH and nucleic acid extraction for GEP.
iFISH
Analysis was conducted on CD138-purified plasma cells using probes for numerical changes of the chromosome regions 1q21, 5p15, 5q31 or 5q35, 8p21, 9q34, 11q22.3 or 11q23, 13q14.3, 15q22, 17p13, and 19q13 and translocations t(4;14) (p16.3;q32.3), t(11;14) (q13;q32.3), and t(14;16) (q32.3;q23) or any other immunoglobulin H (IgH)-rearrangement with unknown translocation partner, according to the manufacturer's instructions (Kreatech, Amsterdam, the Netherlands and MetaSystems, Altlussheim, Germany). Data were analyzed as published.28
Analysis of Gene Expression
RNA was extracted using the Qiagen AllPrep DNA/RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Quality control and quantification of total RNA was performed using an Agilent 2100 bioanalyzer (Agilent, Frankfurt, Germany).
GEP using U133 2.0 plus arrays (Affymetrix, Santa Clara, CA) was performed as published.16,24,25 Expression data are deposited in ArrayExpress under accession numbers GSE19784 and E-MTAB-2299.
Statistical Analysis
Patients were split into training (TG, n = 536) and validation groups (VG, n = 121). The TG consisted of 102 patients treated within the HOVON65/GMMG-HD4 trial,3,18 386 from the GMMG-MM5 trial,14,15 and 48 nontrial patients. The primary endpoint of both trials was progression-free survival (PFS). For validation, 66 patients from the GMMG-MM5 extension cohort and 55 nontrial patients were used.
The primary endpoint of this study was OS, defined as time from the start of induction therapy or random assignment to death from any cause. The following known risk factors were considered for building the prognostic model: age (in years), ISS stage (I/II/III),29 LDH level above upper limit (yes/no), creatinine level >2 g/dL, heavy chain type IgA (yes/no), del17p13 (yes/no), t(4;14) (yes/no), gain 1q21 (no gain/3 copies/>3 copies), GEP-based risk stratification, ie, the UAMS GEP70-score11 (UAMS70), and our GEP-based proliferation index (GPI50).16 GEP-scores were analyzed as continuous predictors. R-ISS7 and two suggested R-ISS-modifications, ie, R2-ISS by the European Myeloma Network10 and a model by the Mayo-clinic (Mayo-2022-score),17 served as comparators.
Plots of martingale residuals were examined to check the linearity of continuous covariates. The proportional hazards assumption for the Cox models was examined graphically and by the proportional hazards test.30 No strong collinearities were detected when checking variance inflation factors. We screened for pairwise interactions between variables. Since models with interaction terms were not superior and to avoid overfitting, we restricted ourselves to simple additive models. Missing values in clinical and cytogenetic parameters in the TG (maximal approximately 2% of missing values per variable) were imputed using the mice R-package31 on the basis of B = 50 imputation runs. A nonstringent backward variable selection procedure with significance level for staying in the model of P = .5 was applied to remove only the surely noninformative predictors. The final Cox model with the remaining predictors was used to build a nomogram for estimating survival probabilities at 3 and 5 years.
The nomogram was validated on the validation cohort and subjected to discrimination and calibration as described.32 We report Harrell's c-index of concordance and AUC from time-dependent receiver operating characteristic curve after 3 years analysis as measures of discrimination. The proposed prognostic model is compared with R-ISS, R2-ISS, and Mayo-2022-score, by testing for difference in respective c-indices.33 Good discrimination is also indicated if the regression coefficient of the linear predictor (prognostic index) as only regressor is close to 1 in the VG data. For visual inspection of discrimination, Kaplan-Meier curves for exemplary risk groups are compared. Calibration, reflecting the accuracy of the estimated survival times, was assessed by smoothed calibration plots of expected versus observed survival probabilities, both on TG data on the basis of bootstrap and on VG data. Another way of exploring calibration is to compare predicted and observed survival curves for exemplary patient risk groups.
Analyses were carried out with software R, and model selection and validation were performed mainly with the rms R-package.34
The Fisher exact test and Wilcoxon test were used to compare distribution of categorical and quantitative parameters.
RESULTS
Quantitative Integrative Prediction of Survival Probability
Six hundred fifty-seven patients were included in this study, split into a training (n = 536) and validation cohort (n = 121). All patients had GEP and iFISH data available at the time of study inclusion, ie, before start of therapy, and were treated with bortezomib-based induction regimen and intended to undergo high-dose chemotherapy, followed by ASCT. One hundred ninety deaths were observed in the TG and 22 in the VG, with a median follow-up time of 5.4 and 3.5 years, respectively. Distribution of risk factors and OS were similar in both cohorts with 3-year OS rates of 80% versus 86%, respectively (Table 1).
TABLE 1.
Patient Characteristics in Training and Validation Group
| Level | Training Group (n = 536) | Validation Group (n = 121) | P |
|---|---|---|---|
| ISS, % | |||
| 1 | 195 (37.3) | 41 (33.9) | .6842 |
| 2 | 174 (33.3) | 45 (37.2) | |
| 3 | 154 (29.4) | 35 (28.9) | |
| R-ISS, % | |||
| 1 | 145 (27.3) | 28 (23.1) | .6581 |
| 2 | 312 (58.6) | 75 (62.0) | |
| 3 | 75 (14.1) | 18 (14.9) | |
| R2-ISS, % | |||
| 1 | 111 (21.4) | 17 (14.0) | .06 |
| 2 | 146 (28.2) | 43 (35.5) | |
| 3 | 227 (43.8) | 48 (39.7) | |
| 4 | 34 (6.6) | 13 (10.7) | |
| Mayo-2022-score, % | |||
| I | 170 (33.1) | 38 (31.7) | .54 |
| II | 179 (34.8) | 48 (40.0) | |
| III | 165 (32.1) | 34 (28.3) | |
| Elevated LDH, % | |||
| No | 432 (81.5) | 92 (76.0) | .203 |
| Yes | 98 (18.5) | 29 (24.0) | |
| Gain 1q21, % | |||
| No | 312 (59.1) | 72 (59.5) | .965 |
| 3 copies | 162 (30.7) | 38 (31.4) | |
| >3 copies | 54 (10.2) | 11 (9.1) | |
| del17p13, % | |||
| No | 475 (88.6) | 107 (88.4) | 1 |
| Yes | 61 (11.4) | 14 (11.6) | |
| t(4;14), % | |||
| No | 473 (88.4) | 111 (91.7) | .3367 |
| Yes | 62 (11.6) | 10 (8.3) | |
| t(14;16), % | |||
| No | 516 (97.0) | 118 (98.3) | .5499 |
| Yes | 16 (3.0) | 2 (1.7) | |
| IgA, % | |||
| No | 423 (78.9) | 94 (77.7) | .8059 |
| Yes | 113 (21.1) | 27 (22.3) | |
| Creatinine >2 g/dL, % | |||
| No | 472 (88.1) | 107 (88.4) | 1 |
| Yes | 64 (11.9) | 14 (11.6) | |
| Age, years, median (IQR) | 58.00 (52.27 to 64.00) | 58.00 (54.00 to 66.00) | .2391 |
| UAMS70, median (IQR) | −0.15 (−0.58 to 0.27) | −0.28 (−0.65 to 0.26) | .3915 |
| GPI50, median (IQR) | 148.94 (102.01 to 210.16) | 140.18 (98.58 to 198.46) | .4454 |
Abbreviations: IgA, immunoglobulin A; LDH, lactate dehydrogenase; (R-) ISS, (revised) International Staging System.
