Abstract
PURPOSE
Prostate cancer (PCa) represents a highly heterogeneous disease that requires tools to assess oncologic risk and guide patient management and treatment planning. Current models are based on various clinical and pathologic parameters including Gleason grading, which suffers from a high interobserver variability. In this study, we determine whether objective machine learning (ML)–driven histopathology image analysis would aid us in better risk stratification of PCa.
MATERIALS AND METHODS
We propose a deep learning, histopathology image–based risk stratification model that combines clinicopathologic data along with hematoxylin and eosin– and Ki-67–stained histopathology images. We train and test our model, using a five-fold cross-validation strategy, on a data set from 502 treatment-naïve PCa patients who underwent radical prostatectomy (RP) between 2000 and 2012.
RESULTS
We used the concordance index as a measure to evaluate the performance of various risk stratification models. Our risk stratification model on the basis of convolutional neural networks demonstrated superior performance compared with Gleason grading and the Cancer of the Prostate Risk Assessment Post-Surgical risk stratification models. Using our model, 3.9% of the low-risk patients were correctly reclassified to be high-risk and 21.3% of the high-risk patients were correctly reclassified as low-risk.
CONCLUSION
These findings highlight the importance of ML as an objective tool for histopathology image assessment and patient risk stratification. With further validation on large cohorts, the digital pathology risk classification we propose may be helpful in guiding administration of adjuvant therapy including radiotherapy after RP.
We proposed a risk stratification model to combine clinical data with H&E- and Ki-67–stained images.
INTRODUCTION
Prostate cancer (PCa) is the second most commonly diagnosed and sixth deadliest cancer among men worldwide.1 It is a heterogeneous disease with a diverse range of histologic patterns. PCa diagnosis and therapeutic decisions are driven primarily by histologic descriptors of prostate tissue samples obtained by prostate needle biopsy under transrectal ultrasound guidance. The biopsy Gleason score (GS) is currently the most common grading system of prostate adenocarcinoma,2,3 where a grade is determined by a pathologist on the basis of the glandular architectural features observed in hematoxylin and eosin (H&E)–stained tissue samples. Although the GS is the strongest clinicopathologic predictor for clinical outcomes,4 its ability to predict clinical outcome is limited by the spatial heterogeneity of PCa5 and high interobserver variability.6
CONTEXT
Key Objective
To develop a risk stratification model on the basis of Cancer of the Prostate Risk Assessment Post-Surgical (CAPRA-S) and hematoxylin and eosin (H&E)– and Ki-67–stained tissue microarrays to predict the risk scores for biochemical recurrence and overall survival in prostate cancer (PCa).
Knowledge Generated
The comparison of concordance indices for the outcomes of interest revealed that using CAPRA-S, H&E, and Ki-67 features is the most effective method among various models accessing different data. Moreover, reclassifying patients on the basis of risk scores demonstrated the importance of digital pathology in outcome prediction for disease management.
Relevance
Machine learning and image-based pipelines to link digital histopathology images directly to cancer outcomes are of value, as they may provide improved risk classification (as demonstrated here in PCa). Additionally, this approach may be the basis for automatization and reduction of inter-observer variability in pathology.
In addition to GS, a number of other clinicopathologic variables such as age, tumor stage or volume, margin status, and prostate-specific antigen (PSA) levels, as well as their various combinations, have been used to predict PCa outcomes. For example, the Cancer of the Prostate Risk Assessment (CAPRA)7 and its analogous derivative, CAPRA Post-Surgical (CAPRA-S),8 represent well-validated risk stratification solutions by offering easy-to-use and clinically accurate prediction models. However, variance in PCa grading among pathologists can still lead to undertreatment or overtreatment of patients, affecting their survival rates and quality of life, as well as their cost to the health care system.
Recent applications of machine learning (ML) in digital pathology images have shown great potential in a range of tasks, where the majority of the effort has been focused on malignancy detection from needle biopsies and GS prediction.9-15 However, because of the high interobserver variability in GS assessment, the ground truth used to develop image-based prediction algorithms remains elusive. There has been little research on directly linking digital histopathology to PCa outcome or response to therapies.16-19 For example, Nagpal et al16 reported that the GS predicted by the ML algorithm, as opposed to assigned by pathologists, achieved slight improvement in risk stratification (concordance index [CI], improved from 0.63 to 0.65). In a follow-up study by the same group on a larger cohort, Wulczyn et al17 achieved a CI value of 0.82 for PCa-specific mortality using ML-based Gleason grading warranting further evaluation for improving disease management.
Building on this body of work, we propose CCHEK (CAPRA-S + CNN H&E & Ki-67), an ML-driven PCa risk assessment method that combines clinicopathologic data with computer-generated features from H&E- and Ki-67–stained tissue microarrays. Compared with the state-of-the-art CAPRA-S model for patient risk stratification, CCHEK is more accurate in predicting biochemical recurrence (BCR), as well as overall survival (OS), as evaluated on our radical prostatectomy (RP) patient data set.
MATERIALS AND METHODS
Cohort Construction and Clinical Data
A total of 502 treatment-naïve PCa patients who underwent RP between 2000 and 2012 were included in this study (Table 1). The study was approved by the University of British Columbia institutional Clinical Research Ethics Board (CREB H15-01064). As previously described,20 tissue microarray (TMA) blocks (one to four cores per patient) constructed from RP and stained by H&E and Ki-67 markers were scanned at 40× magnification using an SCN400 Slide Scanner (Leica Microsystems, Wetzlar, Germany). The cores were scored on the basis of the currently recommended International Society of Urological Pathology GS standards.21
TABLE 1.
