Summary
Intracellular trafficking of fatty acids (FAs) between organelles is critical for cells to adjust their metabolism in response to stimuli such as exercise, fasting, and cold exposure. Here, we describe a protocol to monitor trafficking of FAs from lipid droplets to mitochondria. We describe the labeling of organelles in cultured C2C12 myoblasts with transfection and dyes. We detail a pulse-chase labeling paradigm using a fluorescent FA analog, live-cell imaging to visualize trafficking of FAs, and steps to quantify FA trafficking.
For complete details on the use and execution of this protocol, please refer to Miner et al.1
Subject areas: Cell Biology, Metabolism, Microscopy
Graphical abstract

Highlights
-
•
Guidance on culturing and transfection of cells with a fluorescent mitochondria marker
-
•
Steps for pulse-chase labeling cells with BODIPY 558/568 C12 (Red C12)
-
•
Live imaging of starvation-induced lipid droplet to mitochondria fatty acid trafficking
-
•
Guidance on organelle segmentation and quantification of fatty acid trafficking
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Intracellular trafficking of fatty acids (FAs) between organelles is critical for cells to adjust their metabolism in response to stimuli such as exercise, fasting, and cold exposure. Here, we describe a protocol to monitor trafficking of FAs from lipid droplets to mitochondria. We describe the labeling of organelles in cultured C2C12 myoblasts with transfection and dyes. We detail a pulse-chase labeling paradigm using a fluorescent FA analog, live-cell imaging to visualize trafficking of FAs, and steps to quantify FA trafficking.
Before you begin
The protocol below describes the specific steps for monitoring fatty acid trafficking from lipid droplets to mitochondria in C2C12 myoblast cells. However, we have also used this protocol in mouse embryonic fibroblasts and U-2 OS cells. For the use of other cell lines, we recommend optimizing experimental conditions. The protocol can also be adapted to monitor fatty acid trafficking to other organelles, such as endoplasmic reticulum or peroxisomes.
Culture C2C12 cells
Timing: 2 days
-
1.Passage Cells.
-
a.Passage C2C12 myoblasts when 80% confluent. Culturing in a 10 cm cell culture dish is used as an example here.
-
b.Aspirate to remove media from cell culture dish.
-
c.Wash cells with 2 mL of PBS to remove remaining media.
-
d.Aspirate PBS and add 2 mL of 0.05% trypsin-EDTA solution to cell.
-
e.Incubate the cells at 37°C for 5 min until cells detach from the surface of the dish.
-
f.Add 8 mL of complete medium (CM: DMEM supplemented with 10% fetal bovine serum and 1% L-glutamine) with phenol red and 1% penicillin-streptomycin, to neutralize the trypsin.
-
g.Triturate cells to fully disperse cells and ensure detachment from the surface of the dish.
-
h.Seed cells into a new 10 cm dish containing 9.5 mL of complete medium, with phenol red and 1% penicillin-streptomycin, with a split ratio of 1:20.
-
i.Incubate the cells at 37°C with appropriate humidity and 5% CO2.
-
a.
