Abstract
In metazoans mitochondrial DNA (mtDNA) or retrotransposon cDNA released to cytoplasm are degraded by nucleases to prevent sterile inflammation. It remains unknown whether degradation of these DNA also prevents nuclear genome instability. We used an amplicon sequencing-based method in yeast enabling analysis of millions of DSB repair products. In non-dividing stationary phase cells, Pol4-mediated non-homologous end-joining increases, resulting in frequent insertions of 1–3 nucleotides, and insertions of mtDNA (NUMTs) or retrotransposon cDNA. Yeast EndoG (Nuc1) nuclease limits insertion of cDNA and transfer of very long mtDNA ( >10 kb) to the nucleus, where it forms unstable circles, while promoting the formation of short NUMTs (~45–200 bp). Nuc1 also regulates transfer of extranuclear DNA to nucleus in aging or meiosis. We propose that Nuc1 preserves genome stability by degrading retrotransposon cDNA and long mtDNA, while short NUMTs originate from incompletely degraded mtDNA. This work suggests that nucleases eliminating extranuclear DNA preserve genome stability.
Subject terms: Genetics, Microbiology
Mitochondrial DNA or retrotransposon cDNA released into the cytoplasm is degraded to prevent sterile inflammation. In this study, the authors demonstrate that nucleolytic degradation of these DNA species in a yeast model organism prevents their transfer to the nucleus and genome instability.
Introduction
Mitochondria and chloroplasts carry multiple copies of a small genome, and transfer of DNA from these organelles to the nuclear genome is a ubiquitous and ongoing process. Genomes of most eukaryotes contain many fragments from these organelles1,2. Insertions of these DNA species within the nuclear genome can inactivate genes, form new exons, act as new or modify existing origins of replication, modify gene expression, and impact adaptation and genome evolution3–7. Insertions of mtDNA and cDNA form by their capture at spontaneous, programmed or induced DNA double-strand breaks (DSB) via nonhomologous end joining (NHEJ) (e.g.,8–12). The frequency of these insertion events increases with stress, aging, and tumorigenesis8,13–17, yet little is known about the enzymes that regulate mtDNA or cDNA transfer to the nucleus. While it is established that nucleases such as EndoG and TREX1 can degrade cytoplasmic DNA in humans to prevent activation of the immune response18,19, it remains unclear whether these mechanisms could also play a role in preventing extranuclear DNA from causing harmful genome instability. A major barrier to addressing this question is the difficulty of detecting these rare insertion events. Here we introduce a high-throughput amplicon sequencing pipeline that enables detection and analysis of many thousands of these events in yeast, and we provide evidence that the yeast EndoG homolog prevents genome instability in stressed cells by degrading extranuclear DNA species.
Results
Break-Ins method for insertion analysis
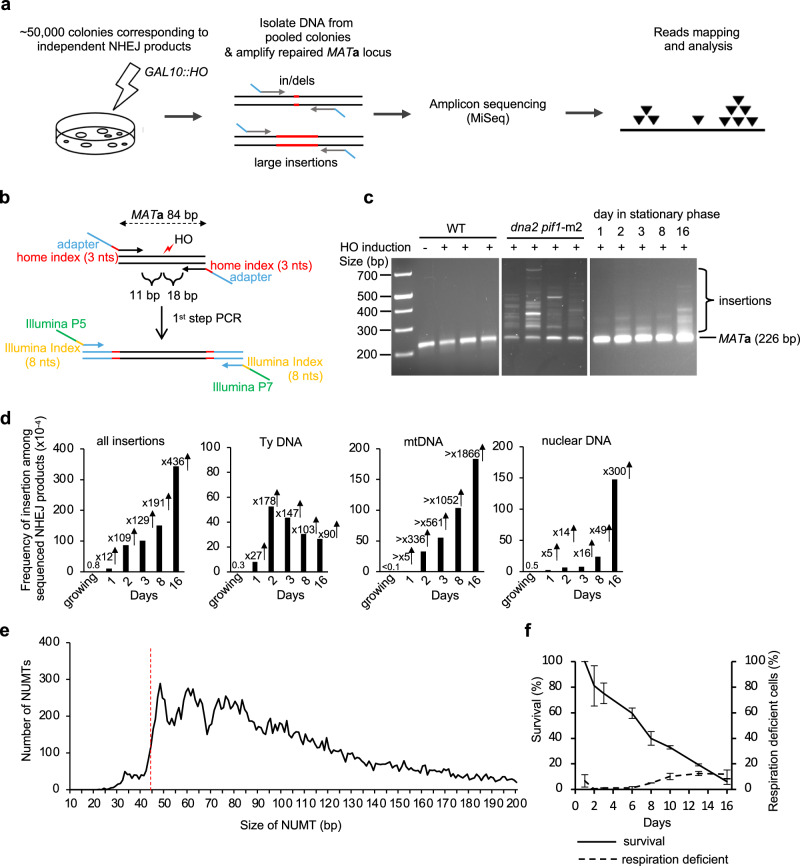
To study nuclear genome instability caused by insertion of mtDNA and retrotransposon cDNA, we developed a high-throughput amplicon sequencing based method called Break-Ins (Break Insertions) which allows screening of hundreds of thousands of NHEJ products simultaneously for the events carrying NUMTs or cDNA insertions (Fig. 1a). This method is suitable for testing many mutant strains or conditions simultaneously. We used yeast haploid cells carrying a galactose-inducible HO endonuclease that generates a single double-strand break (DSB) per genome at the MATa locus. In this strain, homologous sequences for DSB repair, HML and HMR, were deleted. Therefore, this DSB is repaired mostly by NHEJ; only the cells that repaired the break imprecisely, altering the HO recognition site, can survive continuous HO induction and form colonies. Most common sequence changes during DSB repair by NHEJ are small insertions and deletions of a few nucleotides (indels) that were previously characterized in detail (e.g.,20–22). To identify rare long > 10 bp insertion events, repaired MATa loci were amplified by PCR using primers located upstream and downstream from the HO cut site. Libraries of PCR fragments were prepared using a modified Illumina procedure and subjected to amplicon sequencing using the MiSeq platform (Fig. 1a, b; Supplementary Fig. 1a–e). To test this method, we initially compared wild-type cells in which insertions are very rare and Dna2-deficient cells in which insertions are common23. Over 100,000 colonies corresponding to independent NHEJ repair events were pooled in wild-type or Dna2-deficient cells. Amplification of the MATa locus with NHEJ products from Dna2-deficient cells, but not from wild-type cells, showed a smear of products above the band corresponding to the normally repaired MATa fragment size, suggesting many insertions in mutant but not wild-type cells (Fig. 1c). Only reads carrying insertions of > 10 bp, hereafter called “insertions”, were subjected to analysis. In wild-type growing cells, only 8 unique insertions were identified, three of which were derived from Ty retrotransposons, two from rDNA, and three from other parts of the nuclear genome. Overall, a ~ 280-fold increase in insertions was observed in dna2∆ pif1-m2 cells (Supplementary Fig. 2a). The remaining events were small indels (Supplementary Data 1). Repeating amplicon sequencing of the same DNA but using primers located further from the DSB led to identification of ~25% new insertions (Supplementary Fig. 2b). Nearly all unique insertions observed only with one pair of primers were represented by a lower read number, likely representing events more difficult to sequence. The distribution of inserted DNA within the genome, features of DNA inserted, and insertion junctions were comparable to previous Sanger analysis of individual NHEJ products (Supplementary Fig. 2c–e)23. Together we conclude that Break-Ins produces high-quality data and can be applied to study the transfer of extranuclear DNA to nucleus.
Fig. 1. Insertions of extranuclear DNA species in stationary phase cells.
a Schematic of the Break-Ins method. b Schematic of PCR amplification of the repaired MAT locus with two sets of indexes. Two sets of indexes were used to limit incorrect sample assignment by index hopping. c PCR amplification of uncut and repaired MAT locus in WT, dna2Δ pif1-m2 mutant, and WT stationary phase cells. The experiments were repeated 3 times with similar results. d Frequency and types of DNA inserted at DSBs in wild-type stationary phase cells among cells that repaired DSB by NHEJ. “All insertions” includes single and complex events. There was no single mtDNA insertion in growing cells. n–number of NHEJ products tested is shown in Supplementary Data 1. e Insertion size analysis of NUMTs. The insertions originating from mtDNA were from 16 days stationary phase cells (n = 21,348 independent NUMTs). Red dashed line indicates minimal size of DNA for efficient Ku70/80 heterodimer binding28. f Survival and respiration proficiency of cells plated on rich media after 1–16 days in stationary phase, (mean ± SD; n = 3. n represents biological repeats). Source data are provided as a Source Data file.
