Abstract
Purpose
Globally, colorectal cancer (CRC) is among the most prevalent cancers. One distinctive feature of colorectal cancer is its close relationship to the gut microbiota, which is a crucial component of the tumor microenvironment. Over the last ten years, research has demonstrated that colorectal cancer is accompanied with dysbiosis of gut bacteria, fungi, viruses, and Archaea, and that these alterations may be causal. Objectives: This study aimed to evaluate the disruption of the microorganism composition in the intestine, especially bacteria and to determine their relationship with colorectal cancer.
Methods
An evaluation system for determining colorectal cancer (CRC) risk and prognosis can be established more easily with the help of accurate gut microbiota profiling. Stool samples from 14 CRC patients and 13 controls were collected and the flora relative abundance was measured using targeted quantitative PCR (qPCR) assays to evaluate diagnostic potential of selected biomarkers: Streptococcus gallolyticus and Enterococcus faecalis. Culture and MALDI-TOF mass spectrometry were coupled to identify the gut microbiota in both colorectal cancer and control groups.
Results
Compared with controls, the gut microbiota of CRC patients showed an increase in the abundance of Enterococcus, Fusobacterium and Streptococcus. At the species level, the CRC enriched bacterium including Escherichia coli, Enterococcus faecalis, Fusobacterium nucleatum, Streptococcus gallolyticus, Flavoni fractorplautii and Eggerthella lenta acted as promising biomarkers for early detection of CRC.
Conclusion
This study highlights the potential of gut microbiota biomarkers as a promising non-invasive tool for the accurate detection and distinction of individuals with CRC.
Keywords: Colorectal cancer, Gut microbiota, Risk factors, Culturomics, MALDI TOF mass spectrometry
Background
Colorectal cancer (CRC) is one of the cancers that has a major global impact on human life. It is the third most common cancer in the world after breast and lung cancer, responsible for more than 0.57 million deaths worldwide [1]. The crucial involvement of the Gut microbiota in initiating and advancing various cancer types, especially gastrointestinal tumors, is widely recognized. Specifically, bacteria have the capability to encourage persistent inflammation in the stomach lining, leading to permanent alterations in the cells lining the intestines. Consequently, individuals become more susceptible to developing cancer [1].
The human gut is colonized by dynamic and highly competitive microorganisms. Bacteria represent the most studied part of the human microbiota, with more than 1014 bacterial cells [2]. In addition to bacteria, archaea, viruses, and fungi, their genomes and environmental conditions constitute the entire habitat in the gut microbiome. The dysfunction of this flora can be induced by chronic inflammation and stimulated by the production of carcinogenic metabolites leading to CRC [2]. Over the past few years, the use of metagenomics has significantly enhanced our understanding of the connections between the human microbiome, health, and diseases. However, it has also produced a vast number of sequences that cannot be attributed to any known microorganism. However, the potential of microbial culture techniques has been overlooked, resulting in limited knowledge of the microbial population in the human gut [3].
Increasingly studying the gut microbiota has the potential to provide a novel approach in the battle against colon cancer. Numerous studies have indicated that specific bacteria residing in the colon may facilitate the progression of cancer [2, 4–6]. For several decades, researchers have identified a correlation between the abundance of certain bacterial species and CRC [7–10]. One of the most significant associations is with Streptococcus gallolyticus, previously known as Streptococcus bovis, which has been linked to cases of bacterial endocarditis and CRC as described in some studies [11, 12].
The development of colorectal adenoma or cancer may result in the distortion of the colonic mucosa, thereby providing an opportunity for bacteria like S. gallolyticus to access the collagen fibers in the basement membrane that were previously inaccessible [11]. In addition, Enterococcus faecalis infective endocarditis has also been associated with CRC [13]. The aim of this study was to describe the bacterial composition of the gut microbiota in CRC patients and detect the disproportion that exists compared to controls. Comprehensive consideration of demographic and clinical factors is essential for achieving reliable and applicable results in CRC research.
The relationship between the gut microbiota and the development of various diseases, including colorectal cancer (CRC), has garnered increasing attention in global biomedical research. Previous studies have extensively explored this relationship across various populations, primarily in North America, Europe, and Asia, revealing significant variations in microbiota composition based on geographical and demographic factors [14, 15]. However, our study is the first to examine this relationship within a Tunisian population, providing a novel and essential contribution to the existing literature. Tunisia, as a North African country with distinct dietary habits and environmental factors, offers a unique context for studying specific characteristics of the gut microbiota in CRC patients. The data obtained from this study can provide valuable insights not only for the region but also for the global understanding of geographical variations in gut microbiota composition. By comparing our findings with those from international studies, we hope to identify crucial similarities and differences that can illuminate the underlying mechanisms of CRC and potentially guide prevention and treatment strategies tailored to each region.
Materials and methods
Study design
This was a case–control study, and participants were prospectively enrolled between March 23 and August 26, 2021.Fecal samples were collected from colorectal cancer patients and a control group in the gastroenterology department at Charles Nicolle Hospital of Tunis, then transported to the Microbiology laboratory where they were immediately aliquoted and frozen at −80 °C before being transported to the laboratory of the Institute of Mediterranean Infection (IHU), Microbes, Evolution, Phylogeny, and Infection Unit for analysis.
Patients
The cases were CRC patients with a confirmed histological diagnosis of malignant tumors who were newly diagnosed without any previous treatment. Patients who had taken antibiotics during the 4 weeks leading up to the endoscopy were not included in the study. Controls were also recruited from the gastroenterology department. They were patients who consulted for gastro-intestinal symptoms but were not diagnosed with CRC without acute or chronic inflammation.
Inclusion criteria
Patients included in our study were those with colorectal cancer (CRC) who had a histologically confirmed diagnosis of newly diagnosed malignant tumors, had received no prior treatment, and had not taken antibiotics within the four weeks preceding the endoscopy.