Patient Cohort Summary: Clinical and Surgical Parameter Distribution
| Clinical and Pathologic Parameter | Distribution |
|---|---|
| Treatment | 35 patients underwent adjuvant radiation; 47 patients underwent salvage radiation |
| Age, years | 64.9 (mean) ± 7.5 (SD) |
| Biopsy GS (total 494), No. | |
| 6 | 228 |
| 3 + 4 | 140 |
| 4 + 3 | 61 |
| 8 | 42 |
| 9-10 | 23 |
| Preop PSA, ng/mL | 7.2797 (mean) ± 4.8766 (SD) |
| Clinical T stage (total 494), No. | |
| T1c | 233 |
| T2a | 180 |
| T2b/c | 70 |
| T3 | 11 |
| Pathologic T stage (total 499), No. | |
| T0 | 1 |
| T2a | 40 |
| T2b/c | 307 |
| T3/T4 | 151 |
| GS from RP (total 495), No. | |
| 6 | 127 |
| 3 + 4 | 211 |
| 4 + 3 | 94 |
| 8 | 25 |
| 9-10 | 38 |
| Surgical margins (total 499), No. | |
| Positive | 140 |
| Negative | 359 |
| Lymph node status (total 496), No. | |
| N1 | 16 |
| N0 | 258 |
| Nx | 222 |
Abbreviations: GS, Gleason score; PSA, prostate-specific antigen; RP, radical prostatectomy; SD, standard deviation.
CAPRA and CAPRA-S
The CAPRA method generates a pretreatment score. Cooperberg et al22 introduced a new postsurgical score (CAPRA-S) that is improved over CAPRA via incorporation of pathologic data. On the basis of PSA, pathologic GS, surgical margins, extracapsular extension, seminal vesicle invasion, and lymph node invasion, the CAPRA-S score was developed and validated as a predictor of downstream oncologic end points including BCR and systemic progression.8 The CAPRA-S score divides the patients into three groups of low-risk (CAPRA-S ≤2), intermediate-risk (CAPRA-S 3-5), and high-risk (CAPRA-S ≥6).
Proposed Risk Stratification Pipeline
A schematic representation of our proposed risk stratification method CCHEK is shown in Figure 1. We first extracted up to 48 768 × 768-pixel patches from each TMA core image. In addition to manually engineered feature sets,23 we trained a CNN to extract the most relevant features from individual image patches. A survival regression model on the basis of the Cox proportional hazards model24 with elastic net regularization was then trained to predict a risk score for every image patch. Finally, in an aggregation step, a smooth maximum function estimated a patient’s overall risk score from all their patch-wise risk scores.
FIG 1.
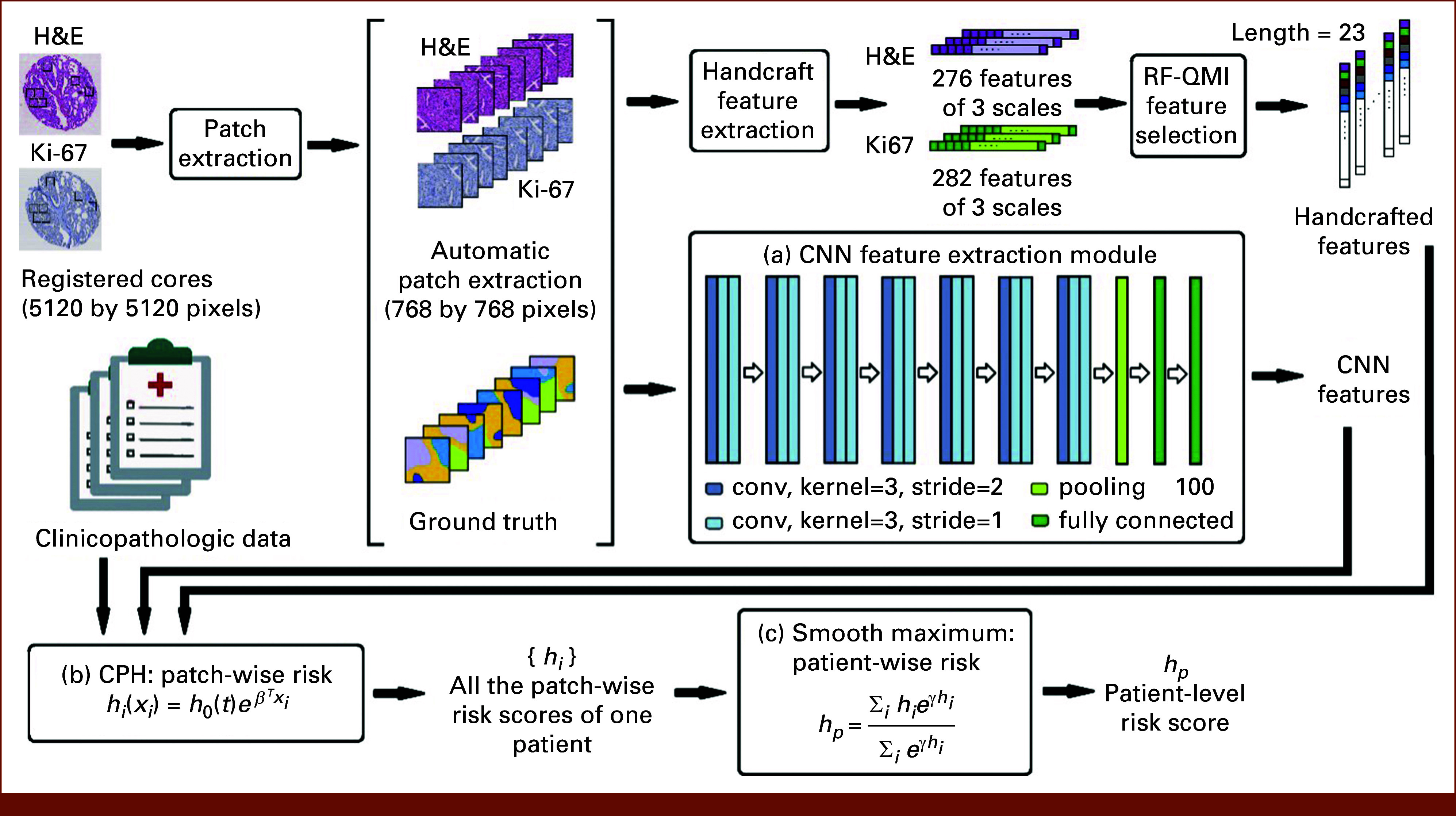
Overview of the proposed ML method to link histopathology to outcome (eg, BCR or OS). The signal processing pipeline takes as input the H&E and Ki-67 images and produces as output the risk score (eg, BCR) for a given patient. BCR, biochemical recurrence; CNN, convolutional neural network; CPH, Cox proportional hazards; H&E, hematoxylin and eosin; ML, machine learning; OS, overall survival; RF-QMI, Random Forest Quadratic Mutual Information.