Frequent extranuclear DNA insertions in stationary phase cells
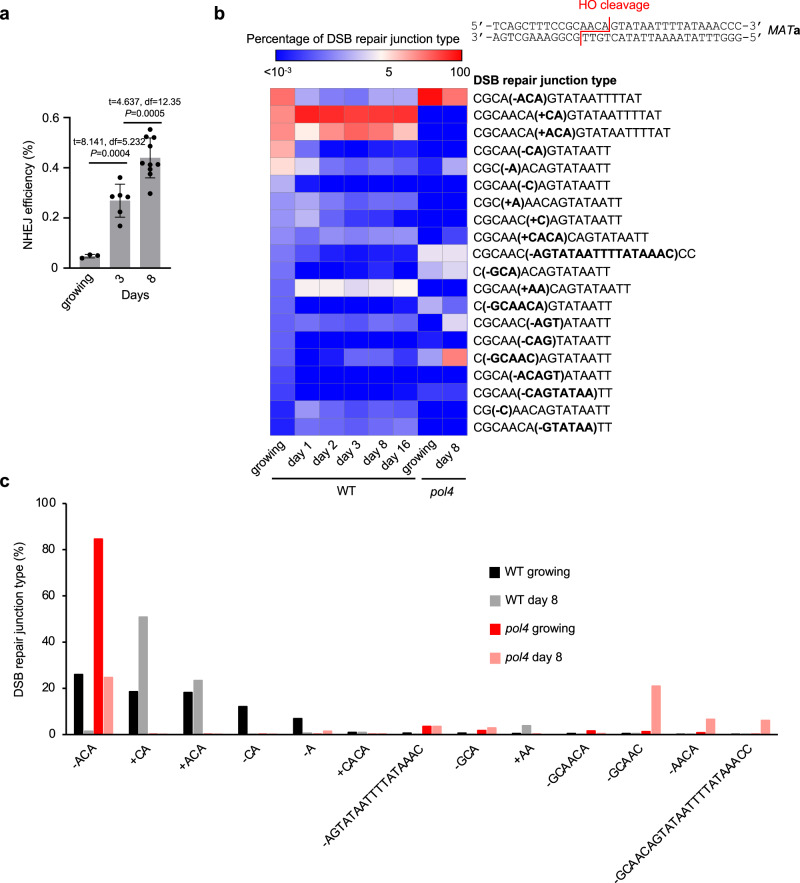
Analysis of insertions in wild-type cells suggests that the transfer of mtDNA or cDNA is rare in dividing cells grown in optimal conditions (Fig. 1d). To study the transfer of extranuclear DNA to the nucleus, we decided to test NHEJ products from nondividing, stationary phase cells where transfer of mtDNA was observed in fission yeast8 and where retrotransposon transcription increases24. NHEJ products were analyzed in nondividing cells incubated in synthetic complete media for 1, 2, 3, 8 or 16 days followed by DSB induction on YEPGal plates. The time of survival after cells enter stationary phase due to the exhaustion of nutrients is often referred to as chronological lifespan25. PCR amplification of the MATa locus suggests a high level of inserted DNA (Fig. 1c). Indeed, a large increase in insertions was observed in early 1-, 2- or 3-days stationary phase wild-type cells (Fig. 1d). At these earliest time points (1-3 days), inserted DNA originated mostly from mtDNA (~340–560-fold increase at 2-3 days) and Ty retrotransposons (~150-fold increase at 2-3 days). At later 8- and 16-day timepoints, NUMTs further increased up to over 1900-fold compared to growing cells and pieces of the nuclear genome were inserted as well (~50–300-fold increase). Insertions from all DNA sources (Ty, mtDNA, nuclear genome) were dependent on Lig4, essential for NHEJ. They were independent of Rad51 or a nonessential component of Polδ, Pol32, which makes it unlikely that break-induced replication-like events were involved (Supplementary Fig. 3a, b). Very rare insertions were observed in pol4Δ cells, implying a major role of Pol4 known to promote NHEJ26. (Supplementary Data 1). Genetic requirements and microhomology at DSB ends (Supplementary Fig. 3c) suggest that most of the insertions occurred by capture of DNA fragments at DSB by NHEJ. Additional analysis of ~0.5 mln NHEJ products from 16-day stationary phase wild-type cells revealed together over 20,000 NUMTs. The size of NUMTs ranged from 22 to 492 bp, comparable to natural yeast NUMTs (Supplementary Fig. 3d)27. Interestingly, the minimal size of mtDNA for efficient insertion is 44–46 bp (Fig. 1e) which likely reflects the DNA length-dependent cooperative binding of Ku70/Ku80 heterodimer to DNA, where ~45 bp-long dsDNA is the minimal DNA size for optimal binding28. The size of insertions originating from Ty retrotransposons was comparable to NUMTs while the size of insertions from the nuclear genome was about twice as long (Supplementary Fig. 3d). Among NHEJ products that have altered the HO cleavage site (~0.2–0.4% of cells plated), about 1% carried NUMTs, which corresponds to about 2-4 NUMTs per 105 cells plated. Inserted fragments of the nuclear genome often originated from fragile regions of the genome such as telomeric, or repetitive regions. Inserted fragments also originated from R-loop-prone regions that were previously mapped in growing cells29(Supplementary Fig. 3e). Thus, insertions at DSBs can occur in stressed stationary phase wild-type cells and are not limited to mutant cells such as Dna2-deficient yeast or human cells, or cancer cells23,30,31. We also noted an increase of error-prone NHEJ and altered distribution of NHEJ junctions in stationary phase cells. The proportion of “+CA” or “+ACA” nucleotide insertions that are mediated by Pol426,32 increased at the expense of “-ACA” mediated by Pol2 (Fig. 2a, b), suggesting an increased role of Pol4-mediated error-prone NHEJ in stationary phase cells. In growing pol4Δ cells, most junctions are represented by “-ACA” deletions while in pol4Δ stationary phase cells, additional longer deletions are common (Fig. 2c).
Fig. 2. The role of Pol4 in NHEJ increases in stationary phase cells.
a NHEJ efficiency during chronological aging. Mean values are plotted, with error bars representing standard deviation (SD). P-values were determined using an unpaired two-tailed Welch’s t-test. n = 3 for growing, 6 for 3 days, and 10 for 8 days. n represents biological repeats. b Analysis of top indel junctions among DSB repair products in growing and stationary phase wild-type or pol4Δ cells. Junctions are compared to the top 20 indels in growing wild-type cells. c Comparison of five most frequent junctions among DSB repair products in growing and stationary phase wild-type or pol4Δ cells. Source data are provided as a Source Data file.
Insertions of mtDNA at DSBs could originate from other cells in culture, dead or alive, or from loss of respiration coupled with loss of mtDNA. The level of respiration deficient petite colonies that either lost (rho0) or severely mutated mtDNA (rho-) does not change in day 1–3 of stationary phase as measured by ability to grow on a nonfermentable carbon source (Fig. 1f). Also, viability in days 1–3 is only slightly decreased while the number of NUMTs increases a lot (Fig. 1d, f). Thus, NUMT formation in early stationary phase cells is not associated with overall loss of mtDNA or loss of viability. Another argument against dead cells being a source of mtDNA for NUMT formation is that sch9Δ mutant maintains 100% viability even at 8 days stationary phase (Supplementary Fig. 3f)33, yet the number of insertions is similar when compared to wild-type. Finally, mtDNA fragments could originate from a cell’s own mitochondria, including connected mother-daughter pairs, or from other live cells in culture. We favor the first possibility because a previous study demonstrated that mtDNA can transfer to the nucleus but not between two yeast strains34. Overall, we conclude that NUMT formation increases in stationary phase cells and that mtDNA inserted at DSBs likely originates from within the same cell.
Nuc1 nuclease regulates cDNA and mtDNA insertions
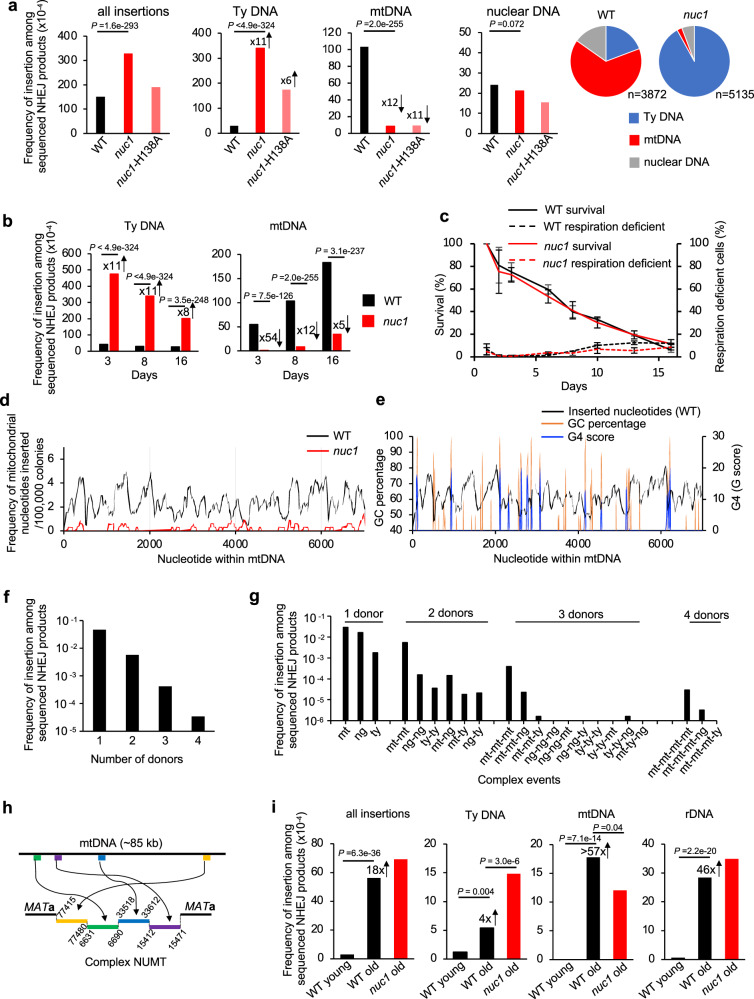
In humans, nucleases such as EndoG eliminate cytoplasmic DNA to prevent inflammation (reviewed in ref. 35) but it remains unknown if they impact nuclear genome stability. To test this in yeast, we followed genomic insertions of mtDNA and cDNA in cells lacking Nuc1, homolog of human EndoG. The primary localization of Nuc1 is in mitochondria but during stress or sporulation it can relocate to the nucleus or to the cytoplasm, respectively36,37. EndoG nucleases have DNA endonuclease and 5’ exonuclease activities with preference for ssDNA over dsDNA38. NUC1 deletion does not affect DNA insertions at DSBs in growing cells (Supplementary Data 1); however, we found it has a profound impact in stationary phase cells. In nuc1Δ cells, the overall level of insertions is higher due to ~11-fold increase of retrotransposon cDNA insertions. This is consistent with the possibility that Nuc1 degrades Ty1 cDNA that forms in cytoplasm. Unexpectedly, NUMTs were reduced by ~12-fold in nuc1Δ, suggesting a more complex role of this nuclease in regulation of mtDNA transfer to nucleus that is addressed below. Insertions from the nuclear genome at 8 days remained at the level comparable to wild type (Fig. 3a). Nuc1 nuclease activity is responsible for the changes of NUMTs and Ty cDNA insertions, as evidenced by a similar pattern of insertions in nuc1-H138A nuclease dead mutant and nuc1Δ cells (Fig. 3a). Considering the broad impact of Nuc1 on generation of insertions, we analyzed the pattern of insertions additionally at 3 and 16 days and observed a comparable effect on mtDNA and Ty DNA (Fig. 3b). Low mtDNA insertions in nuc1Δ cells are not related to a change in viability or respiration proficiency (Fig. 3c). By 16 days, NUMTs increased in nuc1Δ cells, indicating the existence of a less efficient Nuc1-independent pathway of NUMT formation of slightly longer size.
Fig. 3. Nuc1 regulates insertions of extranuclear DNA.
a–b Frequency and types of DNA inserted at DSBs in wild-type, nuc1Δ, and nuc1-H138A mutant cells in 8 days (a) or 3, 8 and 16 days (b) stationary phase cells. Pie charts demonstrate the distribution of different insertion donor DNA in wild-type and nuc1Δ cells at 8 days in stationary phase. n represents number of inserted DNA. P values were determined using two-tailed χ2 test. n–number of NHEJ products tested is shown in Supplementary Data 1. c Survival and respiration proficiency of wild-type and nuc1Δ cells plated on rich media after 1–16 days in stationary phase, (mean ± SD; n = 3. n represents biological repeats). d Analysis of sequences inserted at DSB from the first 7 kb of mtDNA in wild-type and nuc1Δ cells (insertions from the complete mtDNA genome are shown in Supplementary Fig. 4). Single donor insertions were analyzed. e Sequences inserted into DSBs and the distribution of G4 forming sequences and GC content within the first 7 kb of mtDNA. Single donor insertions only were analyzed. f Analysis of number of donor DNA per insertion event in wild-type cells at 16 days in stationary phase. Number of insertions involving 1, 2, 3 or 4 DNA pieces are shown. g Distribution of different complex insertion events in wild type at 16 days. Mt-mitochondrial DNA, ty-retrotransposon DNA, ng- nuclear genome DNA. h Example of a complex NUMT. i Frequency and types of insertions in wild-type and nuc1Δ cells in young cells and aged mother cells. P values were determined using two-tailed χ2 test. n-number of NHEJ products tested is shown in Supplementary Data 1. Source data are provided as a Source Data file.
