Abstract
Background and Purpose
Neonatal encephalopathy (NE) is a neurological syndrome that presents with severe neurological impairments and complications. Hypoxic-ischemic encephalopathy is a major contributor to poor outcomes, being responsible for 50%–80% of admissions to neonatal intensive care units. However, some cases of NE accompanied by hypoxic brain damage cannot be solely attributed to hypoxia-ischemia. We aimed to identify diverse pathogenic genetic variations that may be associated with cases of NE accompanied by hypoxic brain damage rather than hypoxia-ischemia.
Methods
We collected data from 34 patients diagnosed with NE accompanied by hypoxic brain damage over a 10-year period. Patients with the following specific conditions were excluded: 1) premature birth (<32 weeks), 2) no history of hypoxic events, 3) related anomalies, 4) neonatal infections, 5) antenatal or perinatal obstetrical complications, 6) severe hypoxia due to other medical conditions, and 7) early death (within 1 week). A comprehensive review of clinical and radiological features was conducted.
Results
A genetic diagnosis was made in 11 (32.4%) patients, with pathogenic variants being identified in the following 9 genes: CACNA1A (n=2), KCNQ2 (n=2), SCN2A (n=1), SCN8A (n=1), STXBP1 (n=1), NSD1 (n=1), PURA (n=1), ZBTB20 (n=1), and ENG (n=1). No specific treatment outcomes or clinical features other than preterm birth were associated with the results of the genetic analyses. Personalized treatments based on the results of genetic tests were attempted, such as the administration of sodium-channel blockers in patients with KCNQ2 or SCN8A variants and the implementation of a ketogenic diet in patients with STXBP1 or SCN2A mutations, which demonstrated some degree of effectiveness in these patients.
Conclusions
Genetic analyses may help in diagnosing the underlying etiology of NE and concurrent hypoxic brain damage, irrespective of the initial clinical features.
Keywords: targeted gene panel sequencing, neonatal encephalopathy, hypoxic brain damage, hypoxic-ischemic encephalopathy
Graphical Abstract
INTRODUCTION
Neonatal encephalopathy (NE) is a neurological syndrome with clinical features such as low levels of consciousness, abnormalities in muscle tone, neonatal seizures, focal neurological deficits, and depressed respiration.1,2 NE reportedly affects 1–6 of every 1,000 full-term live births, with a mortality rate of 15%–20% during the newborn period. In addition, around 25% of affected patients experience permanent neurological deficits.3,4,5,6 Factors such as maternal antepartum/intrapartum comorbidities, placental abnormalities, hypoxia-ischemia, perinatal infection, coagulopathies or neonatal stroke, metabolic disorders, and genetic/epigenetic abnormalities contribute to the onset of NE either alone or in combination. Neonatal hypoxic-ischemic encephalopathy (HIE) is recognized as the predominant risk factor, accounting for 50%–80% of cases that present in neonatal intensive care unit (NICUs).7,8,9,10,11,12,13
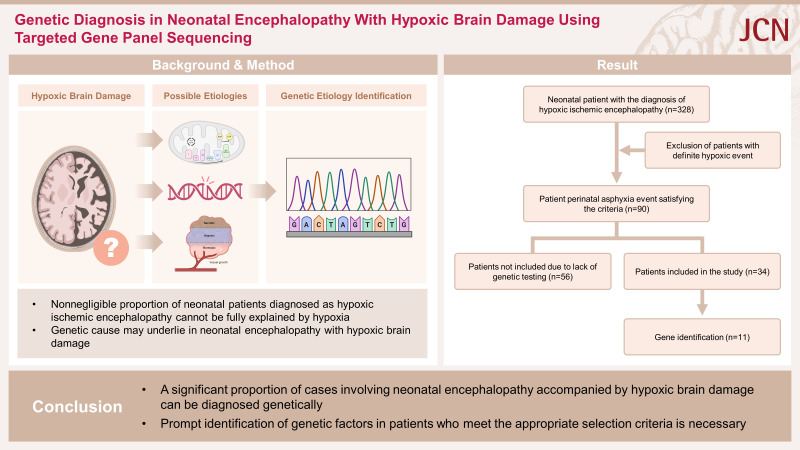
However, a nonnegligible proportion of diagnosed HIE cases cannot be fully explained by a lack of oxygen or blood flow during birth. Moreover, up to 50% of NE cases do not show any signs of perinatal asphyxia, even when neuroimaging findings are abnormal. These patients are often challenging when performing diagnostic workups (including genetic testing), which can lead to delays in them receiving proper treatment and accurate prognostic predictions. For these reasons, previous studies that aimed at identifying the underlying causes of NE have often excluded those with hypoxic brain damage, and the few studies that focused on identifying genetic variations associated with NE accompanied by hypoxic brain damage have lacked the appropriate criteria for patient selection, resulting in inaccuracies in the identified diagnostic rates.14,15,16,17,18
Here we attempted to determine whether various pathogenic genetic variations can be diagnosed in cases of NE accompanied by hypoxic events and brain damage when patients are carefully selected, excluding those with prominent intrapartum and postpartum factors. Moreover, we compared the clinical features of the patients based on the results of genetic testing with the aim of determining whether specific factors are correlated with genetic findings. The findings of this study support the importance of genetic testing methods such as targeted gene panel sequencing and clinical exome sequencing in properly identifying patients with NE accompanied by hypoxic brain damage.
