Abstract
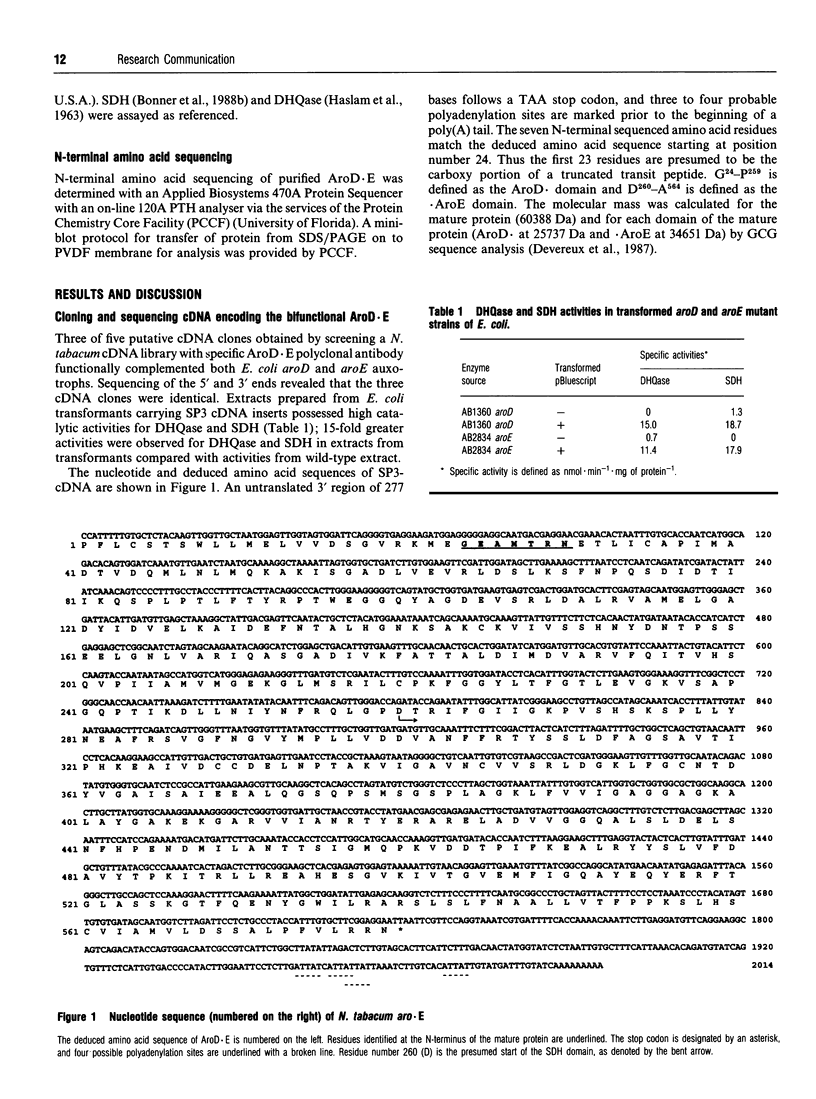
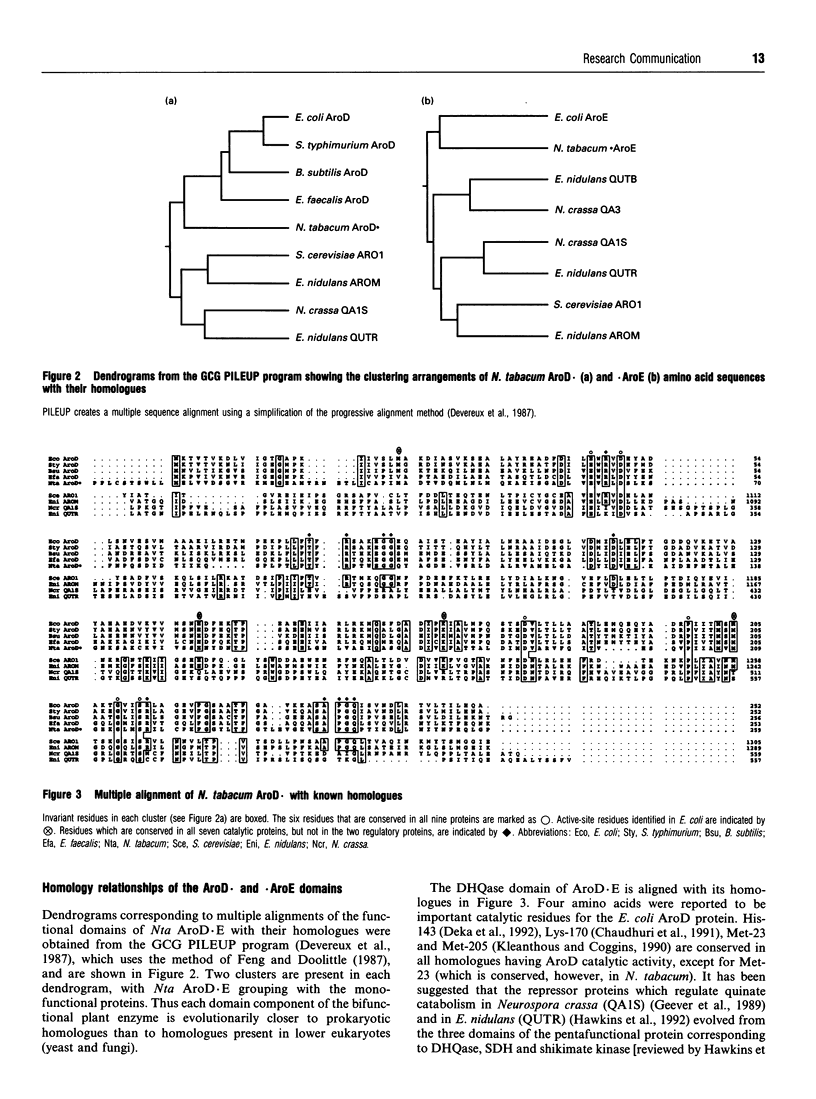
Nicotiana tabacum cDNA encoding a bifunctional protein having catalytic domains for dehydroquinase and shikimate dehydrogenase was cloned and sequenced. Complementation of Escherichia coli aroD and aroE auxotrophs was successful. Amino acid sequencing located the N-terminus of the mature protein. The two catalytic domains exhibited greater amino acid identity with prokaryote homologues than with yeast and fungal homologues.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anton I. A., Coggins J. R. Sequencing and overexpression of the Escherichia coli aroE gene encoding shikimate dehydrogenase. Biochem J. 1988 Jan 15;249(2):319–326. doi: 10.1042/bj2490319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B. A., Dunny G. M. Cloning and molecular analysis of genes affecting expression of binding substance, the recipient-encoded receptor(s) mediating mating aggregate formation in Enterococcus faecalis. J Bacteriol. 1993 Nov;175(22):7421–7429. doi: 10.1128/jb.175.22.7421-7429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Duncan K., Graham L. D., Coggins J. R. Identification of the active-site lysine residues of two biosynthetic 3-dehydroquinases. Biochem J. 1991 Apr 1;275(Pt 1):1–6. doi: 10.1042/bj2750001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka R. K., Kleanthous C., Coggins J. R. Identification of the essential histidine residue at the active site of Escherichia coli dehydroquinase. J Biol Chem. 1992 Nov 5;267(31):22237–22242. [PubMed] [Google Scholar]