CRITICAL: Ensure C2C12 myoblasts do not reach confluency to prevent myotube formation.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Lipofectamine 2000 transfection reagent | Thermo Fisher Scientific | Cat#11668019 |
| Fibronectin | MilliporeSigma | Cat#F2006 |
| BODIPY 665/676 | Thermo Fisher Scientific | Cat#B3932 |
| BODIPY 558/568 C12 | Thermo Fisher Scientific | Cat#D3835 |
| DMEM with phenol red | Corning | Cat# 15017CV |
| DMEM without phenol red | Corning | Cat# 15013CV |
| Trypsin | Corning | Cat# 25051CI |
| L-glutamine | Corning | Cat# 25005CI |
| Pen/strep | Gibco | Cat# 15140122 |
| PBS | Corning | Cat# 21031CV |
| HBSS (with calcium, magnesium, and glucose) | Gibco | Cat# 14025092 |
| Fetal bovine serum | Corning | Cat# 35015CV |
| Experimental models: Cell lines | ||
| Mouse: C2C12 cell line | ATCC | CRL 1772 |
| Recombinant DNA | ||
| Mito-GFP | Jennifer Lippincott-Schwartz2 | N/A |
| mEmerald-Mito-7 | Michael Davidson3 | Addgene 54160 |
| Software and algorithms | ||
| ImageJ | NIH4 | https://imagej.net/ij/index.html |
| Zen 2 | Carl Zeiss | https://www.micro-shop.zeiss.com/en/us/softwarefinder/software-categories/zen-blue/ |
| Other | ||
| 8-well chamber slide | Cellvis | C8-1.5H-N |
Materials and equipment
-
•
BODIPY 558/568 C12 is critical for this assay. Additional fluorescent fatty acid analogs are available; however, their trafficking and metabolism by the cell may differ and therefore may not be directly compatible with this method.
-
•
All other reagents, software, and equipment can be substituted for equivalent alternatives.
Step-by-step method details
Culturing and transfection of cells
Timing: 2 days
To monitor trafficking of fatty acids to mitochondria, cells need to be transfected with a fluorescent mitochondria marker. An 8-well glass coverslip bottom chamber slide is used as an example here.
-
1.Seed Cells.
-
a.Coat wells with 200 μL fibronectin (diluted 1:10 with sterile PBS) and incubate chamber slide at 37°C for 15 min.
-
b.Remove fibronectin from wells.
-
c.Seed cells into chamber slide wells (3–5 × 104 cells/well). The total culture medium volume should be 200 μL.
-
d.Incubate the cells at 37°C with appropriate humidity and 5% CO2 for 18–24 h.
-
a.
CRITICAL: Optimize the cell number for your experimental setup. Cells should be 60%–70% confluent the next day.
-
2.Plasmid Transfection.
-
a.Prepare DNA dilution: Dilute 50 ng of Mito-GFP or mEmerald-Mito-7 plasmid in 50 μL Opti-MEM. Mix by tapping the tube 3–4 times rather than repeated pipetting, to prevent shearing of DNA.
-
b.Prepare Lipofectamine dilution: Dilute 0.1 μL of Lipofectamine 2000 in 50 μL Opti-MEM.Note: DNA and Lipofectamine dilutions are for a single well. Reactions can be scaled for the number of experimental wells. It is recommended to scale up to avoid pipetting amounts smaller than 0.2 μL. If transfecting additional plasmids, ensure DNA: Lipofectamine ratio is maintained at 1 μg DNA: 2 μL Lipofectamine.
-
c.Incubate DNA dilution and Lipofectamine dilution at 20°C–25°C for 5 min.
-
d.Pipette diluted DNA into diluted Lipofectamine and combine by tapping the tube to mix. Incubate at 20°C–25°C for 15 min.
-
e.Aspirate culture medium from well and replace with 100 μL Opti-MEM.
-
f.Add DNA-Lipofectamine mixture to well, to make a total volume of 200 μL/well.
-
g.Gently swirl the chamber slide to mix and incubate cells at 37°C with appropriate humidity and 5% CO2 for 4–6 h.
-
a.
Pulse-chase labeling cells with BODIPY 558/568 C12 (red C12)
Timing: 1 day
Cells are pulse-chased with a fluorescent fatty acid analog, BODIPY 558/568 C12 (Red C12), resulting in accumulation in lipid droplets. Lipid droplets are concurrently labeled with BODIPY 665/676 to monitor initial Red C12 localization.
-
3.Pulse cells with Red C12 and label lipid droplets with BODIPY 665/676.
-
a.Prepare labeling medium: Complete medium containing 5 μM Red C12 and 50 ng/mL BODIPY 665/676.
-
b.Aspirate culture medium and replace with freshly prepared labeling medium.
-
c.Incubate the cells at 37°C with appropriate humidity and 5% CO2 for 16 h.
-
a.