METHODS
Patient selection
This retrospective study identified patients diagnosed with NE between June 2012 and June 2022 at Severance Children’s Hospital. From these patients we specifically selected those diagnosed as NE and exhibiting neuroimaging findings consistent with hypoxic brain damage before reaching a postconceptual age of 1 month. We subsequently excluded patients with the following specific conditions: 1) born extremely (<28 weeks) or very (<32 weeks) preterm, 2) no hypoxic event requiring oxygen support within 24 hours after birth, 3) having related anomalies that could cause hypoxic events, such as cardiac or pulmonary congenital disease, 4) neonatal infections including sepsis and bacterial meningitis, 5) antenatal or perinatal obstetrical complications, such as placental abruption, uterine rupture, cord prolapse, or severe preeclampsia, 6) experiencing severe hypoxia due to other medical conditions, such as emergency operations, or 7) dying within 1 week after birth. The last exclusion criterion was applied due to the difficulty of performing genetic tests under such circumstances. Laboratory tests for inborn metabolic errors, such as plasma amino acids and urine organic acids, were performed to exclude patients with metabolic disorders.
Ultimately 90 patients met the eligibility criteria, of which only 34 had undergone targeted gene panel sequencing. The parents/guardians of the participants provided informed consents, and this study was approved by the Institutional Review Board of Severance Hospital (4-2023-0865).
Clinical and radiological evaluations
Upon arrival at the NICU, clinical data on each patient were carefully collected. The baseline data collection included antenatal details such as the presence of fetal distress (e.g., decreased fetal tone/activity and fetal deceleration/bradycardia) and obstetrical complications (e.g., preterm labor, prolonged labor, premature rupture of membranes, breech presentation, or cephalopelvic disproportion), delivery method, birthweight, neonatal resuscitation procedures (e.g., noninvasive oxygen support, invasive ventilator, or chest compression), sex, presence of neonatal seizures, age at seizure onset (in days), accompanying anomalies, muscle tone, and head circumference.
The patients were subsequently followed up for at least 1 year, during which additional information was obtained such as the tube feeding status due to poor sucking ability, gastrostomy placement status, tracheostomy status, presence of a ventriculoperitoneal shunt, epilepsy development, epilepsy intractability, intellectual disability, neurological motor dysfunction, and mortality.
Motor disability was assessed using the Gross Motor Function Classification System, which is a well-established, five-level, evidence-based tool designed to measure gross motor function in children with cerebral palsy.19 Furthermore, intellectual disability was detected and quantified based on the criteria established by the American Association of Intellectual and Developmental Disorders. We also collected intelligence quotient (IQ) scores on the Wechsler Intelligence Scale for Children (WISC) and the Bayley Scale of Infant Development (BSID) for each patient, and categorized them into the following groups of intellectual disability: mild (WISC: 50≤IQ<70; BSID: 70≤IQ<85), moderate (WISC: 35≤IQ<50; BSID: 55≤IQ<70), severe (WSIC: 20≤IQ<35; BSID: 20≤IQ<55), and profound (WISC: IQ<20; BSID: IQ<20). When developmental examinations could not be performed due to inadequate cooperation, patients were classified as either severe or profound intellectual disability.
Neuroimaging that included cranial ultrasound and brain magnetic resonance imaging (MRI) was performed within 3 days of encephalopathy onset or upon the patient’s arrival at the NICU. The timing for subsequent imaging follow-up was determined based on clinical judgment, with brain MRI scanning typically performed at the postconceptual age of 1 month regardless of the status of previous imaging studies. The MRI protocol encompassed T1-weighted, T2-weighted, diffusion-weighted, and FLAIR (fluid-attenuated inversion recovery) sequences. The presence of hypoxic brain damage was determined based on clinically documented hypoxic events accompanied by radiological findings, taking into consideration relevant information from previous studies.20,21,22 Primary radiographic features of hypoxic brain damage included bilateral or multifocal chances in signal intensity in subcortical regions such as the basal ganglia or thalamus, white matter, cerebral cortex, cerebellum, and brainstem. All imaging findings were carefully reviewed by specialized pediatric neurologists and neuroradiologists.
Targeted gene panel sequencing
We included 4,872 candidate genes associated with neurodevelopment in our gene panel to identify pathogenic variants in patients diagnosed with NE accompanied by hypoxic brain damage. These genes were selected after performing an extensive review of relevant literature and information in the Online Mendelian Inheritance in Man (OMIM) database. A complete list of the genes included in the panel is provided in Supplementary Table 1 (in the online-only Data Supplement).
The DNA library was prepared in accordance with the manufacturer’s protocol. Genomic DNA was extracted from leukocytes in whole blood samples using a QIAamp Blood DNA Mini Kit (QIAGEN, Hilden, Germany) for DNA target preparation. The DNA from each patient was subsequently fragmented and amplified using the polymerase chain reaction to prepare the library. This pooled DNA library was then subjected to massive sequencing using a MiSeq sequencer (Illumina, San Diego, CA, USA) and the MiSeq Reagent Kit (version 2) (300 cycles). The sequenced data were analyzed using BaseSpace (Illumina) and NextGENE (SoftGenetics, State College, PA, USA), and cross-referenced using our custom analysis pipeline. Copy-number variants were assessed using a custom analysis pipeline. Various databases were used for the analysis and variant annotation, including the OMIM, Human Gene Mutation Database, ClinVar, dbSNP, 1000 Genomes, Exome Aggregation Consortium, Exome Sequencing Project, and Korean Reference Genome Database. In four patients, trio sequencing was performed using Sanger sequencing on a 3730 DNA Analyzer with the BigDye Terminator (version 3.1) Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
The sequence variants were interpreted using the American College of Medical Genetics and Genomics Guidance Classification System. We selected variants classified as “pathogenic” or “likely pathogenic” causative variants, which were further confirmed using Sanger sequencing.23
Statistical analysis
To identify the associated factors, we compared the clinical and radiological variables between patients with pathogenic or likely pathogenic variants and those with negative test results from targeted gene panel sequencing. We applied Student’s t-test and the Mann–Whitney U test to continuous variables that did and did not conform to a normal distribution, respectively. Categorical variables were compared between two groups using the chi-square test or Fisher’s exact test. Statistical analyses were performed using R software (version 4.3.1, R Foundation for Statistical Computing, Vienna, Austria), and p values of <0.05 were considered statistically significant. Data were expressed as numbers and percentages, mean and standard-deviation values, or median and interquartile range (IQR) values.