Non-inclusion criteria
Individuals with chronic inflammatory bowel diseases (IBD), those who had taken antibiotics in the last six months, and those who had undergone a colonoscopy at least one month prior were not included.
Exclusion criteria
Patients with other diagnosed gastrointestinal disorders, such as irritable bowel syndrome or chronic gastrointestinal infections during the study were excluded, those regularly using laxatives, as they can influence the composition of the gut microbiota and those taking other medications known to impact the gut microbiota, such as immunosuppressants and treatments for autoimmune disorders.
Ethical considerations + + +
All participants were informed about the purpose of the study and have given their written consent. All collected information and data analysis were confidential and anonymous during and after data collection. The study was approved by the local Ethics Committee of Charles Nicolle Hospital in Tunis, Tunisie (OHRP number: IRB00013338).
Samples
During the study period, 27 different stool samples were collected in sterile collection containers from the two groups: CRC patients and controls. The main characteristics of the participants, including weight, height, and body mass index (BMI), are summarized in Table 1.
Table 1.
Main characteristics of the participants from the two groups: CRC group and controls
| Groupes | Sample number | Weight (kg) | Height (m) | BMI |
|---|---|---|---|---|
| Groupe de cancer colorectal | 1 | 59 | 1,71 | 20,18 |
| 2 | 49 | 1,65 | 18,00 | |
| 3 | 70 | 1,69 | 24,51 | |
| 4 | 50 | 1,78 | 15,78 | |
| 5 | 80 | 1,8 | 24,69 | |
| 6 | 82 | 1,76 | 26,47 | |
| 7 | 87 | 1,81 | 26,56 | |
| 8 | 67 | 1,68 | 23,74 | |
| 9 | 70 | 1,72 | 23,66 | |
| 10 | 58 | 1,64 | 21,56 | |
| 11 | 60 | 1,69 | 21,01 | |
| 12 | 62 | 1,76 | 20,02 | |
| 13 | 67 | 1,78 | 21,15 | |
| 14 | 65 | 1,65 | 23,88 | |
| Groupe de contrôles | 15 | 62 | 1,68 | 21,97 |
| 16 | 76 | 1,75 | 24,82 | |
| 17 | 65 | 1,65 | 23,88 | |
| 18 | 83 | 1,7 | 28,72 | |
| 19 | 70 | 1,61 | 27,01 | |
| 20 | 75 | 1,7 | 25,95 | |
| 21 | 64 | 1,67 | 22,95 | |
| 22 | 70 | 1,8 | 21,60 | |
| 23 | 55 | 1,6 | 21,48 | |
| 24 | 65 | 1,68 | 23,03 | |
| 25 | 68 | 1,75 | 22,20 | |
| 26 | 67 | 1,6 | 26,17 | |
| 27 | 69 | 1,74 | 22,79 |
Fecal DNA isolation
Caution is imperative when interpreting current studies, as the efficiency of DNA extraction varies widely among stool samples. Here is why different fecal DNA extraction protocols were tested: The QiAMP power DNA extraction kit directly on fecal samples without mechanical lysis of stool samples, the second protocol of DNA extraction designed as previously described [16], and the third protocol used for the extraction of Archeae DNA is the protocol that was chosen [17]. DNA extraction was performed by mixing 0.2 g of each feces sample with 500 µL of G2 buffer (QIAGEN, Hilden, Germany) in an Eppendorf tube (Fisher Scientific, Illkirch,France). Then, 0.3 g of acid-washed beads ≤ 106 µm (Sigma-Aldrich, Saint-Quentin Fallavier, France) was added in each tube and shaken in a FastPrep BIO 101 device (MP Biomedicals, Illkirch, France) for 45 s for mechanical lysis before 10 min incubation at 100 °C. A 180 µL volume of the mixture was then incubated with 20 µL of proteinase K (QIAGEN) at 56 °C overnight before a second mechanical lysis was performed. Total DNA was finally extracted with the EZ1 advanced XL extraction kit (QIAGEN) and a 200 µL eluted volume. Sterile phosphate-buffered saline (PBS) was used as a negative control in each DNA extraction run [17].
Real-time quantitative PCR assays
The relative abundance of S.gallolyticus and E.faecalis in fecal samples was measured by real-time quantitative PCR. The target genes, probes, primer sequences, and PCR product sizes for the two real-time PCR assays used in this study are summarized in Table 2. The PCR amplification program was 95 °C for 15 min, followed by 45 cycles of 95 °C for 30 s and 60 °C for 1 min. All DNA samples were tested in duplicate. Results are expressed as the number of 16S rRNA and RecN copies per gram of feces.
Table 2.
