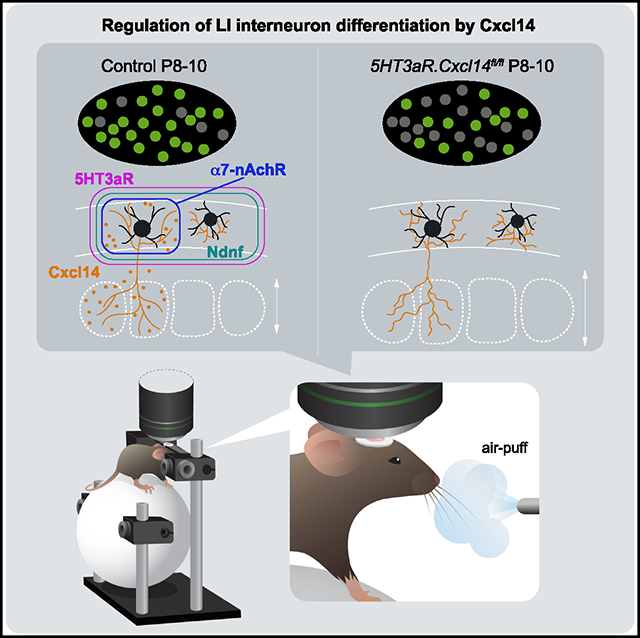
SUMMARY
Spontaneous and sensory-evoked activity sculpts developing circuits. Yet, how these activity patterns intersect with cellular programs regulating the differentiation of neuronal subtypes is not well understood. Through electrophysiological and in vivo longitudinal analyses, we show that C-X-C motif chemokine ligand 14 (Cxcl14), a gene previously characterized for its association with tumor invasion, is expressed by single-bouquet cells (SBCs) in layer I (LI) of the somatosensory cortex during development. Sensory deprivation at neonatal stages markedly decreases Cxcl14 expression. Additionally, we report that loss of function of this gene leads to increased intrinsic excitability of SBCs—but not LI neurogliaform cells—and augments neuronal complexity. Furthermore, Cxcl14 loss impairs sensory map formation and compromises the in vivo recruitment of superficial interneurons by sensory inputs. These results indicate that Cxcl14 is required for LI differentiation and demonstrate the emergent role of chemokines as key players in cortical network development.
In brief
Iannone et al. characterize the development of single-bouquet cells (SBCs) and neurogliaform cells (NGFCs) in LI. They show that Cxcl14 expression in SBCs is crucial for morphological and electrophysiological maturation. In addition, loss of Cxcl14 impairs sensory-evoked responses in LI and the proper development of the barrel map in LIV.
Graphical Abstract
INTRODUCTION
Perinatal activity patterns are prominent in the neocortex and profoundly shape neuronal differentiation.1–5 However, it is unclear whether neuronal activity exerts a permissive or an instructive influence on the emergence of functionally distinct neuronal subtypes.
In cortical layer I (LI), reelin (Re)-expressing interneurons receive transient inputs from the thalamus at postnatal days 6 and 7 (P6 and P7).6,7 Passive whisker stimulation in mouse pups triggers LI network synchronization required for the refinement of somatosensory topography in LIV barrel fields and for sensory discrimination in adult mice.6 This early engagement of LI interneurons highlights the key developmental role of this layer as a recipient of bottom-up inputs. However, whether these inputs contribute to the diversification of LI interneurons into distinct functional subtypes is unknown. Transcriptomic and functional analyses of adult LI interneurons reveal substantial cell diversity in this layer, satisfying the circuit demands of complex cortical computations.8 For instance, LI integrates thalamocortical axonal (TCA) inputs, originating in both first-order and higher-order sensory nuclei, carrying salient features of primary sensory experience while mediating TCA feedback.6,7,9 LI additionally receives corticocortical inputs relaying both cross-modal sensory and higher-order associative information into the cortical column.10,11
Recent evidence identified four LI subtypes in adult mice: canopy cells, alpha 7-nicotinic acetylcholine receptor (α7-nAChR)-expressing single-bouquet cells (SBCs),12–14 vasoactive intestinal peptide (Vip) cells, and neurogliaform cells (NGFCs).13–16 Among these subtypes, only LI NGFCs have been well characterized at a functional level. These cells control the integration of thalamic (bottom-up) and top-down (intracortical, modulatory) inputs during associative sensory learning7,17–19 and show significant expansion in the human brain.20,21 However, a lack of genetic tools to parse out LI subtypes has limited both the study of their functional diversity and an understanding of how this diversity arises through developmental and sensory-dependent processes.
C-X-C motif chemokine ligand 14 (Cxcl14), a chemokine family member with a poorly understood function in the nervous system, is expressed in superficial cortical layers.16,22–24 Other C-X-C motif chemokines are present in LI during development, such as Cxcl12, which regulates neuronal maturation and survival.25,26 While the role of these molecules in chemoattraction has been well characterized,27 their function in activity-dependent circuit assembly is unknown. Converging evidence suggests that expression of Cxcl14 is modulated by activity in adult inhibitory neurons,28,29 but whether this expression is driven by transient sensory inputs that regulate interneuron development has not been investigated. In this study, we thus set out to determine the role of Cxcl14 in superficial interneuron differentiation and network function.
We found that Cxcl14 expression delineates LI SBC, but not NGFC, interneurons by P8 in the barrel cortex. LI NGFCs and Cxcl14-expressing SBCs can already be distinguished based on electrophysiological and morphological properties at this early stage. Furthermore, we uncovered a dependency of Cxcl14 expression on ascending sensory inputs, demonstrating that whisker plucking results in significantly reduced Cxcl14 expression and impaired morphological development of SBC interneurons. In addition, Cxcl14 loss of function leads to increased excitability in SBC, but not NGFC, interneurons. Finally, this manipulation alters single-cell dynamics, revealed by longitudinal in vivo assessment, and impairs sensory map formation, ultimately compromising LI interneuron recruitment after sensory stimulation. Altogether, these results reveal that chemokine expression can be regulated by perinatal sensory experience and can modulate developmental programs for functional diversification of superficial neuronal subtypes.
RESULTS
Cxcl14 expression delineates interneuron subtypes during development
To reveal Cxcl14 interneuron subpopulations, we utilized a Cxcl14.eGFP mouse line and characterized transgene expression in pups at P8 (Figure S1). Sparse expression of GFP could be detected in superficially positioned cells in Cxcl14.eGFP pups, which displayed interneuron morphology (Figures 1A and S1A–S1E). These cells often lay within LI and could be found across the somatosensory, auditory, visual, motor, and prefrontal cortices (Figures S1A–S1E). Subcortically, strong GFP expression was noted in the olfactory tubercle, particularly the GABAergic granule cells of the islands of Calleja (Figures S1A and S1B). Labeling was also detected in the CA1 region of the hippocampus, including the stratum lacunosum-moleculare, but not in the somatosensory thalamus (Figures S1D and S1E). Together, these labeled populations suggested prominent expression of Cxcl14 by various GABAergic populations across both cortical and subcortical regions.
Figure 1. Cxcl14 expression delineates a subtype of layer I interneurons.
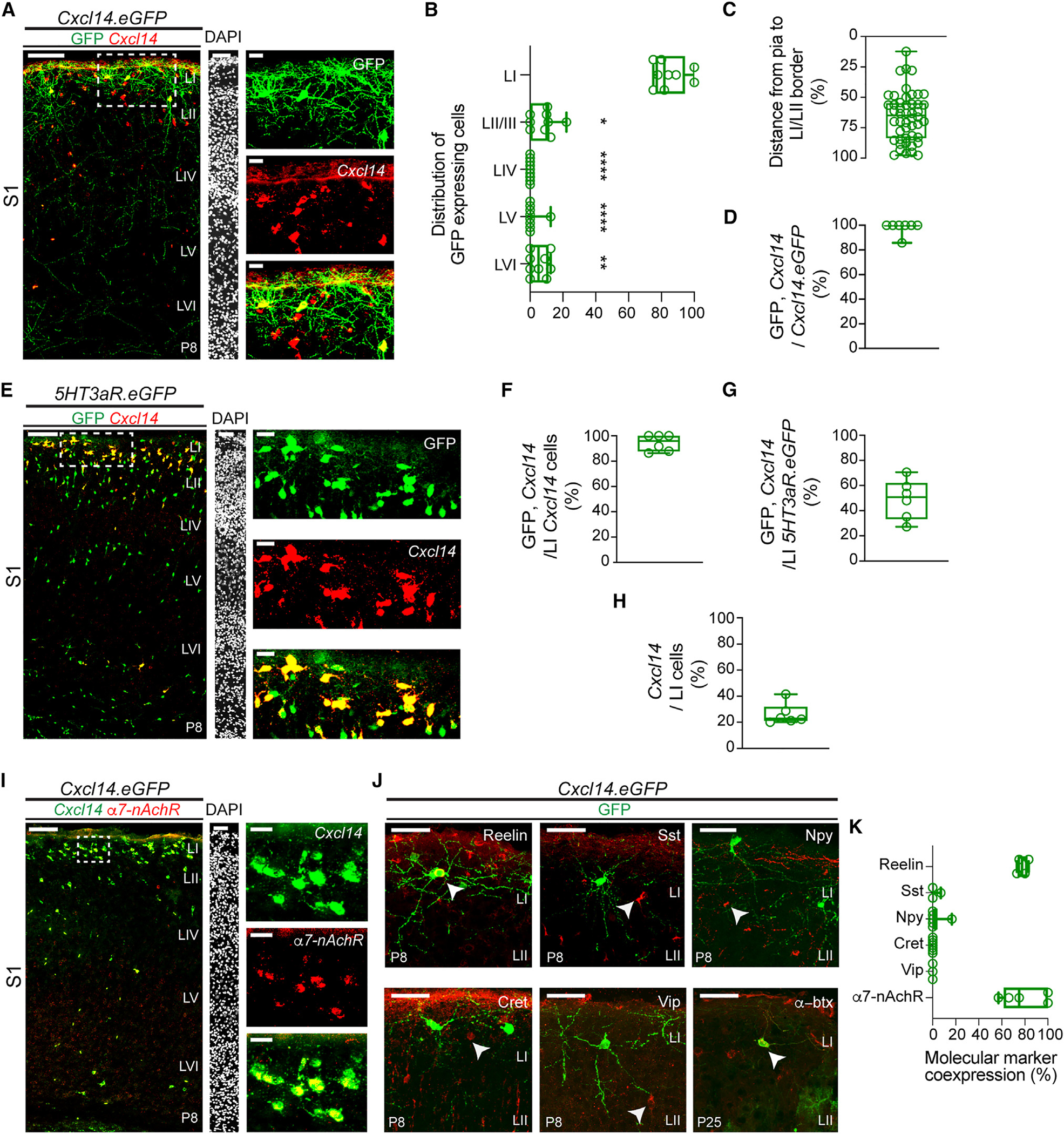
(A) Overlay of Cxcl14 in situ hybridization and GFP fluorescent signals in Cxcl14.eGFP mice at P8 in primary somatosensory cortex (S1); scale bar, 100 μm (left). Layer boundaries were determined by DAPI labeling; scale bar, 50 μm (middle). Enlarged detail of area delineated by the dotted box shows expression of GFP (top right), Cxcl14 transcript (middle right), and merged (bottom right); scale bars, 20 μm.
(B) Percentage distribution by layer of GFP-expressing cells in Cxcl14.eGFP mice at P8 in S1 (n = 10 mice). Kruskal-Wallis test (****p < 0.0001) followed by Dunn’s multiple comparisons test of laminar distribution; LI vs. LII/III, *p = 0.0443; LI vs. LIV, ****p < 0.0001; LI vs. LV, ****p < 0.0001; LI vs. LVI, **p = 0.008. All other comparisons are non-significant.
(C) Distribution of individual GFP-expressing cells in LI as a percentage of the depth from the pial border (0%) to the LI/LII border (100%). Cxcl14.eGFP n = 48 cells, 7 mice.
(D) Percentage of cells that co-express Cxcl14 and GFP over the total number of GFP-expressing cells in LI of Cxcl14.eGFP mice at P8. n = 7 sections, 2 mice.
(E) Expression of Cxcl14 transcript in 5HT3aR.eGFP mice at P8 in S1; scale bar, 100 μm (left). DAPI; scale bar, 50μm (middle). Enlarged detail of area delineated by the dotted box shows expression of GFP (top right), Cxcl14 transcript (middle right), and merged (bottom right); scale bars, 20 μm.
(F) Percentage of cells that co-express GFP and Cxcl14 over the total number of Cxcl14-expressing cells in LI of 5HT3aR.eGFP mice at P8. n = 6 sections, 2 mice.
(G) Percentage of cells that co-express GFP and Cxcl14 over the total number of 5HT3aR.eGFP-expressing cells in LI at P8. n = 6 sections, 2 mice.
(H) Percentage of Cxcl14-expressing cells over the total number of LI cells revealed by DAPI at P8. n = 6 sections, 2 mice.
(I) Overlap of Cxcl14 and α7-nAChR transcripts in LI of Cxcl14.eGFP mice (GFP expression omitted for clarity) at P8 in S1; scale bar, 100 μm (left). DAPI; scale bar, 50 μm (middle). Enlarged detail of area delineated by the dotted box shows expression of Cxcl14 transcript (top right, pseudocolored green), α7-nAChR transcript (middle right), and merged (bottom right); scale bars, 20 μm. n = 6 sections, 1 mouse.
(J) Immunohistochemical staining for reelin (top left), somatostatin (Sst, top middle), neuropeptide Y (Npy, top right), calretinin (Cret, bottom left), and vasoactive intestinal peptide (Vip, bottom middle) in Cxcl14.eGFP mice at P8 in S1. Expression of the α7 subunit of the nicotinic acetylcholine receptor (α7-nAChR) revealed through fluorescently labeled α-bungarotoxin (α-btx, bottom right) in Cxcl14.eGFP mice at P25; scale bars, 20 μm.
(K) Molecular marker expression in GFP+ LI interneurons of Cxcl14.eGFP mice at P8 (reelin, Sst, Npy, Cret, Vip) or P25 (α7-nAChR). Reelin, n = 4 mice; Sst, n =3 mice; Npy, n = 6 mice; Cret, n = 7 mice; Vip, n = 3 mice; α7-nAChR, n = 5 sections, 2 mice.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile ranges (box).
See also Figures S1 and S2.
We next used the Cxcl14.eGFP mouse line in tandem with fluorescence in situ hybridization (FISH) to assess laminar distribution in the primary somatosensory cortex (S1) (Figures 1A–1C). We found that 85% of Cxcl14.eGFP cells lay within LI, significantly more than in any other layer (Figure 1B). The remainder appeared split between LII (typically at the border between LI and LII) or deep within LVI. Cxcl14.eGFP-labeled cells were predominately located within the bottom half of LI (LIb) (Figure 1C) and extended translaminar axons (Figure 1A), suggestive of the previously described morphology of the SBC.14 Finally, we found that 98% of GFP-expressing cells co-expressed Cxcl14 (Figure 1D) in Cxcl14.eGFP mice, validating the specificity of this previously poorly characterized mouse line.
To determine if Cxcl14-expressing cells were derived from the caudal ganglionic eminence (CGE), we employed a 5HT3aR. eGFP mouse line30 in combination with FISH against Cxcl14 (Figures 1E–1H). Our analysis revealed that nearly all Cxcl14-expressing cells in LI co-express 5HT3aR (Figure 1F), in agreement with previous adult analysis.31 Conversely, ~50% of CGE-derived interneurons (Figure 1G) and ~20% of all LI cells (Figure 1H) expressed Cxcl14, suggesting that Cxcl14 expression is subtype specific.