The CCHEK risk stratification models were trained on three categories of data: clinicopathologic data, manually engineered image features consisting of 276 features from H&E images and 282 features from Ki-67 images,23 and CNN-extracted features from the 48 patches of each core image. The details of the proposed ML pipeline can be found in the Data Supplement Text.
Statistical Methods
To evaluate the proposed models, we used a five-fold cross-validation strategy. CI was used to evaluate the performance of various risk stratification models, and the Student's t-test25 was used to evaluate whether the differences between the performance of various models were statistically significant. Statistical tests were performed on the bootstrap sets of 100 held-out samples.
RESULTS
Table 2 shows the CI results for the risk stratification models on the basis of (1) time to BCR and (2) OS after treatment (surgery/radiation). The median follow-up time was 65.9 months after surgery/radiation. Of the 502 patients, 102 had experienced BCR.
TABLE 2.
CI Results and the Associated SDs (100 bootstrap sets) for Risk Stratification Models on the Basis of Various Clinicopathologic and ML-Driven Features
| Data | Row | Risk Score | Follow-Up After Surgery/Radiation. Censor: BCR | Follow-Up After Surgery/Radiation. Censor: Death |
|---|---|---|---|---|
| Clinicopathologic data | 1 | Biopsy GS | 0.63 ± 0.07 | 0.68 ± 0.10 |
| 2 | Clinical T stage | 0.56 ± 0.07 | 0.64 ± 0.10 | |
| 3 | Preop PSA | 0.57 ± 0.06 | 0.60 ± 0.08 | |
| 4 | Preop PSA, biopsy GS | 0.64 ± 0.07 | 0.68 ± 0.09 | |
| 5 | Preop PSA, biopsy GS, clinical T stage | 0.66 ± 0.07 | 0.71 ± 0.10 | |
| 6 | Pathologic T stage | 0.69 ± 0.05 | 0.79 ± 0.09 | |
| 7 | Pathologic T stage, clinical T stage | 0.70 ± 0.06 | 0.81 ± 0.08 | |
| 8 | GS from TMA cores | 0.66 ± 0.07 | 0.74 ± 0.10 | |
| 9 | GS from RP | 0.70 ± 0.06 | 0.82 ± 0.07 | |
| ML | 10 | Handcrafted H&E | 0.69 ± 0.08 | 0.68 ± 0.09 |
| 11 | CNN H&E | 0.72 ± 0.06 | 0.72 ± 0.10 | |
| 12 | Handcrafted Ki-67 | 0.66 ± 0.07 | 0.67 ± 0.09 | |
| 13 | CNN Ki-67 | 0.68 ± 0.07 | 0.70 ± 0.09 | |
| 14 | Handcrafted H&E, Ki-67 | 0.71 ± 0.07 | 0.70 ± 0.10 | |
| 15 | CNN H&E, Ki-67 | 0.74 ± 0.08 | 0.75 ± 0.09 | |
| Postsurgical | 16 | CAPRA-S | 0.68 ± 0.06 | 0.80 ± 0.09 |
| 17 | CAPRA-S, handcrafted H&E | 0.71 ± 0.07 | 0.82 ± 0.09 | |
| 18 | CAPRA-S, CNN H&E | 0.73 ± 0.07 | 0.83 ± 0.09 | |
| 19 | CAPRA-S, handcrafted Ki-67 | 0.69 ± 0.06 | 0.81 ± 0.08 | |
| 20 | CAPRA-S, CNN Ki-67 | 0.71 ± 0.06 | 0.82 ± 0.10 | |
| 21 | CAPRA-S, handcrafted H&E, Ki-67 | 0.73 ± 0.07 | 0.83 ± 0.08 | |
| 22 | CAPRA-S, CNN H&E, Ki-67 | 0.76 ± 0.07 | 0.85 ± 0.09 |
NOTE. Preop represents measurements before RP. Bold values indicate highest numbers in each column.
Abbreviations: BCR, biochemical recurrence; CAPRA-S, Cancer of the Prostate Risk Assessment Post-Surgical; CI, concordance index; CNN, convolutional neural network; GS, Gleason score; H&E, hematoxylin and eosin; ML, machine learning; PSA, prostate-specific antigen; RP, radical prostatectomy; SD, standard deviation; TMA, tissue microarray.
GS From RP Remains the Most Significant Risk Predictor for PCa
We evaluated the contribution of various clinicopathologic factors in the risk stratification of our PCa cohort. In line with previous findings,16,23 GS from RP (CI = 0.70 and 0.82 on row 9) remains the most significant postsurgical risk predictor for both BCR and OS. This provides evidence that our patient cohort was representative of the entire PCa population.