RESULTS
Patient demographics and antenatal features
NE was diagnosed in 428 patients, among which 100 who did not experience any hypoxic events that required oxygen support within 24 hours after birth and 58 with normal radiological findings were excluded from the study. Another 180 patients were further excluded from the study due to the following indications: 1) 58 born extremely or very preterm, 2) 41 with related anomalies, 3) 14 with neonatal infections, 4) 49 with obstetrical complications, 5) 7 who had experienced severe hypoxia due to other medical conditions, and 6) 11 who died early. An additional 56 patients were excluded from our cohort because they had not undergone genetic testing, leading to the final inclusion of 34 patients (20 males and 14 females) (Fig. 1).
Fig. 1. Eligibility criteria and study enrollment flow for patients with neonatal encephalopathy accompanied by hypoxic brain damage.
The diagnostic yield of targeted gene panel sequencing was 32.4% (n=11) in our patient cohort. Notably, no cases of moderate to late preterm birth were observed in those with pathogenic genetic variants (henceforth referred to as the genetic group), while eight (34.8%) patients were not genetically diagnosed (henceforth the nongenetic group) (p=0.034). Vaginal delivery was performed in 4 (36.4%) of the 11 patients in the genetic group and in 7 (30.4%) of the 23 patients in the nongenetic group. In addition, antenatal fetal distress was observed in 11 (32.4%) of the 34 patients in our cohort, with meconium aspiration in 8 (23.5%) patients and accompanying obstetrical complications in 14 (41.2%) patients; the proportions were similar in the genetic and nongenetic groups (Table 1).
Table 1. Baseline demographic and clinical characteristics of patients (<1 month after birth) with neonatal encephalopathy accompanied by hypoxic brain damage (n=34).
| Characteristic | Genetic group (n=11) | Nongenetic group (n=23) | p | ||
|---|---|---|---|---|---|
| Sex, male | 8 (72.7) | 12 (52.2) | 0.295 | ||
| Seizure onset age (days) | 1.00 [2.00] | 1.50 [1.75] | 0.835 | ||
| Vaginal delivery | 4 (36.4) | 7 (30.4) | >0.999 | ||
| Prematurity | 0 (0.0) | 8 (34.8) | 0.034* | ||
| Birthweight (g) | 2,836±588 | 3,126±551 | 0.174 | ||
| Meconium aspiration | 1 (9.1) | 7 (30.4) | 0.228 | ||
| Antenatal fetal distress† | 3 (27.3) | 8 (34.8) | >0.999 | ||
| Antenatal obstetrical complications‡ | 4 (36.4) | 10 (43.5) | >0.999 | ||
| Accompanying anomaly | 0 (0.0) | 3 (13.0) | 0.535 | ||
| Neonatal seizure | 10 (90.9) | 18 (78.3) | 0.638 | ||
| Hypothermia | 1 (9.1) | 3 (13.0) | >0.999 | ||
| Neonatal resuscitation | - | - | - | ||
| Oxygen support | 11 (100.0) | 23 (100.0) | >0.999 | ||
| Chest compression | 0 (0.0) | 2 (8.7) | >0.999 | ||
| Invasive ventilator | 7 (63.6) | 19 (82.6) | 0.388 | ||
| Head circumference | - | - | 0.179 | ||
| Normal | 9 (81.8) | 16 (69.6) | - | ||
| Microcephaly | 1 (9.1) | 7 (30.4) | - | ||
| Macrocephaly | 1 (9.1) | 0 (0.0) | - | ||
| Tonicity | - | - | 0.302 | ||
| Normotonicity | 4 (36.4) | 6 (26.1) | - | ||
| Hypotonicity | 6 (54.5) | 9 (39.1) | - | ||
| Hypertonicity | 1 (9.1) | 8 (34.8) | - | ||
Data are n (%), mean±standard-deviation, or median [interquartile range] values.
*Statistical significance; †Antenatal fetal distress, including decreased fetal tone/activity and fetal deceleration/bradycardia; ‡Antenatal obstetrical complications included preterm labor, prolonged labor, premature rupture of membranes, breech presentation, and cephalopelvic disproportion.
Clinical characteristics based on genetic results
No significant differences in baseline clinical characteristics were detected between the genetic and nongenetic groups. The 31 patients with seizure history in the genetic and nongenetic groups had median ages at seizure onset of 1.00 and 1.50 days (IQR=2.00 and 1.75 days), respectively. Neonatal seizures within 1 month after birth were observed in 10 (90.9%) and 18 (78.3%) patients in the genetic and nongenetic groups, respectively. All 34 patients received at least oxygen support, with 26 (76.5%) receiving this via an invasive ventilator. Hypothermia was induced in four patients: 1 (9.1%) in the genetic group and 3 (13.0%) in the nongenetic group. At birth, the head circumference was within the normal range in most patients, and varying features of muscle tonicity were observed in both the genetic and nongenetic groups. Furthermore, 3 (13.0%) patients in the nongenetic group had coexisting anomalies, comprising clubfoot, amniotic band syndrome, and duodenal atresia in one patient each (Table 1).
Follow-up clinical features were also similar in the two groups. Supportive treatments such as persistent nasogastric tube feeding, gastrostomy placement, and tracheostomy were found to be similarly prevalent in the two groups. Mortality within 1 year of birth was observed in only one patient in the nongenetic group. Moreover, only 3 (8.8%) patients were not diagnosed with epilepsy during the follow-up period, with 16 (47.1%) diagnosed with epilepsy syndrome and 22 (64.7%) experiencing intractable epilepsy. Most of the patients in both groups had severe intellectual and motor disabilities (Table 2).