Real-time PCR primers and probes sequences for detecting Streptococcus gallolyticus and Enterococcus faecalis 16 S rRNA and RecN genes
| Organism | Assay | Primer/probe name | Primer/probe sequence (5ʹ—3ʹ) | Product size (bp) |
|---|---|---|---|---|
| Streptococcus gallolyticus (ex bovis) | 16S rRNA | Sgallo_16S_F | TTTAACMCATGTTAGATGCTTGAAAGR | 165 pb |
| Sgallo_16S_R | GTAGGAGTCTGGGCCGTGTC | |||
| Sgallo_16S_P | 6FAM- GGGTGATCGGCCACACTGGGA | |||
| Enterococcus faecalis | RecN | EFS_recN_F2 | CACAGGCGTATCAAGAGTATCG | 90 pb |
| EFS_recN_R2 | GCATGTCCATTCTTTGGGCAA | |||
| EFS_recN_P2 | 6FAM- CACTCGAAGCCAAAGTCAGAAAGCGACA |
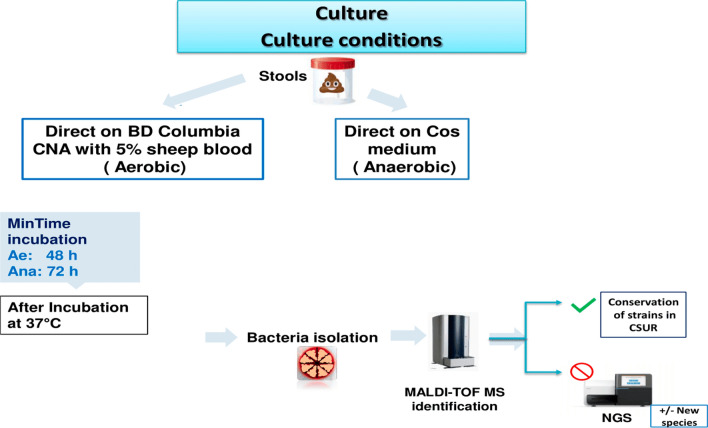
Culture and identification using MALDI-TOF mass spectrometry method
Bacterial culture at different conditions coupled with high-throughput identification by MALDI TOF mass spectrometry was employed to refine the isolation methodology for oxygen-intolerant bacteria and medically relevant species within colorectal cancer (CRC).Culture was conducted by direct plating of cascade dilutions of each stool sample on Columbia CNA medium (Becton Dickinson, Le Pont de Claix, France) supplemented with 5% Gram-negative selective sheep blood under aerobic conditions. Simultaneously Simultaneously, culture was performed on COS medium Columbia Blood Agar (Biomerieux, Marcy l’Etoile, France) for Streptococci under anaerobic conditions. The media plates were incubated aerobically at 37 °C and monitored at intervals of 24, 48, and 72 h. Furthermore, the Columbia with 5% sheep blood are then incubated at 37 °C under anaerobic condition, acilitated by a Zip bag (Oxoid, Dardilly, France) enclosing an anaerobic GasPak (Becton Dickinson, Le Pont de Claix, France) for 48 h [3].
Bacterial isolation and identification were accomplished using MALDI-TOF mass spectrometry. Unidentified bacteria underwent further scrutiny via high-throughput Next-Generation Sequencing (NGS) utilizing MiSeq technology (Illumina, San Diego, CA, USA). The methodology employed for characterizing the cultivable aerobic and anaerobic bacterial communities is illustrated in Fig. 1 [3]. MALDI-TOF MS is an identification device that has allowed the expansion of the knowledge of biodiversity, it is used here for the identification of the different bacterial species that were cultured. Three systems, the Andromas database (Andromas SAS, Paris, France), the Vitek-MS platform (bioMérieux, Marcy l’Etoile, France), and the Bruker Biotyper (Bruker Daltonics, Heidelberg, Germany, in collaboration with Becton Dickinson, Franklin Lakes, NJ, USA), are available for MALDI TOF identification of microorganisms [18]. The principle of MALDI-TOF corresponds to a soft ionization process, which is obtained by adding a matrix to bacterial colonies on metal plates. A UV laser beam was used to perform the ionization. Spectra are generated by measuring the time of flight in the tube to reach a detector. The identification was done automatically by comparing the spectra with the data of the defined database available [18].
Fig. 1.
Methodology of bacterial identification [3]
Statistical analyses
Statistical analyses were performed using SPSS software (version 23.0, IBM Corp.). For descriptive statistics, frequency and percentage values were calculated for categorical variables, and median (Interquartile range (IQR)) values were used to describe continuous data. The Pearsonchi-squared test or Fisher’s exact test were used to compare categorical variables. The U Mann–Whitney test was used to compare the medians of continuous variables between cases and controls because quantitative variables had an abnormal distribution. An odds ratio (OR) was calculated to assess the association between body mass index (BMI) categories (BMI > 25 vs. BMI ≤ 25) in colorectal cancer patients compared to controls, yielding an OR of 0.375 with a p value of 0.1. A p value ≤ 0.05 was considered significant.
Results
Sampling and demographic analysis
We studied a total of 14 patients diagnosed with colorectal cancer (CRC). The stages of the disease were classified according to the TNM staging criteria to provide a detailed understanding of the distribution of stages within our cohort. The detailed distribution of stages is as follows:
Stage IIA 2 patients (14.3%).
Stage IIB 1 patient (7.1%).
Stage IIC 3 patients (21.4%).
Stage IIIA 4 patients (28.6%).
Stage IIIB 2 patients (14.3%).
Stage IIIC 2 patients (14.3%).
This distribution shows a majority of patients in the advanced stages (III) with a significant proportion also in the intermediate stages (II). This information provides crucial context for our subsequent analyses, as it suggests a potential trend in the stages of CRC where gut dysbiosis might be more pronounced.
Among the risk factors studied, male gender, tobacco and alcohol consumption, diabetes, and family history of CRC are significantly associated with increased odds for the CRC group (OR > 1). There was no statistically significant difference between the studied groups regarding age and gender (P > 0.05), but diabetes showed a significant association with CRC (OR = 0.03, p = 0.03). However, no statistically significant associations were observed for tobacco consumption, alcohol consumption, or family history of CRC (p > 0.05). The analysis included two groups: patients with colorectal cancer and controls. Among the patients with colorectal cancer, 2 had a BMI > 25, while 12 had a BMI ≤ 25. In the control group, 4 individuals had a BMI > 25 and 9 had a BMI ≤ 25. The odds ratio (OR) calculated for the association between a BMI > 25 and colorectal cancer compared to controls was 0.375, indicating an inverse association. The associated p value for this analysis was 0.1, suggesting a trend that, although not statistically significant at the 0.05 level, warrants further exploration in future studies. The detailed demographic characteristics of the whole study are presented in Table 3.
Table 3.