To elucidate the subtype identity of Cxcl14-expressing neurons, we performed analysis of interneuron markers in Cxcl14.eGFP mice (Figures 1I–1K). We found more than 90% co-expression of both Cxcl14 and α7-nAChR transcripts within LI Cxcl14.eGFP neurons at P8 (Figures 1I and S2A) as well as continued expression of α7-nAChR subunits into adulthood (Figures 1J and 1K), as demonstrated by α-bungarotoxin labeling.32,33 By contrast, Cxcl14.eGFP cells rarely co-expressed neuropeptide Y (Npy), a marker for LI NGFCs.19 Furthermore, Vip was not co-expressed with Cxcl14.eGFP at P8 (Figures 1J and 1K). Despite previous findings of Cxcl14 transcript in adult Vip-expressing cells,34 we found that, on average, only 5% of Cxcl14.eGFP cells co-expressed Vip at P25 (Figures S2B and S2C). Notably, we also observed GFP expression in astrocytes at this older age (Figure S2B). In addition, Cxcl14.eGFP cells did not express somatostatin (Sst) or calretinin (Cret) (Figures 1J and 1K). Together, these results indicate that Cxcl14.eGFP delineates an α7-nAChR-expressing population characterized by a translaminar axon, consistent with the SBC subtype.14 Thus, Cxcl14 expression provides a genetically tractable means to access a poorly understood interneuron subtype during development.
Cxcl14.eGFP interneurons exhibit morphological and electrophysiological distinctions of SBCs
We next sought to determine if Cxcl14-expressing interneurons display morphological and electrophysiological identity in early development. To that end, we performed whole-cell patch-clamp recordings paired with morphological reconstructions in Cxcl14.eGFP or in 5HT3aR.Cre mice crossed with RCE mice to achieve Cre-recombinase-directed expression of eGFP within CGE-derived interneurons (hereafter 5HT3aR.RCE) at P8–P10 (Figure 2A). Cxcl14.eGFP interneurons in S1 often displayed a descending axon stretching into the deeper cortical layers with axon collaterals in LI as well as deeper layers (Figures 2A and 2B), consistent with the morphological features of LI SBC interneurons.12,16 We then compared these cells to LI NGFCs6 (Figures 2A and 2B) sampled from either GFP-negative LI interneurons in Cxcl14.eGFP mice or GFP+ LI cells in 5HT3aR.RCE mice. Comparing these two populations, we found that the vertical axonal branch of Cxcl14.eGFP neurons extended significantly deeper than that of NGFCs (Figures 2B and 2C). In addition, Cxcl14.eGFP neurons showed significantly larger axonal coverage compared to NGFCs (Figure 2D). These results indicate that Cxcl14.eGFP-expressing SBCs and NGFCs are morphologically distinct at P8.
Figure 2. Early functional specification of layer I interneuron subtypes.
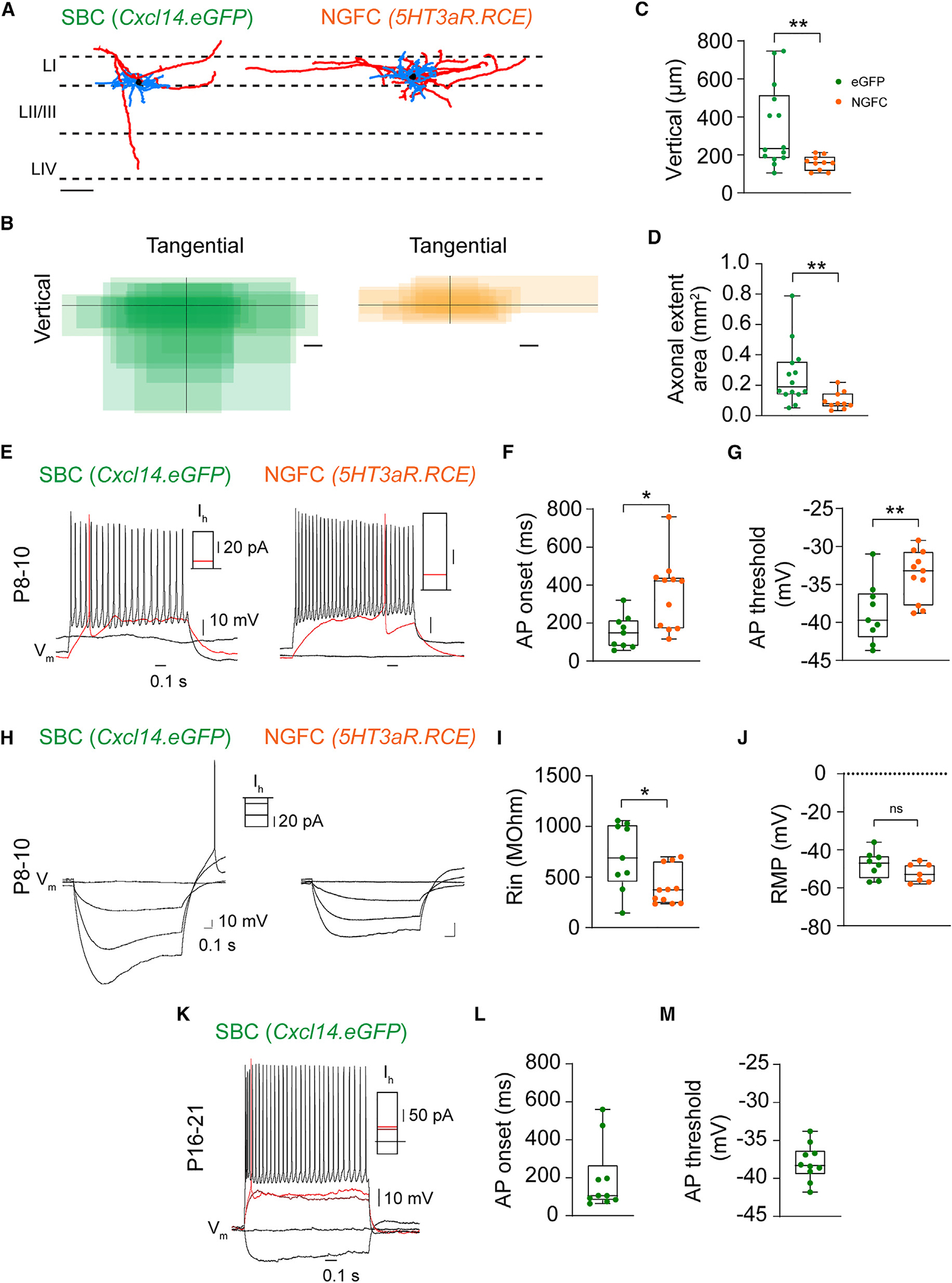
(A) Representative morphological reconstruction of a single-bouquet cell (SBC) (left) and a neurogliaform cell (NGFC, right) in Cxcl14.eGFP and 5HT3aR.RCE mice, respectively, at P8–P10 in S1. Dashed lines demarcate layer borders (LI, LII/III, and LIV). Scale bar, 100 αm. Blue, dendrite; red, axon; black, soma.
(B) Summary of axonal extent in SBC (green) and NGFC (orange) interneurons with respect to the dura. Soma location is represented by the intersection of black lines. SBC, n = 14 cells, 8 mice; NGFC, n = 10 cells, 9 mice; scale bars, 100 μm.
(C) Vertical axonal length. Unpaired t test, n = 14 SBCs, 8 mice, vs. n = 10 NGFCs, 9 mice; **p = 0.0048.
(D) Axonal areal extent. Unpaired t test, n = 14 SBCs, 8 mice, vs. n = 10 NGFCs, 9 mice; **p = 0.0091.
(E) Representative voltage traces (Vm) of an LI SBC (left) and LI NGFC (right) recorded at resting state (~ −65 mV) in Cxcl14.eGFP and 5HT3aR.RCE mice, respectively, at P8–P10. Rheobase trace (red; SBC, 10 pA; NGFC, 10 pA) and action potential (AP) train at ~30 Hz (SBC, 70 pA; NGFC, 50 pA).
(F) AP onset. Mann-Whitney test of SBCs (n = 9 cells, 3 mice) vs. NGFCs (n = 11 cells, 6 mice); *p = 0.013.
(G) AP threshold. Unpaired t test of SBCs (n = 9 cells, 3 mice) vs. NGFCs (n = 11 cells, 6 mice); **p = 0.007.
(H) Representative voltage traces at resting (flat traces) and hyperpolarizing states in Cxcl14.eGFP mice at P8–P10. A single spike was observed in three of seven GFP-expressing interneurons following hyperpolarizing stimulation and was absent in the NGFC group. Current stimulation protocol: −50, −30, −10, and 0 pA.
(I) Input resistance. Mann-Whitney test of SBCs (n = 9 cells, 3 mice) vs. NGFCs (n = 11 cells from 6 mice); *p = 0.038.
(J) Resting membrane potential. Unpaired t test of SBCs (n = 9 cells, 3 mice) vs. NGFCs (n = 11 cells, 6 mice); non-significant (ns).
(K) Representative voltage traces recorded in current clamp at resting state in Cxcl14.eGFP mice at P16–P21. Rheobase trace (red; SBC, 52 pA) and AP train at ~30 Hz (SBC, 200 pA).
(L) AP onset. Cxcl14.eGFP, n = 10 cells, 3 mice.
(M) AP threshold. Cxcl14.eGFP, n = 10 cells, 3 mice.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile range (box).
See also Table S1.
We next analyzed the passive and active intrinsic membrane properties in these cells (Figures 2E–2M; Table S1). Our analysis revealed that SBC interneurons in Cxcl14.eGFP mice and NGFC interneurons in 5HT3aR.RCE mice differed significantly based on action potential (AP) onset fired at rheobase (Figures 2E and 2F), AP threshold (Figures 2E and 2G), AP peak (Table S1), AP rise (Table S1), and input resistance (Rin) (Figures 2H and 2I), whereas the resting membrane potential was not significantly different at P8–P10 (Figure 2J). Furthermore, Cxcl14.eGFP SBC interneurons, unlike NGFCs, occasionally fired a rebound spike after a hyperpolarizing current step (3 of 11 at P8–P10, Figure 2H), suggesting that SBCs might exhibit burst firing in the network.35 In the adult, α7-nAChR-expressing SBC interneurons show early-onset APs and a prefiring membrane bump, distinguishing them from NGFC interneurons.14 Accordingly, we recorded from Cxcl14.eGFP cells at P16–P21 and similarly found that 80% of Cxcl14.eGFP interneurons were early-onset spiking (<300 ms) and exhibited the characteristic depolarized bump before firing their first AP (Figures 2K and 2L); notably, however, the AP threshold remained within a similar range as seen from P8–P10 (Figure 2M).
Together, these results indicate that Cxcl14.eGFP expression delineates LI SBCs and that these cells are intrinsically and molecularly different from NGFCs by the early second postnatal week, suggesting early functional specification.
Sensory inputs regulate Cxcl14 expression and morphological development of LI SBCs
Since thalamic inputs transiently innervate LI during the first postnatal week,6 we next determined if these inputs are necessary for Cxcl14 expression. To that end, we assessed the impact of whisker plucking on Cxcl14 transcript expression (Figures 3 and S3). Effective sensory deprivation was determined by the robust disruption of ascending TCAs, delineated by the vesicular glutamate transporter 2 (VGlut2), into characteristic barrels within LIV of S1 (Figure 3A). In whisker-plucked pups, the density of LI neurons expressing Cxcl14 was significantly decreased compared to unplucked control littermates, as was the density of Cxcl14-expressing neurons located in upper LII (Figure 3B). Importantly, whisker plucking had no impact on Cxcl14 expression in other sensory cortices, such as the primary auditory cortex (Figures S3A and S3B). We found an overall decrease in LI cell density, delineated by DAPI labeling, at P5 after sensory deprivation (Figure S3C), which may explain some loss of Cxcl14 signal. However, as this analysis additionally includes non-neuronal cell types, we cannot rule out an effect of these populations contributing to total density. Importantly, sensory deprivation does not impact neuron-derived neurotrophic factor (Ndnf) interneuron survival at P8 (Figure S3D, see below). Together, these results show that Cxcl14 expression is regulated by passive sensory experience in superficial layers of developing S1.
Figure 3. Cxcl14 expression is reliant on sensory inputs during development.
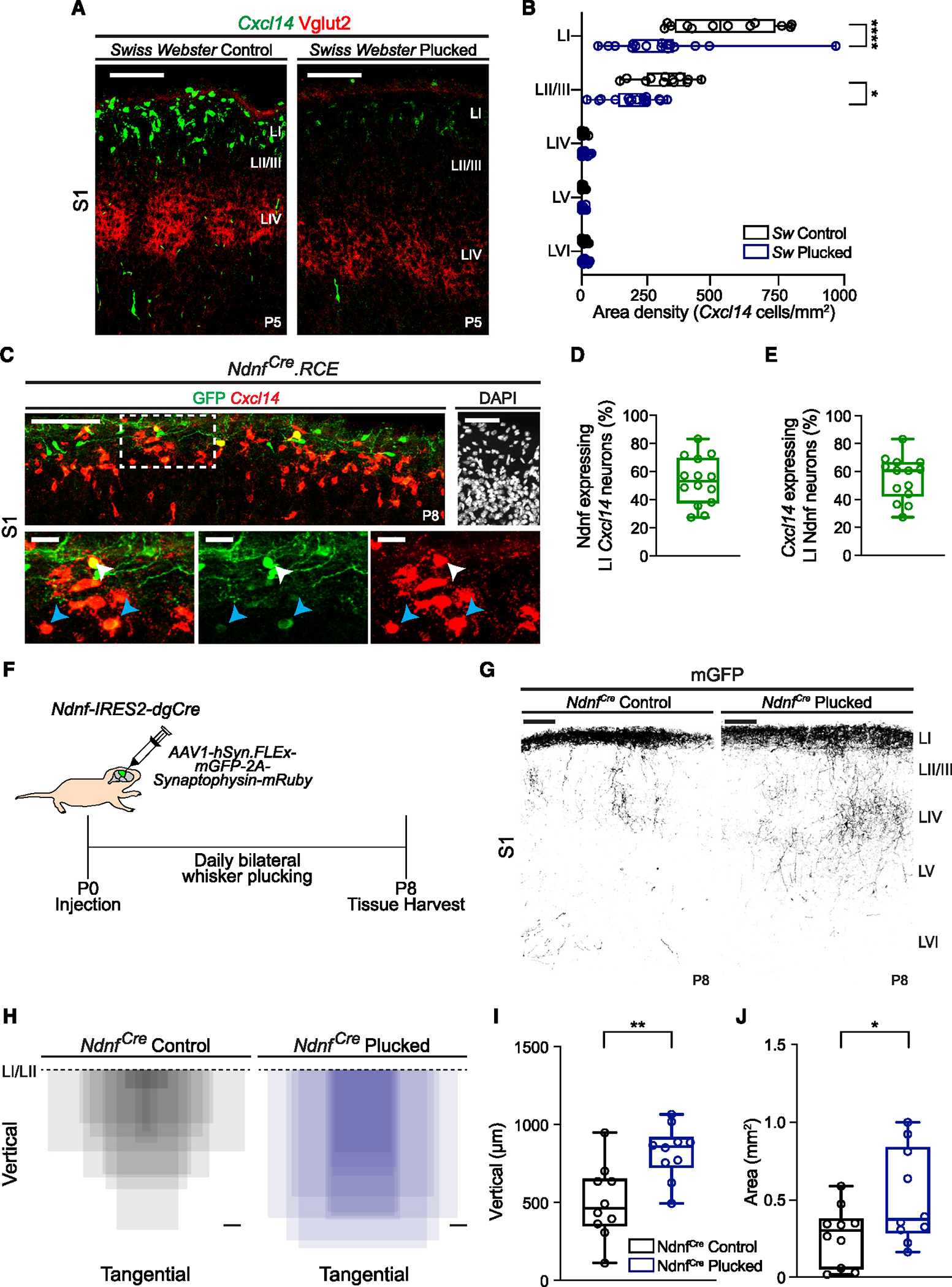
(A) Cxcl14 expression in Swiss Webster (Sw) littermate control (left) and daily whisker-plucked (right) pups at P5 in S1; scale bars, 100 μm. VGlut2 labels thalamocortical axon afferents.