A prognostic model for OS was developed on the basis of established risk factors, that is, age; ISS-stage; LDH-level above upper limit; creatinine-level >2 g/dL; heavy chain type IgA (yes/no); presence of del17p13 (yes/no), t(4;14) (yes/no), or gain 1q21 (no gain/3 copies/>3 copies); and gene expression-based risk stratification, ie, UAMS7011 and GPI50.16 The latter two were analyzed as continuous variables. Owing to the low frequency (3%) in our cohort, t(14;16) was not considered as individual predictor for model building but used to define R-ISS and R2-ISS. Linear effect of continuous predictors was verified. No strong collinearity between predictors was observed. We did not identify any interaction between predictors that would improve model fit. IgA and elevated creatinine were discarded from the model during backward variable selection.
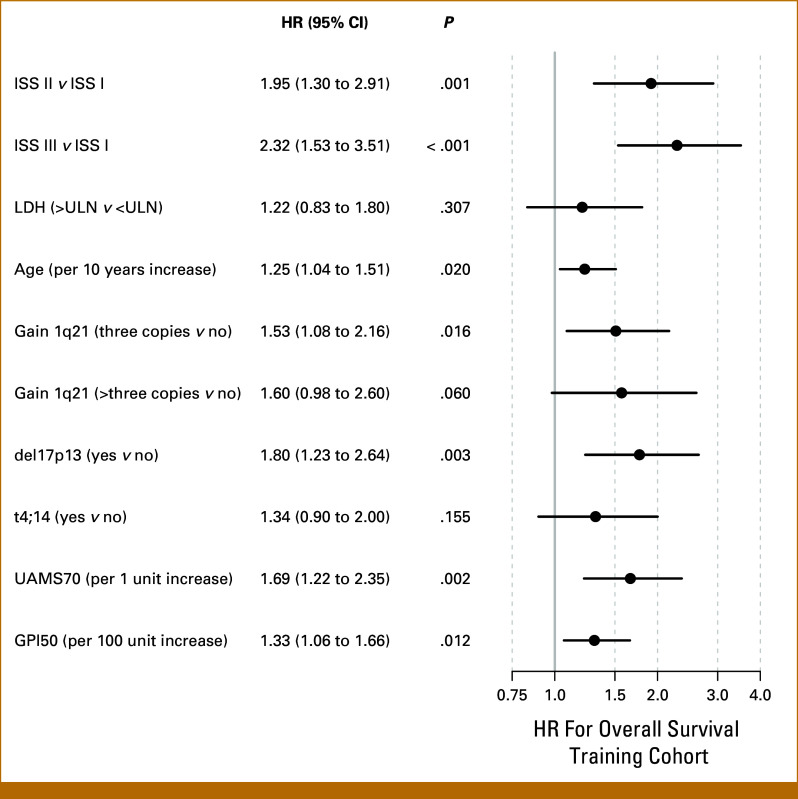
The final Cox model is based on age, ISS-stage, LDH, and molecular prognostic factors, that is, del17p13, t(4;14), gain 1q21, UAMS70, and GPI50; corresponding hazard ratios on training data are shown in Figure 1. This model was then used to build a nomogram for estimating survival probabilities (Fig 2A). Points are attributed to each of the prognostic factors and summed up. Total points translate into estimated 3-/5-year OS probabilities on a continuous scale. Example is given for an actual patient (Fig 2B): The patient's risk profile is translated into points for each characteristic indicated by different colors and then totaled. One hundred seventy total points correspond to an estimated 3- and 5-year OS probability of 51% and 26%, respectively.
FIG 1.
Cox model on training cohort. HRs for the final Cox model based on age, ISS stage, LDH, and molecular prognostic factors, that is, del17p13, t(4;14), gain 1q21, the UAMS GEP70-score (UAMS70), and the gene expression-based proliferation index (GPI50). HR, hazard ratio; ISS, international staging system; LDH, lactate dehydrogenase; ULN, upper limit of normal.
FIG 2.