Demographic data recorded for each study participant for each group colorectal cancer and controls
| Characteristics | Colorectal cancer group (N = 14) | Control group (N = 13) | OR | P |
|---|---|---|---|---|
| Median Age (years-IQR) | 58.0 (51—67.2) | 59.0 (50.5 -66.0) | – | 0.9 |
| Gender (Male/female) | 0.69 | |||
| Male | 10 | 8 | 1.56 | |
| Female | 4 | 5 | ||
| Median weight (Kg-IQR) | 66.0 (58.7–66.0) | 68.0 (64.5–72.5) | – | 0.4 |
| Tobacco consumption | 0.16 | |||
| Yes | 8 | 4 | 3.0 | |
| No | 6 | 9 | ||
| Alcohol consumption | 1.0 | |||
| Yes | 3 | 2 | 1.5 | |
| No | 11 | 11 | ||
| Diabetes | 0.03 | |||
| Yes | 7 | 1 | 12 | |
| No | 7 | 12 | ||
| Family history of colorectal cancer | 0.18 | |||
| Yes | 5 | 2 | 3.05 | |
| No | 9 | 11 | ||
| Median BMI (kg/m2–IQR) | 22.61 (24.35–20.38) | 23.03 (25.95–22.20) | 0.375 | 0.1 |
Real-time quantitative PCR assays
Two bacterial CRC candidate biomarkers were selected for qPCR quantification, including Streptococcus bovis/gallolyticus involved in the infection in patients with CRC as well as Enterococcus faecalis causing infective endocarditis.The results are summarized in Fig. 2, which shows that S.gallolyticus is more prevalent in the group with CRC than in the control group. Similarly, E.faecalisis more prevalent in the group with CRC than in the control group.
Fig. 2.
Distribution of candidate bacteria based on the number of isolates in the two groups: colorectal cancer group and controls
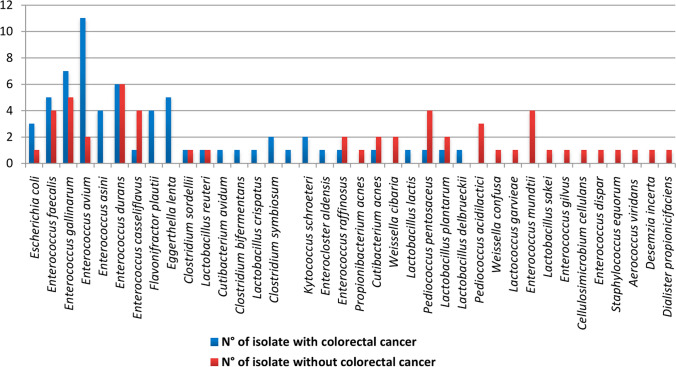
Culturomics results
A total of 117 bacterial strains were isolated from 27 stool samples, with 63 isolates (53.85%) identified among patients with CRC and 54 isolates (46.15%) identified among controls. Overall, 38 bacterial species were identified, with 24 bacterial species among CRC cases and 26 bacterial species among controls (Fig. 3). Finally, 14 out of the 26 bacterial species (53.8%) identified among controls were specific for this group, whereas12 out of the 24 bacterial species (50.0%) identified among patients with CRC were specific for this group, although this difference was not significant (P = 0.7).A number of 10 bacterial species (26.31%) (Flavonifractor plautii, Eggerthella lenta, Clostridium bifermentans, Lactobacillus crispatus, Clostridium symbiosum, Clostridium sordellii, Paraclostridiumbifermentans, Lactobacillus reuteri, Clostridium tertium, and Enteroclosteraldensis) were strict anaerobic bacteria from the two groups.The median number of different bacterial species per stool sample was not significantly different (p = 0.69) between CRC cases (median = 5.5; IQR = 4.75) and controls (median = 7; IQR = 3.5).In the control group, too many species were identified, such as Pediococcusa cidilactici, Weissella confusa, Lactococcus garvieae, Enterococcus mundtii, Lactobacillus sakei, Enterococcus gilvus, Cellulosi microbium cellulans, Enterococcus dispar, Staphylococcus equorum, Aerococcus viridans, Desemzia incerta, Dialister propionicifaciens, Weissella cibaria, Propionibacterium acnes.Of the 38 bacterial isolates analyzed, 37(97.5%) yielded an accurate identification using MALDI-TOF mass spectrometry (Fig. 3).
Fig. 3.
Representation of number of isolates for each bacteria identified between the two groups: colorectal cancer group and control group
Discussion
Our findings are important because they contribute to the growing body of research on the intestinal microbiota and its link with colorectal cancer, specifically in a Tunisian context. The significance of incorporating geographical and demographic variables in microbiome research is underscored by the regional specificity of our data. Implementing this approach can lead to more precise and effective public health interventions. Furthermore, our research highlights the importance of increasing the representation of different populations in scientific research to ensure the inclusion and implementation of health strategies internationally.
Our results should be interpreted taking into account certain limitations. The size of our sample was insufficient due to the challenges posed by the collection of samples in the midst of the COVID-19 pandemic. Demand for stool samples from patients with colorectal cancer was difficult, as these immunosuppressed patients were at high risk and many unfortunately succeeded to the virus during this period. We also acknowledge the absence of a validation cohort, which introduces some uncertainty in our conclusions. A validation cohort is essential to confirm the robustness and generalization of our results. We plan to address this limitation in future research. This could involve collaboration with other research centers or hospitals to recruit a sufficient number of patients with colorectal cancer, following the same conditions and protocols as those used for the initial sampling.
Our study included 14 patients diagnosed with colorectal cancer (CRC), classified according to the TNM staging criteria. Notably, a majority of patients were in the advanced stages (Stage III), with a substantial number in the intermediate stages (Stage II). This distribution indicates that a significant portion of our cohort is in the advanced stages of CRC, providing crucial context for our subsequent analyses. This distribution is consistent with previous findings suggesting that gut dysbiosis is often associated with more advanced stages of tumorigenesis [4, 19]. The observed predominance of advanced-stage CRC patients in our cohort underscores the potential role of gut microbiota alterations in cancer progression. Specifically, the presence of dysbiosis in the later stages of CRC suggests that microbial imbalances might not only be an early event in tumorigenesis but could also contribute to the progression and aggressiveness of the disease. This aligns with the hypothesis that gut dysbiosis could be both a marker and a mediator of cancer development and progression. Further research should focus on longitudinal studies to clarify whether dysbiosis is a driving factor in the transition from early to advanced stages of CRC or primarily a consequence of tumor progression. Additionally, understanding the specific microbial changes associated with each stage could provide insights into potential therapeutic targets for preventing or slowing down CRC progression.