(B) Layer distribution of Cxcl14-expressing cells in control (n = 12 sections, 3 mice) and whisker-plucked (n = 16 sections, 4 mice) pups. Two-way ANOVA (****pwhisker status < 0.0001) with Sidák’s multiple comparisons test of control vs. whisker-plucked by layer; LI, ****p < 0.0001; LII/III, *p = 0.011. All other comparisons are non-significant.
(C) Co-expression of GFP and Cxcl14 in NdnfCre.RCE mice at P8. Scale bar, 100 μm. DAPI; scale bar, 50 μm (top right). Detail of the area in the dotted box shows expression of merged (bottom left), GFP (bottom middle), and Cxcl14 transcript (bottom right); scale bars, 20 μm. White arrowheads, Ndnf cells that robustly co-express Cxcl14; blue arrowheads, Ndnf cells with weak Cxcl14 expression.
(D) Percentage of LI Cxcl14 cells that co-express GFP in NdnfCre.RCE mice at P8 in S1. n = 14 sections, 4 mice.
(E) Percentage of LI GFP cells that co-express Cxcl14 in NdnfCre.RCE mice at P8 in S1. n = 14 sections, 4 mice.
(F) Schematic of experimental approach for the anatomical tracing of axonal terminals in NdnfCre mice undergoing daily bilateral whisker plucking.
(G) Representative images of mGFP-labeled axonal collaterals of Ndnf LI interneurons at P8 (inverted images). Scale bars, 100 μm.
(H) Axonal extent of Ndnf LI interneurons with respect to the LI/LII border (dashed line). Control, n = 10 mice; plucked, n = 10 mice; scale bars, 100 μm.
(I) Vertical axonal depth. Unpaired t test of controls (n = 10 mice) vs. plucked (n = 10 mice); **p = 0.003.
(J) Axonal extent in S1. Unpaired t test of controls (n = 10 mice) vs. plucked (n = 10 mice); *p = 0.046.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile ranges (box).
See also Figure S3.
Our previous work indicates that sensory inputs are fundamental for the morphological development of LII/III interneurons.36 Therefore, we determined if this manipulation also impacts LI interneuron maturation. We utilized the NdnfCre knockin mouse line to target Ndnf-expressing LI interneurons that do not co-express Vip17,22 and crossed it with RCE mice. While evidence in adult animals suggests little Ndnf expression in LI SBCs,14 this gene was expressed in Cxcl14 populations at P8 (Figures 3C–3E). At this age, nearly half of Cxcl14-expressing interneurons co-express Ndnf in LI (Figure 3D). To visualize LI interneuron processes, we carried out a daily bilateral whisker plucking paradigm in NdnfCre pups from P0 to P8, while additionally injecting low-volume, superficially targeted AAV1-hSyn. FLEx-mGFP-2A-Synaptophysin-mRuby (hereafter AAV1-hSyn-mGFP) virus into the S1 at P0 (Figure 3F). This allows for robust labeling of neuronal processes of LI interneurons due to the membrane targeting of GFP.37 Our results revealed an exuberance of axonal spread into deeper layers in sensory-deprived mice at P8 (Figures 3G and 3H). The depth of axonal traversal increased (Figure 3I), as did the area of axonal coverage (Figure 3J). Importantly, this increase in axonal coverage cannot be explained by differences in viral infection, as both plucked and unplucked littermates showed similar densities of infected cells by AAV1-hSyn-mGFP (Figure S3D). In agreement with these observations, sensory deprivation led to a decrease in the number of intersections proximal to the soma and a strong trend toward longer branches, reaching deeper in the cortex of Cxcl14.eGFP-expressing SBC interneurons (Figures S3E–S3G). More generally, these findings revealed a cell-type-specific impact of sensory deprivation on neuronal morphology, as it limits axonal elaboration in LII/III CGE-derived interneurons,36 yet promotes it in CGE-derived interneurons in LI.
Abrogation of Cxcl14 in development does not impact interneuron migration
Given that the chemokine Cxcl12 is a key regulator of interneuron migration,38–41 we evaluated the impact of Cxcl14 loss on lamination of interneuron subtypes in S1. To this end, we utilized a Cxcl14 fl/fl mouse line42,43 and crossed it to 5HT3aR.Cre mice. To ensure the effectiveness of this genetic strategy (Figures S4A and S4B), we performed a PCR with two custom-designed sets of primers flanking the conditional exon 2, which contains the C-X-C motif essential for the stabilization of the complex between this family of chemokines and their cognate receptors43,44 (Figures S4C and S4D). This analysis indicated effective deletion of the exon in 5HT3aR.Cxcl14fl/fl mice (Figure S4D). In addition, we carried out FISH with probes specifically against Cxcl14 exon 2, which also demonstrated substantial loss of signal within superficial layers of S1 in 5HT3aR.Cxcl14fl/fl animals (Figure 4A). We confirmed the specificity of the Cxcl14 fl/fl line in recombining only in the presence of Cre recombinase by evaluating Cxcl14 expression in the islands of Calleja, which robustly express Cxcl14 (Figure S1A) but not 5HT3aR (Figure 4B). As expected, we observed no loss of Cxcl14 expression within this region in either controls or mutants (Figure 4C).
Figure 4. Normal laminar migration after loss of Cxcl14 function in superficial interneurons.
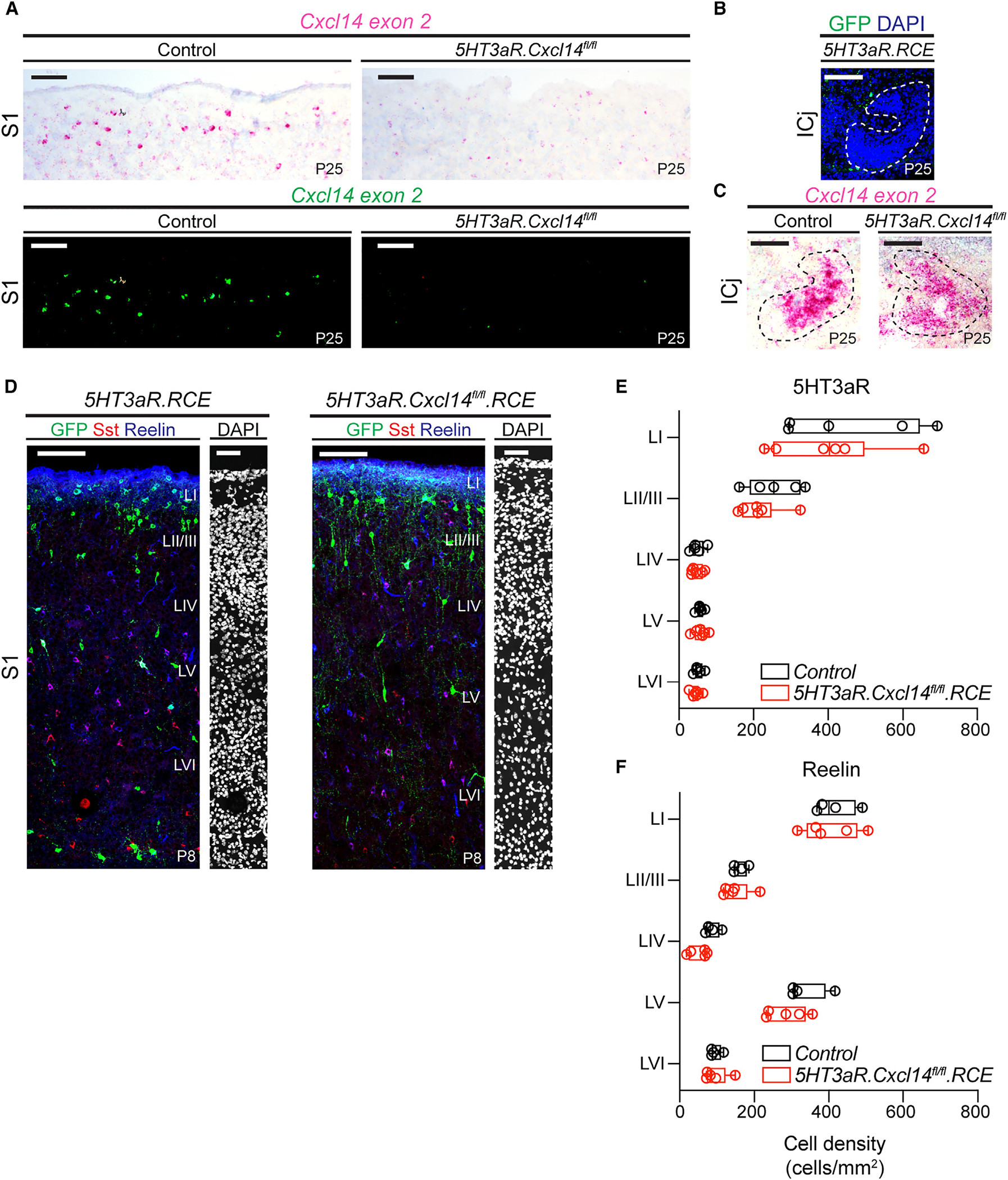
(A) Representative images of LI in 5HT3aR.RCE (left, control) and 5HT3aR.Cxcl14fl/fl.RCE (right, mutant) mice at P25, following in situ hybridization with a probe specific for Cxcl14 exon 2 (top). Tissue counterstained with hematoxylin. Images with probe labeling isolated and color inverted (bottom). Control, n = 4 sections, 2 mice; mutant, n = 3 sections, 2 mice; scale bars, 100 μm. S1, primary somatosensory cortex.
(B) GFP expression in a major island of Calleja (ICj; dotted outline) in a 5HT3aR.RCE mouse at P25; scale bar, 100 μm.
(C) Cxcl14 expression in an ICj (dotted outline) from a control (left) and a mutant (right) mouse at P25; scale bar, 100 μm.
(D) Representative images of interneuron distribution in 5HT3aR.RCE (control, left) and 5HT3aR.Cxcl14fl/fl.RCE (mutant, right) mice at P8 in S1. Left images, scale bars, 100 μm; DAPI, scale bars, 50 μm.
(E) Layer distribution of GFP-expressing interneurons (cell density [cells/mm2] across cortical layers) in 5HT3aR.RCE and 5HT3aR.Cxcl14fl/fl.RCE mice at P8.Two-way ANOVA with Sidák’s multiple comparisons test of control (n = 5 mice) vs. mutant (n = 6 mice) by layer; all genotype comparisons by layer are non-significant.
(F) Layer distribution of reelin-expressing interneurons in 5HT3aR.RCE and 5HT3aR.Cxcl14fl/fl.RCE mice at P8. Two-way ANOVA with Sidák’s multiple comparisons test of control (n = 4 mice) vs. mutant (n = 5 mice) by layer; all genotype comparisons by layer are non-significant.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile ranges (box).
See also Figures S4 and S5.
To assess differences in lamination, we determined medial ganglionic eminence (MGE)- and CGE-interneuron density and distribution across layers in 5HT3aR.Cxcl14fl/fl.RCE and control mice at P8 (Figures 4D–4F and S5). We did not observe any significant impact on the density of 5HT3aR-expressing interneurons (Figure S5A) nor the more specific subtypes of Re-, Sst-, Vip-, or Npy-expressing interneurons in 5HT3aR.Cxcl14fl/fl.RCE mice compared to controls (Figures S5B–S5E). Similarly, the distribution of these interneuron subtypes into their cortical laminae appeared unaffected, nor was the cortical laminar area (Figures 4E, 4F, and S5F–S5I). These results indicate that, unlike Cxcl12, Cxcl14 is not required for the laminar position of cortical interneurons.
Cxcl14 is necessary for the morphological development of LI interneurons
As activity-dependent Cxcl14 expression in LI SBCs predominates at a time when these neurons are acquiring subtype-specific features (Figure 2), we hypothesized that this chemokine may regulate functional differentiation. Thus, we performed whole-cell patch-clamp recordings of LI interneurons in control and 5HT3aR.Cxcl14fl/fl.RCE mice from P8 to P10 and reconstructed their neuronal morphology (Figure 5A). We classified the cells as LI SBC or NGFC based on the laminar allocation (LIb vs. LIa), spike onset, and presence of a translaminar axon (see STAR methods; Figure 2). SBC reconstructions were analyzed for axonal complexity and length (Figures 5B and S6A), vertical axonal length and areal coverage (Figures 5C–5E), and number of axonal nodes (Figure S6B). Our analysis revealed an increase in axonal complexity in mid-deep layers of the cortex (350–500 μm) in 5HT3aR.Cxcl14fl/fl.RCE compared to 5HT3aR.RCE (control) mice (Figures 5A and 5B). Yet, despite the increase in axonal complexity, neither the total length nor the number of nodes was different between genotypes at this developmental stage (Figures S6A and S6B). Similarly, the vertical axonal length and coverage were not significantly different between the controls and the mutants (Figures 5C–5E). Interestingly, however, two SBC reconstructions revealed axons that aberrantly crossed the entire cortical column to reach the white matter in mutant mice (Figure 5A). In addition, LI SBCs exhibited a significant increase in dendritic complexity in 5HT3aR.Cxcl14fl/fl.RCE compared to control mice, with an increased number of dendritic intersections (Figures 5F and 5G). Dendritic length also showed a trend toward larger values in mutant mice (Figure S6C), yet the number of dendritic nodes was not significantly different between the groups (Figure S6D). This suggested to us that tortuosity of the dendritic tree, as opposed to branching per se, was increased in SBCs following loss of Cxcl14. Note that differences in SBC axons and dendrites may become more apparent at later developmental stages as CGE-derived interneurons continue to grow into the third postnatal week.45 Altogether, these results demonstrate that Cxcl14 is required in SBC interneurons to shape the tortuosity of axonal and dendritic processes.
Figure 5. Cxcl14 loss leads to increased tortuosity in neuronal processes of single-bouquet cells.