NUMTs in wild-type cells originated from throughout the 86 kb-long yeast mitochondrial chromosome (Fig. 3d, Supplementary Fig. 4a). We have not observed insertions of spliced mtDNA fragments, indicating that reverse transcribed RNA is not a source of inserted mtDNA. Interestingly, G-rich sequences often mark the boundaries of the peaks of inserted mtDNA (Fig. 3e). Indeed, insertions from wild-type cells (p-value < 2.2e-16, one-sided Wilcoxon test) but not nuc1Δ (p-value = 1, one-sided Wilcoxon test) have an inverse relationship with G4- or GC-rich sequences. Nuc1 homolog EndoG preferentially binds and cuts poly-G rich sequences including G4 structure forming sequences39–41. Additionally, yeast Nuc1 and human EndoG are responsible for mtDNA deletions, which often occur within or at the boundaries of G sequence clusters (e.g.,39). Thus, possible preferential mtDNA degradation of G rich sequences by EndoG nucleases could reflect a slight insertion bias toward AT rich sequences. However, the nuclease activity of yeast Nuc1 toward G4 structures has not yet been studied.
Some of the sequenced NUMTs within eukaryotic genomes are complex, with multiple fragments joined together or in combination with pieces of nuclear genome or Ty retrotransposons (e.g.,13,42,43). Among ~33,000 insertions obtained from 16-day stationary phase wild-type cells, nearly 4000 contain complex insertions of two to four different fragments joined together (Fig. 3f). The frequency of complex NUMTs, but not of other multi-insertions, is higher than expected by chance (p-value = 0.0002, Fisher’s exact test), a phenomenon similar to the higher than expected level of multiple de novo NUMTs observed in cancer cells13. A possible explanation for the high level of multi-insertions is unequal release of mtDNA among cells. Most of these complex events carried multiple fragments of mtDNA, while the rest included only Ty DNA or nuclear DNA or were the mixture of multiple DNA sources (Fig. 3g). An example of a complex NUMT is shown in Fig. 3h. Complex mitochondrial rearrangements are reminiscent of chromothripsis events, in which fragments of nuclear chromosomes are subjected to clustered rearrangements44. As suggested previously, the shattering of the mitochondrial genome followed by random assembly of DNA fragments could constitute a mito-chromothripsis13. Our data provide compelling evidence for such a possibility with Nuc1 being responsible for mtDNA fragmentation.
NUMTs increase during aging across many species14,45,46. To test the transfer of extranuclear DNA to the nucleus during aging and the possible role of Nuc1, we first analyzed the lifespan of the strains used to capture DNA sequences at DSB using a microfluidic device. Wild-type and nuc1Δ showed comparable lifespans (Supplementary Fig. 4e). We then isolated old mother cells corresponding to ~60–80% of their lifespan. Old mother cells were plated on YEPGal plates to induce a DSB and repair products were followed using Break-Ins analysis as described above. An 18-fold increase of insertions was observed compared to young cells. Inserted DNA originated from mtDNA (>57-fold increase), Ty retrotransposon (4-fold increase) and the genome, mostly from rDNA (Fig. 3i). Further, we tested the possible impact of Nuc1 on NUMTs and Ty insertions in aged cells. In nuc1Δ cells, we observed a small (less than 2-fold) reduction of NUMTs and 3-fold increase of Ty cDNA insertions (Fig. 3i). A total of 158 NUMTs were observed in aged wild-type or nuc1Δ cells and they originated from throughout the mtDNA genome (Supplementary Fig. 4b). Thus, Nuc1 regulates transfer of mtDNA and cDNA also during aging, but likely additional nuclease(s) are involved. Finally, NHEJ efficiency was slightly increased in old mother cells while the NHEJ junctions remained comparable to young cells (Supplementary Fig. 5a, b).
Nuc1 degrades long mtDNA to prevent its transfer to nucleus
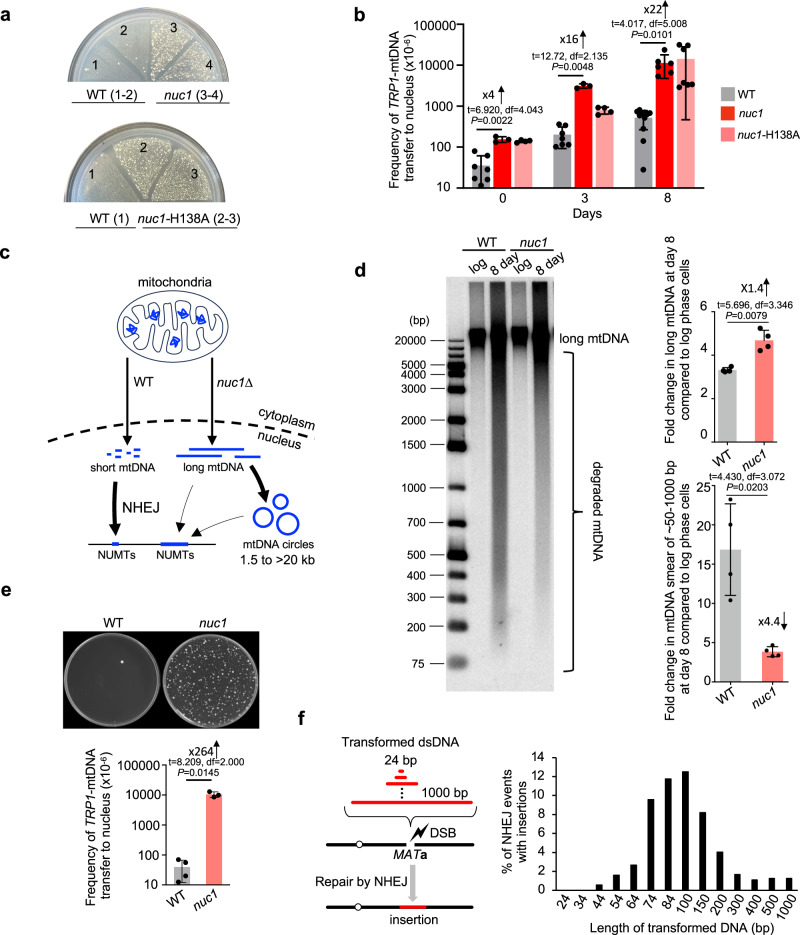
Break-Ins method can only monitor transfer/insertions of short mtDNA. To test a possible role of Nuc1 in the transfer of long mtDNA, we used a previously established assay47. In this assay, the transfer of long mtDNA fragments carrying a TRP1 reporter gene, which is not expressed in mitochondria, to the nucleus generates cells expressing TRP1 to form Trp+ colonies. Nuclear mtDNAs are maintained as circular fragments of a few to >30 kb that are easily lost and thus do not represent stable NUMTs47. These nuclear mtDNA circles are likely formed by ligation of linear DNA ends by NHEJ or other DSB repair pathways as was previously demonstrated for other types of circles (e.g.,48). The multitude of putative origins of replication within the AT-rich mitochondrial genome allows propagation of mtDNA as circular DNA5,7. Unexpectedly, and in contrast to short NUMTs that are nearly eliminated in nuc1Δ mutant cells, transfer of large mtDNA was greatly increased upon loss of NUC1 (Fig. 4a, b). A 16- to 22-fold increase in mtDNA transfer frequency was observed in nuc1Δ stationary phase cells when compared to wild-type cells. Thus, the release of mtDNA from mitochondria is independent of Nuc1 status. None of ~300 Trp+ tested colonies from wild-type or nuc1Δ cells carried stable TRP1-mtDNA.We confirmed that nuclear TRP1-mtDNA forms circles by testing these DNA by Southern blot in cells from which mtDNA was eliminated by EtBr treatment as described previously (Supplementary Fig. 6a, b)47. As expected, a comparable increase was noted in nuc1-H138A nuclease dead cells (Fig. 4a, b). At 8 days ~1% of nuc1Δ cells carried nuclear TRP1-mtDNA, which is likely an underestimate of total mtDNA transfer, considering that we can only follow TRP1-marked mtDNA fragments. There is a ~ 250-fold higher level of mtDNA transfer to the nucleus forming large circles in nuc1Δ when compared to number of short stable NUMTs at HO breaks in wild-type cells. Overall, Nuc1 limits the transfer of mtDNA to the nucleus. We propose that Nuc1 degrades long mtDNA, while incomplete degradation of long mtDNA by Nuc1 may result in shorter mtDNA fragments that form NUMTs (model in Fig. 4c). In support of such a possibility, we observed increased levels of long mtDNA in nuc1Δ and more degraded short mtDNA in wild-type stationary phase cells (Fig. 4d). These results also suggest that having long mtDNA transferred to the nucleus is not a common prerequisite of short stable NUMTs, as the number of NUMTs is greatly decreased in nuc1Δ, yet long mtDNA transfers more frequently. It is also possible that in nuc1Δ cells, very long mtDNA is occasionally inserted at DSB but these insertions cannot be sequenced using MiSeq platform. It was previously shown that Kap123 karyopherin mediates the transfer of Nuc1 to the nucleus in stressed cells36. Here, we tested the possible role of Kap123-mediated transfer of Nuc1 to the nucleus in the insertions of cDNA or mtDNA and the transfer of long TRP1-mtDNA to the nucleus, all of which are controlled by Nuc1. Unlike in nuc1Δ mutant cells, kap123Δ cells almost entirely lost insertions of retrotransposon cDNA. Additionally, the elimination of KAP123 reduced insertions of retrotransposon cDNA in nuc1Δ cells. These results likely reflect the known role of Kap123 in promoting transposition49,50. Also, kap123Δ reduced NUMTs formation by 3-fold compared to the wild type, which is less than the reduction observed in nuc1Δ single mutants. The kap123Δ nuc1Δ double mutant showed much lower levels of mtDNA insertions compared to each single mutant suggesting independent role of Kap123 and Nuc1 in promoting NUMTs. Insertions from the nuclear genome decreased in kap123Δ as well. Finally, kap123Δ, unlike nuc1Δ, showed no increase but rather a decrease in long mtDNA transfer to the nucleus, and this decrease was entirely dependent on Nuc1 (Supplementary Fig. 7). These results suggest that less transfer of Nuc1 to the nucleus in kap123Δ may help degrade extranuclear mtDNA, or alternatively, that Kap123 plays a positive role in insertions of all types of DNA by transporting protein(s) needed for efficient capture of DNA fragments. We conclude that the degradation of extranuclear DNA by Nuc1 likely occurs before its transfer to the nucleus.