ML-Based H&E Image Features Outperform Most Clinicopathologic Predictors
We next sought to evaluate the performance of the CCHEK model in predicting BCR and OS from H&E- and Ki-67–stained histopathology slides. Although ML models built upon handcrafted features did not improve over GS from RP, deep learning (DL)–based models applied to H&E- and Ki-67–stained TMA slides (Table 2, rows 11 and 15) improved CIs associated with BCR and OS. It is important to note that although the DL-based features were extracted from TMA cores, they improved upon risk stratification on the basis of GS that was assessed by pathologists through the inspection of the entire RP specimen. This suggests that DL models extract subtle information in the TMA images that are not captured by conventional and previously characterized morphologic features and Gleason grading.
ML Improves Upon CAPRA-S
We next sought to investigate whether the CCHEK risk scores could complement CAPRA-S.
In a multivariate Cox regression model, when ML-based H&E features were added, the CIs improved from 0.68 in BCR and 0.80 in OS (CAPRA-S, Table 2 row 16) to 0.73 and 0.83, respectively (combined CAPRA-S and image features, row 18; P: .00998 and .0112). Furthermore, when Ki-67 was added, the CIs further increased to 0.76 and 0.85 (row 22; P: .00013 and .00016, respectively). The Ki-67 protein is a cellular marker of cell proliferation. Its prognostic value has been investigated in various cancer types, including breast, soft tissue, lung, and prostate.26-29 Overall, these results indicate that integrating clinicopathologic data with ML-based digital histopathology in our complete CCHEK model could enhance patient risk stratification in comparison with existing state-of-the-art models.
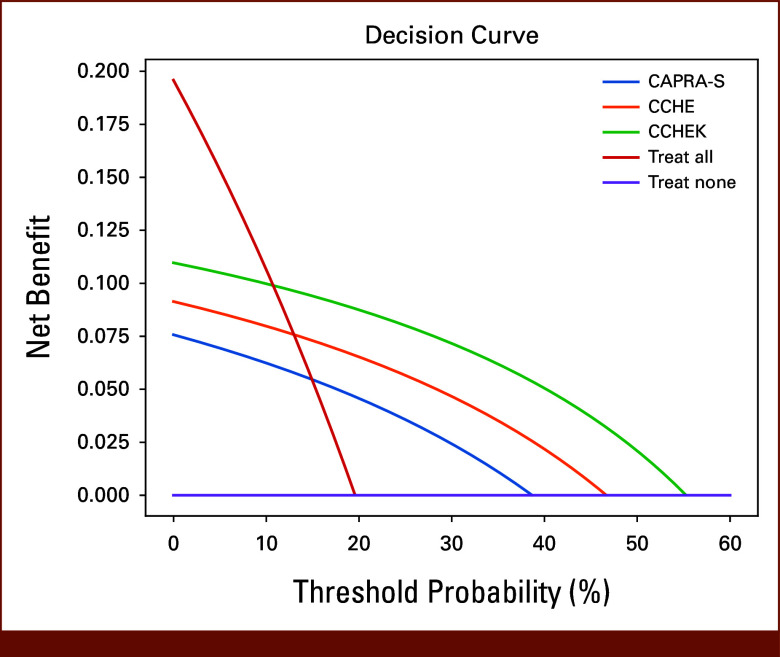
Furthermore, we experimented the classification power of CAPRA-S, CCHE (CAPRA-S + CNN H&E), and CCHEK in estimating whether a patient might have a BCR in 5 years. We censored patients whose last follow-up was <5 years. We used the risk scores and set the threshold in a way to predict BCR for the same ratio of patients who had BCR in 5 years. 19.58% of the patients had recurrence in the 5-year period. After sorting the risk scores, we picked the threshold for the methods on the basis of the patient who had lower risk score than 19.58% of the patients. We predicted that those patients are going to have BCR in 5 years. The macro averaged F1 score, precision, and recall was 62% for CAPRA-S, 67% for CCHE, and 73% for CCHEK. Moreover, decision curve analysis to compare the net benefit of CAPRA-S, CCHE, and CCHEK on the basis of BCR in 5 years suggested that CCHEK has the most net benefit compared with other methods (Fig 2).
FIG 2.
Decision curve analysis for CAPRA-S, CCHE, and CCHEK. CCHEK has the most net benefit compared with other methods. CAPRA-S, Cancer of the Prostate Risk Assessment Post-Surgical; CCHE, CAPRA-S + CNN H&E; CCHEK, CAPRA-S + CNN H&E & Ki-67.
We next divided patients into three risk groups on the basis of CAPRA-S scores as suggested in the previous study8: low-risk (CAPRA-S ≤2), intermediate-risk (CAPRA-S 3-5), and high-risk (CAPRA-S ≥6). The risk groups are used in determining patient management. To facilitate comparison between our proposed AI-based risk models and CAPRA-S through Kaplan-Meier survival analysis, we also divided the cohort into the low-risk, intermediate-risk, and high-risk groups of the same ratio as the CAPRA-S groups, but with patient grouping determined on the basis of our two ML-based risk stratification models: (1) combination of CAPRA-S– and CNN-based features from H&E images (Table 2 row 18), and (2) combination of CAPRA-S– and CNN-based features from H&E and Ki-67 images (Table 2 row 22). CAPRA-S predicts 52.23% of patients as low-risk, 38.26% of patients as intermediate-risk, and the rest 9.51% are grouped as high-risk. Therefore, after sorting risk scores, we chose the risk-score threshold for low-risk group on the basis of the patient who has higher risk score than 52.23% of patients. We also chose the risk-score threshold for high-risk group on the basis of the patient who has lower risk score than 9.51% of the patients. The patients who have a risk score between these two values are grouped as the intermediate-risk group. With this process, we would end up having the same ratio of patients as in CAPRA-S groups.