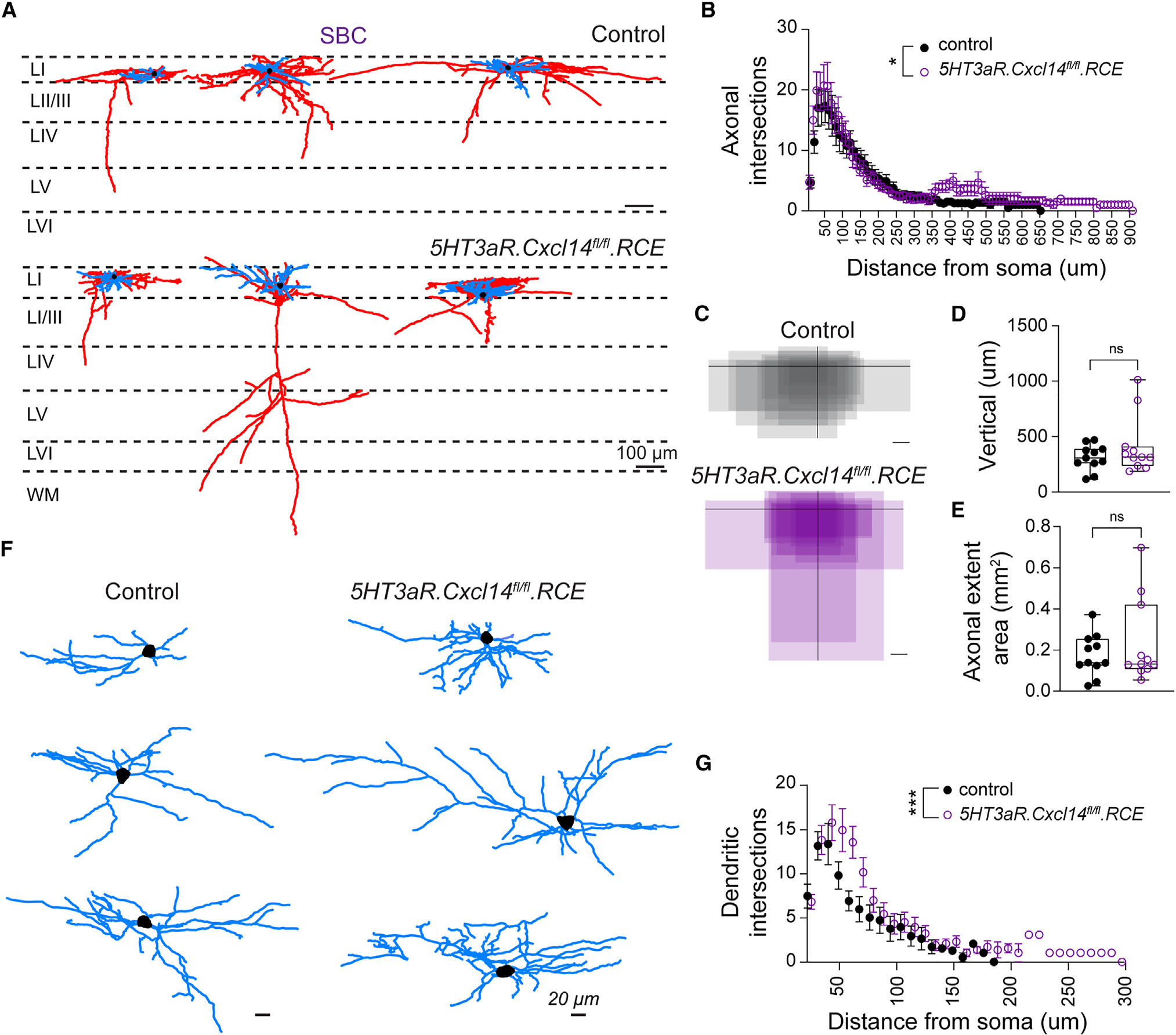
(A) Representative reconstructions of SBCs from 5HT3aR.RCE (control, top) and 5HT3aR.Cxcl14fl/fl.RCE (mutant, bottom) mice at P8–P10 in S1. Dashed lines demarcate layer borders; scale bars, 100 μm. Blue, dendrite; red, axon; black, soma.
(B) Scholl analysis for axonal complexity. Two-way ANOVA of control (n = 11 cells, 7 mice) vs. mutant (n = 11 cells, 8 mice); *p = 0.037.
(C) Summary of axonal extent in 5HT3aR.RCE (control, top) and 5HT3aR.Cxcl14fl/fl.RCE (mutant, bottom) with respect to the dura. Soma location is represented by the intersection of black lines. Control, n = 11 cells, 7 mice; mutant, n = 11 cells, 8 mice; scale bars, 100 μm.
(D) Vertical axonal length. Unpaired t test of control (n = 11 cells, 7 mice) vs. mutant (n = 11 cells, 8 mice); non-significant (ns).
(E) Axonal areal extent. Unpaired t test of control (n = 11 cells, 7 mice) vs. mutant (n = 11 cells, 8 mice); non-significant (ns).
(F) High-magnification depiction of dendritic trees from cells shown in (A); scale bars, 20 μm.
(G) Dendritic Scholl analysis. Two-way ANOVA of control (n = 11 cells, 7 mice) vs. mutant (n = 11 cells, 8 mice); ***p < 0.0008.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile ranges (box).
See also Figures S6 and S7.
To determine if Cxcl14 loss impacts other neuronal populations, we analyzed the morphological development of LI NGFCs in 5HT3aR.Cxcl14fl/fl.RCE mice (Figure S7). As expected, we found that axonal and dendritic trees of these cells were restricted primarily to LI in both 5HT3aR.Cxcl14fl/fl.RCE and control mice (Figures S7A and S7B). Notably, NGFCs showed increased axonal length and complexity in mutant mice (Figures S7C and S7D). However, like SBCs, axonal nodes in NGFCs were not significantly different between genotypes (Figure S7E). Finally, the dendritic length and complexity were comparable in control and 5HT3aR.Cxcl14fl/fl.RCE mice (Figures S7F–S7H), in contrast to what we observed for SBCs. Altogether these results mirror the overelaboration of neurites seen following knockdown of Cxcl14 in the chick cornea,46 suggesting a consistent role for Cxcl14 in constraining neuronal outgrowth.
SBCs exhibit increased excitability upon Cxcl14 loss
As our previous work revealed a strong link between intrinsic excitability and morphology,47 we next determined if loss of Cxcl14 impacts the maturation of intrinsic electrophysiological properties. To investigate this, we carried out whole-cell patch-clamp recordings from randomly selected LI interneurons (Figures 6A–6C) that expressed GFP in control (5HT3aR.RCE) and 5HT3aR.Cxcl14fl/fl.RCE mice from P8 to P10 in S1 to gather passive and active membrane properties (Table S2).
Figure 6. Developmental loss of Cxcl14 leads to increased excitability of SBCs.
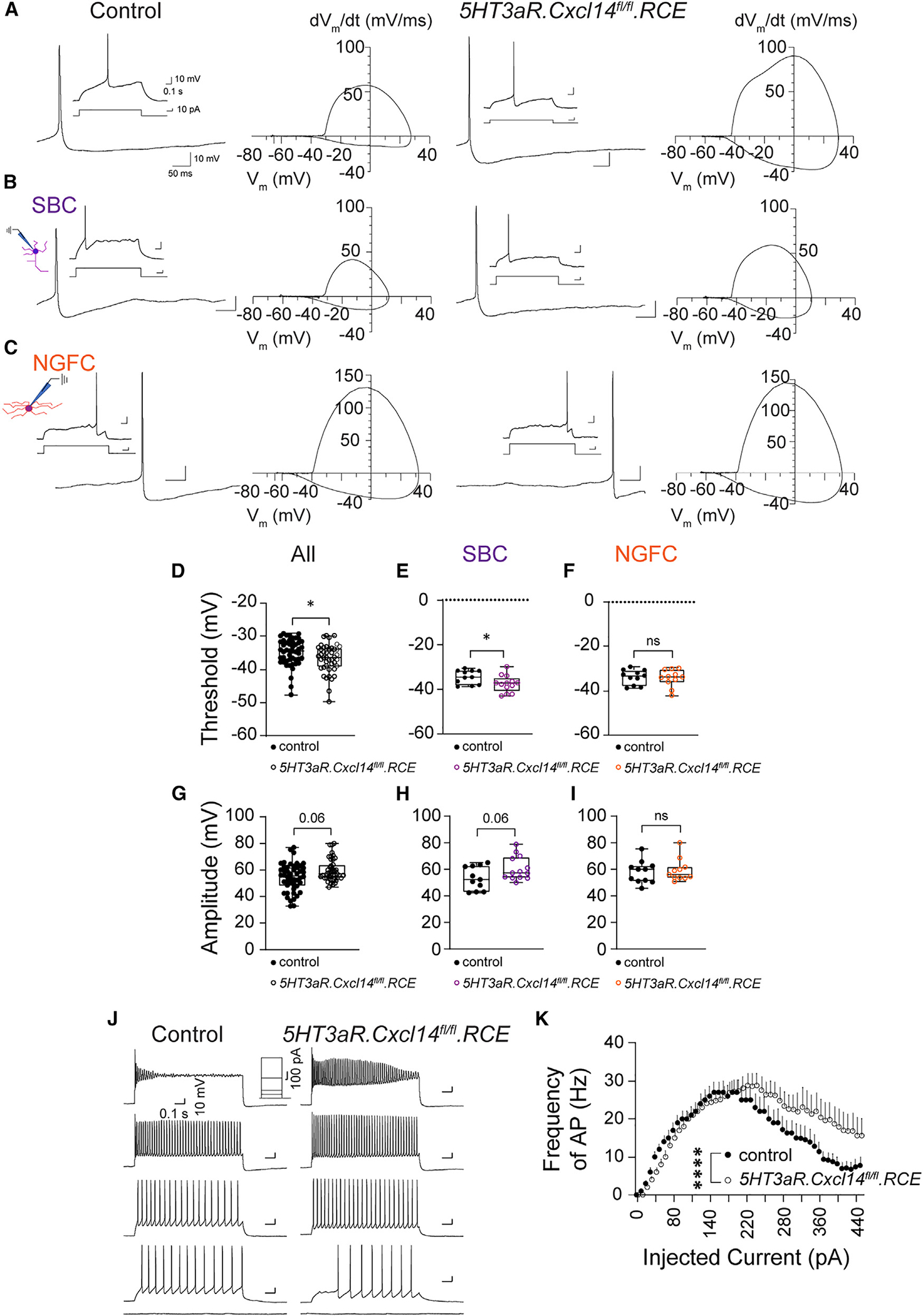
(A) Representative voltage trace of a single action potential (AP) fired at rheobase by an LI interneuron in 5HT3aR.RCE (control) or 5HT3aR.Cxcl14fl/fl.RCE (mutant) mice at P8–P10 in S1. Insets show the 1.5 s recording window with 1 s stimulation eliciting the AP wave. Phase plots are shown to highlight differences between groups identified by genotype. Control, n = 48 cells, 14 mice; mutant, n = 44 cells, 10 mice.
(B) Representative voltage trace of a single AP fired at rheobase by an LI SBC, identified by the presence of a descending axon. Phase plots are shown to highlight differences between groups identified by genotype and morphology. Control, n = 11 cells, 9 mice; mutant, n = 13 cells, 7 mice.
(C) Representative voltage trace of a single AP fired at rheobase by an LI NGFC interneuron. Control, n = 11 cells, 6 mice; mutant, n = 11 cells, 7 mice.
(D) AP threshold (all cells). Mann-Whitney test of control (n = 48 cells, 14 mice) vs. mutant (n = 44 cells, 10 mice); *p = 0.035.
(E) AP threshold (SBC). Unpaired t test of control (n = 11 cells, 9 mice) vs. mutant (n = 13 cells, 7 mice); *p = 0.044.
(F) AP threshold (NGFC). Unpaired t test of control (n = 11 cells, 6 mice) vs. mutant (11 cells, 7 mice); non-significant (ns). The control dataset (black) is replotted from the NGFC cohort in Figure 2G.
(G) AP amplitude (all cells). Mann-Whitney test of control (n = 48 cells, 14 mice) vs. mutant (n = 44 cells, 10 mice); non-significant, p = 0.06.
(H) AP amplitude (SBC). Unpaired t test of control (n = 11 cells, 9 mice) vs. mutant (n = 13 cells, 7 mice); non-significant, p = 0.06.
(I) AP amplitude (NGFC). Mann-Whitney test of control (n = 11 cells, 6 mice) vs. mutant (11 cells, 7 mice); non-significant (ns).
(J) Representative voltage traces in current clamp in response to no stimulation and current steps (50, 100, 200, and 440 pA) in 5HT3aR.RCE (control, left) and 5HT3aR.Cxcl14fl/fl.RCE (mutant, right) mice at P8–P10 in S1.
(K) Input-output relationship. Two-way ANOVA of control (n = 30 cells, 10 mice) vs. mutant (n = 36 cells, 10 mice); ****pgenotype < 0.0001.
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile range (box). See also Tables S2–S4.
As a whole, we found that the LI interneuron population exhibited lower AP threshold in mutants compared to controls (Figures 6A and 6D; Table S2). This indicated increased excitability following developmental loss of Cxcl14. After classifying recorded neurons as SBCs or NGFCs, we found that LI SBCs exhibited a more hyperpolarized AP threshold in 5HT3aR.Cxcl14fl/fl.RCE mice compared to controls (Figure 6E; Table S3). In contrast, the NGFC cohort exhibited no difference in AP threshold (Figure 6F; Table S4). AP amplitude in both the LI interneuron cohort (Figure 6G; Table S2) and the SBC subgroup (Figure 6H; Table S3) trended toward an increase in mutants in comparison with controls, but not in NGFCs (Figure 6I; Table S4). Furthermore, the AP afterhyperpolarization peak was significantly different between mutant and controls (Table S2). Finally, we observed faster AP entrainment at high current stimulations (Figures 6J and 6K), further supporting the presence of an intrinsically hyperexcitable SBC population in the absence of Cxcl14 expression. Altogether, these results reveal that Cxcl14 is both a molecular marker for LI SBC interneurons and a crucial regulator of their cellular physiology.
Dysregulated activation of LI interneurons in 5HT3aR.Cxcl14 mice in vivo
To determine if recruitment of LI neurons is compromised in Cxcl14 mutants, we performed longitudinal two-photon calcium imaging in control and 5HT3aR.Cxcl14fl/fl pups. We crossed these mice to a GCaMP6s conditional reporter line and opened cranial windows over the barrel cortex at P56 (Figures 7A and 7B). We carried out longitudinal imaging in unanesthetized pups at P7 and P14 (Figures 7C–7F; Videos S1 and S2). Our analysis revealed a decreased number of spontaneously active cells in 5HT3aR.Cxcl14fl/fl.GCaMP6s compared to control mice at both stages (Figure 7G). In addition, the event duration of single-cell events decreased in mutant—but not control—mice from P7 to 14 (Figure 7H). In agreement with the developmental maturation of intrinsic properties,37 the frequency of single-cell calcium events increased with age but was not different between genotypes (Figure 7I). However, the single-cell analyses suggested bimodal distributions. Indeed, the duration distribution showed a high percentage of low-duration events and a low percentage of very-long-duration events in mutant mice (Figures S8A and S8B). In addition, the frequency distribution showed a cluster of events with low frequency as well as a cluster of events with a frequency distribution similar to that of the control data (Figures S8C and S8D). This smaller, low-frequency cluster may represent cells that infrequently burst-spike but for longer duration than the dominant cohort of frequently firing, short-duration cells. Finally, the average amplitude of single-cell events decreased from P7 to P14 in controls but not in 5HT3aR.Cxcl14fl/fl.GCaMP6s mice (Figure 7J). Together, these data support our findings that Cxcl14 loss leads to persistence of burst-spiking within a cohort of LI interneurons, but not the population en masse, which may reflect increased intrinsic excitability in SBC, but not NGFC, LI interneurons in cortical slices.
Figure 7. Cxcl14 is necessary for the assembly of thalamocortical circuits in the developing somatosensory cortex.