Fig. 4. Nuc1 suppresses transfer of long mtDNA to nucleus.
a Analysis of TRP1-mtDNA transfer to nucleus. Wild-type, nuc1Δ and nuc1-H138A mutant cells replica plated from rich YEPD media onto plates with media lacking tryptophan. Trp+ colonies represent transfer of TRP1-mtDNA to nucleus. b Frequency of Trp+ colonies in wild-type, nuc1Δ, and nuc1-H138A mutant cells, (mean ± SD; n = 7 for 0 or 3 days and 11 for 8 days for wild type. n = 3 for 0 or 3 days, and 6 for 8 days for nuc1Δ mutant. n = 4 for 0 or 3 days, and 7 for 8 days for nuc1-H138A mutant. n represents biological repeats. P values determined using unpaired two-tailed Welch’s t-test). c Model of Nuc1 mediated control of mtDNA transfer to the nucleus. In stationary phase cells, mtDNA is released from mitochondria in Nuc1-independent manner. MtDNA is degraded by Nuc1 nuclease. Incomplete degradation leaves small mtDNA pieces that can be integrated at DSBs in nuclear genome. In the absence of Nuc1, mtDNA is not degraded and large mtDNA pieces are transferred to the nucleus where they either form circular mtDNA of different sizes or rarely integrate in the genome. d Southern blot analysis and quantification of long mtDNA and mtDNA smear in wild-type and nuc1Δ at 8 days stationary phase cells, (mean ± SD; n = 4; n represents biological repeats. P values determined using unpaired two-tailed Welch’s t-test). e Random spore analysis on plates with media lacking tryptophan. Trp+ colonies represent transfer of TRP1-mtDNA to nucleus. Frequency of spores carrying nuclear TRP1-mtDNA is shown. (mean ± SD; n = 3 for wild type and 4 for nuc1Δ mutant; n represents biological repeats. P values determined using unpaired two-tailed Welch’s t-test). f Scheme and analysis of insertion originating from transformed DNA fragments of different sizes. Frequency of insertions of annealed oligonucleotides (24–74 bp) or PCR fragments (84–1000 bp) are shown. 480–1680 independent NHEJ products were tested by PCR. Source data are provided as a Source Data file.
Next, we decided to test whether Nuc1 function in limiting mtDNA transfer to the nucleus also applies to yeast stationary phase diploid cells that undergo meiotic recombination and sporulation. Wild-type and nuc1Δ/nuc1Δ diploids were sporulated and subjected to random spore analysis on plates lacking tryptophan. As shown in Fig. 4e, spores produced in Nuc1-deficient cells showed a large > 250-fold increase of Trp+ colonies corresponding to mtDNA transfer to the nucleus. About 1% of all spores carried nuclear TRP1 marked mtDNA in Nuc1-deficient cells. Thus, Nuc1 controls the frequency of mtDNA transfer to nuclei in diploid cells during meiosis. 180 independent Trp+ spores were tested for the maintenance of nuclear TRP1-mtDNA and only one carried a stable TRP1 marker integrated in the nuclear genome (Supplementary Fig. 6c). In summary, Nuc1 appears to prevent the transfer of long mtDNA to the nucleus during sporulation or starvation that can lead to rare (below 1%) stable genomic integration.
In nuc1Δ cells, long mtDNA is transferred to the nucleus, yet this DNA is not frequently inserted at HO breaks or anywhere else in the genome. This means that the short size of NUMTs is not likely related to the inability of longer DNA to cross the nuclear membrane. Also, MiSeq read length of up to 600 bp cannot explain a clear bias toward ~100 bp NUMTs and far fewer NUMTs of 200–600 bp. One possibility is that NHEJ has a preference for inserting shorter DNA. To test this, we transformed an equal molar amount of 14 DNA fragments ranging from 24 bp to 1 kb, each carrying the same sequences on 5’ and 3’ ends matching HO cut overhangs. Each fragment was transformed independently, and cells were spread on YEPGal to induce DSB. To avoid any Illumina sequencing bias toward DNA fragments of a particular size, we tested 400 to > 1000 NHEJ products individually by PCR. The shortest fragment inserted was 44 bp long and fragments of ~60 bp to ~150 bp inserted most frequently (Fig. 4f). The length of transformed DNA optimal for insertion at DSB is comparable to the size of all types of inserted DNA (mtDNA, Ty DNA, nuclear genome); however, NUMTs and Ty DNA insertions are on average slightly shorter while insertions from the nuclear genome are longer when compared to the size of insertions of transformed DNA (Supplementary Fig. 3d). Thus, the bias toward short insertions ( ~ 45 to ~300 bp) could stem from easier insertion of short DNA fragments at DSBs by NHEJ. In general, the size of insertions observed in different experimental systems and species is within the range observed here9,12,23,43.
Nuc1 degrades retrotransposon cDNA
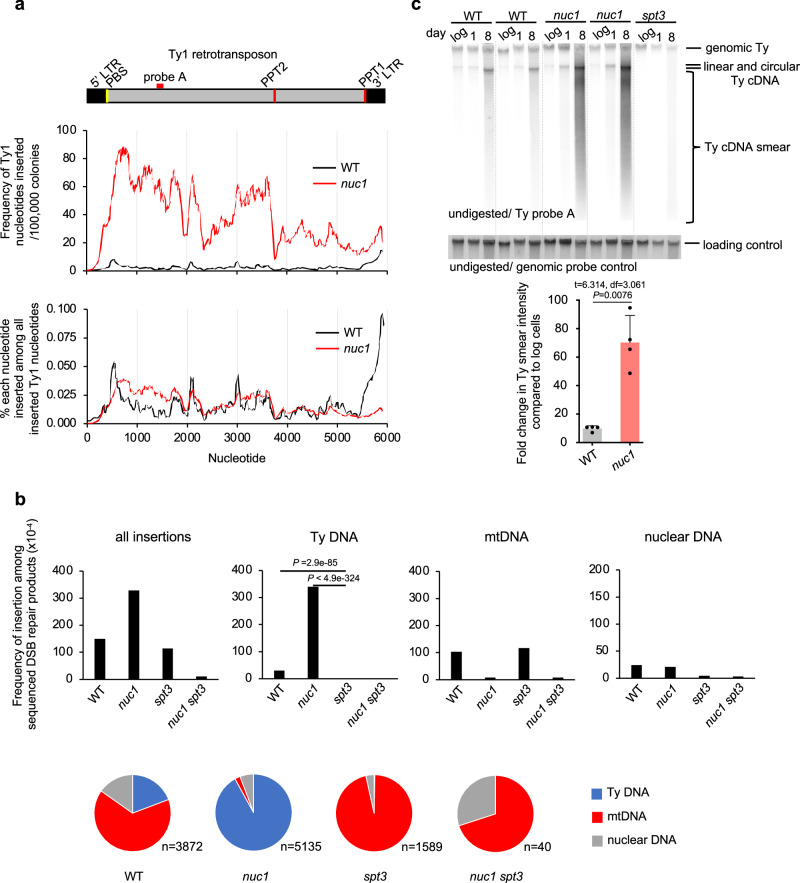
In stationary phase cells, insertions of Ty DNA increase by ~90–180-fold, and Nuc1 absence leads to a further increase by ~11-fold (Figs. 1d, 3a). To analyze transposon insertions in detail, we selected the ones originating from Ty1, the most common retrotransposon in yeast. All regions of Ty1 contributed to insertions, and as previously shown the 3’ LTR region is a clear hotspot in wild-type cells (Fig. 5a, Supplementary Fig. 4c)10,11,51. In nuc1Δ cells, a large increase of inserted Ty1 fragments was observed at the region outside of LTR ( ~ 10 to 40-fold), particularly from the primer binding site (PBS) to the central priming site (PPT2), while the increase of the 3’ LTR region was very modest ( ~ 2-fold). A large fraction of 3’ LTR insertions in wild-type cells (40/56) carried extra nucleotides present only in cDNA, consistent with intermediates of Ty1 replication being inserted at DSBs11,51. Also, insertions of retrotransposon DNA, but not mtDNA, were largely dependent on Spt3, required for the transcription of the Ty1 encoding reverse transcriptase52, suggesting that cDNA fragments were inserted rather than pieces of genomic Ty1 (Fig. 5b). Increased insertions of Ty DNA in stationary phase cells are consistent with greatly increased Ty transcription24. However, high Ty1 expression is not associated with a change in transposition in wild-type or nuc1Δ old stationary phase cells as measured using a previously described Ty1 transposition reporter assay53 (Supplementary Fig. 4d). The full-length Ty cDNA amount was measured by Southern blot and shows no major difference between wild-type and nuc1Δ cells that could account for the large increase of cDNA insertions observed in nuc1Δ (Fig. 5c). As expected, no cDNA was observed in the absence of Spt3 needed for Ty1 expression. Besides complete linear or circular cDNA of Ty1, another possible source of fragments inserted at DSBs could be intermediates of Ty replication. ssDNA minus strand intermediates of Ty1 replication can be inserted at DSBs and can recombine with the yeast genome54, and nucleocapsids are known to carry Ty1 replication intermediates observed as a smear of Ty1 cDNA55. We found ~10-fold increase of the smear of Ty1 cDNA in 8-day-old stationary phase wild-type cells when compared to growing cells and ~70-fold increase in nuc1Δ, which may explain the higher level of Ty1 insertions in nuc1Δ cells (Fig. 5c). Together, the increase of partially replicated retrotransposons in wild-type and particularly in nuc1Δ cells may contribute to inserted cDNA at DSBs. Enzymatic activities of purified EndoG nucleases show a preference for cleavage of ssDNA over dsDNA and the ability to degrade RNA in DNA:RNA hybrids38,56,57, which could explain the strong impact of Nuc1 on intermediates of Ty replication, many of which are ssDNA or RNA:DNA hybrids. We propose that Nuc1 nuclease limits cDNA insertions by degrading incomplete Ty1 replication intermediates in stationary phase cells. Besides dsDNA and ssDNA that can be inserted at DSBs, it is possible that DNA:RNA hybrids are also captured by NHEJ.