The gut microbiota plays a pivotal role in the maintenance of human health and the pathogenesis of diseases. The recognition of this significance has been facilitated by the emergence of omics technologies, which have been instrumental in advancing our understanding of the gut microbial ecosystem. Notably, metagenomics has unveiled the vast diversity of the gut microbiota, concurrently highlighting that a significant proportion of gut bacteria elude cultivation. Culturomics, conceived as part of the resurgence of culture techniques in microbiology, was devised to culture and identify previously unknown bacteria inhabiting the human gut. This approach provides a novel perspective on comprehending the knowledge about the human gut microbiota, encompassing contemporary clinical implications. However, our current constraint lies in the restricted quantity of fecal samples amassed thus far. Within a nationwide sample encompassing 14 colorectal cancer (CRC) cases and 13 controls, we have discerned statistically significant associations between the CRC group and certain factors. Specifically, male gender, tobacco and alcohol consumption, diabetes, and a familial history of colorectal cancer exhibit elevated odds within the CRC group. The P value pertaining to diabetes is 0.03 (< 0.5), indicating a statistically significant association between diabetes and CRC. Previous investigations have substantiated the correlation between diabetes and an augmented risk of developing colorectal cancer [20, 21].
Body Mass Index (BMI) has been positively associated in many observational studies with risk of CRC, especially with colon cancer and in men [22–24]. In this study, we observed an intriguing association between body mass index (BMI) and colorectal cancer (CRC), with a calculated odds ratio of 0.375 suggesting that individuals with a BMI > 25 may have lower odds of being diagnosed with CRC compared to those with a BMI ≤ 25. However, as highlighted in previous studies [20, 21], BMI is often assessed shortly before diagnosis in case–control studies, which can introduce bias. Indeed, prediagnostic weight loss may result from the underlying disease, potentially leading to an underestimation or even reversal of the association between BMI and CRC risk. Thus, while our findings are relevant, they emphasize the need to consider these potential biases to avoid erroneous conclusions regarding the relationship between body weight and cancer risk.
Our results reveal unique characteristics of the gut microbiota in Tunisian patients compared to other studied populations. Although Streptococcus bovis/gallolyticus and Enterococcus faecalis have been identified as potential biomarkers for CRC in several international studies, their significantly higher prevalence in CRC patients compared to controls in our Tunisian cohort highlights regional specificities. This observation may be influenced by environmental, cultural, and dietary factors specific to Tunisia, such as the Mediterranean diet rich in vegetables and olive oil. Additionally, the bacterial diversity identified in the samples shows marked differences between CRC patients and controls, with certain species being specific to each group. For instance, 53.8% of bacterial species in the controls were specific to that group, compared to 50.0% in CRC patients, although this difference is not significant. Comparing our results with international studies, we observe both similarities and differences. For example, certain bacterial species associated with CRC in studies conducted in Europe or North America are also present in our Tunisian cohort, although their relative abundance may vary. These comparisons allow us to place our study within a global context and contribute to a more nuanced understanding of microbiotic diversity and its health implications.
We used qPCR as a robust method to detect and quantify Streptococcus gallolyticus (Sgg) in stool samples of Tunisian patients with colorectal cancer and controls. This approach allowed us to observe a significantly higher prevalence of Sgg in patients with colorectal cancer, corroborating the findings of previous studies using qPCR to analyze this association. The results of a recent molecular analysis of normal and adjacent tumor tissues in unselected patients with CRC by quantitative PCR using Sgg-specific primers revealed that 74% of tumor tissues and 47% of these tissues were positive for Sgg [10]. In contrast, another study on non-selected CRC patients found a much lower prevalence using real-time quantitative PCR. Only six of the 190 patients included (3.2%) tested positive for Sgg [11]. These divergent results may be attributed either to variations in the populations sampled, but more likely to methodological discrepancies in the detection of Sgg. These methodological differences include detection site, number of samples, sample processing and storage, Sgg enrichment in specific medium, and the primers and qPCR techniques employed. A study conducted by sheikh et al. 2020 [25] explored the presence of Streptococcus gallolyticus subsp. gallolyticus (Sgg) in fecal samples collected in Ahvaz, Iran, from patients with colorectal cancer (CRC), inflammatory bowel disease (IBD), and a control group of healthy individuals. They found that Sgg was detected by culture in 13.6% of patient samples, with higher prevalences in IBD patients (15.9%) compared to CRC patients (9%). By PCR, Sgg was found in 36.4% of patient samples, with higher rates in CRC patients (40.9%) than in IBD patients (34.1%). These results suggest a potential pathogenic role of Sgg in CRC and IBD, prompting further research to confirm and expand these observations in other geographic and clinical settings.Streptococcus bovis has long been associated with CRC [26–31]. However, not all genospecies are as closely related to CRC [12], this study indicates that Streptococcus bovis should no longer be regarded as a single species in clinical practice, because S. gallolyticus (S. bovis biotype I) infection, in particular, has an unambiguous association with CRC. S. gallolyticus is a typical resident of various mammalian herbivores and birds' gastrointestinal tracts. According to several studies [32–34], S. gallolyticus can also be detected as a saprophyte outside of its animal host. S. gallolyticus was found in the human digestive tract as well; however, it is still a rare species [35]. Because only S. gallolyticus subsp gallolyticus seems to have virulence characteristics by comparative bacterial virulence analysis, that clearly associate endocarditis with underlying colon malignancies, the specific diagnosis of S. gallolyticus subsp gallolyticus infection might become a valuable tool for the early detection of CRC [11].