- Duncan K., Chaudhuri S., Campbell M. S., Coggins J. R. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem J. 1986 Sep 1;238(2):475–483. doi: 10.1042/bj2380475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Edwards R. M., Coggins J. R. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem J. 1987 Sep 1;246(2):375–386. doi: 10.1042/bj2460375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Roberts C. F. Structure of the Aspergillus nidulans qut repressor-encoding gene: implications for the regulation of transcription initiation. Gene. 1992 Jan 2;110(1):109–114. doi: 10.1016/0378-1119(92)90452-u. [DOI] [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Smith M., Keyte J. W., Roberts C. F. Molecular organisation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. Mol Gen Genet. 1988 Oct;214(2):224–231. doi: 10.1007/BF00337715. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Coggins J. R. Reversible alkylation of an active site methionine residue in dehydroquinase. J Biol Chem. 1990 Jul 5;265(19):10935–10939. [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966 Apr;91(4):1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servos S., Chatfield S., Hone D., Levine M., Dimitriadis G., Pickard D., Dougan G., Fairweather N., Charles I. Molecular cloning and characterization of the aroD gene encoding 3-dehydroquinase from Salmonella typhi. J Gen Microbiol. 1991 Jan;137(1):147–152. doi: 10.1099/00221287-137-1-147. [DOI] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin A., Zumstein E., Azevedo V., Ehrlich S. D., Serror P. The organization of the Bacillus subtilis 168 chromosome region between the spoVA and serA genetic loci, based on sequence data. Mol Microbiol. 1993 Oct;10(2):385–395. doi: 10.1111/j.1365-2958.1993.tb02670.x. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]