Note: Following transfection with Lipofectamine 2000, complete medium without antibiotics and without phenol red should be used.
-
4.Cell Chase.
-
a.Aspirate labeling medium from wells.
-
b.Wash cells three times with complete medium.
-
c.Aspirate wash and replace with complete medium containing 50 ng/mL BODIPY 665/676 but no Red C12.
-
d.Transfer slide to a microscope environment chamber and incubate the cells at 37°C with appropriate humidity and 5% CO2 for 1 h. This step allows the remaining free Red C12 to be incorporated into neutral lipids within lipid droplets.
-
a.
CRITICAL: Cells must be thoroughly washed to remove excess Red C12. Additionally, BODIPY 665/676 must be maintained in media, to retain sufficient signal in lipid droplets for downstream image analysis.
Note: Before transferring cells to the microscope environment chamber, turn on the temperature and CO2 controllers for ∼30 min to allow conditions to stabilize.
Live cell confocal microscopy
Timing: 5 h
Time-lapse images of live cells are acquired using a confocal microscope to observe intracellular trafficking of Red C12 (Figure 1). Use of a Zeiss 800 confocal microscope is described here.
-
5.Microscopy setup using an inverted Zeiss 800/Airyscan laser scanning confocal microscope equipped with 405, 488, 561 and 647 nm diode lasers, and Galium Arsenide Phosphid (GaAsP) and Airyscan detectors, and Focus Controller.2. Confocal images were acquired using a 63x/1.4 NA objective lens and Zen 2 (Blue) software, at 37°C and 5% CO2.
-
a.Track 1: BODIPY 665/676, Excitation 640 nm, Emission 647–700 nm.
-
b.Track 2: Red C12, Excitation 561 nm, Emission 561–600 nm.
-
c.Track 3: Mito-GFP, Excitation 488 nm, Emission 491–536 nm.
-
a.
Note: BODIPY 665/676 shows a change in emission spectrum in response to lipid peroxidation, with a shift towards 600 nm. Therefore, the emission wavelength used when detecting Red C12 should be limited to 600 nm to avoid spectral overlap.
-
6.Set up imaging positions for time-lapse imaging.
-
a.Identify Mito-GFP transfected cells and save as positions; this should be completed during the 1 h cell chase.
-
a.
Note: The number of cells that can be imaged is limited by the time interval chosen for the time-lapse imaging and the acquisition time for each image. We have found imaging at 30 min time intervals sufficient to accurately observe and quantify Red C12 trafficking. In addition, cells may migrate significantly during the time-lapse imaging, therefore it is advisable to utilize a zoom setting that allows resolution of organelle structure while avoiding cells moving out of the field of view.
-
7.Set up Definite Focus using Focus Controller.2.
-
a.Establish a position marker for 10 cells in each well; this allows the same cells in each condition to be imaged throughout the time-lapse imaging.
-
b.Establish a definite focus reference for each position to reduce z-drift during imaging using the Mito-GFP channel as a reference.
-
c.Verify definite focus positions immediately prior to imaging.
-
a.
CRITICAL: A focus strategy must be used to ensure the same z-plane is imaged throughout the time lapse. Failure to do so commonly leads to mitochondria no longer remaining in focus and therefore Red C12 accumulation cannot be accurately assessed.
-
8.
Acquire images of all cells at initial time point (T = 0); this is immediately following the 1 h cell chase.
-
9.Starvation.
-
a.Aspirate medium from wells.
-
b.Wash cells three times with Hank’s balanced salt solution (HBSS) containing calcium, magnesium, and glucose, but without phenol red or sodium pyruvate.
-
c.Aspirate wash and replace with HBSS containing 50 ng/mL BODIPY 665/676.
-
a.
Note: HBSS is used for these starvation studies as it has been shown to induce lipolysis-mediated trafficking of lipids from lipid droplets to mitochondria.5
Note: Keep the slide on the microscope during the starvation step to maintain cell positions for time-lapse imaging.
-
10.Time-lapse image acquisition.