Table 2. Follow-up (>1 year) clinical characteristics of patients with neonatal encephalopathy accompanied by hypoxic brain damage (n=34).
| Clinical characteristic | Genetic group (n=11) | Nongenetic group (n=23) | p | |
|---|---|---|---|---|
| Poor eye contact | 5 (45.5) | 8 (34.8) | 0.709 | |
| Poor sucking with tube feeding | 6 (54.5) | 11 (47.8) | 0.714 | |
| Gastrostomy | 4 (36.4) | 6 (26.1) | 0.692 | |
| Tracheostomy | 3 (27.3) | 1 (4.3) | 0.089 | |
| Ventriculoperitoneal shunt | 0 (0.0) | 2 (8.7) | >0.999 | |
| Mortality | 0 (0.0) | 1 (4.3) | >0.999 | |
| Epilepsy | - | - | >0.999 | |
| No diagnosis | 1 (9.1) | 2 (8.7) | - | |
| Focal seizure only | 5 (45.5) | 10 (43.5) | - | |
| Focal and generalized seizure | 5 (45.5) | 11 (47.8) | - | |
| ASM | - | - | 0.850 | |
| None | 1 (9.1) | 2 (8.7) | - | |
| Single | 2 (18.2) | 7 (30.4) | - | |
| Multiple | 8 (72.7) | 14 (60.9) | - | |
| Intellectual disability | - | - | 0.523 | |
| Normal to mild | 1 (9.1) | 4 (17.4) | - | |
| Moderate to profound | 10 (90.9) | 19 (82.6) | - | |
| Motor disability | - | - | 0.850 | |
| GMFCS levels I–III | 3 (27.3) | 7 (30.4) | - | |
| GMFCS level IV or V | 8 (72.7) | 16 (69.6) | - | |
Data are n (%) values.
ASM, antiseizure medication; GMFCS, Gross Motor Function Classification System.
Demonstration of patients with genetic variations
Pathogenic variants were identified in 9 genes in the 11 patients in the genetic group, with no copy-number variation identified. The following five genes were found to be associated with developmental and epileptic encephalopathy (DEE): CACNA1A (voltage-gated calcium channel alpha subunit 1A, with mutations c.1841T>C and c.4177G>A in two cases), KCNQ2 (voltage-gated potassium channel subfamily Q member 2, with mutation c.601C>T in two cases), SCN2A (voltage-gated sodium channel alpha subunit 2, with mutation c.5308A>G in one case), SCN8A (voltage-gated sodium channel alpha subunit 8, with mutation c.4398C>A in one case), and STXBP1 (syntaxin-binding protein 1, with mutation c.1216C>T in one case). Three genes were associated with neurodevelopmental disorder: NSD1 (nuclear-receptor-binding SET domain protein 1, with mutation c.3549dupT in one case), PURA (purine-rich element-binding protein A, with mutation c.72dup in one case), and ZBTB20 (zinc finger and BTB-domain-containing 20, with mutation c.1492C>T in one case). In addition, ENG (endoglin, with mutation c.154G>C) was identified as the primary cause of hereditary hemorrhagic telangiectasia in one case. All patients had documented hypoxic events within 24 hours after birth, with radiographically features consistent with hypoxic brain damage (Fig. 2).
Fig. 2. Magnetic resonance imaging findings of patients genetically diagnosed with hypoxic brain damage. A and B: T2- and diffusion-weighted sequences of patients with the SCN8A variant, revealing diffuse excessively large signals and diffusion-restriction foci in the white matter. C and D: T1- and diffusion-weighted sequences of patients with the SCN2A variant, revealing T1 hyperintensities and diffusion restriction in the deep white matter and periventricular area. E and F: T1- and diffusion-weighted sequences of patients with the STXBP1 variant, revealing multifocal T1-weighted hyperintensities and diffusion-restriction areas in the frontal periventricular white matter, basal ganglia, and corpus callosum. G and H: T2- and T1-weighted sequences of patients with the ZBTB20 variant, revealing multifocal hemorrhages and a prominent T2 hyperintensity in the deep white matter.
All patients except the one carrying an NSD1 mutation were diagnosed with epilepsy. Five of the seven patients diagnosed with genetic variants related to DEE had intractable epilepsy with profound intellectual disability, with the other two patients having a CACNA1A mutation. Neonatal seizures were observed in all patients, and phenobarbital was initially selected as the medication. However, little or no response to phenobarbital resulted in multiple antiseizure medications (ASMs) being administered, and a continuous infusion of midazolam was used in one patient with an SCN2A variant. Following a genetic diagnosis, patients received treatments tailored to their specific genetic variants, which demonstrated some effectiveness in controlling seizures.
Three of the four patients diagnosed with genetic mutations associated with neurodevelopmental disorders and hereditary hemorrhagic telangiectasia experienced neonatal seizures, with the exception being the patient with an NSD1 variant. However, the symptoms of all patients except those with the ZBTB20 mutation were effectively controlled with either one or two ASMs, with phenobarbital being the initial treatment in all cases. Abnormal muscle tone was observed in all patients, but the head circumference was normal except for the patient with the NSD1 variant, who exhibited macrocephaly. Two patients with PURA-related neurodevelopmental disorder and hereditary hemorrhagic telangiectasia were diagnosed with profound intellectual disability, while the other two were diagnosed with moderate (NSD1) or borderline (ZBTB20) intellectual disability. All patients were finally treated using a multidisciplinary approach and family consultations (Table 3).