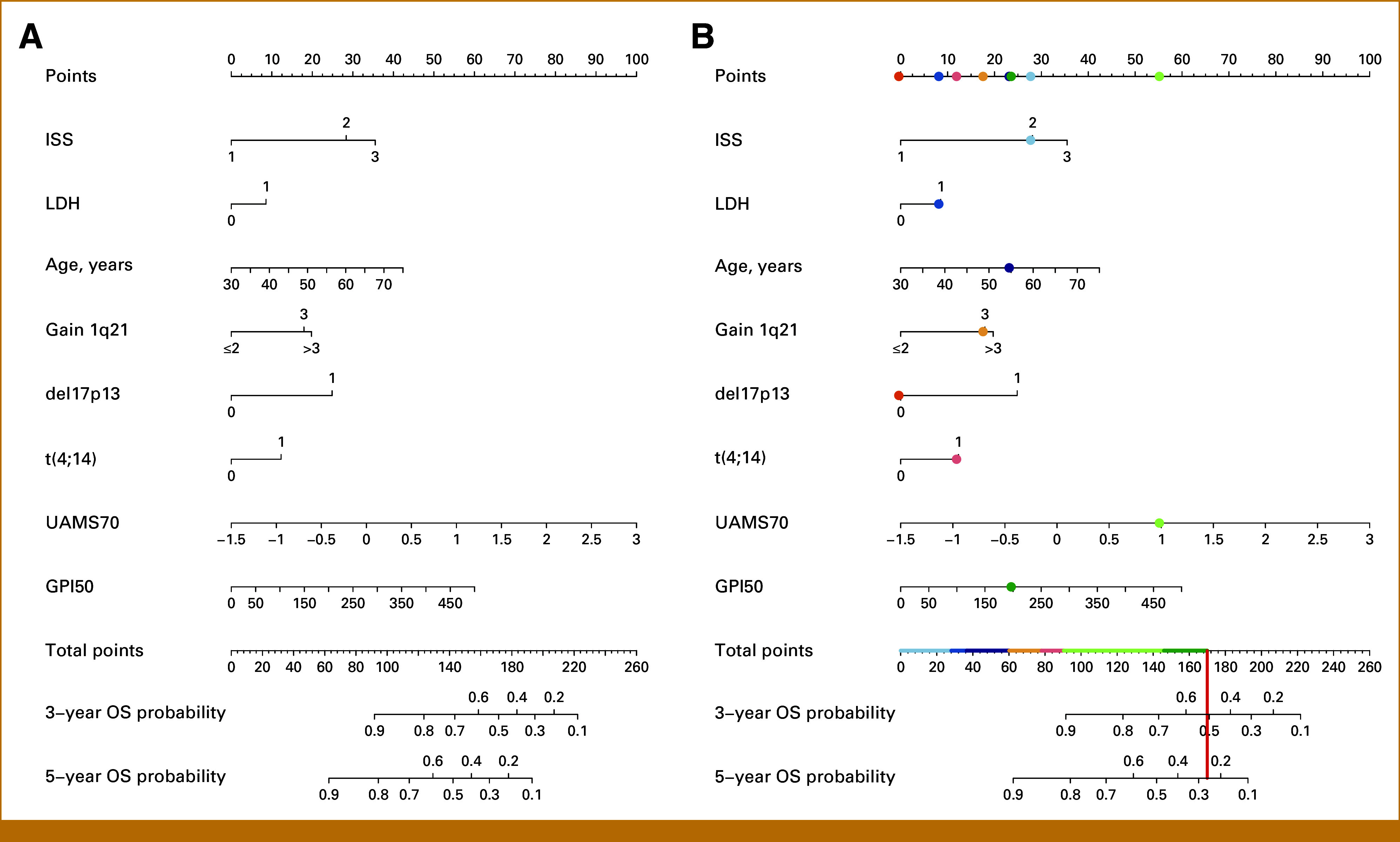
Nomogram. (A) Nomogram for estimating survival probabilities. (B) Exemplary patient. Here, 170 total points correspond to a 3-/5-year OS probability of 51% and 26%, respectively. Contribution of each risk factor is visualized by different colors. OS, overall survival.
The continuous scale allows also to group patients subsequently in low, intermediate, and high risk, eg, a sum of <123/123-171/>171 and <94/94-142/>142 points corresponds to 3-/5-year OS probabilities of >80 versus 50-80 versus <50% respectively.
Validation and Comparison With R-ISS, R2-ISS, and Mayo-2022-Score
The nomogram was validated on the VG regarding discrimination and calibration.32 The prognostic index was highly significant in the VG (P < .001) and with a regression coefficient of 1.04 very close to the optimal value of 1, indicating no obvious bias or overfitting.
Discrimination signifies the ability of the model to distinguish patients with poor and good prognosis. The model showed equally good discrimination in TG with a c-index of 0.76 and VG with a c-index of 0.75. The time-dependent AUC at 3 years was 0.74 in the VG.
In comparison with the nomogram score, the c-index for R-ISS was 0.65 in TG and 0.56 in VG, ie, in both groups significantly lower (P < .001). For R2-ISS, the c-index was 0.70 in TG (P < .001) and 0.63 in VG (P < .01). Regarding the Mayo-2022-score, the c-index was 0.70 in TG (P < .001) and 0.66 in VG (P = .07). The 3-year AUC of R-ISS/R2-ISS/Mayo-2022-score was 0.57/0.65/0.69 in the VG.
Subsequently, we assessed the distribution of continuous nomogram score values with R2-ISS and Mayo-2022-score (Figs 3A and 3B). Boxplots show a significant association between nomogram score and both scores (Kruskal-Wallis test, P < .001). This figure likewise exemplifies one of the most relevant aspects of continuous risk assessment: within each of the risk groups, high variation of risk is evident. For example, for R2-ISS3, nomogram scores between 50 and 200 are found, corresponding to predicted 5-year survival probabilities between >90% and <10%, respectively. While patients within each R-ISS group or its modifications are considered to have a similar risk, the quantitative nomogram score allows to further discriminate between those patients. For example, within VG patients with R-ISS II, the largest R-ISS subgroup (n = 75/121), the prognostic index was a significant predictor for OS (P < .001).