-
a.Re-center imaging positions and reset definite focus positions.Note: Allow 15 min for temperature to stabilize following HBSS treatment before resetting definite focus positions.
-
b.Acquire images every 30 min for 4 h.Note: Length of time-lapse may need to be adjusted depending on assay conditions and cell type.
-
a.
-
11.
Save images for downstream analysis. Save images in a format that maintains image meta data (e.g., .czi for images acquired on a Zeiss microscope).
Figure 1.
Time-lapse micrographs of Red C12 trafficking
Representative images of Red C12 trafficking in complete medium or in response to starvation. Following Red C12 pulse-chase labeling of myoblasts, cells were imaged to measure initial Red C12 incorporation into lipid droplets and mitochondria (T = 0). Cells were then maintained in complete medium (CM) or incubated in HBSS for 4 h with imaging every 30 min to monitor FA trafficking from lipid droplets to mitochondria. Scale bar, 10 μm.
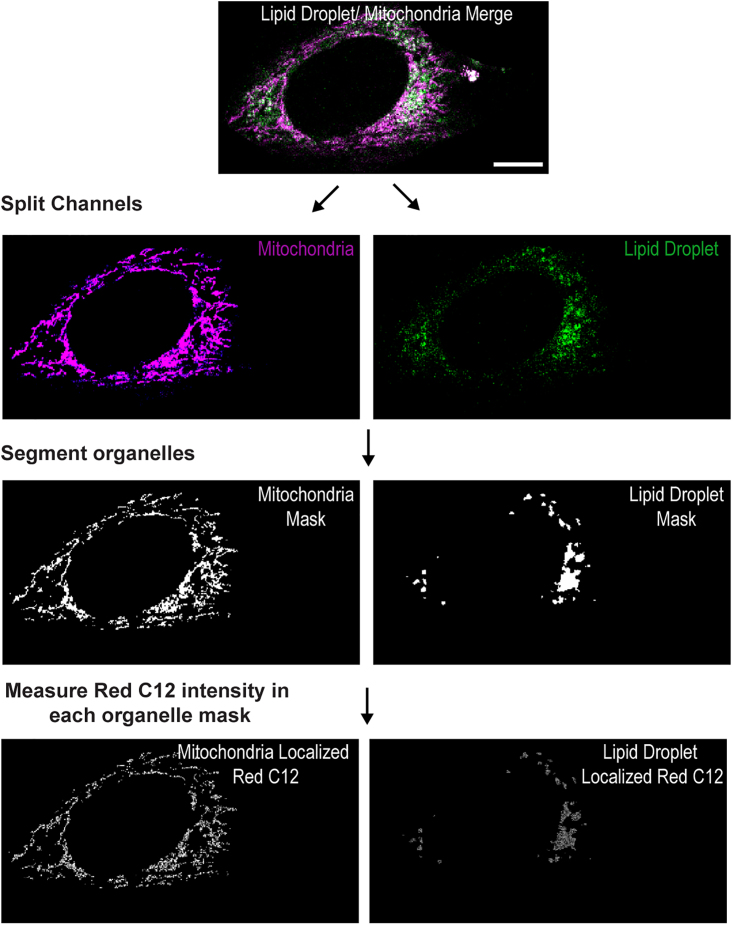
Image analysis
Timing: 2 h
Mito-GFP and BODIPY 665/676 are used to segment mitochondria and lipid droplets respectively. The total Red C12 intensity within mitochondria is measured at each time point to quantify Red C12 trafficking over time. Values are normalized to initial Red C12 accumulation in lipid droplets at T = 0 to account for variation in FA accumulation during pulse-chase labeling (Figure 2).
-
12.Open Files in Fiji.
-
a.Channel 1: Lipid Droplets, BODIPY 665/676.
-
b.Channel 2: Red C12.
-
c.Channel 3: Mitochondria, Mito-GFP.
-
a.
-
13.Set Measurements (Figure 3A).