Table 3. Clinical and radiological features of NE accompanied by hypoxic brain damage, with diagnosed pathogenic genetic variants.
| Patient | Gene | ACMG classification | Nucleotide change | Amino-acid change | Sex | Ventilator duration (days) | Seizure onset age (days) | ASM | Epilepsy syndrome | Eye contact | Ketogenic diet | MRI findings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | ENG | Likely pathogenic | c.154G>C | p.Gly52Arg | M | 191 | 1 | Single | - | Y | N | Diffuse excessively large signals in WM ICH of posterior cingulate |
| P2 | CACNA1A | Likely pathogenic | c.1841T>C | p.Ile614Thr | F | - | 2 | Multiple | - | Y | N | T2 hyperintensity bilaterally in anterior temporal lobes |
| P3 | CACNA1A | Likely pathogenic | c.4177G>A | p.Val1393Met | M | - | 1 | Multiple | - | Y | E | T1 hyperintensity bilaterally in globus pallidum |
| P4 | KCNQ2 | Pathogenic | c.601C>T | p.Arg201Cys | M | 27 | 3 | Multiple | EIEE → IS | N | N/E | Diffuse excessively large signals in periventricular WM with mild brain atrophy |
| P5 | KCNQ2 | Pathogenic | c.601C>T | p.Arg201Cys | M | 4 | 1 | Multiple | EIEE | N | N/E | CC thinning Diffuse excessively large signals in WM |
| P6 | NSD1 | Likely pathogenic | c.3549dupT | p.Glu1184Ter | M | - | - | N | - | Y | N | CC thinning Diffuse excessively large signals in WM |
| P7 | PURA | Likely pathogenic | c.72dup | p.Gly25ArgfsTer176 | M | 21 | 3 | Multiple | - | N | N | T2 hyperintensity bilaterally in periventricular WM and BG |
| P8 | SCN2A | Pathogenic | c.5308A>G | p.Met1770Val | M | 34 | 1 | Multiple | EIEE → IS | N | E | Multiple T1 hyperintensities in deep WM |
| P9 | SCN8A | Likely pathogenic | c.4398C>A | p.Asn1466Lys | F | 2 | 2 | Multiple | LGS | N | N/E | Diffusion restriction bilaterally in parietal WM |
| P10 | STXBP1 | Likely pathogenic | c.1216C>T | p.Arg406Cys | M | - | 7 | Multiple | IS | Y | E | Diffusion restriction bilaterally in frontal periventricular WM |
| P11 | ZBTB20 | Pathogenic | c.1492C>T | p.Gln498Ter | F | 25 | 1 | Single | - | Y | N | Multifocal hemorrhage T2 hyperintensity in WM |
ACMG, American College of Medical Genetics; ASM, antiseizure medication; BG, basal ganglia; CC, corpus callosum; E, effective; EIEE, early infantile epileptic encephalopathy; F, female; ICH, intracerebral hemorrhage; IS, infantile spasm; LGS, Lennox-Gastaut syndrome; M, male; MRI, magnetic resonance imaging; N, no; NE, neonatal encephalopathy; N/E, not effective; WM, white matter; Y, yes.
DISCUSSION
Neonatal-onset HIE is the predominant cause of death or persistent neurological impairment in infants born at or close to term. The reported mortality rate ranges from 20% to 50%, and 25%–60% of surviving infants experience long-term neurological complications such as cerebral palsy, epilepsy, intellectual disability, and learning disabilities, which necessitates the accurate identification of the pathogenesis in order to improve treatment outcomes.20,24,25,26 Similar demographic characteristics were observed in our patient cohort, signifying that our study had little or almost no patient selection bias when performing the genetic analyses.
Few studies have investigated the genetics of NE, particularly in cases of hypoxic brain damage. However, 32% of NE cases with hypoxic brain damage are identified through genetic diagnosis. Even accounting for the 56 patients who did not undergo genetic testing as a nongenetic group, a minimum of 12% of our cohort displayed genetic susceptibility. Various genes linked to distinct molecular functions and biological processes were identified as the genetic basis in our patients, highlighting the diverse genetic nature of NE. Our results underscore the importance of genetic investigations, not only for neonatal patients without hypoxic events and brain damage but also for those with such damage when appropriate criteria are met.
Furthermore, no notable clinical differences were observed between the patients with and without pathogenic mutations, except for a higher incidence of prematurity in the latter group. Nonetheless, the respiratory failure experienced by patients with a moderately to late preterm birth is generally mild, resulting in low rates of complications.27,28,29 Consequently, excluding these patients from the genetic analyses might oversimplify the problem. Therefore, we suggest performing DNA sequencing in patients with an initial presentation of NE accompanied by hypoxic brain damage regardless of the baseline clinical findings.
Previous studies have established that genetic factors play a significant role in a considerable proportion of individuals diagnosed with epileptic syndromes such as early-onset epileptic encephalopathy with burst suppression (EOEE-BS), with genetic variations accounting for approximately 65% of cases.30,31,32 Therefore, if these conditions were prevalent within the dataset, they might have influenced the findings of the genetic analyses of our cohort. However, only a small proportion (14.7%, n=5) of the 34 patients were identified as having EOEE-BS during the neonatal phase, and most patients presented with nonspecific electroencephalogram (EEG) patterns, which consisted of multifocal epileptic discharges accompanied by slow and disorganized background rhythms. Consequently, it is essential to conduct genetic analyses promptly in these patients, even in the presence of nonspecific EEG findings.