Kaplan-Meier survival curves30 associated with BCR and OS corresponding to the three groups (Fig 3) suggest that the CCHEK risk stratification model performed better than CAPRA-S and CCHE by correctly identifying more low- and high-risk patients.
FIG 3.
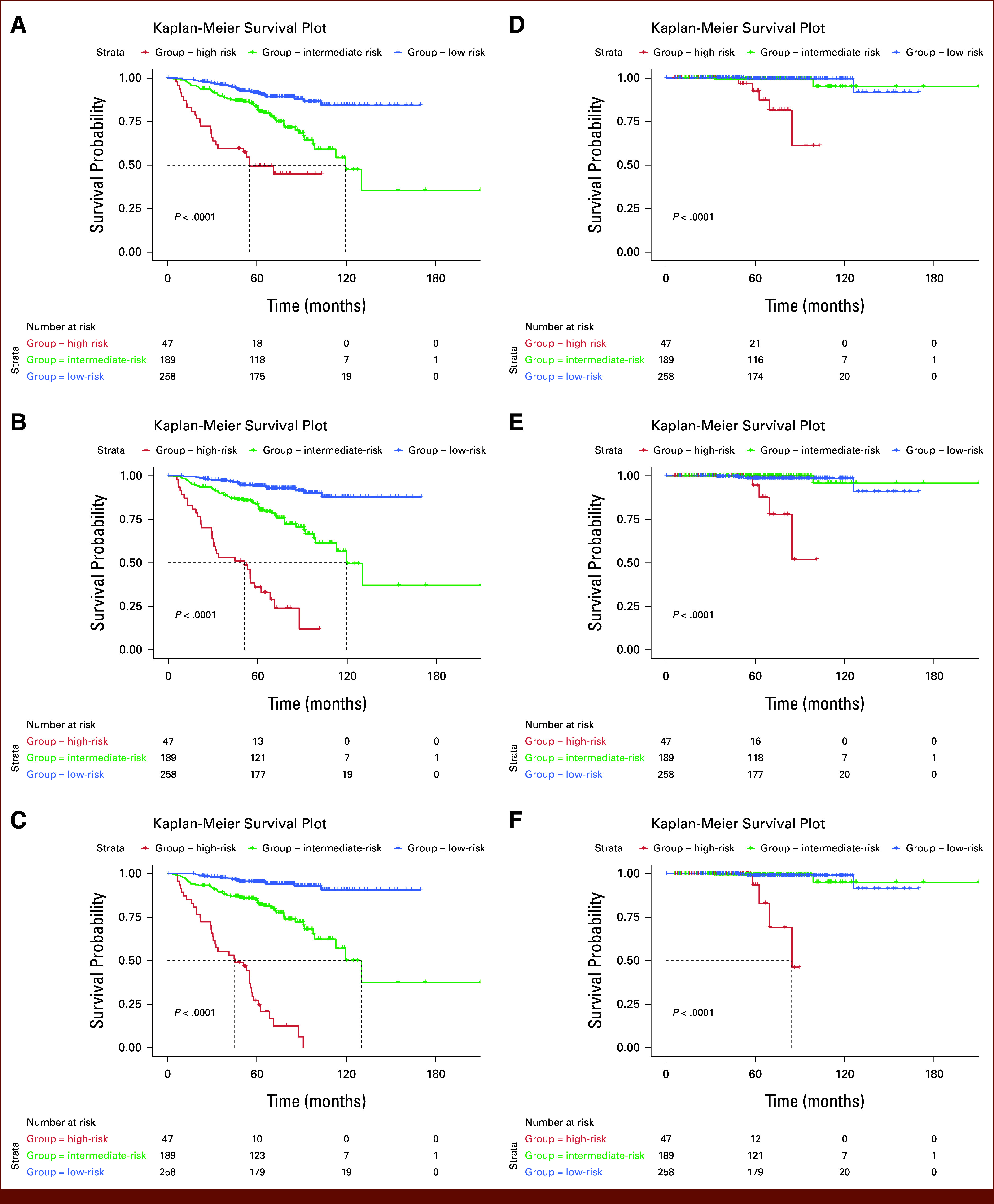
Kaplan-Meier plots associated with the time to BCR for (A) CAPRA-S, (B) CCHE (CAPRA-S + CNN H&E), and (C) the proposed CCHEK risk stratification model (CAPRA-S + CNN H&E, Ki-67 model) and with OS for (D) CAPRA-S, (E) CCHE, and (F) CCHEK. Patients are divided into three risk groups on the basis of CAPRA-S scores: low-risk (CAPRA-S ≤2), intermediate-risk (CAPRA-S 3-5), and high-risk (CAPRA-S ≥6). The cohort is also divided into similar-sized groups on the basis of the ML-based risk stratification models. BCR, biochemical recurrence; CAPRA-S, Cancer of the Prostate Risk Assessment Post-Surgical; CCHE, CAPRA-S + CNN H&E; CCHEK, CAPRA-S + CNN H&E & Ki-67; CNN, convolutional neural network; H&E, hematoxylin and eosin; ML, machine learning; OS, overall survival.
Although 84.3% of low-risk patients on the basis of CAPRA-S were BCR-free 10 years after surgery/radiation, the CCHE model (row 18) identified a low-risk group in which 88.0% were BCR-free after 10 years. Interestingly, when we added Ki-67 to the model (row 22), we identified a low-risk group in which 90.7% were free of BCR 10 years after surgery/radiation. Investigation of the Kaplan-Meier curves associated with OS also revealed similar findings. More specifically, while 95% of low-risk patients on the basis of CAPRA-S survived beyond 10 years, the CCHE (row 18) and CCHEK (row 22) models identified a low-risk group in which 100% were alive after 10 years. Furthermore, when we added Ki-67 to the model (row 22), all the patients in our identified low-risk group survived beyond 10 years after surgery/radiation.
