Abstract
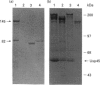
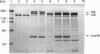
The catalytic domain of the cell-envelope proteinase from Lactococcus lactis SK11 has various inserts, situated in external loops of the catalytic domain, compared with the related subtilisins. Protein engineering was employed to analyse the necessity and function of one of these extra loops (residues 205-219), that is predicted to be located in close proximity to the substrate-binding region and is susceptible to autoproteolysis. We constructed a deletion mutant which lacks 14 residues of this surface loop and subsequently introduced various insertion cassettes coding either for the original loop with three mutations (E205S/E218T/M219S: triple-mutant proteinase) or for neutral spacers (1, 4, 7 and 16 serine residues). Engineered proteinases were analysed for activity, (auto)processing, and cleavage specificity. The presence of residues 205-219 is shown to be essential for proteolytic activity, as only triple-mutant proteinase retained activity towards casein substrates. The triple-mutant proteinase was found to be defective in C-terminal autoprocessing, and subsequent release from the lactococcal cell envelope in a calcium-free medium, indicative of either an altered proteolytic specificity or altered accessibility of the processing site. The specificity change appears to be subtle, as only small differences were found between wild-type and triple-mutant proteinase in the breakdown of casein substrates.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruinenberg P. G., Doesburg P., Alting A. C., Exterkate F. A., de Vos W. M., Siezen R. J. Evidence for a large dispensable segment in the subtilisin-like catalytic domain of the Lactococcus lactis cell-envelope proteinase. Protein Eng. 1994 Aug;7(8):991–996. doi: 10.1093/protein/7.8.991. [DOI] [PubMed] [Google Scholar]
- Bruinenberg P. G., Vos P., De Vos W. M. Proteinase overproduction in Lactococcus lactis strains: regulation and effect on growth and acidification in milk. Appl Environ Microbiol. 1992 Jan;58(1):78–84. doi: 10.1128/aem.58.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini E. A., Hughes J. V., Perlow D. S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985 Sep;55(3):836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., Alting A. C., Slangen C. J. Specificity of two genetically related cell-envelope proteinases of Lactococcus lactis subsp. cremoris towards alpha s1-casein-(1-23)-fragment. Biochem J. 1991 Jan 1;273(Pt 1):135–139. doi: 10.1042/bj2730135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. H., Yang R. C., Wu R. An improved strategy for rapid direct sequencing of both strands of long DNA molecules cloned in a plasmid. Nucleic Acids Res. 1983 Aug 25;11(16):5521–5540. doi: 10.1093/nar/11.16.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A. J., Meesters R., Laan H., Konings W. N., Kok J., Venema G. Processing of the lactococcal extracellular serine proteinase. Appl Environ Microbiol. 1991 Jul;57(7):1899–1904. doi: 10.1128/aem.57.7.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz D. W., Priestle J. P., Rahuel J., Wilson K. S., Grütter M. G. Refined crystal structures of subtilisin novo in complex with wild-type and two mutant eglins. Comparison with other serine proteinase inhibitor complexes. J Mol Biol. 1991 Jan 20;217(2):353–371. doi: 10.1016/0022-2836(91)90549-l. [DOI] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Autoproteolysis of the Extracellular Serine Proteinase of Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1991 Sep;57(9):2586–2590. doi: 10.1128/aem.57.9.2586-2590.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Mechanism of Proteinase Release from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1989 Dec;55(12):3101–3106. doi: 10.1128/aem.55.12.3101-3106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Sletten K. Purification and characterization of the free form of the lactococcal extracellular proteinase and its autoproteolytic cleavage products. J Gen Microbiol. 1991 Jul;137(7):1611–1618. doi: 10.1099/00221287-137-7-1611. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R. J., Bruinenberg P. G., Vos P., van Alen-Boerrigter I., Nijhuis M., Alting A. C., Exterkate F. A., de Vos W. M. Engineering of the substrate-binding region of the subtilisin-like, cell-envelope proteinase of Lactococcus lactis. Protein Eng. 1993 Nov;6(8):927–937. doi: 10.1093/protein/6.8.927. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Bauw G., Puype M., Van Damme J., Van Montagu M. Protein-blotting on Polybrene-coated glass-fiber sheets. A basis for acid hydrolysis and gas-phase sequencing of picomole quantities of protein previously separated on sodium dodecyl sulfate/polyacrylamide gel. Eur J Biochem. 1985 Oct 1;152(1):9–19. doi: 10.1111/j.1432-1033.1985.tb09157.x. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Boerrigter I. J., Buist G., Haandrikman A. J., Nijhuis M., de Reuver M. B., Siezen R. J., Venema G., de Vos W. M., Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991 Apr;4(4):479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Vos P., de Haard H., Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989 Dec 21;85(1):169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- van Asseldonk M., Rutten G., Oteman M., Siezen R. J., de Vos W. M., Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990 Oct 30;95(1):155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]