FIG 3.
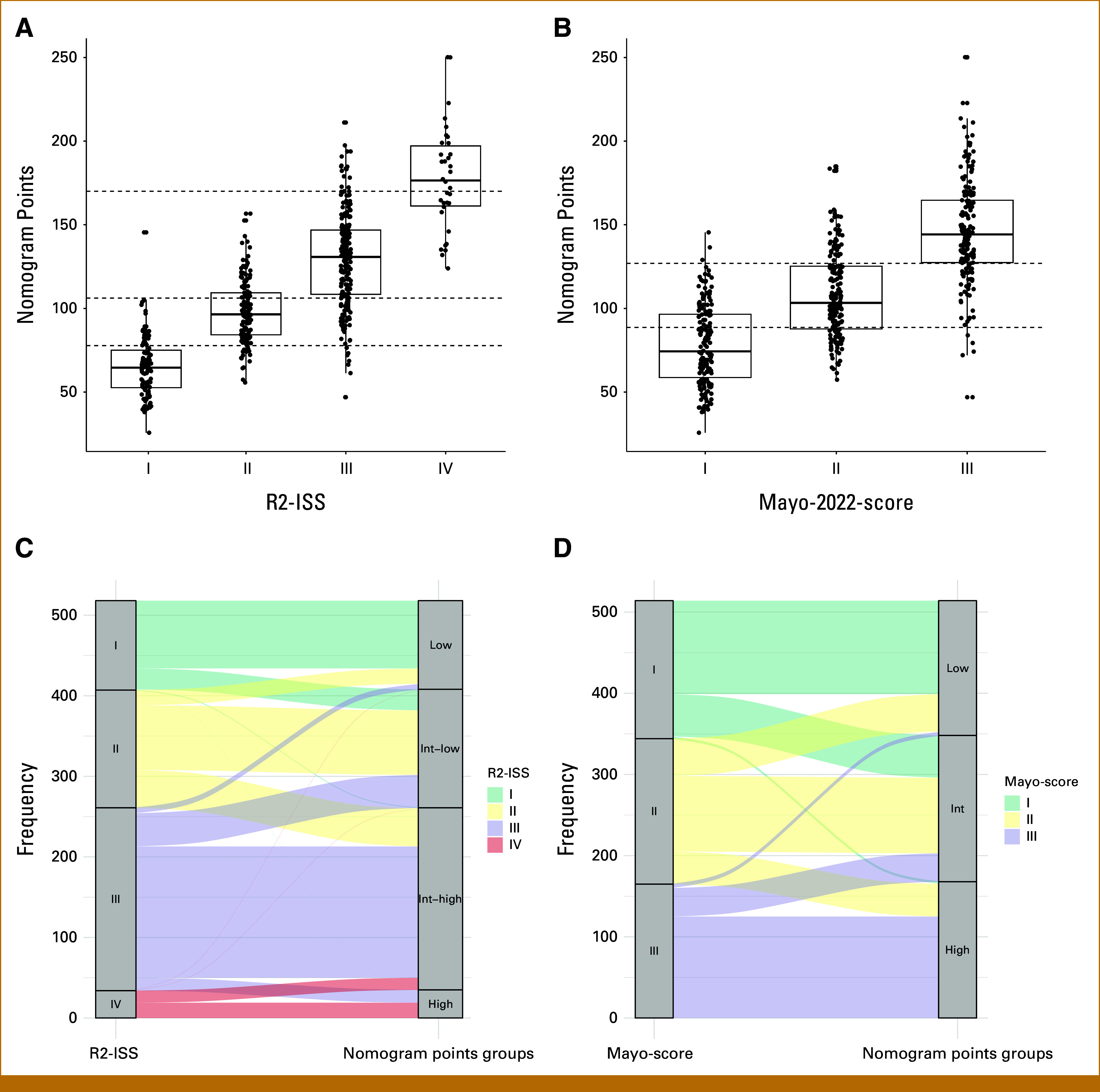
Continuous risk assessment (nomogram-core) versus grouped assessment regarding R2-ISS and Mayo-2022-score and transitions plots. The median nomogram score (continuous predicted survival probability, y-axis) is significantly different between groups for (A) R2-ISS and (B) Mayo-2022-score (Kruskal-Wallis-test, P < .001). At the same time, within each of the R2-ISS or Mayo-2022-score risk groups, score values vary up to four-fold, eg, R2-ISS3 from 50 to 200 points. Continuous risk assessment thus allows substratification within each of the risk groups. Dashed lines depict cut-points to match number and size of R2-ISS and Mayo-2022-groups, respectively. Alluvial plot for (C) R2-ISS and (D) Mayo-2022-score versus nomogram score. The nomogram score is grouped for both scores so that the number/size of the groups corresponds to the respective comparison score. Transitioning patients represent extreme scores within each of the risk groups.
We next depicted the overlap of the continuous nomogram score values with R2-ISS and Mayo-2022-score in a transition (alluvial) plot. To do so, we categorized the nomogram score to match the number and size of the groups in the comparator scores. Patient transitions occur in all risk groups, but rarely across multiple risk categories (Figs 3C and 3D).
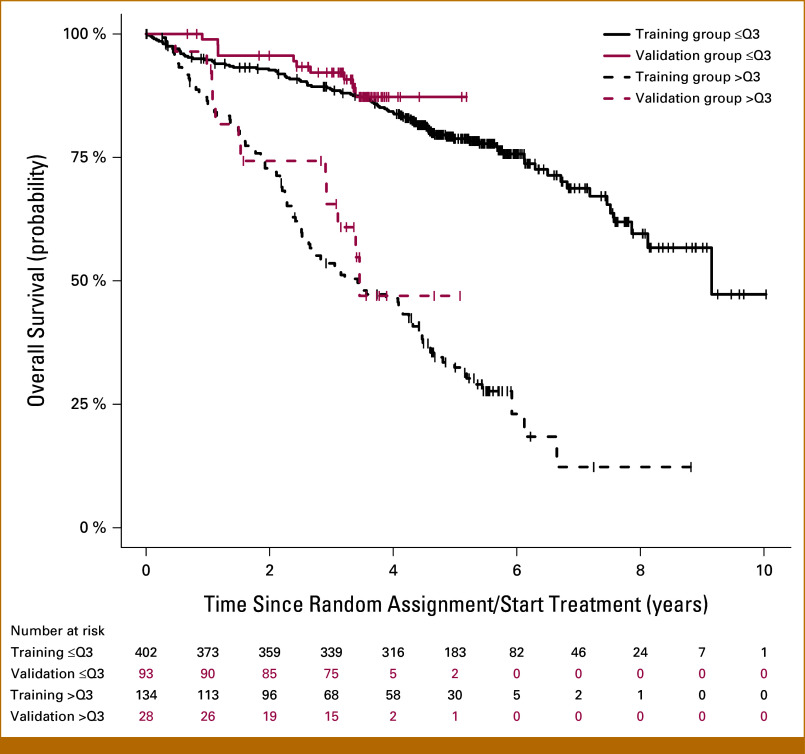
For explorative purpose, we defined two risk groups using the upper quartile of the linear predictor from the training model as cutoff. The same cutoff was applied to the prognostic index calculated for the validation data on the basis of the training model. Kaplan-Meier curves for the resulting patient groups confirmed good discrimination in both cohorts and good agreement between cohorts (Fig 4).
FIG 4.
Kaplan-Meier curves. Kaplan-Meier curves for the patient groups using the upper quartile of the prognostic index from the training group as cutoff confirmed good discrimination in training (black curve) and validation data (red curve).
Calibration, reflecting accuracy of the estimated survival times, was assessed by smoothed calibration plots of expected versus observed survival probabilities on VG and TG (bootstrap). Figure 5 shows calibration plots for both TG and VG data. Resampling based evaluation (TG) showed very good calibration, with tendency of too pessimistic predictions for high-risk patients in the VG as the more recent patient cohort.
FIG 5.