Fig. 5. Nuc1 degrades retrotransposon cDNA replication intermediates.
a Analysis of Ty1 sequences inserted at DSBs in wild-type and nuc1Δ cells. Frequency of each Ty1 nucleotide insertion per 106 NHEJ products or among all Ty1 nucleotides inserted. Schematic of Ty1 shown at top indicating long terminal repeats (LTR), primer binding site (PBS) and polypurine tracts (PPT1/2). b Frequency and types of DNA inserted at DSBs in wild-type, nuc1Δ, spt3Δ, and nuc1Δ spt3Δ mutant cells in 8 days stationary phase cells. Pie charts below demonstrate distribution of different insertion donor DNA. n represents number of inserted DNA. P values were determined using two-tailed χ2 test. n–number of NHEJ products tested is shown in Supplementary Data 1. c Southern blot analysis of Ty1 cDNA in wild-type, nuc1Δ and control spt3Δ cells. Quantification of Ty1 cDNA smear is shown (mean ± SD; n = 4; n represents biological repeats. P values were determined using unpaired two-tailed Welch’s t-test). The position of the probe used is shown in (a). Source data are provided as a Source Data file.
Discussion
Cytoplasmic DNA must be degraded to limit sterile inflammation in metazoans. We provide evidence that degradation of long mtDNA and retrotransposon cDNA is important for nuclear genome instability. In wild-type cells grown in optimal conditions, the transfer of cDNA or mtDNA to nuclei is very rare. However, in aging or in stationary phase when cells experience stress and limited nutrients, transfer of these DNA species to the nucleus increases. Insertions of extranuclear DNA fragments at DSBs are mediated by NHEJ and require Pol4. High activity of Pol4 in stationary phase cells is evident based on increased error-prone NHEJ repair with most junctions specific for Pol4 activity.
Yeast EndoG is a major regulator of extranuclear DNA transfer to the nucleus. Elimination of Nuc1 increases partially replicated retrotransposon cDNA fragments and their insertions in the genome. In stressed cells, transcription of transposons is upregulated24; however, with limited nutrients it is likely that neither complete cDNA nor nucleocapsids are formed efficiently, leaving cDNA intermediates vulnerable for Nuc1 mediated degradation. Nuc1 absence also increases transfer of very long extrachromosomal mtDNA to the nucleus that form unstable circles. Up to ~20 or 250-fold increase of the transfer of long TRP1 marked mtDNA in nuc1Δ cells was observed during starvation in haploid cells or during sporulation in diploids, respectively. In yeast, any circular DNA devoid of centromeres shortens the lifespan of the cells58; therefore, elimination of mtDNA released from mitochondria during sporulation or starvation produces healthier cells. While nuc1Δ cells show an increased level of nuclear circular mtDNA, these circles are present in a small fraction of cells and therefore do not impact nuc1Δ population lifespan. MtDNA carries many repetitive sequences including microsatellite DNA, direct and inverted repeats, sequences forming G4 structures and sequences that may serve as efficient origins of replication in the nucleus. Therefore, nuclear mtDNA circles that are often present at a high copy number could instigate genome instability, recruit proteins needed for regular genome replication, and likely integrate into the genome as was shown for other circular DNA fragments in yeast and humans59,60. Nuclear circular mtDNA has not yet been studied in a systematic way in mammalian systems. MtDNA is excluded from sequencing analysis of circular DNA because of the inability to distinguish it from hundreds/thousands of regular mtDNA per cell. However, several pieces of evidence suggest that very long mtDNA can transfer to nuclei. Recent studies demonstrated many cases of multicopy mega-NUMTs inserted in the human genome61,62. Long NUMTs are also common in cancer cells13. Finally, nuclear extrachromosomal circular mtDNA was observed in nuclei of human pluripotent and embryonic stem cells63. Opposite to transfer of long mtDNA, formation of short NUMTs decreases in nuc1Δ cells. However, the highest frequency of stable short NUMTs in the presence of Nuc1 is at least 250-fold lower when compared to the spontaneous transfer of long mtDNA to the nucleus in the absence of Nuc1. Therefore, while the Nuc1 nuclease generates small mtDNA fragments that can be integrated into DSBs, it prevents the far more frequent nuclear transfer of very long mtDNA that also occasionally integrates into the genome (model in Fig. 4c). We propose that nucleases such as EndoG and TREX1 in humans and other organisms, in addition to their role in immune response, also prevent genome instability caused by free DNA species.
Methods
Yeast strains
All yeast strains used here to study insertions at HO endonuclease induced DSB are derivatives of JKM139 strain, a gift from James Haber, and are listed in Supplementary Table 1. All strains used to study the transfer of TRP1-mtDNA to the nucleus are derivatives of PTY44 strain, a gift from Peter Thorsness. In this system, a TRP1 marker was inserted upstream of the COX2 gene and TRP1 marker carries no homology to the nuclear genome where TRP1 is entirely deleted (trp1-Δ1). The nuc1-H138A mutants were constructed using the plasmid pCORE-UH (a gift from Francesca Storici). First, the KlURA3-hphMX double maker were integrated into NUC1 locus. Then a fragment of nuc1 with a point mutation was PCR amplified from the plasmid pESC-nuc1-H138A (a gift from Frank Madeo) and transformed into cells to replace KlURA3-hphMX. All mutations were confirmed by Sanger sequencing.
Yeast growth, media and DSB induction
For amplicon sequencing analysis in normal conditions, cells were grown in YEPD (1% yeast extract, 2% peptone, 2% dextrose) o/n to saturation. Cells were washed twice in YEP-Raffinose (1% yeast extract, 2% peptone, 2% raffinose) media and inoculated in 10 ml of YEP-Raffinose and grown overnight. Once cells reached the density of 2 × 107/ml, they were plated on YPE-Gal plates (1% yeast extract, 2% peptone, 2.5% agar, 2% galactose. Galactose was filter sterilized and added to the autoclaved media). Control plating on YEPD plates provided information on the number of viable cells. Old yeast mother cells isolated as described below were plated on YEPGal plates the same way. For all analyses in stationary phase cells, we followed an established protocol for yeast growth64. Yeast cells were initially streaked out on YEPD plates and individual colonies were inoculated in 3 ml SC media (1.72 g/L yeast nitrogen base without amino acids (USBiological, cat# Y2030), 2 g/L amino acid mix (Bufferad, cat#S0051), 0.5% ammonium sulfate, 2% dextrose) and incubated overnight at 30 °C. The culture was diluted into fresh 50 mL SC media in 250 mL flasks to an OD600 of 0.1. Logarithmic growth phase control cells were plated when the culture reached 1 × 107 cells/ml. Cells were incubated with shaking for 24 hrs (day 1), 3 days, 8 days, 16 days or as specified in individual experiments and plated on YEPGal plates for HO induction. Colonies were collected after 5 days of incubation for analysis of NHEJ products.
Measurement of survival, respiration and NHEJ efficiency
To measure yeast cell survival (CFU), the cells were diluted and spread on YEPD plates. Colonies were counted 5 days after plating. To check the respiration capacity, colonies on YEPD plates were replica plated to YPEGlycerol plates, where fermentation is not possible. The number of colonies that failed to grow on YPEG out of all colonies replica plated from YEPD is the proportion of respiration-deficient cells. To analyze the efficiency of DSB repair by NHEJ, cells were plated on YEP-Gal (GAL10-HO induction) and YEPD (no HO induction). As only a small fraction of cells survive constant HO induction, typically 100,000 cells were plated on YEPGal plates and 100 cells on YEPD plates. The efficiency of NHEJ was calculated as the percentage of cells growing on YEPGal out of all cells plated on YEPD plates.
Isolation of old yeast cells
Populations of old yeast mother cells were isolated as described65. 3 × 108 yeast cells from a fresh log-phase culture in YEPD were treated with 3 mg of Sulfo-NHS-LC-Biotin (Thermo Scientific) and were used to seed a 1 L culture in YEP-Raffinose. Seeded cultures were incubated with shaking at 30 °C for 15 hours to age biotin-labeled cells. The aged biotin-labeled cells were isolated using 1 mL Dynabeads Biotin Binder (Thermo Scientific). Three rounds of aging and sorting were conducted to obtain old mother cells with an average replicative age of around 20. Control young cells were obtained from last sorting and wash.
Yeast replicative lifespan measurement
The yeast replicative lifespan was determined in microfluidic devices (AD-Chips, obtained from Innovative Biochips) with time-lapse imaging as described66,67. Briefly, filter-sterilized YEPD was loaded into the AD-Chip channels at 20 µL/min using 20 mL syringes (BD Biosciences) driven by a KDS-230 pump (KD Scientific); medium flow was subsequently set to 1 µL/min prior to cell loading. Yeast strains were grown to mid-log phase in filter-sterilized YEPD and diluted 1:20 in the same prior to manually loading cells into the AD-Chip. After cell loading, the medium flow was set to 5 µL/min for the duration of the experiment. The AD-Chip was placed in an environmental chamber set to 30 °C for the lifespan analysis. Multi-position time-lapse imaging was performed with an EVOS FL Auto system (Thermo Fisher), using a 20× objective and transmitted light optics. Images (4 images encompassing 480 traps per microfluidic channel) were taken every 15 minutes for 65 hours. The timelapse image series were analyzed with ImageJ (National Institutes of Health) and cell divisions were manually counted for a minimum of 50 cells from each strain.
Analysis of Ty1 transposition
Rates of Ty1 retrotransposition were estimated using strains carrying a Ty1-270his3-AI reporter53. Individual colonies of wild-type and nuc1Δ strains were inoculated in 5 mL YEPD media overnight at 24 °C. Overnight cultures were then diluted and cells were grown to 1 × 107 cells/mL before plating ~20–30 cells on YEPD plates. After 3–5 days of incubation at 24 °C, ten individual colonies per strain were inoculated to 8 mL SC media and incubated at 24 °C overnight. 2 × 108 cells of each culture were plated on 150 × 15mm plates of synthetic complete medium lacking histidine, and 100 cells were plated on YEPD plates to measure viability. The remaining Day 0 culture was used to inoculate fresh 8 mL SC media to an OD of 0.1. The plating procedure was repeated on Day 3 and Day 8 without dilution to fresh SC. After growth at 24˚C for ~7–10 days, His+ colonies were counted. Statistical analysis of retrotransposition rates was performed with Drake estimator68, normalizing the number of cells plated by multiplying 2 × 108 cells by the number of cells that grew on YEPD plates/100. P-values were determined using the bootstrap resampling approach.
Analysis and quantification of mtDNA, Ty1 linear, circular, and smear cDNA amounts
For analyses of Ty1 cDNA or mtDNA in stationary phase cells, yeast cultures were grown as described above. Cells from log phase, saturated, and 8 days stationary phase were collected by centrifugation, washed in sterile water and frozen. Genomic DNA was extracted as described69 with modifications. Cells were resuspended in 500uL extraction buffer (2% SDS, 100 mM Tris-HCl pH 8.0, 50 mM EDTA) and 5 μL β-mercaptoethanol and 10 μL 100 T Zymolyase (5 mg/mL) were added. Cells were lysed by 30-60 minutes incubation with shaking at 37 °C. DNA was extracted using a standard phenol/chloroform extraction and digested with RNaseA (0.1 mg/mL) overnight at 37 °C. For analysis of Ty cDNA, undigested DNA was run on a 1 % agarose gel and subjected to Southern blot analysis using a Ty1 specific DNA probe amplified by PCR using primers Typeak fw and Typeak rv. For analysis of mtDNA amount, the same DNA was probed with a mtDNA specific DNA probe amplified by PCR using primers mtDNA F1 and mtDNA R2. See Supplementary Table 2 for primer sequences.