Numerous genes linked to channelopathies, including KCNQ2, SCN1A, SCN2A, SCN8A, KCNT1, and CACNA1A, are known to contribute to the onset of NE and potentially constitute a significant proportion of the causative genes.14,15,33 Our study produced equivalent findings, with more than half of the patients diagnosed with pathogenic variants of these genes. Given that the molecular functions of these genes do not align with the pathological mechanisms linked to hypoxic brain damage, we assumed that the frequent identification of variations in these genes in cases of neonatal hypoxic brain damage may stem from either the early onset of severe neurological dysfunctions that trigger hypoxic events or the resemblance of neuroimaging findings to those of HIE when variations are present in these genes. In addition to the genes related to channelopathy, one patient was diagnosed with STXBP1 encephalopathy, which is also concordant with previous studies.15
Individualized treatments were administered to the patients based on which specific pathogenic genetic mutations were identified. Sodium-channel blockers such as oxcarbazepine or phenytoin were administered to patients with KCNQ2 variants, lamotrigine was initiated in patients with CACNA1A mutations, and a ketogenic diet was implemented in patients with SCN2A, SCN8A, and STXBP1 variants.34,35,36,37,38 While exhibiting some variability, these treatments were effective in managing intractable epilepsy among these patients, except in a case involving SCN8A encephalopathy, where a ketogenic diet showed no effect and so a sodium-channel blocker was employed. Nonetheless, the cessation of recurrent seizures did not translate into sustained favorable outcomes over the long term, such as the amelioration of intellectual disabilities or neurological motor impairments.
In addition to the genes associated with DEE, our patient cohort carried several other gene mutations that could make individuals susceptible to NE, which is accompanied by hypoxic brain damage. We identified a specific case with a pathogenic NSD1 variant that exhibited clinical features closely resembling those of Sotos syndrome, including hypotonia, macrocephaly, intellectual impairment, and motor dysfunction.39 Our findings are further supported by a few cases in which early respiratory difficulty was observed at birth, similar to the situation in our case.40,41 Two other cases associated with neurodevelopmental disorder consisted of Primrose syndrome with pathogenic ZBTB20 variants and PURA-related neurodevelopmental disorder. Although corpus callosum abnormality was not identified in our patient with the ZBTB20 mutation, as in previous studies, there were other accompanying clinical characteristics such as intellectual disability, behavioral issues, and epilepsy.42,43 In the case of the PURA variant, recurrent central apnea, hypotonia, and early-onset seizures with progression to profound mental retardation were observed, which is consistent with previously reported findings.44,45
Finally, we detected a pathogenic variant of a gene associated with the vascular endothelium glycoprotein endoglin (i.e., ENG). Given its recognized involvement in the regulation of angiogenesis, it is conceivable that a pathogenic mutation in this gene directly contributes to the occurrence of hypoxic brain damage. Furthermore, a few studies examining the molecular pathogenesis underlying cerebral ischemia have suggested a connection to this gene.46,47 Therefore, future research into the contribution of this gene to the development of hypoxic brain damage is strongly recommended. In our patient, severe perinatal asphyxia with MRI findings of diffuse excessively large signals in the white matter and intracerebral hemorrhage of the posterior cingulate were observed. The patient had a family history of cerebral hemorrhage, and genetic analyses of both parents revealed that the pathogenic variant identified in the patient was identical to the maternal variant. This finding prompted family counseling for genetic screening during prenatal care as part of the preparation for future pregnancies.
Our study had certain limitations, including the smallness of the sample of 34 patients, since many patients were excluded due to the lack of genetic testing. In addition, the study design was retrospective and further genetic examinations such as whole-exome or whole-genome sequencing were not conducted. Nonetheless, the demographic characteristics of the patient cohort resembled those of previously reported clinical cohorts, implying that selection bias might have been negligible. Furthermore, given the scarcity of studies in this area, we believe that our study represents a significant contribution to the literature.
In summary, this study has demonstrated that a significant proportion of cases involving NE accompanied by hypoxic brain damage can be diagnosed genetically, regardless of the early manifestation of clinical characteristics. Therefore, we strongly advise the prompt identification of genetic factors in patients who meet the appropriate selection criteria.
Footnotes
- Conceptualization: Sangbo Lee, Ara Ko, Hoon-Chul Kang.
- Data curation: Sangbo Lee, Se Hee Kim, Joon Soo Lee, Heung Dong Kim.
- Formal analysis: Sangbo Lee, Ara Ko, Hoon-Chul Kang.
- Methodology: Sangbo Lee.
- Resources: Se Hee Kim, Joon Soo Lee, Heung Dong Kim.
- Supervision: Ara Ko, Hoon-Chul Kang.
- Validation: Ara Ko, Hoon-Chul Kang.
- Writing—original draft: Sangbo Lee.
- Writing—review & editing: all authors.
Conflicts of Interest: Hoon-Chul Kang, a contributing editor of the Journal of Clinical Neurology, was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
Funding Statement: This research was supported by the grant of the MD-Phd/Medical Scientist Training Program through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea, funded by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2022R1A2C1012522), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI21C1659), and the Team Science Award of Yonsei University College of Medicine (6-2021-0007).
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Supplementary Materials
The online-only Data Supplement is available with this article at https://doi.org/10.3988/jcn.2023.0500.