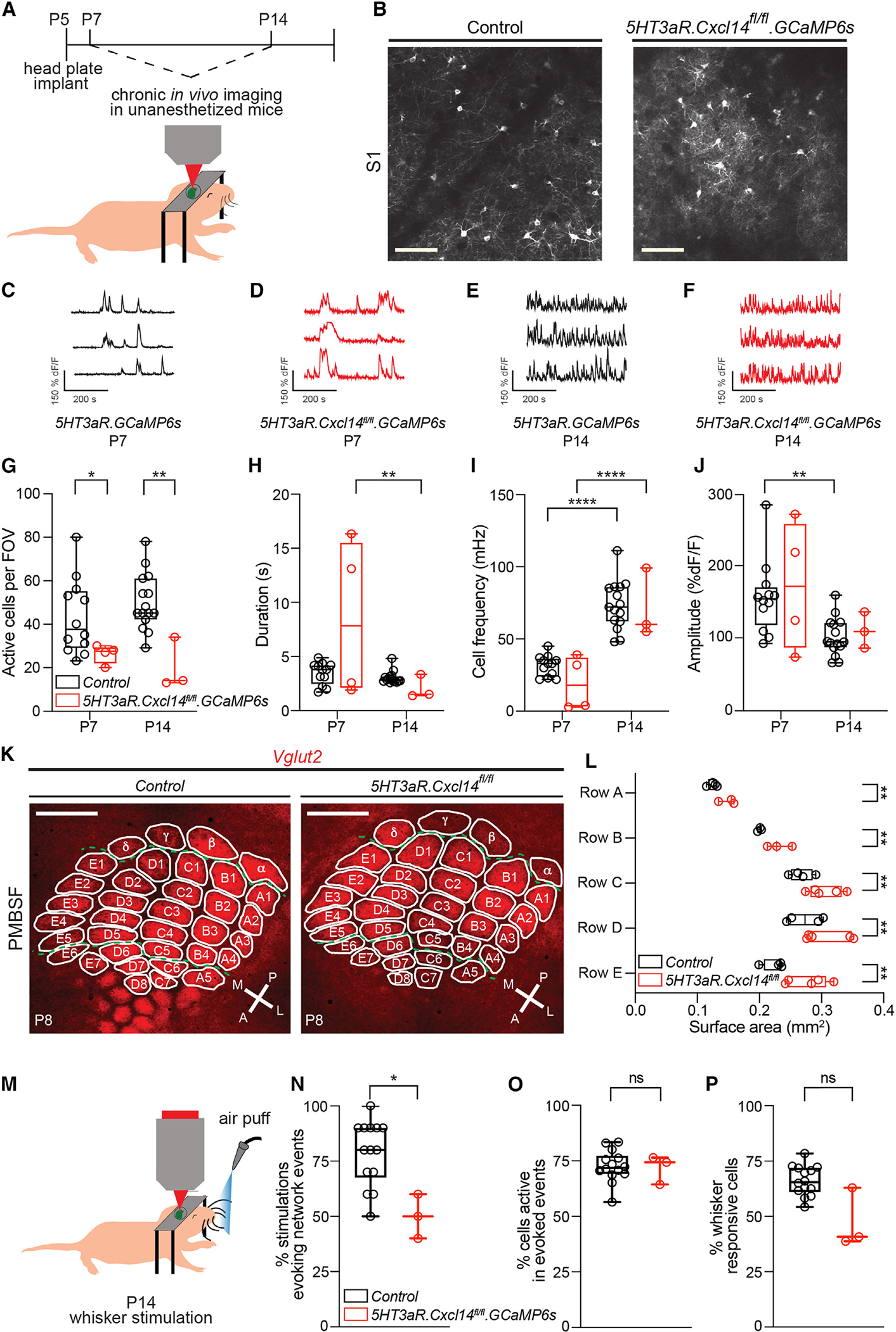
(A) Schematic for longitudinal two-photon imaging through a cranial window in S1.
(B) Representative images of fields of view (FOVs) from 5HT3aR.GCaMP6s (control, left) and 5HT3aR.Cxcl14fl/fl.GCaMP6s (mutant, right) mice at P7. Scale bars, 100 μm.
(C–F) Representative dF/F traces of three 5HT3aR.GCaMP6s LI interneurons at P7 (C and D) and P14 (E and F). Control: P7, n = 12 FOVs, 9 mice; P14, n = 15 FOVs, 10 mice. Mutants: P7, n = 4 FOVs, 3 mice; P14, n = 3 FOVs, 3 mice (C–J).
(G) Average number of spontaneously active cells per FOV. Two-way ANOVA, ***pgenotype < 0.001; Tukey’s multiple comparison test, control vs. mutant at P7, *p = 0.046, and P14, **p = 0.002.
(H) Average single-cell event duration. Two-way ANOVA, **page < 0.01; Tukey’s multiple comparisons test, mutant at P7 vs. P14, **p = 0.001.
(I) Average single-cell event frequency. Two-way ANOVA, ****page < 0.0001; Tukey’s multiple comparisons test, control at P7 vs. P14, ****p < 0.0001, and mutant at P7 vs. P14, ****p < 0.0001.
(J) Average single-cell event amplitude. Two-way ANOVA, **page < 0.01; Tukey’s multiple comparisons test, control at P7 vs. P14, **p = 0.004.
(K) Representative images of posteromedial barrel subfield (PMBSF) in 5HT3aR.Cre (control) and 5HT3aR.Cxcl14fl/fl (mutant) mice at P8. Thalamic afferents were visualized by VGlut2 immunoreactivity. Individual barrels are outlined and labeled. A, anterior; P, posterior; M, medial; L, lateral. Scale bar, 500 μm.
(L) Quantification of barrel areas as the sum of individual barrel areas per row (rows A to E). Two-way ANOVA, ****pgenotype < 0.0001; Tukey’s multiple comparisons test of control (n = 4 mice) vs. mutant (n = 3 mice), rows A–E, **p = 0.001.
(M) Schematic for calcium imaging in S1 after whisker stimulation. 5HT3aR.GCaMP6s (control), n = 14 FOVs, 10 mice, and 5HT3aR.Cxcl14fl/fl.GCaMP6s mice, n = 3 FOVs, 3 mice (M–P).
(N) Average percentage of whisker stimulations that evoked network events. Unpaired t test of control vs. mutant, *p = 0.04.
(O) Average percentage of cells active in whisker-evoked network events, normalized to the percentage of total active cells per FOV. Unpaired t test of control vs. mutant, non-significant (ns).
(P) Average percentage of cells responsive to whisker stimulation. Unpaired t test of control vs. mutant, non-significant (ns).
Data are represented as box-and-whisker plots from minimum to maximum range (whiskers) with interquartile ranges (box).
See also Figure S8.
Since LI interneurons regulate sensory map formation in response to sensory inputs,6 we determined whether this process is compromised in 5HT3aR.Cxcl14fl/fl mutants. To this end, we assessed barrel map cytoarchitecture by measuring TCA elaboration in flat-mount preparations of the primary somatotopic representation of murine vibrissae in S1, the posteromedial barrel subfield (PMBSF), in control and 5HT3aR.Cxcl14fl/fl mice at P86,48 (Figure 7K). Our results revealed an expansion of TCAs across all principal rows of the PMBSF in mutant pups compared to controls (Figure 7L), suggesting that Cxcl14 expression in LI interneurons is necessary for the appropriate restriction of TCAs into whisker-aligned cortical columns. The overall cortical area devoted to the PMBSF was not significantly affected by loss of Cxcl14 (Figure S8E), consistent with the strong influence of sensory-independent processes in cortical regionalization.49–51 Interestingly, however, we did not observe a significant increase in the total area of TCA elaboration within the PMBSF despite detecting an impact on a row-by-row basis (Figure S8F), perhaps signifying a role for Cxcl14 in the distribution of ascending TCAs among individual neighboring barrels within and across rows of the PMBSF.
To determine the functional impact of disrupted barrel structure, we recorded cortical responses to whisker stimulation in 5HT3aR.GCaMP6s and 5HT3aR.Cxcl14fl/fl.GCaMP6s mice at P7 and P14, during a critical window for sensory circuit assembly (Figures 7M–7P and S8G–S8K). To mimic passive whisker deflections resulting from interactions with the dam and littermates,52 we applied air puffs to repetitively stimulate the whisker pad, causing a displacement of all vibrissae. This strategy allowed for the quantification of cortical responses after stimulation. Our results revealed a significant decrease in the total number of active cells and stimulations that triggered network events at P14, but not P7 (Figures 7N, S8G, and S8I). In contrast, neither the amplitude of single-cell events nor the percentage of LI interneurons active in network events or the percentage of cells responding to whisker stimulation were different between genotypes (Figures 7O, 7P, S8H, S8J, and S8K), indicating that these cells are still able to respond to whisker stimulation after Cxcl14 abrogation, but these inputs are less effective in triggering network activation. These results demonstrate that Cxcl14 is required for proper assembly of sensory maps and processing during postnatal development.
DISCUSSION
The role of sensory experience in circuit assembly has been increasingly appreciated.52–54 However, the mechanisms that translate sensory-evoked activity into programs for interneuron development still need elucidating. Here, we demonstrate that Cxcl14 is expressed in LI SBC interneurons in response to sensory experience at neonatal stages and that loss of its function compromises the functional properties of this layer.
Neuronal activity shape interneuron differentiation during development.36,47,55–61 Within superficial circuits, thalamic afferents reach LI as early as P6 and regulate network events across this layer.6 We find that SBC and NGFC interneurons are apparent from P8 onward, shortly after the arrival of thalamic terminals. Furthermore, our experiments indicate that sensory inputs and Cxcl14 regulate the maturation of intrinsic electrophysiological and morphological properties of LI SBCs, which are fundamental signatures of subtype specification.14
Although loss of Cxcl14 function did not impair the intrinsic maturation of NGFCs, both SBCs and NGFCs showed morphological deficits after Cxcl14 abrogation. The morphological deficits in LI SBCs may be a consequence of abnormal excitability or reflect an independent requirement of Cxcl14 for the elaboration of dendrites and axons.62–65 Cxcl14 may act in both a paracrine and an autocrine manner. In other systems, Cxcl14 is secreted66; however, the cellular allocation in neurons is not known.67 Based on transcriptomic analysis of adult cells,68 NGFCs appear to lack Cxcl14 expression. Therefore, this cytokine may shape morphology via paracrine action on these cells, consistent with the known mechanism of related chemokine signaling within the developing cortex.27,69,70 Altogether, these findings support a role for sensory inputs and Cxcl14 in the emergence of subtype identity.
What is the cellular mechanism by which Cxcl14 regulates subtype differentiation? As the cognate receptor for Cxcl14 has not been identified, efforts to untangle its functional role in the nervous system have been particularly complicated. Nevertheless, experimental evidence supports a role for this chemokine in circuit function. In the adult dentate gyrus niche, Cxcl14 attenuates, while Cxcl12 promotes, GABAergic transmission.71 Similarly, Cxcl14 attenuates,46 while Cxcl12 promotes, axonal growth.72 In contrast to the limited understanding of Cxcl14 signaling, the receptors for Cxcl12 have been well characterized. In the CNS, this chemokine signals through Cxcr4 and Ackr3 (formerly Cxcr7)38,73 and regulates neuronal excitability via the G protein-coupled inwardly rectifying pottasium (GIRK) channels, big potassium (BK) channels, voltage-gated sodium (NaV) channels,74,75 and neurotransmitter receptor expression.76 Cxcl14, therefore, may similarly shape intrinsic excitability by regulating the expression and/or activation of select voltage-gated channels in SBC interneurons.
Is LI functionally specialized during the first week of development? In the barrel cortex, the basic functional unit is the column, which represents the receptive field for a single whisker.77 From P0 to P4, TCA inputs trigger columnar-restricted activity.78–80 Furthermore, these cortical columns show whisker selectivity long before active whisking.81 We found that LI SBC axons reach deep layers already at P8, suggesting a role in barrel column maturation. Indeed, loss of Cxcl14 rendered LI interneurons less spontaneously active and impaired their recruitment by whisker stimulation in vivo and caused an expansion of barrel columns. Because the ratio of interneuron subtypes in the cortex did not change upon Cxcl14 loss, it is unlikely that impediments in network activity are due to cell death or migratory deficits. Instead, the reduced number of active LI neurons after Cxcl14 loss in vivo may reflect an augmented inhibitory synaptic drive from LI NGFCs,12,82,83 which show increased axonal coverage in Cxcl14 mutants. Connectivity between NGFCs and SBCs is observed in the adult and may also be present in early development.84 Thus, abnormal inhibitory output from NGFCs may decrease the probability of whisker-evoked events in mutant mice.85 Furthermore, our previous work and that of others show that LI interneurons receive thalamic connectivity and respond to whisker stimulation at P6–P12.6,86 Our current results indicate that this recruitment is compromised in Cxcl14 mutants, suggesting decreased excitatory/inhibitory synaptic balance in SBCs. In addition, LI SBCs in the mutant group may undergo homeostatic downscaling of excitatory synapses to counteract intrinsic hyperexcitability (Figures 6E, 6J, and 6K), rendering them less active in the network.87 Together, these changes in the SBC and the NGFC cohorts might impede the recruitment of LI interneurons in early spontaneous and evoked networks.
How do superficial LI SBCs sharpen the barrel map in LIV? During the first postnatal week, thalamic inputs recruit LV Sst interneurons to inhibit LIV spiny stellate cells.88,89 Similarly, LI SBC activation in response to sensory inputs may provide further spatially constrained feedforward inhibition onto LIV and LV excitatory cells, facilitating columnar segregation and refinement of the sensory map. Such a translaminar function of SBCs may be conserved across sensory cortical areas. In the auditory cortex, superficial interneurons with vertically descending axons, reminiscent of SBCs, relay sensory inputs to LIV excitatory cells.18,90 Future efforts to uncover the downstream neuronal targets of LI SBCs in S1 would thus be illuminating and aid in underscoring a stereotyped role for early sensory experience in tailoring subtype-specific interneuron circuits to promote the development of columnar topography across cortical domains.
In summary, these results provide mechanistic insight into how early sensory experience is translated into cell-type-specific programs for circuit assembly and uncover an unexpected role for chemokines in activity-dependent differentiation of cortical interneurons.
Limitations of the study
While our study advances understanding of the molecular diversity of LI interneurons, the ability to genetically manipulate and independently parse between the four currently described LI neuronal subtypes is not currently possible. While we report the regulatory role of Cxcl14 in the development of SBCs and NGFCs in LI, we were unable to manipulate either cell type independent of the other nor able to explore impacts of Cxcl14 loss of function on other LI populations, such as canopy cells.14 Isolation of specific interneuron types with an intersectional approach through the use of multiple genetic drivers, viral infections, and/or in utero electroporation will enable the study of the contribution to LI cell types in the future.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Natalia V. De Marco García (nvd2018@med.cornell.edu).