Band intensities corresponding to probed DNA fragments were analyzed using ImageQuant software. All quantifications were done with a normalized amount of DNA in each lane. For undigested DNA, normalization was done with a genomic mixture of three probes amplified using primers ACT1-fw and ACT1 rv, TRA1 fw and TRA1 rv, TOM1 fw and TOM1 rv. All probes were gel purified using the Nucleospin Gel and PCR Clean-up kit (Macherey-Nagel, cat# 740609). The pixel intensities of the bands corresponding to linear, circular, or smear of Ty cDNA were subtracted by the corresponding background intensity observed in spt3Δ mutant cells that do not generate cDNA. The pixel intensities of cross-hybridizing bands observed in the spt3Δ mutant cells that do not generate cDNA were subtracted from the bands corresponding to linear, circular, or smear of Ty cDNA. For smear, all DNA below linear cDNA was measured. The amounts observed in saturated culture cells (day 0) and day 8 were compared to cDNA levels observed in wild-type growing cells. Three to four repeats were done. T-test was used to quantify P-values.
Amplicon sequencing of repaired MAT locus by “Break-Ins”
Cells from an overnight saturated YEPD culture were washed twice with YEP-Raffinose, inoculated into 10 ml YEP-Raffinose and incubated overnight at 30 °C. When the density of the culture was ~2 × 107 cells/ml, ~1 × 107 cells were spread on a YEP-galactose plates and incubated at 30 °C for 5 days. Each culture was spread on 5–7 150 × 15 mm plates. Colony number was counted. All colonies were collected, pooled together and mixed vigorously. ~80 μl cells were spun down and the genomic DNA was extracted using standard glass beads and phenol/chloroform. The genomic DNA was treated with RNase A overnight. The genomic DNA was dissolved with water and adjusted to a concentration of 10 ng/μl. The construction of the sequencing library was adapted from Illumina’s amplicon sequencing protocol #15044223 Rev. B. Two rounds of PCR were performed to construct the sequencing library. For each sample, 3.2 μl genomic DNA, 0.5 μl 10 μM primers and 12.5 μl KAPA HiFi HotStart ReadyMix (Roche, 7958927001) was used for the first round of PCR for a total volume of 25 μl. The forward primers are 66-mers that contain (5’ to 3’): 33 bases Illumina adapter sequence, 3 bases unique home index, and 30 bases targeting the MATa locus which is located 11 bp upstream of the HO cleavage site. The reverse primers are 66-mers that contain (5’–3’): 34 bases Illumina adapter sequence, 3 bases unique home index, and 29 bases targeting the MATa locus which is located 18 bp downstream the HO cleavage site. The following conditions were used for the first round of PCR: 95 °C for 5 min; 22 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 3 min; 72 °C for 10 min. 18 μl PCR product was purified with 18 μl AMPure XP beads (Beckman Coulter, A63880) and eluted with 52.5 μl 10 mM Tris pH 8.5. For the second round of PCR, 5 μl purified PCR product, 5 μl Nextera XT V2 Index (Illumina, FC-131-2001) primer N7xx, 5 μl index primer S5xx, and 25 μl KAPA HiFi HotStart ReadyMix were used. The total volume of PCR is 50 μl. The following conditions were used for PCR: 95 °C for 5 min; 8 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 3 min; 72 °C for 10 min. 40 μl PCR product was purified with 48 μl AMPure XP beads and eluted with 27.5 μl 10 mM Tris pH 8.5. The DNA concentration of each sample was determined with Qubit dsDNA BR Assay Kit (ThermoFisher, Q32850) and the average size of the DNA was determined with TapeStation. The DNA was diluted to 4 nM using 10 mM Tris pH 8.5. An equal amount of DNA was pooled from ~20 samples into the library. The pooled library and PhiX library (Illumina, FC‐110‐3001) were denatured with 0.2 M NaOH separately and diluted with pre-chilled HT1 to 12 pM. 540 μl pooled library and 60 μl PhiX were mixed, incubated at 96 °C for 2 min and immediately placed in an ice-water bath for 5 min. The denatured combined library was loaded into the MiSeq Reagent Kit v3 (600 cycle) (Illumina, MS-102-3003). The cluster density was ~1100 K/mm2.
Analysis of TRP1-mtDNA transfer to the nucleus
To analyze the frequency of transfer of TRP1-mtDNA to the nucleus, cells were grown as described above in SC media and plated on Trp- plates. Control plating on YEPD informed about the number of viable cells plated. Trp+ colonies were scored after 5 days incubation at 30 °C. Analysis was repeated 3 to 8 times. The frequency of nuclear TRP1-mtDNA was calculated as the number of Trp+ colonies divided by the number of viable cells plated. To analyze the stability of the nuclear TRP1-mtDNA, an initial Trp+ colony was streaked onto Trp- plates and grown for 2-3 days at 30 °C. Resulting single Trp+ colonies were streaked on nonselective YEPD plates and grown for 2 days at 30 °C. Once colonies formed, the plates were replica plated on Trp- plates and the number of Trp+ colonies were counted. To analyze the size of nuclear TRP1-mtDNA, we streaked the Trp+ colonies for singles on Trp- plates and inoculated in 3 ml Trp- minimal liquid media containing 25 μg/ml ethidium bromide and incubated overnight at 30 °C on a roller drum as described70. The growth in Trp- minimal liquid media with ethidium bromide media was repeated once more, and the cells were streaked on Trp- plates. A single colony was used to inoculate 5 ml Trp- minimal media, and cells were grown to saturation at 30 °C. Cells were pelleted, and DNA was isolated using standard glass beads and phenol/chloroform procedure. Isolated DNA was digested with PstI restriction enzyme, separated on 0.8% agarose gel, and subjected to Southern blot analysis using PCR amplified 3’ TRP1 fragment (~300 bp on each side of the PstI cleavage site). Probe labelling and hybridization was done as described above. Size was estimated based on marker DNA.
Analysis of insertion of transformed DNA
To obtain the set of 84 bp, 100 bp, 150 bp, 200 bp, 300 bp 400 bp, 500 bp and 1 kb dsDNA, we PCR amplified the DNA using Q5 polymerase (NEB, M0491) and Lambda DNA (NEB, N3011S) as a template. All of the ends of this set of PCR products have AACA. DNA was purified using NucleoSpin PCR clean-up kit (Macherey-Nagel, cat# 740609), dissolved with ddH2O and adjusted to a concentration of 0.6 μM. To obtain the set of 24 bp, 34 bp, 44 bp, 54 bp, 64 bp, 74 bp, dsDNA, the reverse-phase cartridge-purified oligos ordered from Sigma-Aldrich were dissolved with annealing buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 50 mM NaCl) to 200 μM. Equal amounts of complementary oligos were mixed, heated at 95 °C for 5 min and cooled down at room temperature. Every 10 bp of this set of dsDNA has AACA. To analyze the insertions of transformed DNA, the cells were collected when the density reached ~2 × 107 cells/mL in YEP-Raffinose and washed twice with ddH2O. For each transformation, 2 × 108 cells were mixed with 240 μl 50% PEG3350, 36 μl 1 M lithium acetate, water, and 20 μl 100 μM 24 bp-74 bp dsDNA or 64 μl 0.6 μM 84 bp-1kb dsDNA to total 360 μl. The mixture was incubated at 30 °C for 30 min followed by 42 °C for 30 min. The cells were centrifuged and resuspended in ddH2O, spread on YEP-Galactose plates and incubated at 30 °C for 5 days. To test insertion of transformed DNA, the amfiSure PCR Master Mix (GenDEPOT, P0311) was used for colony PCR with the following conditions: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 2 min 30 s. To test the insertion of short DNA (<100 bp), the primers MATa-F3 and mat-Rw3 were used for PCR, and the PCR products were visualized on EtBr containing 3% agarose gel. To test the insertion of long DNA (>100 bp), the primers MATa-F and mat-Rw were used for PCR, and the PCR products were analyzed by electrophoresis (1.2% agarose in 1x TBE buffer) at 120 V for 30 min.
Processing of raw reads from Break-Ins analysis
Paired-end reads were assigned to samples based on the Illumina indexes. Then the custom indexes were used to discharge reads for which the Illumina indexes and the matched custom indexes were not mapped to the same sample. Thereafter, the software BBDuk v38.46 at the BBMap website (https://sourceforge.net/projects/bbmap/) was used to remove PhiX reads with default parameters. Subsequently, low-quality reads were removed while high-quality reads were retained with the average sequencing quality score (Q score) of each read to be at least 25 for bases in the MATa region and 15 for bases in the rest of the region. To calculate the average Q score in each region of a read, the quality symbol for each base in the FASTQ file was converted to the Q-score based on the Quality Score Encoding table provided at the Illumina web site (https://support.illumina.com/help/BaseSpace_OLH_009008/Content/Source/Informatics/BS/QualityScoreEncoding_swBS.htm). We then calculated the mean value of the scores in each region of each read.
Detection of reads carrying insertions of 10 bp or longer
We used the software PEAR v0.9.1171 to merge the forward and reverse reads with default parameters. The merged reads were aligned to the yeast genome (S288C, 2μ plasmid sequence was included). For each pair of reads that could not be merged, e.g., because of no overlap between sequences of the two reads, we aligned the two reads to the genome separately. We used the software BLASTN v2.8.172 to perform the alignment. A pair of merged reads was defined as having an insertion if the merged read were aligned to the MATa sequence but with at least 10 bp insertion at the break point of the MATa sequence. For a pair of reads that could not be merged, we considered it as having an insertion if the insertion portions of the two reads were at the 3’ ends and aligned to different loci that are at least 10 bp and at most 3 kb away from each other on the same chromosome.
Defining unique insertions
If there was no sequencing error, two pairs of sequencing reads derived from a unique insertion event would have the identical sequence, and thus be recognized as a sequencing duplication. To eliminate insertion duplicates that differ in sequence due to sequencing error, we performed additional analysis. We used a 60 bp region (11 bp in the MATa region followed by 19 bp in the insertion donor sequence region at both junctions) of each insertion to identify the sequencing duplications of a unique insertion. The reads that had identical or nearly identical junctions were considered as originating from the same insertion. Specifically, two pairs of reads that aligned to each other based on the 60 bp region with at least 95% identity, at least 95% coverage, and no more than 6 bp mismatches and indels were defined as duplications of one unique insertion.