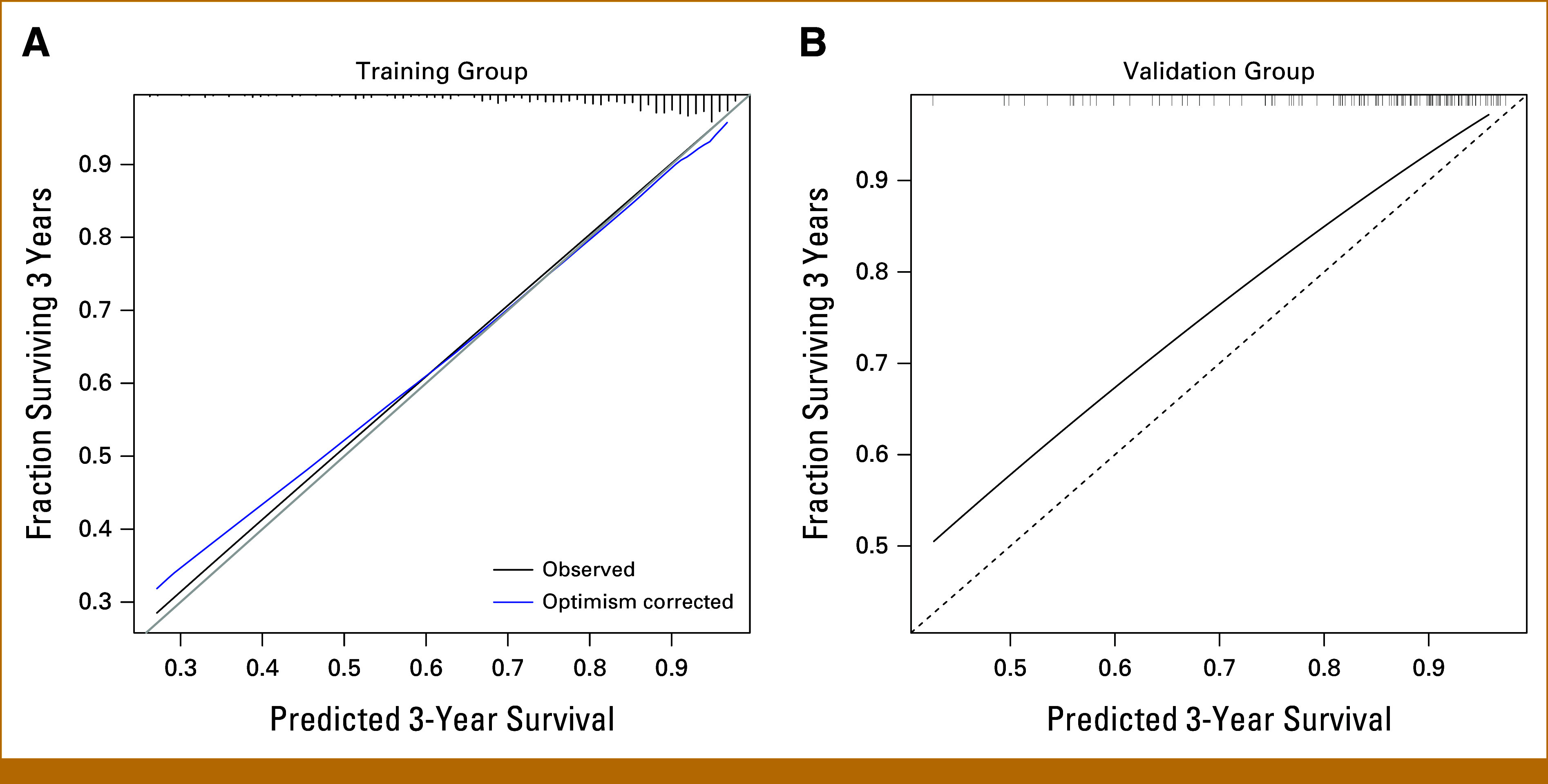
Calibration plots for training group and validation group data. Shown are smoothed calibration plots of observed versus estimated survival probabilities after 3 years for (A) training data and (B) validation data. Bootstrapping is used for training group (A) to get bias-corrected estimates.
DISCUSSION
Many different prognostic factors and prognostic scores attributing patients to two to four arbitrary groups, ie, (ultra-) high, intermediate, and low risk, have been proposed in myeloma, and the discussion of which factor to include is ongoing.4,6,10,17,35,36 This is further complicated by only partial overlap between patients identified as high risk by different scores.4,6,16 The variety of clinical and molecular prognostic factors necessitates integrating different factors into a single prognostic information.4,6,23 Widely accepted standard is the R-ISS.7 Prognostic power can be increased by suggested R-ISS-modifications, that is, R2-ISS10 or Mayo-2022-score,17 including 1q21-gains and integration of gene expression-based risk scores or proliferation, for example, UAMS GEP70-score4,6,11 and GPI.6,16
We integrated clinical and molecular prognostic factors, on the basis of iFISH and GEP analyses, into a comprehensive model followed by developing a quantitative prediction of individual patient's OS probability.
In addition to del17p13 and t(4;14) being part of R-ISS, we included 1q21-gains into our model. Associated with significantly higher proliferation,16 1q21-gain has been shown by others and us to be (copy number dependently) associated with adverse PFS and OS.37,38 1q21-gain was also included in two recent modifications of the R-ISS score.10,17 By contrast, t(14;16) was not considered as individual predictor for model building because of the low frequency (3% in our cohort). Boyd et al38 have shown t(14;16) and 1q21-gain cosegregating in about two thirds of patients. Most risk classifications, including R-ISS,7 do not take into account that it is not (only) the single adverse aberration impacting on patient's outcome but also their combination. This limitation is overcome by using quantitative prediction models as in our approach. Here, points are attributed to each of the prognostic factors and summed up. The contribution of each factor represented by different colors is shown in Figure 2B.
Continuous assessment allows more differentiated assessment for patients throughout the risk groups and better risk prediction (significantly higher c-index): Within each of the risk groups in R-ISS, R2-ISS, or Mayo-2022-score, risk varies widely if assessed continuously. For example, the nomogram score varies for R2-ISS3 from 50 to 200 points and corresponding predicted 5-year survival probability of >90% versus <10% (Figs 2 and 3). Methodologically, continuous risk assessment implies that for any given cut-point, risk changes gradually as opposed to step-wise for attribution to risk groups. This especially holds true for patients with risk scores at the border of cutoffs. An evident example is that it would be very difficult to suggest a pathophysiological explanation why a β2-microglobulin-level of 5.49 mg/L (below the threshold for ISS3) would convey a different prognosis compared with a β2-microglobulin-level of 5.5 mg/L (above this threshold). If comparing a grouping of the continuous nomogram score with R2-ISS or Mayo-2022-score matching the size and number of risk groups of the respective score, transitioning patients represent extreme scores within each of the risk groups, in agreement with above discussed argument.