-
a.Select: Area, Integrated Density, Limit to Threshold, Redirect to = None, Decimal = 3.
-
a.
-
14.Trace Mito-GFP positive cells and clear outside signal (Figure 3B).
-
a.Use the Mito-GFP channel to visualize transfected cells.
-
b.Use the “polygons selection” tool to trace the outline of the Mito-GFP positive cell.
-
c.Use “Clear Outside” to remove signal outside the Mito-GFP positive cell.
-
a.
Note: The “Brightness/Contrast” tool can be used to adjust the brightness to more easily define the cell area for tracing without altering intensity values.
-
15.Segment organelles.
-
a.Use “Split Channels” (Figure 4).
-
b.Segment mitochondria channel (Figure 5).
-
i.Convert channels to 8-bit.
-
ii.Use “Subtract Background” with “rolling = 50”.
-
iii.“Auto Threshold” using “method = Max Entropy white”.
-
i.
-
c.Segment lipid droplet channel using segmentation steps detailed above for mitochondria.
-
d.Remove overlapping lipid droplet-mitochondria pixels from mitochondria segmentation. This step is necessary to avoid double counting Red C12 intensity at co-localized pixels which will confound the results (Figure 6).
-
i.Use “Image Calculator” with “And” function to select mitochondria and lipid droplet co-localized pixels.
-
ii.“Make Binary”.
-
iii.“Create Selection”.
-
iv.Use “Image Calculator” with “Subtract” function to select mitochondria pixels that are not colocalized with lipid droplets.
-
v.“Make Binary”, ensure Black Background is selected.Note: Settings/values required for proper organelle segmentation will be dependent on imaging settings and experimental conditions.
-
i.
-
a.
-
16.Measure Red C12 intensity in each organelle.
-
a.Select segmented organelle channels and add each to the region of interest (ROI) Manager (Figure 7A).
-
i.“Create Selection”.
-
ii.“Add” to “ROI Manager”.
-
i.
-
b.Measure organelle ROIs in Red C12 Channel (Figure 7B).
-
i.Select Red C12 Channel.
-
ii.Select lipid droplet ROI in “ROI Manager”.
-
iii.Select “Measure” in “ROI Manager”.
-
iv.Select mitochondria ROI in “ROI Manager”.
-
v.Select “Measure” in “ROI Manager”.
-
i.
-
a.
-
17.Quantify Change in Mitochondrial Red C12 over time.
-
a.(Mito Red C12t=x - Mito Red C12t=0) / LD Red C12 t=0.
-
i.Subtract the mitochondrial Red C12 intensity at T = 0 from the Red C12 intensity at each subsequent time point to measure change in Red C12 intensity over time.
-
ii.Divide change in Red C12 intensity values at each time point by the lipid droplet Red C12 intensity value at T = 0 to normalize intensity values to initial Red C12 accumulation.
-
i.
-
b.Data can be represented as a scatter plot or bar graph with representative images (Figure 8).
-
a.
Note: Change in Mitochondrial Red C12 over time can also be quantified as fold change by dividing mitochondrial Red C12 at each time point by mitochondrial Red C12 intensity at T = 0.
Figure 2.
Image analysis of Red C12 trafficking
Representative images of Red C12 trafficking analysis. Merged images are split into respective channels. Split channels are processed through FIJI to segment the organelles of each cell and create an organelle mask. Finally, Red C12 intensity within each organelle mask is measured. Scale bar, 10 μm.
Figure 3.
Fiji analysis steps for clearing signal outside cell of interest
Figure 4.
Fiji Analysis steps for splitting lipid droplet, mitochondria, and Red C12 channels
Figure 5.
Fiji Analysis steps for segmenting mitochondria
Figure 6.
Fiji Analysis steps for removal of lipid droplet-mitochondria co-localized pixels from mitochondria segmentation
Figure 7.
Fiji analysis steps for measuring Red C12 intensity within an organelle
Figure 8.