Genes included in the panel of targeted gene sequencing
References
- 1.Leviton A, Nelson KB. Problems with definitions and classifications of newborn encephalopathy. Pediatr Neurol. 1992;8:85–90. doi: 10.1016/0887-8994(92)90026-u. [DOI] [PubMed] [Google Scholar]
- 2.Ferriero DM. Neonatal brain injury. N Engl J Med. 2004;351:1985–1995. doi: 10.1056/NEJMra041996. [DOI] [PubMed] [Google Scholar]
- 3.Brown JK, Purvis RJ, Forfar JO, Cockburn F. Neurological aspects of perinatal asphyxia. Dev Med Child Neurol. 1974;16:567–580. doi: 10.1111/j.1469-8749.1974.tb04176.x. [DOI] [PubMed] [Google Scholar]
- 4.Levene ML, Kornberg J, Williams TH. The incidence and severity of post-asphyxial encephalopathy in full-term infants. Early Hum Dev. 1985;11:21–26. doi: 10.1016/0378-3782(85)90115-x. [DOI] [PubMed] [Google Scholar]
- 5.Volpe JJ. Neurology of the newborn. 4th ed. Philadelphia: WB Saunders; 2001. p. 25. [Google Scholar]
- 6.Kurinczuk JJ, White-Koning M, Badawi N. Epidemiology of neonatal encephalopathy and hypoxic-ischaemic encephalopathy. Early Hum Dev. 2010;86:329–338. doi: 10.1016/j.earlhumdev.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Badawi N, Kurinczuk JJ, Keogh JM, Alessandri LM, O’Sullivan F, Burton PR, et al. Intrapartum risk factors for newborn encephalopathy: the Western Australian case-control study. BMJ. 1998;317:1554–1558. doi: 10.1136/bmj.317.7172.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badawi N, Kurinczuk JJ, Keogh JM, Alessandri LM, O’Sullivan F, Burton PR, et al. Antepartum risk factors for newborn encephalopathy: the Western Australian case-control study. BMJ. 1998;317:1549–1553. doi: 10.1136/bmj.317.7172.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blume HK, Li CI, Loch CM, Koepsell TD. Intrapartum fever and chorioamnionitis as risks for encephalopathy in term newborns: a case-control study. Dev Med Child Neurol. 2008;50:19–24. doi: 10.1111/j.1469-8749.2007.02007.x. [DOI] [PubMed] [Google Scholar]
- 10.Nelson KB, Bingham P, Edwards EM, Horbar JD, Kenny MJ, Inder T, et al. Antecedents of neonatal encephalopathy in the Vermont Oxford network encephalopathy registry. Pediatrics. 2012;130:878–886. doi: 10.1542/peds.2012-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volpe JJ. Neonatal encephalopathy: an inadequate term for hypoxic-ischemic encephalopathy. Ann Neurol. 2012;72:156–166. doi: 10.1002/ana.23647. [DOI] [PubMed] [Google Scholar]
- 12.Aslam S, Strickland T, Molloy EJ. Neonatal encephalopathy: need for recognition of multiple etiologies for optimal management. Front Pediatr. 2019;7:142. doi: 10.3389/fped.2019.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandoval Karamian AG, Mercimek-Andrews S, Mohammad K, Molloy EJ, Chang T, Chau V, et al. Neonatal encephalopathy: etiologies other than hypoxic-ischemic encephalopathy. Semin Fetal Neonatal Med. 2021;26:101272. doi: 10.1016/j.siny.2021.101272. [DOI] [PubMed] [Google Scholar]
- 14.Bruun TUJ, DesRoches CL, Wilson D, Chau V, Nakagawa T, Yamasaki M, et al. Prospective cohort study for identification of underlying genetic causes in neonatal encephalopathy using whole-exome sequencing. Genet Med. 2018;20:486–494. doi: 10.1038/gim.2017.129. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Chen X, Liu X, Dong X, Ye C, Deng D, et al. Clinical features and underlying genetic causes in neonatal encephalopathy: a large cohort study. Clin Genet. 2020;98:365–373. doi: 10.1111/cge.13818. [DOI] [PubMed] [Google Scholar]
- 16.Xiao TT, Yang L, Wu BB, Peng XM, Wang HJ, Cheng GQ, et al. [Genotype and phenotype analysis of neonates with neonatal encephalopathy complicated with perinatal hypoxic event] Zhonghua Er Ke Za Zhi. 2021;59:280–285. doi: 10.3760/cma.j.cn112140-20201130-01065. Chinese. [DOI] [PubMed] [Google Scholar]
- 17.Lenahan A, Mietzsch U, Wood TR, Callahan KP, Weiss EM, Miller DE, et al. Characteristics, genetic testing, and diagnoses of infants with neonatal encephalopathy not due to hypoxic ischemic encephalopathy: a cohort study. J Pediatr. 2023;260:113533. doi: 10.1016/j.jpeds.2023.113533. [DOI] [PubMed] [Google Scholar]
- 18.Woodward KE, Murthy P, Mineyko A, Mohammad K, Esser MJ. Identifying genetic susceptibility in neonates with hypoxic-ischemic encephalopathy: a retrospective case series. J Child Neurol. 2023;38:16–24. doi: 10.1177/08830738221147805. [DOI] [PubMed] [Google Scholar]
- 19.Palisano R, Rosenbaum P, Walter S, Russell D, Wood E, Galuppi B. Development and reliability of a system to classify gross motor function in children with cerebral palsy. Dev Med Child Neurol. 1997;39:214–223. doi: 10.1111/j.1469-8749.1997.tb07414.x. [DOI] [PubMed] [Google Scholar]
- 20.Douglas-Escobar M, Weiss MD. Hypoxic-ischemic encephalopathy: a review for the clinician. JAMA Pediatr. 2015;169:397–403. doi: 10.1001/jamapediatrics.2014.3269. [DOI] [PubMed] [Google Scholar]
- 21.Trivedi SB, Vesoulis ZA, Rao R, Liao SM, Shimony JS, McKinstry RC, et al. A validated clinical MRI injury scoring system in neonatal hypoxic-ischemic encephalopathy. Pediatr Radiol. 2017;47:1491–1499. doi: 10.1007/s00247-017-3893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisnowski JL, Wintermark P, Bonifacio SL, Smyser CD, Barkovich AJ, Edwards AD, et al. Neuroimaging in the term newborn with neonatal encephalopathy. Semin Fetal Neonatal Med. 2021;26:101304. doi: 10.1016/j.siny.2021.101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahearne CE, Boylan GB, Murray DM. Short and long term prognosis in perinatal asphyxia: an update. World J Clin Pediatr. 2016;5:67–74. doi: 10.5409/wjcp.v5.i1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkholy UM, Abdalmonem N, Zaki A, Ali YF, Mohamed SA, Abdelsalam NI, et al. Early predictors of brain damage in full-term newborns with hypoxic ischemic encephalopathy. Neuropsychiatr Dis Treat. 2017;13:2133–2139. doi: 10.2147/NDT.S144225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim GH. Which factors predict outcomes of neonates with hypoxic-ischemic encephalopathy following therapeutic hypothermia? Clin Exp Pediatr. 2021;64:169–171. doi: 10.3345/cep.2020.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H, Xu F, Xiong H, Kang W, Bai Q, Zhang Y, et al. Characteristics of respiratory distress syndrome in infants of different gestational ages. Lung. 2013;191:425–433. doi: 10.1007/s00408-013-9475-3. [DOI] [PubMed] [Google Scholar]