The median 10-year BCR-free survival in the high-risk groups decreased from 69.6 (CAPRA-S) to 51.8 (CCHE) and 48.2 months (CCHEK), suggesting that both CNN-based models identify higher-risk patients compared with CAPRA-S. Further investigation revealed that four (1.6%) of the patients identified by CAPRA-S as low-risk were reclassified as high-risk and three (6.4%) of high-risk patients were reclassified as low-risk by the CCHE model (Fig 4A). Interestingly, all four new high-risk patients experienced BCR and the three new low-risk patients did not. Upon pathologic review of the TMA cores associated with these patients, we did not observe any revision in Gleason grades, suggesting that the ML models identify subtle features in the images that were not captured by Gleason grading. When we investigated the risk groups on the basis of the CCHEK model (row 22: CAPRA-S + CNN H&E, Ki-67), we identified 10 high-risk patients (representing 3.9%) by CAPRA-S who were reclassified as low-risk and 10 low-risk patients (representing 21.3%) who were reclassified as high-risk (Fig 4B). Although all the newly identified high-risk patients experienced BCR, none of the newly classified low-risk patients did. A pathologic review of H&E TMA cores associated with these 20 patients did not result in the revision of GSs. However, Ki-67 scores associated with the newly identified high-risk group were higher than the low-risk group (mean Ki-67 score 15 v 51 per 1 mm2), suggesting that Ki-67 likely contributes to better risk stratification.
FIG 4.
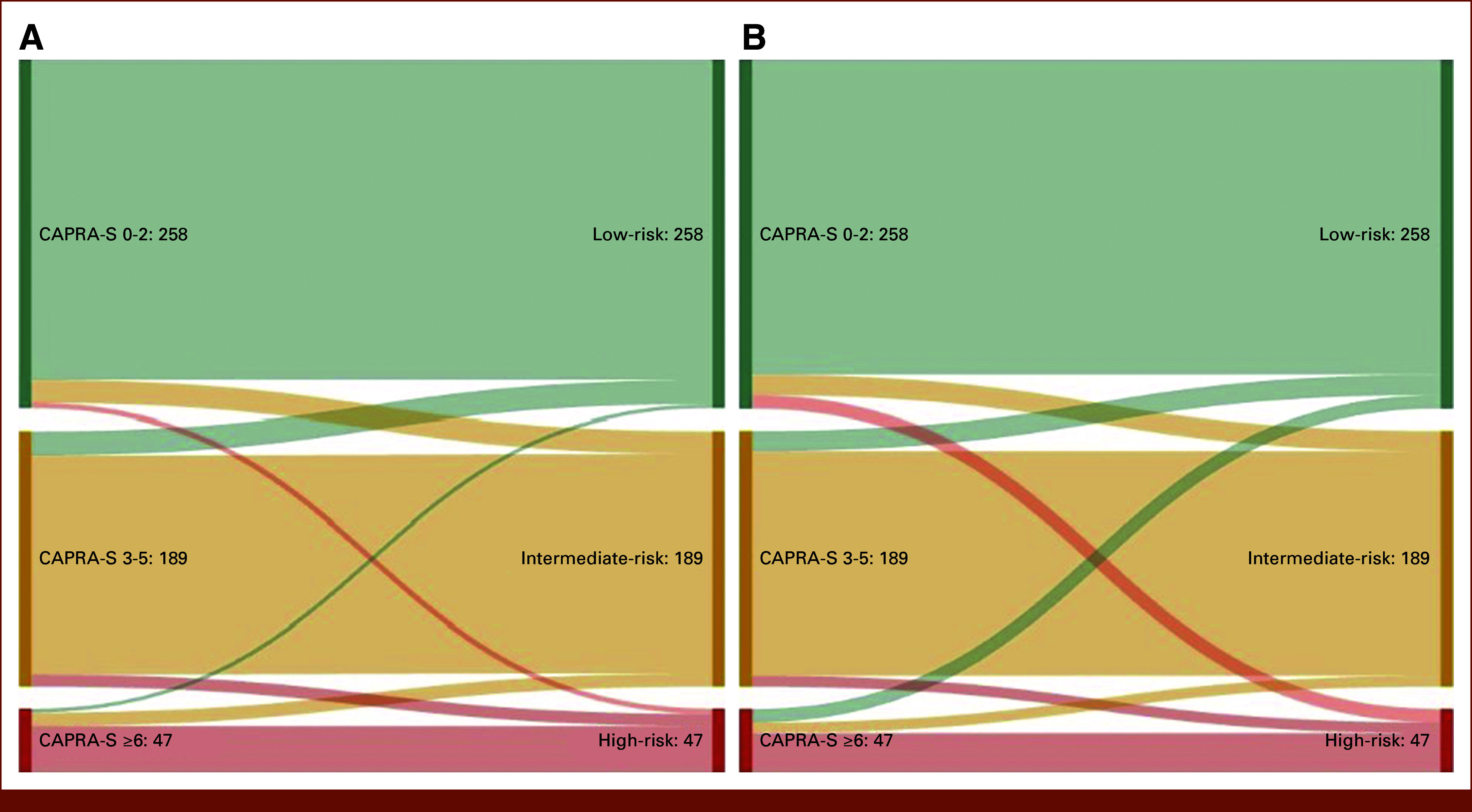
Sankey diagrams showing reassignment of patients from CAPRA-S risk categories to new ML-based risk groups. (A) CAPRA-S (Table 2 row 16) versus CAPRA-S + CNN H&E model (Table 2 row 18), and (B) CAPRA-S (row 16) versus CAPRA-S + CNN H&E Ki-67 model (Table 2 row 22). Each row represents one patient. CAPRA-S, Cancer of the Prostate Risk Assessment Post-Surgical; CNN, convolutional neural network; H&E, hematoxylin and eosin; ML, machine learning.