On the basis of the depicted nomogram, continuous risk assessment in general could be used in clinical routine. For the underlying methods used in our nomogram score, this might be called in question for expression profiling-based approaches so far only been used at specialized centers or as part of clinical trials, with cost and local infrastructure being the main limiting factors: introduced in myeloma research in 200239 and 2011,40 GEP and NGS revolutionized our understanding of myeloma biology, pathogenesis, and risk41,42; however, the standard myeloma workup is still based on morphological bone marrow assessment and iFISH because of several reasons.6 Of these, practical issues can be easily disproven: GEP can be applied in clinical routine in academic (eg, GEP-R,43 UAMS70-score,44 IFM-score45) and commercial settings (eg, MyPRS, Signal Genetics,46 MMprofiler, SkylineDiagnostics47) in most patients within 4 weeks.4 NGS-based techniques can be performed in academic (CoMMpass)48 or private laboratories,49 even within 14 days in a tertiary hospital.50 The same holds true for RNA sequencing48,51-53 in >90% of patients in clinical trials or routine as shown by us.6 However, for rare circumstances, myeloma treatment is not an emergency, and a time interval of for example, 2-4 weeks can be covered with a short course of steroids while waiting for test results.41
From a methodological point of view, continuous nomogram-based risk assessment can in principle be easily translated into RNA sequencing–based assessments. As recently published, we have shown the transferability of GEP-based scores (including GEP70 and GPI50 used in the nomogram) to RNA sequencing data.6
Treatment has significantly improved since the conduct of our GMMG-HD4 and GMMG-MM5 trials, especially including immune-oncological drugs. Effective quadruple combinations including anti-CD38 antibodies followed by ASCT increase response rates from about one-third for single agents54-60 to almost 100% of patients61,62 and are the evolving standard of care. One potential limitation of our study is that these regimens are only accounted for in terms of relapse and salvage, not up-front treatment. This is on the one hand true, as we could not assess the same question in a comparable cohort, for example, the GMMG-HD7 trial63 (including isatuximab-VRd induction treatment) because of nonmature data. On the other hand, most patients with myeloma worldwide are not treated with such four-compound induction regimen because of restrictions in reimbursement and approval. Thus, seemingly outdated regimen and corresponding prognostication are still of significant value. We, therefore, deliberately focused on a homogenous patient cohort treated with bortezomib-based induction therapy and long follow-up. As soon as the data from current studies are mature, they can likewise be the basis for a nomogram-based risk assessment as presented here.
In summary, we developed and validated individual quantitative nomogram-based prediction of survival in multiple myeloma which can in principle be used in clinical routine and methodically be translated to other settings (eg, RNA-sequencing based assessment). Integrating serum and molecular prognostic factors including iFISH- and GEP-based risk scores and proliferation, continuous risk assessment allows superior granular and individual risk stratification. This likewise overcomes heterogeneous grouping of patients to discrete risk groups, that is, high variation of risk within all groups for example, in R-ISS, R2-ISS, or Mayo-2022-score. Our study will hopefully serve as a bridge toward the goal of wider use of continuous risk assessment and inclusion of molecular profiling in clinical routine.
ACKNOWLEDGMENT
The authors thank Maria Dörner, Ewelina Nickel, and Birgit Schneiders for technical assistance in the enrichment of CD138-positive plasma cells; Tine Borowski, Michaela Brough, Michelle Ebentheuer, Marie-Christine Meffert, and Stephanie Pschowski-Zuck for technical assistance in iFISH; and Véronique Pantesco for performing DNA-microarrays, as well as the Transcriptomics Platform at CHU Montpellier.
PRIOR PRESENTATION
Presented in part at the American Society of Hematology Annual Meeting & Exposition, San Diego, CA, December 2, 2018.
SUPPORT
M.H., T.H., A.S., and D.H. contributed equally to this work.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.23.00613.
AUTHOR CONTRIBUTIONS
Conception and design: Manuela Hummel, Thomas Hielscher, Anja Seckinger, Dirk Hose
Financial support: Hartmut Goldschmidt
Provision of study materials or patients: Hans Salwender, Christof Scheid, Hartmut Goldschmidt, Dirk Hose
Collection and assembly of data: Manuela Hummel, Thomas Hielscher, Martina Emde-Rajaratnam, Susanne Beck, Uta Bertsch, Anna Jauch, Jérôme Moreaux, Anja Seckinger, Dirk Hose
Data analysis and interpretation: Manuela Hummel, Thomas Hielscher, Anja Seckinger, Dirk Hose
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Manuela Hummel
Employment: Roche
Stock and Other Ownership Interests: Roche
Patents, Royalties, Other Intellectual Property: Patent application under examination procedure: P36767 - Method for automated quality check of chromatographic and/or mass spectral data WO2023/031447
Hans Salwender
Honoraria: Takeda, Chugai Pharma, Janssen, BMS GmbH & Co KG, Amgen, Abbvie, Stemline Therapeutics, Oncopeptides, AstraZeneca, Sanofi, Genzyme, GlaxoSmithKline, Pfizer, Roche
Consulting or Advisory Role: Pfizer, Janssen Oncology, Sanofi, Oncopeptides, GlaxoSmithKline, Amgen, Abbvie, Bristol Myers Squibb/Celgene, Roche, Genzyme, Stemline Therapeutics, AstraZeneca
Travel, Accommodations, Expenses: Amgen, BMS GmbH & Co KG, Janssen, GlaxoSmithKline
Christof Scheid
Employment: University of Cologne
Honoraria: Amgen, Bristol Myers Squibb/Celgene, Janssen Oncology, Novartis, Pfizer, Takeda, Sanofi/Aventis, GlaxoSmithKline, Stemline Therapeutics, Oncopeptides, Adaptive Biotechnologies
Consulting or Advisory Role: Amgen, Roche, Janssen Oncology, Bristol Myers Squibb/Celgene, GlaxoSmithKline, Sanofi/Aventis
Research Funding: Janssen Oncology (Inst), Takeda (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: Bristol Myers Squibb/Celgene, Janssen Oncology, Amgen
Hartmut Goldschmidt
Honoraria: Janssen-Cilag, Novartis, Bristol Myers Squibb, Chugai Pharma, Sanofi, Amgen, GlaxoSmithKline, Pfizer
Consulting or Advisory Role: Janssen-Cilag (Inst), Bristol Myers Squibb (Inst), Amgen (Inst), Adaptive Biotechnologies (Inst), Sanofi (Inst)
Research Funding: Bristol Myers Squibb (Inst), Janssen (Inst), Novartis (Inst), Celgene (Inst), Amgen (Inst), Sanofi (Inst), Takeda (Inst), Molecular Partners (Inst), MSD (Inst), Incyte (Inst), GlycoMimetics Inc (Inst), GlaxoSmithKline (Inst), Heidelberg Pharma (Inst), Roche (Inst), Karyopharm Therapeutics (Inst), Millennium Pharmaceuticals (Inst), MorphoSys (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: Janssen-Cilag, Sanofi, Amgen, Bristol Myers Squibb/Celgene, GlaxoSmithKline, Novartis, Pfizer
Other Relationship: Amgen (Inst), Celgene/Bristol Myers Squibb (Inst), Chugai Pharma Europe (Inst), Janssen (Inst), Sanofi (Inst), Mundipharma (Inst), Array BioPharma/Pfizer (Inst)
Jérôme Moreaux
Honoraria: Diag2Tec
Anja Seckinger
Employment: LamKap Bio Group