The role of E.faecalis in CRC development still remains controversial. In our study, we found that the prevalence of E.faecalis in stool samples from the CRC group is higher than that of the controls. While some studies suggest an adverse effect attributable to its capacity to inflict damage on the DNA of colonic epithelial cells, alternative scientific literature underscores the considerable potential of E. faecalis as a beneficial probiotic microorganism, especially in the context of its utility in food products [19, 36]. A review of [37] delineates the dual functionality of E. faecalis concerning injury to human intestinal epithelial cells. On one facet, its association with the production of reactive oxygen species is implicated in subsequent cellular damage. Conversely, an observed protective effect is evident in these cells, where in E. faecalis fosters the expression of interleukin-10 (IL-10); thereby diminishing the secretion of proinflammatory cytokines such as interleukin-8 (IL-8).A case report is presented on endocarditis caused by E. faecalis, where the source of the infection remains unidentified. Moreover, the report establishes an association between stage 1 colorectal cancer (CRC) and endocarditis, implying that endocarditis functioned as the origin of E. faecalis bacteremia [38].
We combined culture and MALDI TOF mass spectrometry for the identification of gut microbiota in fecal samples from both the CRC group and controls. We found that several species of enterococci were associated with CRC such as E.faecalis, E.asini, E.avium, and E.gallinarum. The literature reports the virulence and multidrug resistance exhibited by Enterococci, which could pose a potential epidemic risk, especially within healthcare settings. Enterococci can be acknowledged as pathogens. [39]. Enterococci, in particular E.faecalis and E. faecium, are key players in nosocomial infections. They can result in bacteremia, endocarditis, and urinary tract and intra-abdominal infections [40]. Moreover, there are indications suggesting their potential involvement in the pathogenesis of colon tumors. However, certain strains of enterococci have been demonstrated to support immunotherapy in the context of cancer treatment [41]. Moreover, E. faecalis has been recognized as a bacterium associated with colorectal cancer (CRC) due to the detrimental effects of its metabolites on the DNA of intestinal epithelial cells. This capacity to induce harm to cellular DNA may contribute to the initiation of neoplastic changes and potentially impede the expression of genes crucial for the repair of DNA damage, including PMS1 (PMS1 Homolog 1, Mismatch Repair System Component), MSH2 (MutS Homolog 2), MSH3 (MutS Homolog 3), MSH6 (MutS Homolog 6), and the regulation of the cell cycle [42]. Conversely, numerous species of enterococci are recognized for their potential to activate the immune system, playing a crucial role in maintaining intestinal homeostasis. The utilization of enterococci in cancer therapy has become the focus of an expanding body of literature. These microorganisms produce various compounds with antibacterial and/or anticancer properties. Increasing attention is being directed towards understanding this dual mechanism of action, encompassing both bactericidal and tumoricidal effects in certain instances. This dual functionality holds promise for the treatment of immunocompromised cancer patients, who are prone to life-threatening bacterial infections. These compounds could serve a dual purpose by addressing the infection and acting as an adjuvant in fundamental oncological therapy [41]. E.coli was also detected in the CRC group more than the control group; it is often linked to CRC in many studies [43–45]. Furthermore, a bacterial species identified as Flavonifractor plautii, specializing in the degradation of flavonoids, was exclusively identified in the CRC group. A recent investigation highlighted the significance of Flavonifractor plautii as one of the primary bacteria associated with CRC in Indian populations. This bacterium possesses the ability to metabolize flavonoids, compounds obtained by humans through the consumption of plants and fruits, known for their anticancer properties. The researchers propose that, without resorting to invasive techniques, the presence of this bacterium may serve as a non-intrusive biomarker for the early detection of colon cancer [46]. In addition, another pathogen, Eggerthella lenta, was detected in the CRC group. Actually, patients with serious comorbidities are increasingly being diagnosed with Eggerthella lenta because newly developing germs are introduced to oncologic patients [47]. Fecal Clostridium symbiosum was identified in the literature as a biomarker for non-invasive early detection of CRC [46, 47].We found it in the CRC group only.
Our study has certain limitations, such as sample size and the methodologies employed, which could introduce biases. To overcome these limitations, future studies should include larger and more diverse samples, as well as more robust methodological approaches, such as longitudinal studies or clinical trials. This research could deepen our understanding of the interactions between the gut microbiota and colorectal cancer in Tunisia and help develop more effective prevention and treatment strategies.
Conclusions
In conclusion, this study has produced promising results regarding the relationship between colorectal cancer (CRC) and the microbiome. However, the challenges posed by the COVID-19 pandemic have limited the size of our sample and introduced uncertainty in our findings. Collecting stool samples from patients with colorectal cancer has been difficult in this context, significantly impacting the robustness of our results. To address these constraints, we are considering creating a validation cohort in our future research. This initiative will help verify and strengthen the findings of this preliminary study. We plan to collaborate with other research centers or hospitals to recruit a sufficient number of participants, following the same rigorous protocols used in the initial sample collection. Although there has been little research on the relationship between CRC and the microbiome in developing nations, the incidence of CRC is rising there. This study highlights the potential of gut microbiota biomarkers as a promising non-invasive tool for the accurate detection and distinction of individuals with CRC.
Author contributions
Conceptualization, M.Z. and A.F. and I.B.B.B.; methodology, M.Z.; software, M.Z and M.N.; validation, A.F.; formal analysis, M.Z.; investigation, M.Z.; resources, M.Z.; data curation, M.Z Sample provision, L.M. and R.D. and S.H. and N.G.; writing—original draft preparation, M.Z.; writing—review and editing, A.F and I.B.B.B and D.R. All authors have read and agreed to the published version of the manuscript.”