Interestingly, of the seven patients who were reclassified by the H&E-based ML model, six were a subset of the 20 patients identified by the H&E + Ki-67 ML model. These results suggest that H&E images potentially contain subtle features that are not captured by GS but contribute to risk stratification. This is evidenced by new ML model developments in which Ki-67–stained images were virtually constructed from H&E images,31 suggesting that H&E images carry information about the Ki-67 status.
DISCUSSION
We proposed an ML- and image-based pipeline for linking digital histopathology images directly to PCa outcome after RP. Our proposed pipeline achieved superior performance compared with the current GS-based risk stratification and CAPRA-S, which suggests that artificial intelligence (AI)–based risk stratification models could potentially introduce significant changes in PCa management. Our work validates the findings of other studies17 as it confirms the effectiveness of ML in PCa risk stratification. Furthermore, we show the utility of ML in predicting BCR and improving patient stratification relative to well-validated nomograms (ie, CAPRA-S). Finally, our work gleans on the links between Ki-67 status and patient outcome, as the Ki-67 scores in the patients who were reclassified as high-risk by our model were higher than those in the low-risk group.
Using the proposed CCHEK risk stratification model, 21.3% of patients identified to be high-risk by CAPRA-S were reclassified as low-risk and another 3.9% of patients identified to be low-risk by CAPRA-S were reclassified as high-risk. These patients may have been counseled and managed differently had this information been available after RP. For example, the digital pathology risk classification could steer one higher-risk patient with adverse pathologic features toward adjuvant radiation but another lower-risk patient with the same pathologic features toward surveillance.
Accurate risk stratification is critically important also in deciding on initial management of men with newly diagnosed, localized PCa. Men predicted to have indolent disease may undergo active surveillance (AS), while men with high-risk disease may require aggressive multimodal therapy. Current risk stratification tools on the basis of clinical and pathologic parameters have clinical utility but still do not allow optimal distinction of patients best suited for AS versus single or combined modality therapy. Our CCHEK model was developed on RP specimens and not prostate biopsies. If it can be validated on prostate biopsies, it could be evaluated as a biomarker to guide patients to either AS or definitive treatment.
Our CCHEK model was trained on TMA images rather than RP sections. As PCa is a multifocal cancer with different biology in each focus, TMA cores sampled from RP material do not overcome the limitations of spatial heterogeneity and therefore the proposed models may perform even better if the whole RP sections were used. Nonetheless, our models outperform established models even with this limitation. Furthermore, our study only considered H&E and Ki-67 histopathology data with CAPRA-S. Other predictive tools, such as Decipher32 or ArteraAI,18 could be used in future studies.
Although our results suggest that AI-based models have the promise to refine practice in PCa, further training and validation of our methods on larger multi-institutional external data sets are necessary. Nevertheless, our models show the potential of ML analysis of histopathology data to influence disease management on the basis of outcome prediction.
SUPPORT
Supported by the Canadian Institutes of Health Research grant MOP-14239 (S.S.), PJT-175017 (A.B., S.S.). Prostate Cancer Canada grant D2016-1352 (S.S.). The University of British Columbia C.A. Laszlo Chair grant GF001659 (S.S.).
Y.S. and R.B. contributed equally to this work as co-first authors. A.B. and S.S. contributed equally to this work as co-senior authors.
AUTHOR CONTRIBUTIONS
Conception and design: Yanan Shao, Davood Karimi, S. Larry Goldenberg, Martin E. Gleave, Ali Bashashati, Septimiu Salcudean
Financial support: Septimiu Salcudean
Administrative support: Martin E. Gleave
Provision of study materials or patients: Ladan Fazli, S. Larry Goldenberg, Martin E. Gleave
Collection and assembly of data: S. Larry Goldenberg, Ali Bashashati
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Roozbeh Bazargani
Employment: Prenuvo
S. Larry Goldenberg
Stock and Other Ownership Interests: Telix Pharmaceuticals
Martin E. Gleave
Stock and Other Ownership Interests: Sustained Therapeutics
Honoraria: Janssen, Astellas Pharma, Bayer, Pfizer, Tersera, Roche, AstraZeneca, Sanofi
Consulting or Advisory Role: Janssen, Astellas Pharma, Bayer, Sanofi, AstraZeneca, Genova Diagnostics, Pfizer, Tersera, Roche
Research Funding: Janssen, Astellas Pharma, Bayer
Patents, Royalties, Other Intellectual Property: ST-CP, ST-POP
Peter C. Black
Consulting or Advisory Role: Astellas Pharma, Janssen Oncology, Bayer, Merck, Ferring, Bristol Myers Squibb, EMD Serono, AstraZeneca, TerSera, Protara Therapeutics, Verity Pharmaceuticals, Nonagen Bioscience, Pfizer, Tolmar, Prokarium, NanOlogy, Combat Medical, Nanobot, Photocure, Sumitomo Pharma Oncology
Speakers’ Bureau: TerSera, Pfizer, BioSyent, Bayer
Research Funding: iProgen
Patents, Royalties, Other Intellectual Property: (1) PCT/CA2014/000787. Canada. 2014-11-03 Cancer Biomarkers and Classifiers and uses thereof. (2) #61899648 United States. 2013-03-13 Bladder cancer signature
Septimiu Salcudean
Stock and Other Ownership Interests: Sonic Incytes
Research Funding: Philips Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: I received royalty for real-time elastography software that was initially licensed to Ultrasonix. Analogic corporation, then b&k ultrasound have paid annual royalty until 2022
Uncompensated Relationships: Sonic Incytes
No other potential conflicts of interest were reported.