Materials availability
The BaseScope™ BA-MmCxcl14–2zz-st-C1 probe targeting exon 2 of the Cxcl14 gene is available through ACD Bio (see key resources table).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Goat α-GFP | Rockland Immunochemicals | Cat#600–101–215; RRID: AB_218182 |
| Chicken α-RFP | Rockland Immunochemicals | Cat#600–901–379; RRID: AB_10704808 |
| Guinea pig α-Vesicular glutamate transporter 2 | Synaptic Systems | Cat#135404; RRID: AB_887884 |
| Rabbit α-Somatostatin | Abcam | Cat#ab64053; RRID: AB_1143012 |
| Rabbit α-Vasoactive intestinal peptide | ImmunoStar | Cat#20077; RRID: AB_572270 |
| Rabbit α-Neuropeptide Y | ImmunoStar | Cat#22940; RRID: AB_2307354 |
| Rabbit α-Calretinin | Swant | Cat#7697; RRID: AB_2619710 |
| Mouse α-Reelin | MBL International | Cat#D223–3; RRID: AB_843523 |
| Tetramethylrhodamine conjugated α-Bungarotoxin | Molecular Probes | Cat#T1175; RRID: AB_2313931 |
| Alexa Fluor 647 conjugated Streptavidin | Molecular Probes | Cat#S32357 |
| AlexaFluor 488 Donkey α-Goat | Jackson ImmunoResearch Labs | Cat#705–545-003; RRID: AB_2340428 |
| Cy3 Donkey α-Chicken | Jackson ImmunoResearch Labs | Cat#703–165-155; RRID: AB_2340363 |
| Cy3 Donkey α-Rabbit | Jackson ImmunoResearch Labs | Cat#711–165-152; RRID: AB_2307443 |
| AlexaFluor 647 Donkey α-Guinea pig | Jackson ImmunoResearch Labs | Cat#706–605-148; RRID: AB_2340476 |
| AlexaFluor 647 Donkey α-Mouse | Thermo Fisher Scientific | Cat#A32787; RRID: AB_2762830 |
| TSA Plus Cy3 Reagent | Akoya Biosciences | Cat#TS-000202 |
| TSA Plus Cy5 Reagent | Akoya Biosciences | Cat#TS-000203 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAV1-hSyn-FLEx-mGFP-2A-Synaptophysin-mRuby | Generated from UNC viral core from commercially available plasmid (see recombinant DNA) | RRID: SCR_023280 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| GelRed nucleic acid gel stain | Biotium | Cat#41003–1 |
| Bovine serum albumin, 0.5% | Miltenyi Biotec | Cat#130–091–376 |
| Dulbecco’s phosphate-buffered saline with calcium and magnesium | Thermo Fisher Scientific | Cat#14040133 |
| 20x saline sodium citrate buffer | Thermo Fisher Scientific | Cat#AM9770 |
| 4’,6’-Diamidino-2-Phenylindole (DAPI) | Thermo Fisher Scientific | Cat#D-1306 |
| Gill’s Hematoxylin I | Thermo Fisher Scientific | Cat#HXGHE1LT |
| GeneRule 1kb Plus DNA Ladder | Thermo Fisher Scientific | Cat#SM1333 |
| Biocytin | Sigma Aldrich | Cat#B4261 |
| Sodium chloride | Sigma Aldrich | Cat#S7653 |
| Sodium bicarbonate | Sigma Aldrich | Cat#S5761 |
| Sodium phosphate monobasic monohydrate | Sigma Aldrich | Cat#S9638 |
| Potassium chloride, 4M | Sigma Aldrich | Cat#P4504 |
| Potassium D-gluconate | Sigma Aldrich | Cat#G4500 |
| Calcium chloride | Sigma Aldrich | Cat#C3306 |
| Magnesium chloride hexahydrate | Sigma Aldrich | Cat#M2670 |
| Dextrose | Sigma Aldrich | Cat#D9434 |
| Sucrose, 99.5% | Sigma Aldrich | Cat#S9378 |
| Adenosine 5’-triphosphate magnesium salt | Sigma Aldrich | Cat#A9187 |
| Guanosine 5’-triphosphate sodium salt hydrate | Sigma Aldrich | Cat#G8877 |
| Phosphocreatine di(tris) salt | Sigma Aldrich | Cat#P1937 |
| 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) Buffer | Sigma Aldrich | Cat#H3375 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’- tetraacetic acid (EGTA) | Sigma Aldrich | Cat#E3889 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAscope™ Multiplex Fluorescent Reagent Kit v2 | ACD Bio | Cat#323110 |
| BaseScope™ Reagent Kit v2 RED | ACD Bio | Cat#323900 |
| Neural Tissue Dissociation Kit for Postnatal Neurons | Miltenyi Biotec | Cat#130–094-802 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69504 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Swiss Webster: Tac:SW | Taconic Farms | SW-F; SW-M; RRID:MGI:2160840 |
| Mouse: 5HT3aR.eGFP: Tg(Htr3a-EGFP)DH30Gsat/ Mmnc | Mutant Mouse Resource and Research Center (MMRRC) | MMRRC:000273-UNC; RRID: MMRRC_000273-UNC |
| Mouse: Cxcl14.eGFP: Tg(Cxcl14-EGFP)PS15Gsat/ Mmucd | Mutant Mouse Resource and Research Center (MMRRC) | MMRRC:034771-UCD; RRID: MMRRC_034771-UCD |
| Mouse: 5HT3aR.Cre: Tg(Htr3a-cre)NO152Gsat/Mmucd | a gift from N. Heintz, The Rockefeller University | MMRRC:036680-UCD; RRID: MMRRC_036680-UCD |
| Mouse: NdnfCre: B6.Cg-Ndnftm1.1(folA/cre)Hze/J | The Jackson Laboratory | JAX:028536; RRID: IMSR_JAX:028536 |
| Mouse: Cxcl14fl/fl: C57BL/6-Cxcl14tm1/Rbrc | RIKEN BioResearch Resource Center | RBRC:02308; RRID: IMSR_RBRC02308 |
| Mouse: RCE: Gt(ROSA)26Sortm1.1(CAG-EGFP)Fsh/Mmjax | The Jackson Laboratory | JAX:032037; RRID: MMRRC_032037-JAX |
| Mouse: Ai9: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX:007909; RRID: IMSR_JAX:007909 |
| Mouse: GCaMP6s: B6;129S6-Gt(ROSA)26Sortm96(CAG-GCaMP6s)Hze/J | The Jackson Laboratory | JAX:024106; RRID: IMSR_JAX:024106 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Mm-Cxcl14-C1 | ACD Bio | Cat#459741 |
| Mm-Chrna7-C2 | ACD Bio | Cat#465161 |
| BA-Mm-Cxcl14–2zz-st-C1 | ACD Bio | N/A; See in situ hybridization methods |
| S1-sense: 5’-GCCCTCTGTCGCGGTCCTGC-3’ | Eurofins Genomics | N/A |
| S2-sense: 5’-ATGAGGCTCCTGGCGGCC-3’ | Eurofins Genomics | N/A |
| A1-antisense: 5’-GTCllGTCAAAGGCAGAGAG-3’ | Eurofins Genomics | N/A |
| A2-antisense: 5’-GTCCGATCTAACCCTAGGTTG-3’ | Eurofins Genomics | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pAAV-hSyn-FLEx-mGFP-2A-Synaptophysin-mRuby | Generated by Beier et al.91 | Addgene: Cat#71760; http://n2t.net/addgene:71760; RRID: Addgene_71760 |
|
| ||
| Software and algorithms | ||
|
| ||
| SnapGene | SnapGene | https://www.snapgene.com; RRID: SCR_015052 |
| Neurolucida 360 | MBF Bioscience | https://www.mbfbioscience.com/products/neurolucida-360/; RRID: SCR_001775 |
| Neurolucida Explorer | MBF Bioscience | https://www.mbfbioscience.com/products/neurolucida-explorer; RRID: SCR_017348 |
| Fiji (Fiji is just ImageJ) | Developed by Schindelin et al.92 | https://ImageJ.net/software/fiji/; RRID: SCR_002285 |
| Cell counter, ImageJ plugin | Developed by K. De Vos | https://imagej.net/ij/plugins/cell-counter.html |
| Pairwise stitching, ImageJ plugin | Developed by Preibisch et al.93 | https://imagej.net/Image_Stitching#Pairwise_Stitching |
| IsoData thresholding, ImageJ plugin | Developed by Ridler and Calvard94 | https://imagej.net/plugins/auto-threshold |
| AxoGraph | Developed by J. Clements | https://www.axograph.com/; RRID: SCR_014284 |
| MATLAB version: 8.5.0 (R2015a) | MathWorks | https://www.mathworks.com/products/MATLAB.html; RRID: SCR_001622 |
| CalciumDx | Developed by Ackman et al.95 | https://github.com/ackman678/CalciumDX |
| GraphPad Prism 10 | GraphPad | https://www.graphpad.com/; RRID: SCR_002798 |
|
| ||
| Other | ||
|
| ||
| Olympus confocal microscope | Olympus | Model#FV1000; RRID: SCR_016840 |
| Olympus multiphoton laser scanning microscope | Olympus | Model#FVMPE-RS |
| Mai Tai Deepsee Ti:Sapphire ultrafast laser | Spectra-Physics | https://www.spectra-physics.com/en/f/mai-tai-ultrafast-laser |
| HybEZ™ II Oven | ACD Bio | Cat#321710 |
| C&B Metabond Quick Adhesive Cement System | Parkell | Cat#S380 |
| 3M Vetbond Tissue Adhesive | Thermo Fisher Scientific | Cat#NC9259532 |
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines
All animal care and procedures were performed according to the Weill Cornell Medicine Research Animal Resource Center guidelines. Animals were housed in a controlled environment on a 12-hour light/dark cycle with food and water ad libitum. The following lines have been described previously: Swiss Webster (Tac:SW; Taconic Farms), 5HT3aR.eGFP (Htr3a-EGFP; MMRRC 000273-UNC), NdnfCre (Ndnf-IRES2-dgCre-D; JAX 028536), RCE (Gt(ROSA)26Sortm1.1(CAF-EGFP)Fsh/Mmjax; JAX 032037), Ai9 (RCL-tdT; JAX 007909), GCaMP6s (Ai96, JAX 024106). The 5HT3aR.Cre mouse was a gift from N. Heintz, The Rockefeller University in New York City, NY, United States; this line targets most LI interneurons during developmental timepoints.6 Cxcl14.eGFP (Tg(Cxcl14-EGFP)PS15Gsat/Mmucd, MMRRC 034771-UCD) was made available through the Gene Expression Nervous System Atlas (GENSAT) project. Cxcl14fl/fl (Cxcl14<tm1>, RIKEN RBRC02308) was made available through R. Hata, Kanagawa Dental College, Yokosuka, Kanagawa, Japan.
For most experiments investigating the functional roles of Cxcl14 in developing interneurons (interneuron lamination and migration, electrophysiology, neuronal morphology), controls used were 5HT3aRCre+.Cxcl14+/+.RCE, as Cre-dependent GFP expression was needed to identify LI interneuron populations for analyses. 5HT3aRCre+.Cxcl14+/+.GCaMP6s were used in the calcium imaging experiments as controls as expression of GCaMP6s was necessary. Mutants were defined as 5HT3aRCre+.Cxcl14fl/fl with RCE or GCaMP6s as appropriate. Similarly, NdnfCre+ mice were used for viral injection experiments in both the unplucked and plucked condition to ensure Cre-dependent mGFP expression. Controls for barrel morphology analysis contained either 5HT3aRCre+.Cxcl14+/+.RCE or 5HT3aRCre−.Cxcl14+/+.RCE, as Cre-dependent GFP expression was not necessary for the analysis. For experiments involving whisker plucking, only mice reflecting littermate sets were included in analysis, and unplucked control mice were similarly anesthesized and manipulated. For Cxcl14fl/fl validation experiments, controls used were 5HT3aRCre+.Cxcl14fl/+.RCE, as the presence of both a mutant and wildtype band in heterozygotes was utilized in PCR experiments. Mice of both sexes were used for analysis. As all experimental animals used in these experiments were carried out at ages before reaching sexual maturity, the influence of sex was not analyzed.
METHOD DETAILS
Immunohistochemistry
For immunohistochemistry (IHC) experiments, the protocol was performed as previously described.6,37 The following primary antibodies were used at the indicated dilutions made in 5% normal donkey serum blocking buffer: 1:1000 Goat α-GFP (Rockland Immunochemicals, #600–101-215); 1:1000 Chicken α-RFP (Rockland Immunochemicals, #600–901-379); 1:1000 Guinea pig α-Vglut2 (Synaptic Systems, #135404); 1:500 Rabbit α-Sst (Abcam, #ab64053); 1:500 Rabbit α-Vip (ImmunoStar, #20077); 1:500 Rabbit α-Npy (ImmunoStar, #22940); 1:500 Rabbit α-Cret (Swant, #7697); 1:500 Mouse α-Reelin (MBL International, #D223–3); 1:1000 Streptavidin-Alexa Fluor 647 Conjugate (Molecular Probes, #S32357). To label the a7-subunit of nicotinic acetylcholine receptors, a tetramethylrhodamine conjugated α-Bungarotoxin (Molecular Probes, #T1175) diluted 1:500 in the appropriate primary antibody solution as above was used. Incubation was carried out as for other primary antibodies.
The following secondary antibodies were used, all at a dilution of 1:500 made in 5% normal donkey serum blocked buffer: AlexaFluor 488 Donkey α-Goat (Jackson Immunoresearch, #705–545-003); Cy3 Donkey α-Chicken (Jackson ImmunoResearch Labs, #703–165-155); Cy3 Donkey α-Rabbit (Jackson ImmunoResearch Labs, #711–165-152); AlexaFluor 647 Donkey α-Guinea pig (Jackson ImmunoResearch Labs, #706–605-148); AlexaFluor 647 Donkey α-Mouse (Thermo Fisher Scientific, #A32787). Tissue sections were incubated in a 1:1000 solution of 4’,6’-Diamidino-2-Phenylindole (DAPI; Thermo Fisher Scientific, D-1306) in 1x PBS for 2 minutes at room temperature following secondary incubation.
In situ hybridization and analysis
Fluorescent
The manufacturer’s protocol for the RNAscope™ Multiplex Fluorescent Reagent Kit v2 Assay (ACD Biosciences, #323110) for fixedfrozen tissue was largely followed. All hybridization steps were carried out in a HybEZ™ II oven (ACD Biosciences). The following adjustments were made to the protocol to account for the fragility of tissue samples due to younger ages: The target retrieval step was carried out at room temperature by immersing the tissue in solution, instead of steaming the tissue at 99°C. The manufacturer’s DAPI solution was not used. Instead DAPI labeling was carried out as described in the previous section (Immunohistochemistry) to determine layer boundaries (Figures 1A, 1E, 3A, 3C and 3G).
The protocol was split into two days. Samples were stored in a 5x saline sodium citrate (SSC) buffer made fresh from a 20x SSC stock (Thermo Fisher Scientific, #AM9770) overnight at 4°C between days. Hybridization was carried out using the Mm-Cxcl14-C1 probe (ACD Biosciences, #459741) and Mm-Chrna7-C2 probe (ACD Biosciences, #465161) according to manufacturer’s specifications. Hybridization signal was developed using either TSA Plus Cy3 Reagent (Akoya Biosciences, TS-000202) or TSA Plus Cy5 Reagent (Akoya Bioscience, TS-000203). If IHC was to additionally be performed for dual labeling experiments, samples were subjected to the protocol as outlined above following completion of the RNAscope protocol.
Chromogenic
The manufacturer’s protocol for the BaseScope Detection Reagent Kit v2 – RED (ACD Biosciences) was followed. The protocol was completed on fixed frozen tissue. Antigen retrieval steps were followed as detailed in the manufacturer’s protocol. Hybridization was carried out using a custom-designed BaseScope probe targeting an ~40bp region of exon 2 of Cxcl14 (BA-Mm-Cxcl14–2zz-st-C1; ACD Biosciences). Samples were counterstained with Gill’s Hematoxylin I (American Master Tech Scientific, #HXGHE1LT) to determine layer boundaries (Figure 4A) and 0.02% Ammonia water prepared from an Ammonium hydroxide, 28–30% stock (Sigma-Aldrich, #320145). For FISH experiments, images were taken from every tissue slice exhibiting successful hybridization.
Cell density and laminar allocation quantifications
Image postprocessing was carried out in Fiji (ImageJ). Images of the full cortical column represent a stitched pair of images, using the deprecated 2D stitching plugin. Any adjustments to image brightness were equally applied across all color channels. For inversion of ISH signal, the Color Negative preset Curves Adjustment was applied in Photoshop (Adobe). For IHC cell density and lamination analyses,37 total cell density was calculated as the ratio between the total number of immunopositive cells across all cortical layers and the total area in 3–5 (3 for Reelin, Vip; 5 for Sst, Npy, Cret) representative sections to result in a single total areal density for the given marker and represents a single N (mouse). For layer specific areal densities (all other density analyses), the same process was followed, except the summed total of immunopositive cells were divided by the summed total area for the given layer only.
