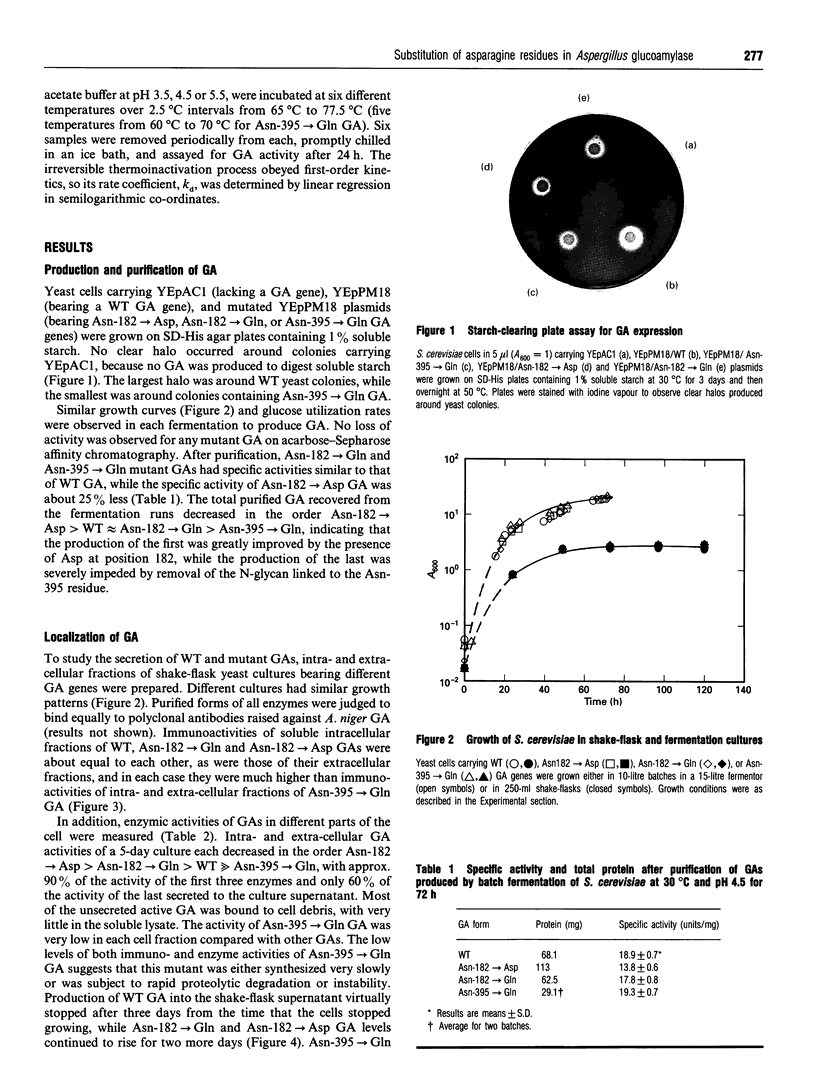
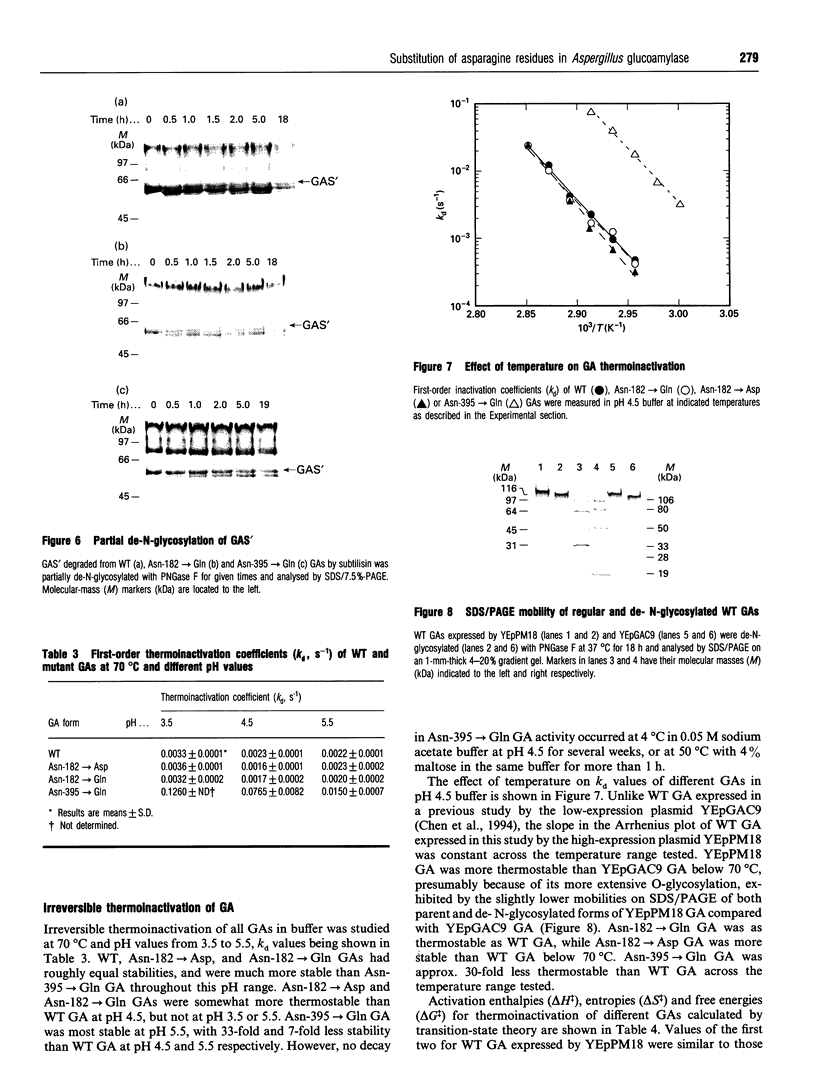
Abstract
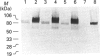
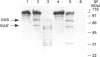
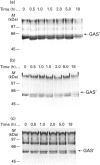
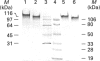
Aspergillus awamori glucoamylase is a secreted glycoprotein containing N-linked carbohydrate recognition sites at Asn-171, Asn-182 and Asn-395. Site-directed mutagenesis was performed at Asn-182 and Asn-395 to determine whether these residues were N-glycosylated by Saccharomyces cerevisiae, to investigate the function of any glycans linked to them, and to determine the effect of their deamidation on glucoamylase thermostability. Asn-171 and Asn-395, but not Asn-182, were N-glycosylated. Deletion of the glycan N-linked to Asn-395 did not affect specific activity, but greatly decreased enzyme secretion and thermostability. The mutant lacking the N-glycan linked to Asn-395 was synthesized very slowly, and was more associated with cell membrane components and susceptible to proteinase degradation than were wild-type or other mutant glucoamylases. Its secreted form was 30-fold less thermostable than wild-type enzyme at pH 4.5. Replacement of Asn-182 by Gln to eliminate deamidation at this site did not change glucoamylase specific activity or thermostability, while replacement by Asp decreased specific activity about 25%, but increased thermostability moderately at pH 4.5 below 70 degrees C. Both mutations of Asn-182 increased glucoamylase production.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aleshin A., Golubev A., Firsov L. M., Honzatko R. B. Crystal structure of glucoamylase from Aspergillus awamori var. X100 to 2.2-A resolution. J Biol Chem. 1992 Sep 25;267(27):19291–19298. doi: 10.2210/pdb1gly/pdb. [DOI] [PubMed] [Google Scholar]
- Alexander S., Elder J. H. Endoglycosidases from Flavobacterium meningosepticum application to biological problems. Methods Enzymol. 1989;179:505–518. doi: 10.1016/0076-6879(89)79151-5. [DOI] [PubMed] [Google Scholar]
- Bakir U., Coutinho P. M., Sullivan P. A., Ford C., Reilly P. J. Cassette mutagenesis of Aspergillus awamori glucoamylase near its general acid residue to probe its catalytic and pH properties. Protein Eng. 1993 Nov;6(8):939–946. doi: 10.1093/protein/6.8.939. [DOI] [PubMed] [Google Scholar]
- Berg D. T., Grinnell B. W. Pro to Gly (P219G) in a silent glycosylation site results in complete glycosylation in tissue plasminogen activator. Protein Sci. 1993 Jan;2(1):126–127. doi: 10.1002/pro.5560020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchinfuso W. P., Ma K. L., Lee W. M., Warmels-Rodenhiser S., Hammond G. L. Selective removal of glycosylation sites from sex hormone-binding globulin by site-directed mutagenesis. Endocrinology. 1992 Nov;131(5):2331–2336. doi: 10.1210/endo.131.5.1425432. [DOI] [PubMed] [Google Scholar]