REFERENCES
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. : Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144:1941-1953, 2019 [DOI] [PubMed] [Google Scholar]
- 2.Gleason DF: Classification of prostatic carcinomas. Cancer Chemother Rep 50:125-128, 1966 [PubMed] [Google Scholar]
- 3.Epstein JI, Zelefsky MJ, Sjoberg DD, et al. : A contemporary prostate cancer grading system: A validated alternative to the Gleason score. Eur Urol 69:428-435, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphrey PA: Gleason grading and prognostic factors in carcinoma of the prostate. Mod Pathol 17:292-306, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Iczkowski KA, Torkko KC, Kotnis GR, et al. : Digital quantification of five high-grade prostate cancer patterns, including the cribriform pattern, and their association with adverse outcome. Am J Clin Pathol 136:98-107, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allsbrook WC Jr, Mangold KA, Johnson MH, et al. : Interobserver reproducibility of Gleason grading of prostatic carcinoma: Urologic pathologists. Hum Pathol 32:74-80, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Cooperberg MR, Pasta DJ, Elkin EP, et al. : The University of California, San Francisco Cancer of the Prostate Risk Assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol 173:1938-1942, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooperberg MR, Hilton JF, Carroll PR: The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer 117:5039-5046, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldenberg SL, Nir G, Salcudean SE: A new era: Artificial intelligence and machine learning in prostate cancer. Nat Rev Urol 16:391-403, 2019 [DOI] [PubMed] [Google Scholar]
- 10.Bulten W, Kartasalo K, Chen P-HC, et al. : Artificial intelligence for diagnosis and Gleason grading of prostate cancer: The PANDA challenge. Nat Med 28:154-163, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nir G, Hor S, Karimi D, et al. : Automatic grading of prostate cancer in digitized histopathology images: Learning from multiple experts. Med Image Anal 50:167-180, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Karimi D, Nir G, Fazli L, et al. : Deep learning-based Gleason grading of prostate cancer from histopathology images—Role of multiscale decision aggregation and data augmentation. IEEE J Biomed Health Inform 24:1413-1426, 2020 [DOI] [PubMed] [Google Scholar]
- 13.Arvaniti E, Fricker KS, Moret M, et al. : Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 8:12054, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazargani R, Fazli L, Gleave M, et al. : Multi-scale relational graph convolutional network for multiple instance learning in histopathology images. Med Image Anal 96:103197, 2024 [DOI] [PubMed]
- 15.Moghadam PA, Bashashati A, Goldenberg SL: Artificial intelligence and pathomics: Prostate cancer. Urol Clin North Am 51:15-26, 2024 [DOI] [PubMed] [Google Scholar]
- 16.Nagpal K, Foote D, Liu Y, et al. : Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2:48, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wulczyn E, Nagpal K, Symonds M, et al. : Predicting prostate cancer specific-mortality with artificial intelligence-based Gleason grading. Commun Med 1:10, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteva A, Feng J, van der Wal D, et al. : Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit Med 5:71, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azadi P, Suderman J, Nakhli R, et al. : ALL-IN: A local global graph-based distillation model for representation learning of gigapixel histopathology images with application in cancer risk assessment, in Greenspan H, Madabhushi A, Mousavi P, Salcudean S, Duncan J, Syeda-Mahmood T, Taylor R (eds), International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham, Switzerland, Springer Nature, 2023, pp 765-775 [Google Scholar]
- 20.Nath D, Li X, Mondragon C, et al. : Abi1 loss drives prostate tumorigenesis through activation of EMT and non-canonical WNT signaling. Cell Commun Signal 17:120, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leenders GJLH, van der Kwast TH, Grignon DJ, et al. : The 2019 International Society of Urological Pathology (ISUP) consensus conference on grading of prostatic carcinoma. Am J Surg Pathol 44:e87-e99, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooperberg MR, Hilton JF, Carroll PR: The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer 117:5039-5046, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nir G, Hor S, Karimi D, et al. : Automatic grading of prostate cancer in digitized histopathology images: Learning from multiple experts. Med Image Anal 50:167-180, 2018 [DOI] [PubMed] [Google Scholar]
- 24.Cox D: Partial likelihood. Biometrika 62:269-276, 1975 [Google Scholar]
- 25.Welch BL: The eneralization of student’s problems when several different population variances are involved. Biometrika 34:28-35, 1947 [DOI] [PubMed] [Google Scholar]
- 26.Ishihara M, Mukai H, Nagai S, et al. : Retrospective analysis of risk factors for central nervous system metastases in operable breast cancer: Effects of biologic subtype and Ki67 overexpression on survival. Oncology 84:135-140, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Sorbye SW, Kilvaer TK, Valkov A, et al. : Prognostic impact of Jab1, p16, p21, p62, Ki67 and Skp2 in soft tissue sarcomas. PloS One 7:e47068, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciancio N, Galasso MG, Campisi R, et al. : Prognostic value of p53 and Ki67 expression in fiberoptic bronchial biopsies of patients with non small cell lung cancer. Multidiscip Respir Med 7:29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josefsson A, Wikstrom P, Egevad L, et al. : Low endoglin vascular density and Ki67 index in Gleason score 6 tumours may identify prostate cancer patients suitable for surveillance. Scand J Urol Nephrol 46:247-257, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457-481, 1958 [Google Scholar]
- 31.Shao Y, Nir G, Fazli L, et al. : Improving prostate cancer classification in H&E tissue micro arrays using Ki67 and P63 histopathology. Comput Biol Med 127:104053, 2020 [DOI] [PubMed] [Google Scholar]
- 32.Jairath NK, Dal Pra A, Vince R Jr, et al. : A systematic review of the evidence for the Decipher genomic classifier in prostate cancer. Eur Urol 79:374-383, 2021 [DOI] [PubMed] [Google Scholar]