Leadership: LamKap Bio Group, Light Chain Bioscience—Novimmune SA
Stock and Other Ownership Interests: LamKap Bio alpha, LamKap Bio beta, LamKap Bio gamma
Patents, Royalties, Other Intellectual Property: Patent: Bispecific antibodies against CEACAM5 and CD47 (WO2022130348A1), Patent: Combination therapy of bispecific antibodies against CEACAM5 and CD47 and bispecific antibodies against CEACAM5 and CD3 (WO2023242351)
Dirk Hose
Employment: LamKap Bio Group
Leadership: LamKap Bio Group
Stock and Other Ownership Interests: LamKap Bio alpha AG, LamKap Bio Beta AG, LamKap Bio gamma AG
Research Funding: BMS
Patents, Royalties, Other Intellectual Property: Patent: Bispecific antibodies against CEACAM5 and CD47 (WO2022130348A1), Patent: Combination therapy of bispecific antibodies against CEACAM5 and CD47 and bispecific antibodies against CEACAM5 and CD3 (WO2023242351)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kyle RA, Rajkumar SV: Multiple myeloma. N Engl J Med 351:1860-1873, 2004 [DOI] [PubMed] [Google Scholar]
- 2.Barlogie B, Mitchell A, van Rhee F, et al. : Curing myeloma at last: Defining criteria and providing the evidence. Blood 124:3043-3051, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldschmidt H, Lokhorst HM, Mai EK, et al. : Bortezomib before and after high-dose therapy in myeloma: Long-term results from the phase III HOVON-65/GMMG-HD4 trial. Leukemia 32:383-390, 2018 [DOI] [PubMed] [Google Scholar]
- 4.Hose D, Beck S, Salwender H, et al. : Prospective target assessment and multimodal prediction of survival for personalized and risk-adapted treatment strategies in multiple myeloma in the GMMG-MM5 multicenter trial. J Hematol Oncol 12:65, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimopoulos MA, Terpos E, Boccadoro M, et al. : Daratumumab plus pomalidomide and dexamethasone versus pomalidomide and dexamethasone alone in previously treated multiple myeloma (APOLLO): An open-label, randomised, phase 3 trial. Lancet Oncol 22:801-812, 2021 [DOI] [PubMed] [Google Scholar]
- 6.Emde-Rajaratnam M, Beck S, Benes V, et al. : RNA-sequencing based first choice of treatment and determination of risk in multiple myeloma. Front Immunol 14:1286700, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palumbo A, Avet-Loiseau H, Oliva S, et al. : Revised International Staging System for Multiple Myeloma: A report from International Myeloma Working Group. J Clin Oncol 33:2863-2869, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neben K, Jauch A, Bertsch U, et al. : Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica 95:1150-1157, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avet-Loiseau H, Durie BG, Cavo M, et al. : Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: An International Myeloma Working Group collaborative project. Leukemia 27:711-717, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Agostino M, Cairns DA, Lahuerta JJ, et al. : Second revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: A European Myeloma Network (EMN) report within the HARMONY project. J Clin Oncol 40:3406-3418, 2022 [DOI] [PubMed] [Google Scholar]
- 11.Shaughnessy JD, Zhan F, Burington BE, et al. : A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109:2276-2284, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Decaux O, Lodé L, Magrangeas F, et al. : Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myélome. J Clin Oncol 26:4798-4805, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Reme T, Hose D, Theillet C, et al. : Modeling risk stratification in human cancer. Bioinformatics 29:1149-1157, 2013 [DOI] [PubMed] [Google Scholar]
- 14.Kuiper R, Broyl A, de Knegt Y, et al. : A gene expression signature for high-risk multiple myeloma. Leukemia 26:2406-2413, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Chng WJ, Chung TH, Kumar S, et al. : Gene signature combinations improve prognostic stratification of multiple myeloma patients. Leukemia 30:1071-1078, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Hose D, Reme T, Hielscher T, et al. : Proliferation is a central independent prognostic factor and target for personalized and risk adapted treatment in multiple myeloma. Haematologica 96:87-95, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdallah NH, Binder M, Rajkumar SV, et al. : A simple additive staging system for newly diagnosed multiple myeloma. Blood Cancer J 12:21, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonneveld P, Schmidt-Wolf IG, van der Holt B, et al. : Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: Results of the randomized phase III HOVON-65/GMMG-HD4 trial. J Clin Oncol 30:2946-2955, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Mai EK, Bertsch U, Durig J, et al. : Phase III trial of bortezomib, cyclophosphamide and dexamethasone (VCD) versus bortezomib, doxorubicin and dexamethasone (PAd) in newly diagnosed myeloma. Leukemia 29:1721-1729, 2015 [DOI] [PubMed] [Google Scholar]
- 20.Merz M, Salwender H, Haenel M, et al. : Subcutaneous versus intravenous bortezomib in two different induction therapies for newly diagnosed multiple myeloma: An interim analysis from the prospective GMMG-MM5 trial. Haematologica 100:964-969, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hose D, Moreaux J, Meissner T, et al. : Induction of angiogenesis by normal and malignant plasma cells. Blood 114:128-143, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Hose D, Rème T, Meissner T, et al. : Inhibition of aurora kinases for tailored risk-adapted treatment of multiple myeloma. Blood 113:4331-4340, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meißner T, Seckinger A, Rème T, et al. : Gene expression profiling in multiple myeloma—Reporting of entities, risk, and targets in clinical routine. Clin Cancer Res 17:7240-7247, 2011 [DOI] [PubMed] [Google Scholar]
- 24.Seckinger A, Delgado JA, Moser S, et al. : Target expression, generation, preclinical activity, and pharmacokinetics of the BCMA-T cell bispecific antibody EM801 for multiple myeloma treatment. Cancer Cell 31:396-410, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Seckinger A, Meißner T, Moreaux J, et al. : Clinical and prognostic role of annexin A2 in multiple myeloma. Blood 120:1087-1094, 2012 [DOI] [PubMed] [Google Scholar]
- 26.Seckinger A, Meissner T, Moreaux J, et al. : Bone morphogenic protein 6: A member of a novel class of prognostic factors expressed by normal and malignant plasma cells inhibiting proliferation and angiogenesis. Oncogene 28:3866-3879, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seckinger A, Hillengass J, Emde M, et al. : CD38 as immunotherapeutic target in light chain amyloidosis and multiple myeloma—association with molecular entities, risk, survival, and mechanisms of upfront resistance. Front Immunol 9:1676, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neben K, Jauch A, Hielscher T, et al. : Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J Clin Oncol 31:4325-4332, 2013 [DOI] [PubMed] [Google Scholar]
- 29.Greipp PR, San Miguel J, Durie BGM, et al. : International staging system for multiple myeloma. J Clin Oncol 23:3412-3420, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Grambsch PM, Therneau TM: Proportional hazards tests and diagnostics based on weighted residuals. Biometrika 81:515-526, 1994 [Google Scholar]