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Bulleid N. J., Bassel-Duby R. S., Freedman R. B., Sambrook J. F., Gething M. J. Cell-free synthesis of enzymically active tissue-type plasminogen activator. Protein folding determines the extent of N-linked glycosylation. Biochem J. 1992 Aug 15;286(Pt 1):275–280. doi: 10.1042/bj2860275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme E., Lorenzini T., Giffin J., Martin F., Jacobsen F., Boone T., Elliott S. Role of glycosylation on the secretion and biological activity of erythropoietin. Biochemistry. 1992 Oct 20;31(41):9871–9876. doi: 10.1021/bi00156a003. [DOI] [PubMed] [Google Scholar]
- Dubé S., Fisher J. W., Powell J. S. Glycosylation at specific sites of erythropoietin is essential for biosynthesis, secretion, and biological function. J Biol Chem. 1988 Nov 25;263(33):17516–17521. [PubMed] [Google Scholar]
- Evans R., Ford C., Sierks M., Nikolov Z., Svensson B. Activity and thermal stability of genetically truncated forms of Aspergillus glucoamylase. Gene. 1990 Jul 2;91(1):131–134. doi: 10.1016/0378-1119(90)90174-p. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990 Apr;3(5):433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabe C. G., Hanover J. A., Lennarz W. J. Glycosylation of ovalbumin nascent chains. The spatial relationship between translation and glycosylation. J Biol Chem. 1980 Oct 10;255(19):9236–9242. [PubMed] [Google Scholar]
- Gunnarsson A., Svensson B., Nilsson B., Svensson S. Structural studies on the O-glycosidically linked carbohydrate chains of glucoamylase G1 from Aspergillus niger. Eur J Biochem. 1984 Dec 17;145(3):463–467. doi: 10.1111/j.1432-1033.1984.tb08578.x. [DOI] [PubMed] [Google Scholar]
- Harris E. M., Aleshin A. E., Firsov L. M., Honzatko R. B. Refined structure for the complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution. Biochemistry. 1993 Feb 16;32(6):1618–1626. doi: 10.1021/bi00057a028. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Holland M. J., McCabe P. C., Cole G. E., Wittman V. P., Tal R., Watt K. W., Gelfand D. H., Holland J. P., Meade J. H. Expression, Glycosylation, and Secretion of an Aspergillus Glucoamylase by Saccharomyces cerevisiae. Science. 1985 Apr 5;228(4695):21–26. doi: 10.1126/science.228.4695.21. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S. M. Preparation of extracts from yeast. Methods Enzymol. 1990;182:154–174. doi: 10.1016/0076-6879(90)82015-t. [DOI] [PubMed] [Google Scholar]