Quantitation of Red C12 trafficking to mitochondria
Quantitation of Red C12 accumulation into mitochondria relative to initial LD incorporation after pulse-chase labeling, followed by incubation in complete medium (CM) or HBSS for 4 h. Values were normalized to HBSS starved cells at t = 4 h. At least 10 cells were analyzed per condition across n = 3 biological replicates. Data are expressed as means, error bars represent ± SEM. ∗p < 0.05. ∗∗p < 0.01∗∗∗p < 0.001. Figure reprinted and adapted with permission from Miner et al., 2023.
Expected outcomes
Following pulse-chase labeling of cells, Red C12 is primarily localized within lipid droplets with minimal accumulation in the mitochondria or other organelles. Continued incubation in complete medium results in loss of Red C12 within mitochondria (Figure 8, CM). In response to starvation however, a rapid accumulation of Red C12 intensity within the mitochondria is expected (Figure 8, 4 h HBSS). The rate of trafficking typically plateaus after ∼4 h in C2C12 myoblast cells. Perturbations that promote lipid droplet-to-mitochondria fatty acid transfer are expected to increase the rate and/or amount of Red C12 accumulation in mitochondria, while perturbations that impair this trafficking are expected to decrease the rate and/or amount of mitochondrial Red C12. For example, treatment with isoproterenol/IBMX increased the flux of Red C12 into mitochondria, while knockdown of the lipid droplet-mitochondria tether proteins PLIN5 or FATP4 decreased Red C12 flux in this assay.1
While our protocol described the analysis of fatty acid trafficking from lipid droplets to mitochondria in C2C12 myoblasts, it can easily be adapted to other cell culture or primary culture systems. For other cell types, the concentration of Red C12, the timing of the initial pulse-chase, or the interval and duration of live-cell imaging may need to be adjusted. In addition, the trafficking of FAs from lipid droplets may differ in other cell types resulting in trafficking to other organelles. FA trafficking to other organelles, such as peroxisomes or the endoplasmic reticulum, can also be visualized. In this case, the Mito-GFP transfection step would be adjusted to transfect cells with an alternative organelle-specific fluorescent marker such as GFP-SKL or GFP-Sec61B to label peroxisomes or the endoplasmic reticulum respectively. Finally, while this protocol specifically uses BODIPY 558/568 C12 (Red C12), fluorescent fatty acid analogues of various lengths and colors are available. These include BODIPY FL-C5, BODIPY FL-C12, and BODIPY FL-C16, all of which are incorporated into the three major classes of complex lipids: phospholipids, triglycerides, and cholesterol esters.6 BODIPY FL-C16 has also been successfully used to assess lipid droplet-to-mitochondria fatty acid trafficking.7 The BODIPY fluorophore is equivalent in length to approximately 4 carbons in a fatty acid chain. Therefore, BODIPY FL-C5 is equivalent to a medium chain C9 fatty acid, BODIPY FL-C12/Red C12 are equivalent to a long chain C16 fatty acid (e.g., palmitic acid), and BODIPY FL-C16 is equivalent to a long chain C20 fatty acid.
Limitations
A primary limitation of this protocol is related to the need for live cell confocal imaging. Cells need to be imaged multiple times throughout the experiment, which may cause bleaching and phototoxicity if high laser powers or long exposure times are required to visualize Red C12 and organelle markers. Drift in the Z-plane can occur due to temperature fluctuations when replacing culture medium with HBSS. Therefore, it is recommended to use a microscope with Z-plane autofocusing.