- 31.Van Buuren S, Groothuis-Oudshoorn K: mice: Multivariate imputation by chained equations in R. J Stat Softw 45:67, 2011 [Google Scholar]
- 32.Royston P, Altman DG: External validation of a Cox prognostic model: Principles and methods. BMC Med Res Methodol 13:33, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang L, Chen W, Petrick NA, et al. : Comparing two correlated C indices with right-censored survival outcome: A one-shot nonparametric approach. Stat Med 34:685-703, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrell FE: rms: Regression Modeling Strategies: R Package Version 5.1-1, 2017. https://CRAN.R-project.org/package=rms [Google Scholar]
- 35.Chng WJ, Dispenzieri A, Chim CS, et al. : IMWG consensus on risk stratification in multiple myeloma. Leukemia 28:269-277, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Fonseca R, Bergsagel PL, Drach J, et al. : International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia 23:2210-2221, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neben K, Lokhorst HM, Jauch A, et al. : Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood 119:940-948, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Boyd KD, Ross FM, Chiecchio L, et al. : A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: Analysis of patients treated in the MRC Myeloma IX trial. Leukemia 26:349-355, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan F, Hardin J, Kordsmeier B, et al. : Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood 99:1745-1757, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Chapman MA, Lawrence MS, Keats JJ, et al. : Initial genome sequencing and analysis of multiple myeloma. Nature 471:467-472, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolli N, Genuardi E, Ziccheddu B, et al. : Next-generation sequencing for clinical management of multiple myeloma: Ready for prime time? Front Oncol 10:189, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lionetti M, Neri A: Utilizing next-generation sequencing in the management of multiple myeloma. Expert Rev Mol Diagn 17:653-663, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Meissner T, Seckinger A, Reme T, et al. : Gene expression profiling in multiple myeloma—reporting of entities, risk, and targets in clinical routine. Clin Cancer Res 17:7240-7247, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Shaughnessy JD Jr, Zhan F, Burington BE, et al. : A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109:2276-2284, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Decaux O, Lode L, Magrangeas F, et al. : Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myelome. J Clin Oncol 26:4798-4805, 2008 [DOI] [PubMed] [Google Scholar]
- 46.van Laar R, Flinchum R, Brown N, et al. : Translating a gene expression signature for multiple myeloma prognosis into a robust high-throughput assay for clinical use. BMC Med Genomics 7:25, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown S, Sherratt D, Hinsley S, et al. : MUKnine OPTIMUM protocol: A screening study to identify high-risk patients with multiple myeloma suitable for novel treatment approaches combined with a phase II study evaluating optimised combination of biological therapy in newly diagnosed high-risk multiple myeloma and plasma cell leukaemia. BMJ Open 11:e046225, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldsmith SR, Fiala MA, Dukeman J, et al. : Next generation sequencing-based validation of the revised international staging system for multiple myeloma: An analysis of the MMRF CoMMpass study. Clin Lymphoma Myeloma Leuk 19:285-289, 2019 [DOI] [PubMed] [Google Scholar]
- 49.Hollein A, Twardziok SO, Walter W, et al. : The combination of WGS and RNA-Seq is superior to conventional diagnostic tests in multiple myeloma: Ready for prime time? Cancer Genet 242:15-24, 2020 [DOI] [PubMed] [Google Scholar]
- 50.Lomas OC, Gooding S, Cabes M, et al. : Validation of clinical-grade whole genome sequencing reproduces cytogenetic analysis and identifies mutational landscape in newly-diagnosed multiple myeloma patients: A pilot study from the 100,000 Genomes Project. EJHaem 2:809-812, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seckinger A, Delgado JA, Moser S, et al. : Target expression, generation, preclinical activity, and pharmacokinetics of the BCMA-T cell bispecific antibody EM801 for multiple myeloma treatment. Cancer Cell 31:396-410, 2017 [DOI] [PubMed] [Google Scholar]
- 52.Seckinger A, Hillengass J, Emde M, et al. : CD38 as immunotherapeutic target in light chain amyloidosis and multiple myeloma-association with molecular entities, risk, survival, and mechanisms of upfront resistance. Front Immunol 9:1676, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bendig S, Walter W, Meggendorfer M, et al. : Whole genome sequencing demonstrates substantial pathophysiological differences of MYC rearrangements in patients with plasma cell myeloma and B-cell lymphoma. Leuk Lymphoma 62:3420-3429, 2021 [DOI] [PubMed] [Google Scholar]
- 54.Richardson PG, Barlogie B, Berenson J, et al. : A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med 348:2609-2617, 2003 [DOI] [PubMed] [Google Scholar]
- 55.O'Connor OA, Stewart AK, Vallone M, et al. : A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clin Cancer Res 15:7085-7091, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar SK, LaPlant B, Roy V, et al. : Phase 2 trial of ixazomib in patients with relapsed multiple myeloma not refractory to bortezomib. Blood Cancer J 5:e338, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singhal S, Mehta J, Desikan R, et al. : Antitumor activity of thalidomide in refractory multiple myeloma. N Engl J Med 341:1565-1571, 1999 [DOI] [PubMed] [Google Scholar]
- 58.Richardson PG, Schlossman RL, Weller E, et al. : Immunomodulatory drug CC-5013 overcomes drug resistance and is well tolerated in patients with relapsed multiple myeloma. Blood 100:3063-3067, 2002 [DOI] [PubMed] [Google Scholar]
- 59.Miguel JS, Weisel K, Moreau P, et al. : Pomalidomide plus low-dose dexamethasone versus high-dose dexamethasone alone for patients with relapsed and refractory multiple myeloma (MM-003): A randomised, open-label, phase 3 trial. Lancet Oncol 14:1055-1066, 2013 [DOI] [PubMed] [Google Scholar]
- 60.Schey SA, Fields P, Bartlett JB, et al. : Phase I study of an immunomodulatory thalidomide analog, CC-4047, in relapsed or refractory multiple myeloma. J Clin Oncol 22:3269-3276, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Voorhees PM, Kaufman JL, Laubach J, et al. : Daratumumab, lenalidomide, bortezomib, and dexamethasone for transplant-eligible newly diagnosed multiple myeloma: The GRIFFIN trial. Blood 136:936-945, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonneveld P, Dimopoulos MA, Boccadoro M, et al. : Daratumumab, bortezomib, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med 390:301-313, 2024 [DOI] [PubMed] [Google Scholar]
- 63.Goldschmidt H, Mai EK, Bertsch U, et al. : Addition of isatuximab to lenalidomide, bortezomib, and dexamethasone as induction therapy for newly diagnosed, transplantation-eligible patients with multiple myeloma (GMMG-HD7): Part 1 of an open-label, multicentre, randomised, active-controlled, phase 3 trial. Lancet Haematol 9:e810-e821, 2022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO.23.00613.