- 28.Debillon T, Tourneux P, Guellec I, Jarreau PH, Flamant C. Respiratory distress management in moderate and late preterm infants: the NEOBS study. Arch Pediatr. 2021;28:392–397. doi: 10.1016/j.arcped.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Smyrni N, Koutsaki M, Petra M, Nikaina E, Gontika M, Strataki H, et al. Moderately and late preterm infants: short- and long-term outcomes from a registry-based cohort. Front Neurol. 2021;12:628066. doi: 10.3389/fneur.2021.628066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko A, Youn SE, Kim SH, Lee JS, Kim S, Choi JR, et al. Targeted gene panel and genotype-phenotype correlation in children with developmental and epileptic encephalopathy. Epilepsy Res. 2018;141:48–55. doi: 10.1016/j.eplepsyres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Kim SH, Kim B, Lee ST, Choi JR, Kim HD, et al. Genetic diagnosis and clinical characteristics by etiological classification in early-onset epileptic encephalopathy with burst suppression pattern. Epilepsy Res. 2020;163:106323. doi: 10.1016/j.eplepsyres.2020.106323. [DOI] [PubMed] [Google Scholar]
- 33.Hayashida T, Saito Y, Ishii A, Yamada H, Itakura A, Minato T, et al. CACNA1A-related early-onset encephalopathy with myoclonic epilepsy: a case report. Brain Dev. 2018;40:130–133. doi: 10.1016/j.braindev.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Numis AL, Angriman M, Sullivan JE, Lewis AJ, Striano P, Nabbout R, et al. KCNQ2 encephalopathy: delineation of the electroclinical phenotype and treatment response. Neurology. 2014;82:368–370. doi: 10.1212/WNL.0000000000000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howell KB, McMahon JM, Carvill GL, Tambunan D, Mackay MT, Rodriguez-Casero V, et al. SCN2A encephalopathy: a major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85:958–966. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardella E, Marini C, Trivisano M, Fitzgerald MP, Alber M, Howell KB, et al. The phenotype of SCN8A developmental and epileptic encephalopathy. Neurology. 2018;91:e1112–e1124. doi: 10.1212/WNL.0000000000006199. [DOI] [PubMed] [Google Scholar]
- 37.Le Roux M, Barth M, Gueden S, Desbordes de Cepoy P, Aeby A, Vilain C, et al. CACNA1A-associated epilepsy: electroclinical findings and treatment response on seizures in 18 patients. Eur J Paediatr Neurol. 2021;33:75–85. doi: 10.1016/j.ejpn.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Thalwitzer KM, Driedger JH, Xian J, Saffari A, Zacher P, Bölsterli BK, et al. Natural history and developmental trajectories of individuals with disease-causing variants in STXBP1. Neurology. 2023;101:e879–e891. doi: 10.1212/WNL.0000000000207550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatton-Brown K, Cole TRP, Rahman N. Sotos Syndrome. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al., editors. GeneReviews® [Internet] Seattle: University of Washington, Seattle; 2022. [cited 2023 Oct 23]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1479/ [Google Scholar]
- 40.Singh P, Suryawanshi P, Garegrat R, Malshe N. Neonatal Sotos syndrome: a novel frameshift mutation of the NSD1 gene associated with neonatal encephalopathy presenting without overgrowth. J Pediatr Neurol. 2024;22:56–59. [Google Scholar]
- 41.Lu DF, Tong XM, Liu YF. A case of Sotos syndrome in a preterm infant with severe bronchopulmonary dysplasia and congenital heart disease. Children (Basel) 2023;10:1111. doi: 10.3390/children10071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alby C, Boutaud L, Bessières B, Serre V, Rio M, Cormier-Daire V, et al. Novel de novo ZBTB20 mutations in three cases with Primrose syndrome and constant corpus callosum anomalies. Am J Med Genet A. 2018;176:1091–1098. doi: 10.1002/ajmg.a.38684. [DOI] [PubMed] [Google Scholar]
- 43.Cleaver R, Berg J, Craft E, Foster A, Gibbons RJ, Hobson E, et al. Refining the Primrose syndrome phenotype: a study of five patients with ZBTB20 de novo variants and a review of the literature. Am J Med Genet A. 2019;179:344–349. doi: 10.1002/ajmg.a.61024. [DOI] [PubMed] [Google Scholar]
- 44.Reijnders MRF, Leventer RJ, Lee BH, Baralle D, Selber P, Paciorkowski AR, et al. PURA-Related Neurodevelopmental Disorders. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al., editors. GeneReviews® [Internet] Seattle: University of Washington, Seattle; 2017. [cited 2023 Oct 23]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK426063/ [PubMed] [Google Scholar]
- 45.Mishra S, Girisha KM, Shukla A. Expanding the phenotype of PURA-related neurodevelopmental disorder: a close differential diagnosis of infantile hypotonia with psychomotor retardation and characteristic facies. Clin Dysmorphol. 2021;30:1–5. doi: 10.1097/MCD.0000000000000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Zhou HS, Chen TB. An integrated analysis of hypoxic-ischemic encephalopathy-related cell sequencing outcomes via genes network construction. Ibrain. 2022;8:78–92. doi: 10.1002/ibra.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes included in the panel of targeted gene sequencing
Data Availability Statement
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.