For FISH experiments, the same analysis process was largely followed37 except each imaged section served as an N (section). This was chosen due to the tissue processing capacity of the RNAscope kits, which are aliquoted to process 20 sections of tissue per kit. Calculation of densities therefore do not involve summing across multiple imaged sections per sample. Note that there are low and high GFP expressing Ndnf and Cxcl14 positive cells (Figure 3C). Both populations were considered Ndnf expressing cells for co-expression analysis.
For assessing the sublamination of Cxcl14.eGFP interneurons within LI (Figure 1), the pial boundary was outlined as was the border between LI and LII using the polygon selection tool in Fiji. Layer boundaries were determined by DAPI (Figures 1I, 1J and 4D). The scale bar of the image was used to set measurement scaling. A GFP expressing cell was identified and a straight line was drawn from the center of the cell body to the outlined pial boundary, stopping immediately upon intersection. The distance of this line was measured as the cell depth within LI. This line was then extended from the other end until it reached the outlined LI/LII border, stopping immediately upon intersection. The distance of this line was measured as the total depth of LI. For each cell, its depth within LI was divided by its corresponding total depth of LI and the result was expressed as a percentage.
Sensory deprivation
Newborn Swiss Webster, NdnfCre, or Cxcl14.eGFP pups underwent a bilateral sensory deprivation paradigm by whisker plucking as previously described.6,36 Maintenance whisker plucking was carried out each subsequent day until sacrifice and analysis at P5 (Swiss Webster) or P8 (NdnfCre and Cxcl14.eGFP). Controls underwent anesthesia by hypothermia, handling, and heating pad recovery, but whiskers were not removed.
Viral injections
Stereotactic injections of AAV1-hSyn.FLEx-mGFP-2A-Synaptophysin-mRuby (AAV1-hSyn-mGFP) virus (UNC Viral core, Addgene plasmid #71760) in the somatosensory cortex were were carried out from P0 to P16,37 and Injections were made at a depth between 70μm and 100μm and 29.4nL of virus was injected in three 9.8nL aliquots spaced apart by 10 seconds.
Axonal distribution analysis
Analysis of the laminar allocation of LI axonal morphology was carried out as previously described,37 with the following modifications. Thresholding was carried out utilizing the Auto Threshold built-in tool in ImageJ and the IsoData thresholding method. Low-volume injections of virus were utilized to minimize infection density within LI of imaged sections and limit the spread of infection beyond the barrel cortex. Mice that had at least two evenly spaced sections (200μm between sections) of tissue demonstrating >90% of infected cells within LI and within S1 were included for analysis. Two to five sections of tissue were analyzed per animal. Thresholded axonal coverage was normalized per mouse by the total number of infected LI interneurons in that mouse, determined by GFP expression. Layer densities were determined by the laminar area delineated by DAPI staining. LI axonal coverage was excluded from all analyses due to the presence of axons, dendrites, and soma all within the layer. Axonal extent was defined as described below (see morphological reconstructions).
Fluorescence activated cell sorting
Acute brain slices from animals were prepared as for electrophysiological recordings and were sliced at 600 μm (see Electrophysiology section below). Tissue slices were transferred to a petri dish containing ice-cold ACSF and bubbled with 95% oxygen/5% carbon dioxide and placed under a dissecting macroscrope. Slices at the coronal plane of the somatosensory cortex were identified and the barrel cortex was carefully dissected away from the surrounding cortical and subcortical tissue using a #11 sterile scalpel blade (Kent Scientific Corporation, #INS750240). Dissected somatosensory cortical tissue was placed into ice cold Dulbecco’s PBS with calcium and magnesium (Thermo Fisher Scientific, #14040133) supplemented with 0.5% bovine serum albumin (BSA, 10% stock, Miltenyi Biotec, #130–091-376). Microdissection of barrel cortex was confirmed by post hoc IHC of leftover, dissected slices for Vglut2. Collected barrel cortical tissue was dissociated into a single cell suspension using the Neural Tissue Dissociation Kit for Postnatal Neurons (Miltenyi Biotec, #130–094-802) using the manual dissociation of brain tissue protocol and 70 μm MACS® SmartStrainers (Miltenyi Biotec, #130–098-462). No deviations from the protocol were made. Single cell suspensions of somatosensory cortical cells from 5HT3aR.RCE mice underwent fluorescence automated cell sorting (FACS) under the expertise of Weill Cornell Medicine’s FACS core facility. GFP+, DAP− Ineurons were sorted into sterile 1x PBS and placed on ice before downstream analysis.
PCR amplification of Cxcl14 exon 2
DNA from cells that underwent FACS was extracted using the DNeasy Blood & Tissue Kit (Qiagen, #69504) according to protocol. No deviations from the protocol were made. To amplify the loxP flanked region of exon 2 of the Cxcl14 gene,43 two separate primer sets were used and evaluated. Primer set 1: S1-sense, 5’-GCCCTCTGTCGCGGTCCTGC-3’, A1-antisense, 5’-GTCTTGTCAAAGGCAG AGAG-3’. Primer set 2: S2-sense, 5’-ATGAGGCTCCTGGCGGCC-3’, A2-antisense, 5’-GTCCGATCTAACCCTAGGTTG-3’. PCR amplification was carried out as following: 1) 120 sec at 94°C, 2) 10 sec at 98°C, 3) 30 sec at 60°C, 4) 60 sec at 68°C, cycling through steps 2 through 4 a total of 30 times. PCR products were separated by electrophoresis on a 2% agarose gel (Denville Scientific, #GR140) with GelRed Nucleic Acid Gel Stain (Biotium, #41003–1) and GeneRuler 1kb Plus DNA Ladder (Fisher Scientific, #SM1333). A Bio-Rad Quantitiy One gel analysis system was used to develop the gel. Gene maps of Cxcl14 exons were created using SnapGene software (www.snapgene.com).
Electrophysiology
Solutions
Sucrose-based artificial cerebral spinal fluid (ACSF) for dissecting (mM): NaCl 83, KCl 2.5, NaH2PO4 1, NaHCO3 26.2, Dextrose 22, Sucrose 72, CaCl2 20.5, MgCl2 3.3, 305mOsm. ACSF buffer for recording (mM): NaCl 125, KCl 3, NaH2PO4 1.25, NaHCO3 25, Dextrose 25, CaCl2 2, MgCl2 1, 305mOsm. Intracellular pipette solution (mM): K-gluconate 125, KCl 10, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) 10, ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA) 0.1, Mg-ATP 4, Na-GTP 0.3, phosphocreatine 10, biocytin 0.1% w/v for a total osmolarity of 293mOsM and pH of 7.43.
Acute slices were prepared for patch clamp electrophysiological recordings as previously described.6,58 Mice were decapitated and brains were coronally sectioned on a Leica V1000s vibratome at 300μm. Slices were recovered for 30–40 minutes at 34°C in sucrose-based ACSF. Slices were maintained at room temperature following recovery.
Whole cell patch clamp electrodes were pulled (Sutter Instrument P-1000) to a measured tip resistance of 3–6 mOhms. An Olympus BX51WI microscope with IR-DIC optics with an Olympus 60x (NA: 1.00W, FN26.5) objective lens and a CMOS camera (ORCA-Flash4.0 LT, Hamamatsu) were used to visualize neurons. In slices from both Cxcl14.eGFP and 5HT3aR.RCE expressing animals, interneurons were identified based on their expression of eGFP and location within layer I.
Patch-clamp recordings were conducted at 32°C to 35°C in the following recording ACSF buffer. All extracellular solutions were buffered to pH 7.4 and recording solutions were bath applied at a perfusion rate of ~3 mL/min. Membrane potentials were amplified with a MultiClamp amplifier and digitized with ITC-18 digitizer using AxoGraph software. Recordings were sampled at 20 kHz (action potentials, AP) and low-pass filtered with a 4-pole Bessel filter at 10 kHz. Membrane potentials were not corrected for liquid junction potentials.
Passive and active membrane properties
When whole cell mode had been achieved, resting membrane potentials (Vm) were measured in current clamp. The amplifier mode was changed to voltage clamp and the baseline holding potential set to −65 mV. We placed all cells at the same Vm so that voltage-dependent channels would be at a similar starting state and rheobase current would be independent of Vm. APs were evoked with a series of depolarizing pulses of 1 sec duration each, with 5 sec between each stimulus. Rheobase (see below) was found by applying 2-pA increments of current when a single AP was not observed during the 10 pA increment recording protocol. Rm is the slope of a line fit to subthreshold voltage responses to current input. Rheobase is the minimum current input over a 1 sec stimulus that generates an AP. The first AP evoked by rheobase was used to characterize AP shape. AP parameters were calculated with built in functions in AxoGraph software. AP rise and decay were used to describe the rising and falling phases of the AP. AP peak is the absolute maximum amplitude. Half-width was measured as the time from the rising phase to the falling phase of the AP at one-half the distance from threshold to peak. Afterhyperpolarization (AHP) was characterized by the AHP peak. AHP peak is the difference between threshold and maximum AHP. The delay to onset of the first AP is measured from the start of a depolarizing stimulus pulse to the peak of the first AP evoked in response to rheobase. AP firing patterns were analyzed in the trace that contained ~30 APs, or equivalently, that fired APs at 30 Hz, the low range of the gamma frequency.96,97
Morphological reconstructions
Biocytin-containing intracellular solution was used to fill cells with biocytin during whole-cell patch clamp recordings for post-hoc morphological analysis. After recording, the sections were placed in a 4% PFA solution for 24 hours at 4°C. After post-fixation, samples were washed with 1x PBS, 3 times at RT. Then, the tissue sections were incubated in a blocking buffer containing Streptavidin-Alexa 647 (1:1000) at RT for 2 hrs. DAPI was applied in between the last two washes with 1xPBS for 5 min at RT. The sections were placed on glass slides, dried at RT for at least 1 hr before mounted with Pro Gold Antifade mounting medium. Fluorescently labeled single neurons were visualized with Olympus Confocal using 10X (to monitor the neuron in the entire cortical lamination) and 40X (to monitor high resolution image of the single neuron for reconstruction). Biocytin filled neurons were imaged at 1.0–1.5 μm steps covering the entire filled morphology. If needed, the multi-area timelapse stitching feature of the Fluoview FV1000 software was used to take composite images (areal survey; morphological reconstructions, not shown). Laser strength and acquisition parameters were kept consistent across all sample images. Recovered cells were reconstructed using Neurolucida 360 and analyzed with Neurolucida Explorer (MBF Bioscience) for process intersections on concentric circles (Scholl), total length, and number of nodes. Dendrites and axons were differentially labeled as blue and red respectively during tracing. Post hoc identification of single bouquet cell or extended neurogliaform cell morphology was determined based on known morphological and electrophysiological distinctions between these two interneuron subtypes.12–14 Briefly, the cells that had axonal processes going into the deeper layers of cortex were categorized as SBCs and the cells that had axonal processes were mostly constrained in the superficial layer were categorized as NGFCs. Cells with incomplete or indeterminate reconstructions were excluded from analysis. Canopy cells identified based on extended axons tangential to pia were not included in the analysis. Prior to manually drawing the boundaries for the axonal extents, cells were first oriented in the same direction with pia at the top. Their somas were labeled with the intersection of a horizontal and vertical black line in Adobe Illustrator. The tangential and vertical axonal extents were then measured after drawing a rectangle that encloses the axonal projections. Axonal extent is calculated as the area of the longest tangential and the longest vertical axon.
Cranial window surgery and chronic in-vivo calcium imaging
Surgery and imaging were performed as previously described.6,37,58 Briefly, mice aged P4–5 were briefly anesthetized by ice-induced hypothermia for 5 minutes and then placed on a Snuggle Safe® warmer (Lenric C21, Ltd.) for the remainder of the surgery. Scalp above the somatosensory cortex was removed, and a custom-made, titanium head plate for small mouse pups was positioned over the area, identified by stereotactic coordinates, and adhered to the skull using a veterinary adhesive (Metabond®). A craniotomy was performed at the center of the circular head plate by gently etching away the skull using the sharp edge of a sterile syringe needle. After achieving homeostasis, a cranial window was lowered on top of the brain. The window was then fixed to the skull using veterinary adhesives (first Vetbond™, then Metabond®). The pup was then placed on a heating pad with bedding for recovery.
Mouse pups were allowed to recover on a heating pad for one hour after surgery was completed. Recordings were performed on unanesthetized mouse pups in which the head was stabilized by attaching the head plate to a fixed fork positioned beneath the objective using two set screws. The pup was kept on a heating pad and bedding was placed loosely around the animal for comfort. During imaging sessions, mouse pups spent the majority of time in a quiet resting state, interrupted by occasional limb or tail twitches. Whisker stimulation37 was performed with a 500ms air puffs were applied every 60 seconds to stimulate whiskers on the left side at P14. Imaging was performed in the right hemisphere.
A FluoView FVMPE-RS multiphoton imaging system (Olympus) was used for detecting brain activity in mouse pups. A Mai Tai® Deepsee Ti:Sapphire laser (Spectra-Physics) was tuned to 920 nm for GCaMP6s excitation and the total laser power delivered to the brain was less than 60 mW. A 25X (1.05 NA) water immersion lens (Olympus) was used for imaging. To ensure that LI was consistently imaged, we performed recordings at less than 60 mm below pia surface. Image frames of 509 × 509 mm (512 × 512 pixels) were imaged with a resonant scanner, and we performed a 3-frame moving average of raw frames to remove fluorescence signal variations and obtained a final frame rate of 5 Hz for 3000 frames. The large field of view (FOV) was used in order to capture as many interneurons as possible in a recording. Each recording consisted of a single, continuous 5 to 10-minute movie. Movies taken from non-overlapping FOVs were used for analysis. The system was controlled by FV30S-SW software (Olympus).
For chronic imaging, mice were returned to their home cage with the dam at the end of the initial imaging session on the day of the surgery or were imaged starting from the day after the implantation. The condition of the animal, head plate, and cranial windows were monitored daily. Subsequent imaging sessions took place on P14. After the last imaging session, the mice were sacrificed and the location of the cranial window was confirmed. Immunohistochemistry was also performed for anti-eGFP to check for expression of GCaMP6s.
Imaging data processing and transient detection
Video processing, event detection and analysis were performed using CalciumDX code95 written in MATLAB (Mathworks) and available in GitHub (https://github.com/ackman678/CalciumDX). Motion correction of movies was performed using the Image Stabilizer plugin of NIH ImageJ, which uses the Lucus-Kanade algorithm (http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html). For each movie, cell contours were manually drawn in the t-stack projections of the average intensity image. Calcium signals as the average intensity inside each cell contour were then measured, and the ΔF/F signal was calculated for each contour. Calcium transients were identified using automatic detection algorithms.6,95 Calcium activity patterns were characterized by quantifying frequency as number of events per second, duration as time between the onset and offset of a calcium and amplitude. The frequency, duration and amplitude were calculated for each contour and then the average was calculated for all contours (cells). From spontaneous activity recordings at P7, we generated histograms as well as cumulative probability plots to visualize the distribution of the data for duration and frequency. To quantify synchronicity of network activity,6,98 the percentage of active cells in each frame were graphed as event histograms. Then, surrogate activity histograms were made by reshuffling the events for each cell 100 times and a distribution of events sizes were created. The significance threshold for network events were set to p<0.01 and then the onset and offset of network events were determined as the percentage of active cells exceeded the threshold or dropped below it. The duration of network events was calculated based on these onset-offset time points. For whisker stimulation,6 network events were calculated as above and the events that peaked within 5 s after the whisker stimulation were deemed as the evoked events.