Another limitation is the use of fluorescent fatty acid analogues. These analogues are powerful tools to visualize intracellular trafficking of fatty acids. Like natural fatty acids, Red C12 incorporates into a variety of complex lipids.8 In the case of trafficking to mitochondria, thin layer chromatography showed that Red C12 is primarily in a free acid form. In addition, trafficking in the absence or presence of the inhibitor etomoxir showed that Red C12 mitochondrial localization is dependent on the carnitine palmitoyltransferase CPT1, and that Red C12 can undergo at least two rounds of β-oxidation52. Nevertheless, incorporation of a fluorophore influences lipid behavior and metabolism. For example, an HPLC-CAD/fluorescence lipidomics platform revealed that the array of complex lipid products formed when BODIPY-C12 is fed to larval zebrafish varies depending on the type of BODIPY label, i.e., the lipid profiles of red vs. green BODIPY-C12 were different.6 Thus, it is recommended to validate key results using alternative approaches such as radioactive pulse-chase assays. While these do not provide spatial information, information on the kinetics of β-oxidation derived from radioactive pulse-chase assays is complementary to the information provided by microscopy-based assays.
Troubleshooting
Problem 1
Low transfection efficiency of C2C12 myoblasts. Step 2.
Potential solution
If transfection efficiencies are low, increase the DNA concentration used or the Lipofectamine 2000: DNA ratio. For this protocol we utilized 50 ng of plasmid with a 2 μl: 1 μg Lipofectamine 2000:DNA ratio, both of which are on the lower end of the manufacturer’s recommendations. Additionally, alternative transfection reagents such as Viafect, Fugene, CellAvalanche, or JetOPTIMUS have been successfully used in C2C12 myoblasts.9 Lentiviral transduction can also be used for expression of fluorescent constructs in C2C12 myoblasts. Alternatively, a mitochondrial dye such as MitoTracker Deep Red can be used in combination with lipid droplet dyes such as BODIPY 493 or Lipi-Blue to visualize organelles without the use of transfection.
Problem 2
Minimal probe accumulation in lipid droplets following Red C12 pulse-chase. Steps 3–4.
Potential solution
It may be necessary to adjust either the cell density or Red C12 concentration. If the cell density is too high, cells will not accumulate sufficient Red C12 for visualization. Try lowering the cell density. Alternatively, the concentration of Red C12 can be increased. However, for C2C12 cells, we have found it is not advisable to increase the Red C12 concentration above 5 μM, due to cytotoxicity (see problem 3).
Problem 3
Cell death following Red C12 pulse-chase. Steps 3–4.
Potential solution
The concentration of Red C12 may be too high. For various cell types, we have found that concentrations between 1-5 μM are optimal to minimize cytotoxicity while maximizing the ability to visualize Red C12 in organelles. Alternatively, the cell density may be too low. If cell density is too low, cells are more sensitive to lipotoxicity. Try increasing the cell density.
Another possibility is that toxicity is due to the transfection reagent. Try lowering the amount of Lipofectamine or using an alternate transfection reagent. Finally, it is crucial to use antibiotic-free medium following transfection. Ensure that for all steps following transfection (Step 2), media free of antibiotics are used.
Problem 4
Minimal probe trafficking to mitochondria in response to starvation. Step 10.
Potential solution
C2C12 myoblasts are sensitive to cell culture confluency. If your cell density is low or approaching confluency, then C2C12 cells will likely respond abnormally to metabolic stressors. This is especially true for C2C12 myoblasts that reach confluency, as they may begin to differentiate and alter their cellular metabolic programs. We have found that C2C12 cells at ∼70%–90% confluency following the pulse-chase behave consistently.
Problem 5
Difficulty segmenting organelles. Step 15.
Potential solution
Segmentation of organelles is highly dependent on the signal-to-noise ratio of each organelle marker and specificity of labeling. If signal is low or hard to distinguish from background, it is recommended to increase the amount of DNA transfected and/or dye used to label. Alternatively, laser power, gain, or exposure time can be increased, however this may cause phototoxicity or bleaching.
When using lipid droplet dyes, it is possible to get non-specific labeling of membranes. If strong signal is detected in non-spherical structures, this indicates non-specific labeling, and the amount of dye should be reduced.