Funding
This research was funded by the Ministry of Higher Education and Scientific Research of Tunisia. The data that support the findings of this study titled ‘‘Diversity in Gut Microbiota among Colorectal Cancer Patients: Findings from a Case–Control Study Conducted at a Tunisian University Hospital’’ are not openly available due to reasons of sensitivity and patient privacy. The data are available from the corresponding author upon reasonable request. Requests for data access should be addressed to zrelli.myriam@gmail.com.
Data availability
The data that support the findings of this study titled ‘‘Diversity in Gut Microbiota among Colorectal Cancer Patients: Findings from a Case–Control Study Conducted at a Tunisian University Hospital’’ are not openly available due to reasons of sensitivity and patient privacy. The data are available from the corresponding author upon reasonable request. Requests for data access should be addressed to zrelli.myriam@gmail.com.
Declarations
Institutional review board statement
The study was conducted in accordance with the Declaration of Helsinki, and it was approved by the local Ethics Committee of Charles Nicolle Hospital in Tunis, Tunisie (OHRP Number: IRB00013338).
Informed consent
Informed consent was obtained from all subjects involved in the study.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mariem Zrelli, Email: zrelli.myriam@gmail.com.
Didier Raoult, Email: didier.raoult@gmail.com.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Liu J, Deng M, Chen X, Jiang L, Zhang J, et al. Enterococcusfaecalis promotes the progression of colorectal cancer via its metabolite biliverdin. J Transl Med. 2023;21(1):72. 10.1186/s12967-023-03929-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;341:16203. 10.1038/nmicrobiol.2016.203 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Gupta A, Dhakan DB, Maji A, Saxena R, et al. Association of Flavonifractor plautii, a flavonoid-degrading bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems. 2017;4(6):00438–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artemev A, Naik S, Pougno A, Honnavar P, Shanbhag NM. The association of microbiome dysbiosis with colorectal cancer. Cureus. 2022;14(2):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu-Ghazaleh N, Chua WJ, Gopalan V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J Gastroenterol Hepatol. 2021;36(1):75–88. 10.1111/jgh.15042 [DOI] [PubMed] [Google Scholar]
- 7.Gut Microbiota for Health. 2019. Two new studies reveal universal gut microbiome signatures in colorectal cancer. https://www.gutmicrobiotaforhealth.com/two-new-studies-reveal-universal-gut-microbiome-signatures-in-colorectal-cancer/
- 8.Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci. 2019;116(48):24285–95. 10.1073/pnas.1912129116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, et al. The Role of the gut microbiome in colorectal cancer development and therapy Response. Cancers. 2020;12(6):1406. 10.3390/cancers12061406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Vogtmann E, Ahlquist DA, Devens ME, Kisiel JB, Taylor WR, et al. Fecal metabolomic signatures in colorectal adenoma patients are associated with gut microbiota and early events of colorectal cancer pathogenesis. mBio. 2020;11(1):03186–19. 10.1128/mBio.03186-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boleij A, Muytjens CMJ, Bukhari SI, Cayet N, Glaser P, Hermans PWM, et al. Novel clues on the specific association of Streptococcusgallolyticus subsp gallolyticus with colorectal Cancer. J Infect Dis. 2011;203(8):1101–9. 10.1093/infdis/jiq169 [DOI] [PubMed] [Google Scholar]
- 12.Boleij A, van Gelder MMHJ, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and Meta-analysis. Clin Infect Dis. 2011;53(9):870–8. 10.1093/cid/cir609 [DOI] [PubMed] [Google Scholar]
- 13.Pericàs JM, Corredoira J, Moreno A, García-País MJ, Falces C, Rabuñal R, et al. Relationship between Enterococcusfaecalis infective endocarditis and colorectal neoplasm: preliminary Results from a cohort of 154 patients. Rev Esp Cardiol Engl Ed. 2017;70(6):451–8. 10.1016/j.recesp.2016.09.055 [DOI] [PubMed] [Google Scholar]
- 14.Guindo CO, Davoust B, Drancourt M, Grine G. Diversity of methanogens in animals’ gut. Microorganisms. 2020;9(1):13. 10.3390/microorganisms9010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–33.e6. 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 16.Raoult D, Audoly G, Angelakis E, Rolain JM. Methode d’extraction d’ADN comprenant un traitement de degradation enzymatique d’un echantillon biologique FR3035409A1. 2016. https://patents.google.com/patent/FR3035409A1/fr
- 17.Guindo CO, Davoust B, Drancourt M, Grine G. Diversity of methanogens in animals gut. Microorganisms. 2020. 10.1128/CMR.00072-12. 10.1128/CMR.00072-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26(3):547–603. 10.1128/CMR.00072-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan Z, Siddiqui N, Saif MW. Enterococcus faecalis infective endocarditis and colorectal carcinoma: case of new association gaining ground. Gastroenterol Res. 2018. 10.4740/gr996w. 10.4740/gr996w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low EE, Demb J, Liu L, Earles A, Bustamante R, Williams CD, et al. Risk factors for early-onset colorectal cancer. Gastroenterology. 2020;159(2):492-501.e7. 10.1053/j.gastro.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gausman V, Dornblaser D, Anand S, Hayes RB, O’Connell K, Du M, et al. Risk factors associated with early-onset colorectal cancer. Clin Gastroenterol Hepatol. 2020;18(12):2752-2759.e2. 10.1016/j.cgh.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang X, Wei J, He X, Lian J, Han D, An P, et al. Quantitative association between body mass index and the risk of cancer: a global meta-analysis of prospective cohort studies. Int J Cancer. 2018;143(7):1595–603. 10.1002/ijc.31553 [DOI] [PubMed] [Google Scholar]