We next further defined additional duplications based on the genomic locations at which the reads aligned. For each insertion detected at the induced break point at the MATa locus, the inserted sequence might come from a single genomic location (single-donor) or multiple genomic locations (multi-donor) in the genome. For two pairs of reads (2 reads for each sequencing pair and thus 4 reads for two pairs) for the same single donor, we define the distance between their mapped locations at the left side of the insertion site (d1a), the distance between their mapped locations at the right side of the insertion site (d1b), the distance between their mapped locations at the left side of the donor site (d1c), and the distance between their mapped locations at the right side of the donor site (d1d). The two pairs of reads are defined as a duplication of one unique insertion if d1a + d1b ≤ 6 bp and d1c + d1d ≤ 6 bp. For a n-donor insertion, we will define the four distances for each donor i as dia, dib, dic, and did (i = 1, 2, …, n). The two pairs are defined as a duplication if (1) the sum of all dia and dib is no more than 10 and (2) the sum of all dic and did is no more than 10.
Furthermore, among the unique insertions defined in the above two steps, we retrieved each insertion that had less than 10 pairs of reads and had more than 300 bp of sequence in the MATa region plus the donor region. For any two of these retrieved insertions that had a sequence identity greater than 80% with each other, we further defined them as a duplication. Finally, we retrieved each unique insertion that had no more than 10 read pairs in one sample but had more than 500 read pairs in another sample sequenced in the same batch. The insertion in the sample that displayed no more than 10 read pairs was recognized as contamination and thus discarded.
At the end, each unique insertion might have multiple pairs of reads that represent sequencing duplications. We selected one pair of reads to represent each unique insertion. For this purpose, we gave high priority to the read pairs for which the forward and reverse reads had overlap and thus could be merged. Thereafter, between different read pairs that each could be merged, we provide high priority to the pairs that have higher sequencing quality. Finally, between different read pairs that have the same sequencing quality, we gave high priority to the pairs that each have large number of identical copies.
Defining the locations of the donor sequences of insertions
For more than 90% of the insertions, the donor locations could be easily defined based on the mapping result of BLASTN. However, it is more complicated to define the donor locations for a few insertions that belong to one of two categories. For the first category, the insertion sequences were not mapped to any donor location by the BLASTN algorithm. We thus used the BLASTN-short algorithm, which allowed us to further map some of these insertions to donor locations. For the second category, each insertion could be mapped to multiple alternative donors due to the great sequence similarity between the alternative donors. In this scenario, each alternative donor sequence covered a large, but not the full, proportion of the insertion sequence. Among the alternative donors, we select the donor that covered the largest proportion of the insertion sequence. Finally, because each insertion sequence might consist of several different donor sequences, our algorithm counts the number of donors in each insertion. We have not analyzed very rare insertions from proximity of DSB.
Analysis of trimming and microhomology around DSB junction
Trimming of MATa sequence near the junction of an insertion was common. The deletion was defined based on the missing nucleotides in the mapping result generated by BLASTN, which aligned the sequencing read to the original MATa sequence. The insertion was defined based on the extra nucleotides that could not be mapped to the MATa sequence and also could not be mapped to any sequence near the original donor location of the insertion. The microhomology between MATa and insertion donor was defined as sequence mapped to both the terminal region of the donor location and the terminal region of the MATa sequence flanking the break site. The microhomology sequence was defined by requiring perfect (100 % identity) match with both the donor sequence and the MATa sequence.
Defining genomic features of the donor sequences of inserted DNA
We used Bedtools v2.253 to retrieve genomic features that overlap with or are located close to the locations of donor sequences in the genome. Locations of the confirmed origins of replication (ARSes) were collected from the OriDB database (http://cerevisiae.oridb.org/). We collected locations of reference R-loops from a published source29. Known tandem repeats were downloaded from UCSC genome browser as a compacted file provided at the website (https://hgdownload.soe.ucsc.edu/goldenPath/sacCer3/bigZips/chromTrf.tar.gz). All other genomic features, i.e., tRNA, telomere, centromere, were acquired from the SGD database (https://downloads.yeastgenome.org/). To evaluate significance of association between a type of genomic feature and the donor locations of insertions, we performed comparison between the observed locations of the insertion sequences and a set of random control locations. For each genotype, random control locations were generated by shuffling the real observed locations of the insertions using the algorithm BEDTools shuffle73. Therefore, the control locations were generated to have the same sizes as for the locations of the observed insertions.
Overlapped events between insertions and genome features are defined as having at least 1-bp overlap. Distance analysis was based on edge distance between an insertion and its closest genome feature. Insertions coming from Ty retrotransposons, mtDNA, rDNA, MATa or 2 µ plasmid were excluded from these analyses. R-loops, ARS, telomeres, centromeres, tRNAs, and tandem repeats were used for comparison between the observed insertion locations and randomized control locations. Randomized control locations were shuffled by 100,000 times bootstrapping. P values for genomic feature overlap or proximity were calculated by one-sided permutation test.
Frequency of Ty1, mtDNA, rDNA sequence insertions
All inserted sequences derived from the LTR retrotransposon (Ty1) were realigned further against the same Ty1 sequence (YPLWTy1-1). Due to the high divergence of Ty1 insertions, we requested a loose parameter for BLASTN realignments (-word_size 11 -evalue 0.01 -gapopen 5 -penalty −1 -perc_identity 60 -dust no -soft_masking false). Among these alternative alignments, we selected the one that covered the largest proportion of the insertion sequence. In the case that Ty1 inserts could map perfectly to both LTR regions, we selected the right LTR alignment.
Based on the aligned positions of Ty1 inserts, we measured the number of times each Ty1 nucleotide was inserted, nt,i. Frequency of nucleotide inserted at each Ty1 position were calculated per 100,000 independent NHEJ products as 100,000*nt,i /n colonies. Percentage of each nucleotide inserted among all Ty1 insertions were measured by 100* n t,i /n t, total. nt,i as the number of insertion events at the i position, whereas n t, total is the total number of insertion times with all the Ty1 positions. And n colonies is the total number of colonies. The same nucleotide frequency methods but without realignments were applied for inserts from mtDNA and rDNA.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Dr. Peter Thorsness, David Garfinkel, James Haber, Frank Madeo for the gift of plasmids or strains. We thank Dr. James Haber for critical reading of the manuscript, Dr. Mitch McVey for helpful suggestions, Alma Papusha for technical help. This work was funded by grants from the National Institute of Health (GM080600 and GM125650 to G.I, R01AG052507 and R01AG081347 to W.D. and R01HL148338, R01CA278832 and R01GM138407 to K.C.).
Author contributions
Y.Y. (BCM) constructed strains, designed experiments, including the amplicon sequencing method, conducted most of the experiments, and analyzed data. X.W. and K.C. developed the software toolkits and bioinformatic pipelines to map and analyze insertions, and conducted most of the statistical data integration; Y.Y. (HMS) performed an independent analysis of the same data sets; J.F. constructed strains, performed an analysis of Ty1 transposition, Ty1 cDNA, mtDNA amount, and analyzed Ty1 cDNA insertions; G.I., Y.Y., and P.T. designed experiments to study TRP1-mtDNA transfer to nucleus; Q.L. helped with mutant strains constructions and Southern blots, P.H. helped with data interpretation; W.D., R.Y., N.N., B.M. provided expertise on yeast aging and helped with isolation of old yeast cells and yeast lifespan analysis; G.I., Y.Y., X. W., and K.C. designed experiments, analyzed the data, and wrote the manuscript. J.F. edited the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The Break-ins data generated in this study have been deposited in NCBI’s Gene Expression Omnibus under GEO series accession number GSE246469. Source data are provided in this paper.
Code availability
All original code has been deposited on GitHub. The code for insertion detection can be accessed at the following GitHub repository: https://github.com/gucascau/iDSBins.git. For short indel detection, the code can be accessed at this GitHub repository: https://github.com/gucascau/iDSBindel.git. The features related to insertion analysis have been uploaded to a dedicated GitHub repository, which can be found here: https://github.com/gucascau/LargeInsertionFeature.git. Any additional information required to reanalyze the data reported in this paper is available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yang Yu, Xin Wang, Jordan Fox.
Contributor Information
Kaifu Chen, Email: Kaifu.Chen@childrens.harvard.edu.
Grzegorz Ira, Email: gira@bcm.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52147-2.