The segmentation steps can also be augmented to the specifics of the cell type being used. For example while “Auto Threshold: Max Entropy” was ideal for segmenting organelles in the method outlined above, other thresholding options may be required in other cell types or with alternative organelle markers. These can be evaluated by using the “Auto Threshold: try all” which will preview all available auto thresholding options in FIJI. In cases of variable labeling between organelle structures, the use of “Auto Local Threshold” in place of “Auto Threshold” may improve segmentation. Additionally, use of a filter such as median or Gaussian blur prior to thresholding can improve segmentation.
Problem 6
Wide margin of error between biological replicates. Step 17.
Potential solution
Large differences between biological replicates most commonly arise during the live-cell imaging portion of the protocol. The most common source of error is ending measurements before FA trafficking has reached its maximum and plateaued. The amount of time required for FA trafficking to occur should be assessed before performing additional experiments.
Another source of error arises from the differential loading of individual cells with Red C12 into lipid droplets. Therefore, it is critical to assess the same cell over the entire time-course. Due to cell migration the entire time-series should be visualized to avoid imaging separate cells.
In addition, it is advisable to remove cells which are undergoing division throughout the time-course from the analysis as only a portion of the initial Red C12 pool will be available for transfer into mitochondria following cell division.
Resource availability
Lead contact
Further information and requests about resources and reagents are available from the corresponding author, Sarah Cohen (sarahcoh@med.unc.edu), upon reasonable request.
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Gregory Miner (gminer2@med.unc.edu), upon reasonable request.
Materials availability
This study did not generate new unique reagents. The recombinant DNA used in this study is available from Addgene or from the lead contact upon request.
Data and code availability
This study did not generate/analyze datasets or code.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R35 GM133460 (S.C.) and F32 GM136027 (G.E.M.).
Author contributions
G.E.M. and S.C. conceptualized, designed, and wrote the protocol. G.E.M. performed research and analyzed the data.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Gregory E. Miner, Email: gminer2@med.unc.edu.
Sarah Cohen, Email: sarahcoh@med.unc.edu.
References
- 1.Miner G.E., So C.M., Edwards W., Ragusa J.V., Wine J.T., Wong Gutierrez D., Airola M.V., Herring L.E., Coleman R.A., Klett E.L., Cohen S. PLIN5 interacts with FATP4 at membrane contact sites to promote lipid droplet-to-mitochondria fatty acid transport. Dev. Cell. 2023;58:1250–1265.e6. doi: 10.1016/J.DEVCEL.2023.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambold A.S., Kostelecky B., Elia N., Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 2011;108:10190–10195. doi: 10.1073/PNAS.1107402108/SUPPL_FILE/PNAS.201107402SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Planchon T.A., Gao L., Milkie D.E., Davidson M.W., Galbraith J.A., Galbraith C.G., Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 2011;8:417–423. doi: 10.1038/NMETH.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty Acid Trafficking in Starved Cells: Regulation by Lipid Droplet Lipolysis, Autophagy, and Mitochondrial Fusion Dynamics. Dev. Cell. 2015;32:678–692. doi: 10.1016/J.DEVCEL.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinlivan V.H., Wilson M.H., Ruzicka J., Farber S.A. An HPLC-CAD/fluorescence lipidomics platform using fluorescent fatty acids as metabolic tracers. J. Lipid Res. 2017;58:1008–1020. doi: 10.1194/JLR.D072918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt E.G., Hurst K.E., Riesenberg B.P., Kennedy A.S., Gandy E.J., Andrews A.M., del Mar Alicea Pauneto C., Ball L.E., Wallace E.D., Gao P., et al. Acetyl-CoA carboxylase obstructs CD8+ T cell lipid utilization in the tumor microenvironment. Cell Metab. 2024;36:969–983.e10. doi: 10.1016/J.CMET.2024.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carten J.D., Bradford M.K., Farber S.A. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 2011;360:276–285. doi: 10.1016/J.YDBIO.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchiararo I., Cornut M., Soldati H., Bonavoglia A., Castets P. Back to basics: Optimization of DNA and RNA transfer in muscle cells using recent transfection reagents. Exp. Cell Res. 2022;421 doi: 10.1016/J.YEXCR.2022.113392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets or code.