- Joao H. C., Scragg I. G., Dwek R. A. Effects of glycosylation on protein conformation and amide proton exchange rates in RNase B. FEBS Lett. 1992 Aug 3;307(3):343–346. doi: 10.1016/0014-5793(92)80709-p. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Li Y., Luo L., Rasool N., Kang C. Y. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993 Jan;67(1):584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Meade J. H., Cole G., Lawyer F. C., McCabe P., Schweickart V., Tal R., Wittman V. P., Flatgaard J. E., Innis M. A. Molecular cloning and characterization of the glucoamylase gene of Aspergillus awamori. Mol Cell Biol. 1984 Nov;4(11):2306–2315. doi: 10.1128/mcb.4.11.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Borchardt R. T. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm Res. 1990 Jul;7(7):703–711. doi: 10.1023/a:1015807303766. [DOI] [PubMed] [Google Scholar]
- Pazur J. H., Knull H. R., Simpson D. L. Glycoenzymes: a note on the role for the carbohydrate moieties. Biochem Biophys Res Commun. 1970 Jul 13;40(1):110–116. doi: 10.1016/0006-291x(70)91053-3. [DOI] [PubMed] [Google Scholar]
- Påhlsson P., Shakin-Eshleman S. H., Spitalnik S. L. N-linked glycosylation of beta-amyloid precursor protein. Biochem Biophys Res Commun. 1992 Dec 30;189(3):1667–1673. doi: 10.1016/0006-291x(92)90269-q. [DOI] [PubMed] [Google Scholar]
- Robinson A. B., Rudd C. J. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr Top Cell Regul. 1974;8(0):247–295. doi: 10.1016/b978-0-12-152808-9.50013-4. [DOI] [PubMed] [Google Scholar]
- Sheares B. T. Site-specific glycosylation in animal cells. Substitution of glutamine for asparagine 293 in chicken ovalbumin does not allow glycosylation of asparagine 312. J Biol Chem. 1988 Sep 5;263(25):12778–12782. [PubMed] [Google Scholar]
- Sierks M. R., Ford C., Reilly P. J., Svensson B. Functional roles and subsite locations of Leu177, Trp178 and Asn182 of Aspergillus awamori glucoamylase determined by site-directed mutagenesis. Protein Eng. 1993 Jan;6(1):75–79. doi: 10.1093/protein/6.1.75. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Stoffer B., Frandsen T. P., Busk P. K., Schneider P., Svendsen I., Svensson B. Production, purification and characterization of the catalytic domain of glucoamylase from Aspergillus niger. Biochem J. 1993 May 15;292(Pt 1):197–202. doi: 10.1042/bj2920197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Cross R., Schirch V. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J Biol Chem. 1991 Nov 25;266(33):22549–22556. [PubMed] [Google Scholar]
- Wright H. T. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26(1):1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]