References
- 1.Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet5, 123–135 (2004). 10.1038/nrg1271 [DOI] [PubMed] [Google Scholar]
- 2.Hazkani-Covo, E., Zeller, R. M. & Martin, W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet6, e1000834 (2010). 10.1371/journal.pgen.1000834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namasivayam, S. et al. Massive invasion of organellar DNA drives nuclear genome evolution in Toxoplasma. Proc. Natl. Acad. Sci. USA120, e2308569120 (2023). [DOI] [PMC free article] [PubMed]
- 4.Kleine, T., Maier, U. G. & Leister, D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev. Plant Biol.60, 115–138 (2009). 10.1146/annurev.arplant.043008.092119 [DOI] [PubMed] [Google Scholar]
- 5.Chatre, L. & Ricchetti, M. Nuclear mitochondrial DNA activates replication in Saccharomyces cerevisiae. PLoS One6, e17235 (2011). 10.1371/journal.pone.0017235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyman, B. C., Cramer, J. H. & Rownd, R. H. The mitochondrial genome of Saccharomyces cerevisiae contains numerous, densely spaced autonomously replicating sequences. Gene26, 223–230 (1983). 10.1016/0378-1119(83)90192-0 [DOI] [PubMed] [Google Scholar]
- 7.Schiestl, R. H., Dominska, M. & Petes, T. D. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol. Cell Biol.13, 2697–2705 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decottignies, A. Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics171, 1535–1548 (2005). 10.1534/genetics.105.046144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricchetti, M., Fairhead, C. & Dujon, B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature402, 96–100 (1999). 10.1038/47076 [DOI] [PubMed] [Google Scholar]
- 10.Teng, S. C., Kim, B. & Gabriel, A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature383, 641–644 (1996). 10.1038/383641a0 [DOI] [PubMed] [Google Scholar]
- 11.Moore, J. K. & Haber, J. E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature383, 644–646 (1996). 10.1038/383644a0 [DOI] [PubMed] [Google Scholar]
- 12.Lebedin, M. et al. Different classes of genomic inserts contribute to human antibody diversity. Proc. Natl Acad. Sci. USA119, e2205470119 (2022). 10.1073/pnas.2205470119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei, W. et al. Nuclear-embedded mitochondrial DNA sequences in 66,083 human genomes. Nature611, 105–114 (2022). 10.1038/s41586-022-05288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou, W. et al. Somatic nuclear mitochondrial DNA insertions are prevalent in the human brain and accumulate over time in fibroblasts. PLoS Biol22, e3002723 (2024). [DOI] [PMC free article] [PubMed]
- 15.Yuan, Y. et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet52, 342–352 (2020). 10.1038/s41588-019-0557-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju, Y. S. et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res25, 814–824 (2015). 10.1101/gr.190470.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner, C. et al. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum. Genet112, 303–309 (2003). 10.1007/s00439-002-0892-2 [DOI] [PubMed] [Google Scholar]
- 18.Kim, J. et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science366, 1531–1536 (2019). 10.1126/science.aav4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature566, 73–78 (2019). 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, J. K. & Haber, J. E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol.16, 2164–2173 (1996). 10.1128/MCB.16.5.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emerson, C. H. et al. Ku DNA end-binding activity promotes repair fidelity and influences end-processing during nonhomologous end-joining in Saccharomyces cerevisiae. Genetics209, 115–128 (2018). 10.1534/genetics.117.300672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, Z., Sunder, S., Nallasivam, S. & Wilson, T. E. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res44, 2769–2781 (2016). 10.1093/nar/gkw013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, Y. et al. Dna2 nuclease deficiency results in large and complex DNA insertions at chromosomal breaks. Nature564, 287–290 (2018). 10.1038/s41586-018-0769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aragon, A. D. et al. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell19, 1271–1280 (2008). 10.1091/mbc.e07-07-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longo, V. D., Shadel, G. S., Kaeberlein, M. & Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab.16, 18–31 (2012). 10.1016/j.cmet.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, T. E. & Lieber, M. R. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem.274, 23599–23609 (1999). 10.1074/jbc.274.33.23599 [DOI] [PubMed] [Google Scholar]
- 27.Richly, E. & Leister, D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol.21, 1081–1084 (2004). 10.1093/molbev/msh110 [DOI] [PubMed] [Google Scholar]
- 28.Ma, Y. & Lieber, M. R. DNA length-dependent cooperative interactions in the binding of Ku to DNA. Biochemistry40, 9638–9646 (2001). 10.1021/bi010932v [DOI] [PubMed] [Google Scholar]
- 29.Wahba, L., Costantino, L., Tan, F. J., Zimmer, A. & Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev.30, 1327–1338 (2016). 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussmann, J. A. et al. Mapping the genetic landscape of DNA double-strand break repair. Cell184, 5653–5669 e5625 (2021). 10.1016/j.cell.2021.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min, J. et al. Mechanisms of insertions at a DNA double-strand break. Mol. Cell83, 2434–2448 e2437 (2023). 10.1016/j.molcel.2023.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng, S. F., Gabriel, A. & Teng, S. C. Proofreading activity of DNA polymerase Pol2 mediates 3’-end processing during nonhomologous end joining in yeast. PLoS Genet4, e1000060 (2008). 10.1371/journal.pgen.1000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabrizio, P., Pozza, F., Pletcher, S. D., Gendron, C. M. & Longo, V. D. Regulation of longevity and stress resistance by Sch9 in yeast. Science292, 288–290 (2001). 10.1126/science.1059497 [DOI] [PubMed] [Google Scholar]
- 34.Shafer, K. S., Hanekamp, T., White, K. H. & Thorsness, P. E. Mechanisms of mitochondrial DNA escape to the nucleus in the yeast Saccharomyces cerevisiae. Curr. Genet36, 183–194 (1999). 10.1007/s002940050489 [DOI] [PubMed] [Google Scholar]
- 35.Riley, J. S. & Tait, S. W. Mitochondrial DNA in inflammation and immunity. EMBO Rep.21, e49799 (2020). 10.15252/embr.201949799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buttner, S. et al. Endonuclease G regulates budding yeast life and death. Mol. Cell25, 233–246 (2007). 10.1016/j.molcel.2006.12.021 [DOI] [PubMed] [Google Scholar]
- 37.Gao, J. et al. Meiotic viral attenuation through an ancestral apoptotic pathway. Proc. Natl Acad. Sci. USA116, 16454–16462 (2019). 10.1073/pnas.1900751116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dake, E., Hofmann, T. J., McIntire, S., Hudson, A. & Zassenhaus, H. P. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem.263, 7691–7702 (1988). 10.1016/S0021-9258(18)68554-0 [DOI] [PubMed] [Google Scholar]
- 39.Dahal, S. et al. Unleashing a novel function of Endonuclease G in mitochondrial genome instability. Elife11, e69916 (2022). [DOI] [PMC free article] [PubMed]
- 40.Cote, J., Renaud, J. & Ruiz-Carrillo, A. Recognition of (dG)n.(dC)n sequences by endonuclease G. Characterization of the calf thymus nuclease. J. Biol. Chem.264, 3301–3310 (1989). 10.1016/S0021-9258(18)94066-4 [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Carrillo, A. & Renaud, J. Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J.6, 401–407 (1987). 10.1002/j.1460-2075.1987.tb04769.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onozawa, M., Goldberg, L. & Aplan, P. D. Landscape of insertion polymorphisms in the human genome. Genome Biol. Evol.7, 960–968 (2015). 10.1093/gbe/evv043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, D., Lloyd, A. H. & Timmis, J. N. Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc. Natl Acad. Sci. USA109, 2444–2448 (2012). 10.1073/pnas.1117890109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leibowitz, M. L., Zhang, C. Z. & Pellman, D. Chromothripsis: a new mechanism for rapid karyotype evolution. Annu Rev. Genet49, 183–211 (2015). 10.1146/annurev-genet-120213-092228 [DOI] [PubMed] [Google Scholar]
- 45.Caro, P. et al. Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion10, 479–486 (2010). 10.1016/j.mito.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 46.Hu, Z. et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev.28, 396–408 (2014). 10.1101/gad.233221.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorsness, P. E. & Fox, T. D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics134, 21–28 (1993). 10.1093/genetics/134.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet52, 331–341 (2020). 10.1038/s41588-019-0576-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dakshinamurthy, A., Nyswaner, K. M., Farabaugh, P. J. & Garfinkel, D. J. BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics185, 1193–1205 (2010). 10.1534/genetics.110.119115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suresh, S. et al. Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon. Mob. DNA6, 22 (2015). 10.1186/s13100-015-0053-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, X. & Gabriel, A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell4, 873–881 (1999). 10.1016/S1097-2765(00)80397-4 [DOI] [PubMed] [Google Scholar]
- 52.Winston, F., Durbin, K. J. & Fink, G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell39, 675–682 (1984). 10.1016/0092-8674(84)90474-4 [DOI] [PubMed] [Google Scholar]
- 53.Sundararajan, A., Lee, B. S. & Garfinkel, D. J. The Rad27 (Fen-1) nuclease inhibits Ty1 mobility in Saccharomyces cerevisiae. Genetics163, 55–67 (2003). 10.1093/genetics/163.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nevo-Caspi, Y. & Kupiec, M. cDNA-mediated Ty recombination can take place in the absence of plus-strand cDNA synthesis, but not in the absence of the integrase protein. Curr. Genet32, 32–40 (1997). 10.1007/s002940050245 [DOI] [PubMed] [Google Scholar]
- 55.Eichinger, D. J. & Boeke, J. D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell54, 955–966 (1988). 10.1016/0092-8674(88)90110-9 [DOI] [PubMed] [Google Scholar]
- 56.Cote, J. & Ruiz-Carrillo, A. Primers for mitochondrial DNA replication generated by endonuclease G. Science261, 765–769 (1993). 10.1126/science.7688144 [DOI] [PubMed] [Google Scholar]
- 57.Foury, F. Endonucleases in yeast mitochondria: apurinic and manganese-stimulated deoxyribonuclease activities in the inner mitochondrial membrane of Saccharomyces cerevisiae. Eur. J. Biochem124, 253–259 (1982). 10.1111/j.1432-1033.1982.tb06585.x [DOI] [PubMed] [Google Scholar]
- 58.Meinema, A. C. et al. DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes. Elife11, e71196 (2022). [DOI] [PMC free article] [PubMed]
- 59.Demeke, M. M., Foulquie-Moreno, M. R., Dumortier, F. & Thevelein, J. M. Rapid evolution of recombinant Saccharomyces cerevisiae for Xylose fermentation through formation of extra-chromosomal circular DNA. PLoS Genet11, e1005010 (2015). 10.1371/journal.pgen.1005010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durkin, K. et al. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature482, 81–84 (2012). 10.1038/nature10757 [DOI] [PubMed] [Google Scholar]
- 61.Lutz-Bonengel, S. et al. Evidence for multi-copy Mega-NUMTs in the human genome. Nucleic Acids Res49, 1517–1531 (2021). 10.1093/nar/gkaa1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, W. et al. Nuclear-mitochondrial DNA segments resemble paternally inherited mitochondrial DNA in humans. Nat. Commun.11, 1740 (2020). 10.1038/s41467-020-15336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider, J. S. et al. Reversible mitochondrial DNA accumulation in nuclei of pluripotent stem cells. Stem Cells Dev.23, 2712–2719 (2014). 10.1089/scd.2013.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu, J., Wei, M., Mirisola, M. G. & Longo, V. D. Assessing chronological aging in Saccharomyces cerevisiae. Methods Mol. Biol.965, 463–472 (2013). 10.1007/978-1-62703-239-1_30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuchiyama, S., Kwan, E., Dang, W., Bedalov, A. & Kennedy, B. K. Sirtuins in yeast: phenotypes and tools. Methods Mol. Biol.1077, 11–37 (2013). 10.1007/978-1-62703-637-5_2 [DOI] [PubMed] [Google Scholar]
- 66.Jo, M. C., Liu, W., Gu, L., Dang, W. & Qin, L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl Acad. Sci. USA112, 9364–9369 (2015). 10.1073/pnas.1510328112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, R., Jo, M. C. & Dang, W. Measuring the replicative lifespan of Saccharomyces cerevisiae using the HYAA microfluidic platform. Methods Mol. Biol.2144, 1–6 (2020). 10.1007/978-1-0716-0592-9_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayle, R. et al. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science349, 742–747 (2015). 10.1126/science.aaa8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zierhut, C. & Diffley, J. F. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J.27, 1875–1885 (2008). 10.1038/emboj.2008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fox, T. D. et al. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol.194, 149–165 (1991). 10.1016/0076-6879(91)94013-3 [DOI] [PubMed] [Google Scholar]
- 71.Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics30, 614–620 (2014). 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye, J., McGinnis, S. & Madden, T. L. BLAST: improvements for better sequence analysis. Nucleic acids Res.34, W6–W9 (2006). 10.1093/nar/gkl164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Break-ins data generated in this study have been deposited in NCBI’s Gene Expression Omnibus under GEO series accession number GSE246469. Source data are provided in this paper.
All original code has been deposited on GitHub. The code for insertion detection can be accessed at the following GitHub repository: https://github.com/gucascau/iDSBins.git. For short indel detection, the code can be accessed at this GitHub repository: https://github.com/gucascau/iDSBindel.git. The features related to insertion analysis have been uploaded to a dedicated GitHub repository, which can be found here: https://github.com/gucascau/LargeInsertionFeature.git. Any additional information required to reanalyze the data reported in this paper is available from the corresponding authors upon request.