- 23.Andreasson A, Hagström H, Sköldberg F, Önnerhag K, Carlsson AC, Schmidt PT, et al. The prediction of colorectal cancer using anthropometric measures: a Swedish population-based cohort study with 22 years of follow-up. United European Gastroenterol J. 2019;7(9):1250–60. 10.1177/2050640619854278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abar L, Vieira AR, Aune D, Sobiecki JG, Vingeliene S, Polemiti E, et al. Height and body fatness and colorectal cancer risk: an update of the WCRF-AICR systematic review of published prospective studies. Eur J Nutr. 2018;57(5):1701–20. 10.1007/s00394-017-1557-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheikh AF, Masjedi Zadeh AR, Saki M, Khani P, Hashemi SJ, Shahin Zadeh S, et al. Detection of Streptococcusgallolyticus in colorectal cancer and inflammatory bowel disease patients compared to control group in southwest of Iran. Mol Biol Rep. 2020;47(11):8361–5. 10.1007/s11033-020-05807-7 [DOI] [PubMed] [Google Scholar]
- 26.Kumar R, Herold JL, Schady D, Davis J, Kopetz S, Martinez-Moczygemba M, et al. Streptococcusgallolyticus subsp gallolyticus promotes colorectal tumor development. PLOS Pathogens. 2017;13(7):1006440. 10.1371/journal.ppat.1006440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andres-Franch M, Galiana A, Sanchez-Hellin V, Ochoa E, Hernandez-Illan E, Lopez-Garcia P, et al. Streptococcus gallolyticus infection in colorectal cancer and association with biological and clinical factors. PLoS ONE. 2017;12(3): e0174305. 10.1371/journal.pone.0174305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aymeric L, Donnadieu F, Mulet C, Du Merle L, Nigro G, Saffarian A, et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci. 2018. 10.1073/pnas.1715112115388. 10.1073/pnas.1715112115388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquereau-Kotula E, Martins M, Aymeric L, Dramsi S. Significance of Streptococcusgallolyticus subsp gallolyticus association with colorectal cancer. Front Microbiol. 2019;9(614):391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–43. 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2017;36(5):757–69. 10.1007/s10096-016-2881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas C, Barnich N, Nguyen HTT. Microbiota inflammation and colorectal cancer. Int J Mol Sci. 2017;18(6):1310. 10.3390/ijms18061310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumke J, Vollmer T, Akkermann O, Knabbe C, Dreier J. Case-control study: Determination of potential risk factors for the colonization of healthy volunteers with Streptococcus gallolyticus subsp gallolyticus. PLoS One. 2017;12(5):e0176515. 10.1371/journal.pone.0176515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong J, Bai T, Zhang L, Qian W, Song J, Hou X. Inhibition effect of Bifidobacterium longum, Lactobacillus acidophilus, Streptococcusthermophilus and Enterococcusfaecalis and their related products on human colonic smooth muscle in vitro. PLoS One. 2017;12(12):0189257. 10.1371/journal.pone.0189257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Habermann W, Zimmermann K, Skarabis H, Kunze R, Rusch V. The effect of a bacterial immunostimulant (human Enterococcusfaecalis bacteria) on the occurrence of relapse in patients with. Arzneimittelforschung. 2001;11:931–7. [DOI] [PubMed] [Google Scholar]
- 36.de Almeida CV, Taddei A, Amedei A. The controversial role of Enterococcusfaecalis in colorectal cancer. Ther Adv Gastroenterol. 2018;11(1756284818783606):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8(217–30):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristich CJ, Rice LB, Arias CA. Enterococcal Infection—Treatment and Antibiotic Resistance. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary. 2014. http://www.ncbi.nlm.nih.gov/books/NBK190420/. [PubMed]
- 39.Grenda A, Grenda T, Domaradzki P, Kwiatek K. Enterococci—involvement in pathogenesis and therapeutic potential in cancer treatment: a mini-review. Pathogens. 2022;11(6):687. 10.3390/pathogens11060687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strickertsson JAB, Desler C, Martin-Bertelsen T, Machado AMD, Wadstrøm T, Winther O, et al. Enterococcusfaecalis infection causes inflammation, intracellular oxphos-independent ROS production, and DNA damage in human gastric cancer cells. PLoS ONE. 2023;8(4):63147. 10.1371/journal.pone.0063147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection: fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528–36. 10.1002/ijc.31011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nouri R, Hasani A, Masnadi Shirazi K, Alivand MR, Sepehri B, Sotoudeh S, et al. Mucosa-associated Escherichiacoli in colorectal cancer patients and control subjects: variations in the prevalence and attributing features. Can J Infect Dis Med Microbiol. 2021. 10.1155/2021/2131787. 10.1155/2021/2131787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu K, Yang X, Zeng M, Yuan Y, Sun J, He P, et al. The role of FecalFusobacterium nucleatum and pks+ Escherichiacoli as early diagnostic markers of colorectal cancer. Dis Markers. 2021;433(1171239):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta A, Dhakan DB, Maji A, Saxena R, et al. Association of Flavonifractor plautii, a Flavonoid-degrading Bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems. 2019;4(6):00438–19. 10.1128/msystems.00438-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woerther PL, Antoun S, Chachaty E, Merad M. EggerthellaLenta bacteremia in solid tumor cancer patients: pathogen or witness of frailty? Anaerobe. 2017;47(70–2):439. [DOI] [PubMed] [Google Scholar]
- 46.Xie YH, Gao QY, Cai GX, Sun XM, Zou TH, Chen HM, et al. Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine. 2017;25(32–40):442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, He Y, Xia L, Yi J, Wang Z, Zhao Y, et al. Expansion of colorectal cancer biomarkers based on gut Bacteria and viruses. Cancers. 2022;14(19):4662. 10.3390/cancers14194662 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study titled ‘‘Diversity in Gut Microbiota among Colorectal Cancer Patients: Findings from a Case–Control Study Conducted at a Tunisian University Hospital’’ are not openly available due to reasons of sensitivity and patient privacy. The data are available from the corresponding author upon reasonable request. Requests for data access should be addressed to zrelli.myriam@gmail.com.