Barrel field analysis
Flat mount tissue preparations of the PMBSF to quantify of TCA axonal surface areas were prepared from perfused littermates of mutant and control pups at P8 6. Hemispheres were flattened between two glass slides separated with glass spacers. Then, cryostat sectioning and immunohistochemistry were carried out. Sections in which the barrels cssomposing the boundaries of the PMBSF (Row A, Row E, arc 1, arc 5) were identifiable and complete were utilized for total PMBSF surface area analysis (Figure S8E). Sections in which barrels A1–4, B1–4, C1–5, D1–5, and E1–5 were identifiable and complete across multiple flat mounted sections for a given animal were utilized for total TCA surface area analyses (Figure S8F). Tissue sections in which each barrel of a given row were identifiable and complete within a single slice of flat mounted tissue were utilized for row-specific TCA surface area analyses, even if other rows were not clearly identifiable in tissue from that same animal (Figure 7). These approaches were used to maximize the utility of flat mount samples in which the full and complete PMBSF was not recovered in a single 50μm slice of tissue.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics and graphs were generated using GraphPad Prism 10 software (www.graphpad.com). The sample size (n), statistical test used, and corresponding p-values are indicated in the appropriate figure legends. For IHC, viral axonal distribution, and barrel field analysis experiments, n refers to the number of mice; for FISH experiments, n refers to the number of sections; for electrophysiological experiments, n refers to the number of neurons recorded; for calcium imaging experiments, n refers to the number of non-overlapping FOVs. Sample sizes were targeted to those previously reported by us and others for experiments of similar approach.6,14,37 Normality testing was performed using the Shapiro-Wilk test and an appropriate parametric or non-parametric test was performed. Outlier testing was performed on completed data sets using the ROUT method in GraphPad Prism with a Q set to 1%. All data points are individually plotted on each graph. Box-and-whisker plots span the minimum and maximum values and denote the median within the box. Other data graphs depict all data points as well as the mean and standard error of the mean (SEM). Significance is indicated on graphs with asterisks and is defined as follows: *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001; ns, p > 0.05. For non-significant findings where p <0.1, the exact p value is reported.
Supplementary Material
Highlights.
Activity-dependent Cxcl14 expression delineates developing SBCs in layer I (LI)
Loss of Cxcl14 causes increased intrinsic excitability and neuronal complexity
5HT3aR.Cxcl14fl/fl mice show abnormal barrel map formation
LI interneurons show decreased network activity in 5HT3aR.Cxcl14fl/fl mice
ACKNOWLEDGMENTS
We are grateful to Mary E. Hatten, Margaret E. Ross, Anjali Rhajadhyaksha, and Rob Machold for helpful discussions. We thank Camilo Ferrer and Seohee Ahn for comments on the manuscript. We thank Ryu-Ichiro Hata and the Gene Expression Nervous System Atlas (GENSAT) Project for making available the Cxcl14 fl/fl and Cxcl14.eGFP mouse lines, respectively. In addition, we recognize the generosity of the Genetically Encoded Neuronal Indicator and Effector (GENIE) Project and the Janelia Research Campus for making GCaMP6s available. We are grateful to Alicia Che for help with pilot SBC slice recordings. We are thankful to Jason McCormick, Tomas Baumgartner, and the Weill Cornell FACS core for their assistance with experimental design and expertise and the Weill Cornell Neuroanatomy EM core for assistance with the in situ hybridization experiments. We thank Julia Kuhl for creating the graphical abstract and additional illustrations. This work was supported by the following grants from the NIH: 2R01MH110553, 1R01NS116137, and 1R01MH125006 to N.V.D.M.G. and F30HD100089 to A.F.I. This work was also supported by the BBRF (29805–01 to G.A.). Additional support for A.F.I. and D.A. (training grant T32GM007739) came from the NIGMS to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interest.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114531.
REFERENCES
- 1.Golshani P, Goncalves JT, Khoshkhoo S, Mostany R, Smirnakis S, and Portera-Cailliau C (2009). Internally mediated developmental desynchronization of neocortical network activity. J. Neurosci. 29, 10890–10899. 10.1523/JNEUROSCI.2012-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egorov AV, and Draguhn A (2013). Development of coherent neuronal activity patterns in mammalian cortical networks: common principles and local hetereogeneity. Mech. Dev. 130, 412–423. 10.1016/j.mod.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Chini M, Pfeffer T, and Hanganu-Opatz I (2022). An increase of inhibition drives the developmental decorrelation of neural activity. Elife 11, e78811. 10.7554/eLife.78811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suarez R, Bluett T, McCullough MH, Avitan L, Black DA, Paolino A, Fenlon LR, Goodhill GJ, and Richards LJ (2023). Cortical activity emerges in region-specific patterns during early brain development. Proc. Natl. Acad. Sci. USA 120, e2208654120. 10.1073/pnas.2208654120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luhmann HJ, Sinning A, Yang JW, Reyes-Puerta V, Stuttgen MC, Kirischuk S, and Kilb W (2016). Spontaneous Neuronal Activity in Developing Neocortical Networks: From Single Cells to Large-Scale Interactions. Front. Neural Circ. 10, 40. 10.3389/fncir.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Che A, Babij R, Iannone AF, Fetcho RN, Ferrer M, Liston C, Fishell G, and De Marco Garcia NV (2018). Layer I Interneurons Sharpen Sensory Maps during Neonatal Development. Neuron 99, 98–116.e7. 10.1016/j.neuron.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim LA, Huang S, Fernandez-Otero M, Sherer M, Qiu Y, Vemuri S, Xu Q, Machold R, Pouchelon G, Rudy B, and Fishell G (2021). Bottom-up inputs are required for establishment of top-down connectivity onto cortical layer 1 neurogliaform cells. Neuron 109, 3473–3485.e5. 10.1016/j.neuron.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S, Wu SJ, Sansone G, Ibrahim LA, and Fishell G (2024). Layer 1 neocortex: Gating and integrating multidimensional signals. Neuron 112, 184–200. 10.1016/j.neuron.2023.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth MM, Dahmen JC, Muir DR, Imhof F, Martini FJ, and Hofer SB (2016). Thalamic nuclei convey diverse contextual information to layer 1 of visual cortex. Nat. Neurosci. 19, 299–307. 10.1038/nn.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon SE, Yang H, Minamisawa G, and O’Connor DH (2016). Sensory and decision-related activity propagate in a cortical feedback loop during touch perception. Nat. Neurosci. 19, 1243–1249. 10.1038/nn.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manita S, Suzuki T, Homma C, Matsumoto T, Odagawa M, Yamada K, Ota K, Matsubara C, Inutsuka A, Sato M, et al. (2015). A Top-Down Cortical Circuit for Accurate Sensory Perception. Neuron 86, 1304–1316. 10.1016/j.neuron.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, and Tolias AS (2015). Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350, aac9462. 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niquille M, Limoni G, Markopoulos F, Cadilhac C, Prados J, Holtmaat A, and Dayer A (2018). Neurogliaform cortical interneurons derive from cells in the preoptic area. Elife 7, e32017. 10.7554/eLife.32017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuman B, Machold RP, Hashikawa Y, Fuzik J, Fishell GJ, and Rudy B (2019). Four Unique Interneuron Populations Reside in Neocortical Layer 1. J. Neurosci. 39, 125–139. 10.1523/JNEUROSCI.1613-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Cui H, Li X, Chen L, Zhang C, Zhao X, Li X, Shao Q, Sun Q, Yan K, and Wang G (2023). Laminar and dorsoventral organization of layer 1 interneuronal microcircuitry in superficial layers of the medial entorhinal cortex. Cell Rep. 42, 112782. 10.1016/j.celrep.2023.112782. [DOI] [PubMed] [Google Scholar]
- 16.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203. 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abs E, Poorthuis RB, Apelblat D, Muhammad K, Pardi MB, Enke L, Kushinsky D, Pu DL, Eizinger MF, Conzelmann KK, et al. (2018). Learning-Related Plasticity in Dendrite-Targeting Layer 1 Interneurons. Neuron 100, 684–699.e6. 10.1016/j.neuron.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Kashi Malina K, Tsivourakis E, Kushinsky D, Apelblat D, Shtiglitz S, Zohar E, Sokoletsky M, Tasaka GI, Mizrahi A, Lampl I, and Spiegel I (2021). NDNF interneurons in layer 1 gain-modulate whole cortical columns according to an animal’s behavioral state. Neuron 109, 2150–2164.e2155. 10.1016/j.neuron.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Machold R, Dellal S, Valero M, Zurita H, Kruglikov I, Meng JH, Hanson JL, Hashikawa Y, Schuman B, Buzsaki G, and Rudy B (2023). Id2 GABAergic interneurons comprise a neglected fourth major group of cortical inhibitory cells. Elife 12, e85893. 10.7554/eLife.85893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD, Baka J, Borde S, Close JL, Diez-Fuertes F, Ding SL, et al. (2018). Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 21, 1185–1195. 10.1038/s41593-018-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, and Kriegstein AR (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587. 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hromas R, Broxmeyer HE, Kim C, Nakshatri H, Christopherson K, 2nd, Azam, M., and Hou, Y.H. (1999). Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem. Biophys. Res. Commun. 255, 703–706. 10.1006/bbrc.1999.0257. [DOI] [PubMed] [Google Scholar]
- 24.Hata R, Izukuri K, Kato Y, Sasaki S, Mukaida N, Maehata Y, Miyamoto C, Akasaka T, Yang X, Nagashima Y, et al. (2015). Suppressed rate of carcinogenesis and decreases in tumour volume and lung metastasis in CXCL14/BRAK transgenic mice. Sci. Rep. 5, 9083. 10.1038/srep09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrosini E, and Aloisi F (2004). Chemokines and glial cells: a complex network in the central nervous system. Neurochem. Res. 29, 1017–1038. 10.1023/b:nere.0000021246.96864.89. [DOI] [PubMed] [Google Scholar]
- 26.Watson AES, Goodkey K, Footz T, and Voronova A (2020). Regulation of CNS precursor function by neuronal chemokines. Neurosci. Lett. 715, 134533. 10.1016/j.neulet.2019.134533. [DOI] [PubMed] [Google Scholar]
- 27.Borrell V, and Marin O (2006). Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat. Neurosci. 9, 1284–1293. 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- 28.Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129. 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wester JC, Mahadevan V, Rhodes CT, Calvigioni D, Venkatesh S, Maric D, Hunt S, Yuan X, Zhang Y, Petros TJ, and McBain CJ (2019). Neocortical Projection Neurons Instruct Inhibitory Interneuron Circuit Development in a Lineage-Dependent Manner. Neuron 102, 960–975.e6. 10.1016/j.neuron.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Hjerling-Leffler J, Zagha E, Fishell G, and Rudy B (2010). The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J. Neurosci. 30, 16796–16808. 10.1523/JNEUROSCI.1869-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei S, Du H, Li Z, Tao G, Xu Z, Song X, Shang Z, Su Z, Chen H, Wen Y, et al. (2019). Transcription factors Sp8 and Sp9 regulate the development of caudal ganglionic eminence-derived cortical interneurons. J. Comp. Neurol. 527, 2860–2874. 10.1002/cne.24712. [DOI] [PubMed] [Google Scholar]
- 32.Colquhoun LM, and Patrick JW (1997). Pharmacology of neuronal nicotinic acetylcholine receptor subtypes. Adv. Pharmacol. 39, 191–220. 10.1016/s1054-3589(08)60072-1. [DOI] [PubMed] [Google Scholar]
- 33.Gotti C, and Clementi F (2004). Neuronal nicotinic receptors: from structure to pathology. Prog. Neurobiol. 74, 363–396. 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Zhao Z, Shi Y, and He M (2022). Cortical VIP(+) Interneurons in the Upper and Deeper Layers Are Transcriptionally Distinct. J. Mol. Neurosci. 72, 1779–1795. 10.1007/s12031-022-02040-8. [DOI] [PubMed] [Google Scholar]
- 35.Aizenman CD, and Linden DJ (1999). Regulation of the rebound depolarization and spontaneous firing patterns of deep nuclear neurons in slices of rat cerebellum. J. Neurophysiol. 82, 1697–1709. 10.1152/jn.1999.82.4.1697. [DOI] [PubMed] [Google Scholar]
- 36.De Marco Garcia NV, Priya R, Tuncdemir SN, Fishell G, and Karayannis T (2015). Sensory inputs control the integration of neurogliaform interneurons into cortical circuits. Nat. Neurosci. 18, 393–401. 10.1038/nn.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babij R, Ferrer C, Donatelle A, Wacks S, Buch AM, Niemeyer JE, Ma H, Duan ZRS, Fetcho RN, Che A, et al. (2023). Gabrb3 is required for the functional integration of pyramidal neuron subtypes in the somatosensory cortex. Neuron 111, 256–274.e10. 10.1016/j.neuron.2022.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Bendito G, Sanchez-Alcaniz JA, Pla R, Borrell V, Pico E, Valdeolmillos M, and Marin O (2008). Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 28, 1613–1624. 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saaber F, Schutz D, Miess E, Abe P, Desikan S, Ashok Kumar P, Balk S, Huang K, Beaulieu JM, Schulz S, and Stumm R (2019). ACKR3 Regulation of Neuronal Migration Requires ACKR3 Phosphorylation, but Not beta-Arrestin. Cell Rep. 26, 1473–1488.e1479. 10.1016/j.celrep.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 40.Squarzoni P, Thion MS, and Garel S (2015). Neuronal and microglial regulators of cortical wiring: usual and novel guideposts. Front. Neurosci. 9, 248. 10.3389/fnins.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venkataramanappa S, Saaber F, Abe P, Schutz D, Kumar PA, and Stumm R (2022). Cxcr4 and Ackr3 regulate allocation of caudal ganglionic eminence-derived interneurons to superficial cortical layers. Cell Rep. 40, 111157. 10.1016/j.celrep.2022.111157. [DOI] [PubMed] [Google Scholar]
- 42.Cao Q, Chen H, Deng Z, Yue J, Chen Q, Cao Y, Ning L, Lei X, and Duan E (2013). Genetic deletion of Cxcl14 in mice alters uterine NK cells. Biochem. Biophys. Res. Commun. 435, 664–670. 10.1016/j.bbrc.2013.04.106. [DOI] [PubMed] [Google Scholar]
- 43.Yajima N, I, K., and Hata, R. (2010). Production of conditional knockout mice for chemokine BMAC/cxcl14 gene by Cre/loxP system. Inflamm. Regen. 30, 536–541. [Google Scholar]
- 44.Wedemeyer MJ, Mahn SA, Getschman AE, Crawford KS, Peterson FC, Marchese A, McCorvy JD, and Volkman BF (2020). The chemokine X-factor: Structure-function analysis of the CXC motif at CXCR4 and ACKR3. J. Biol. Chem. 295, 13927–13939. 10.1074/jbc.RA120.014244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chittajallu R, Wester JC, Craig MT, Barksdale E, Yuan XQ, Akgul G, Fang C, Collins D, Hunt S, Pelkey KA, and McBain CJ (2017). Afferent specific role of NMDA receptors for the circuit integration of hippocampal neurogliaform cells. Nat. Commun. 8, 152. 10.1038/s41467-017-00218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ojeda AF, Munjaal RP, and Lwigale PY (2017). Knockdown of CXCL14 disrupts neurovascular patterning during ocular development. Dev. Biol. 423, 77–91. 10.1016/j.ydbio.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Marco Garcia NV, Karayannis T, and Fishell G (2011). Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature 472, 351–355. 10.1038/nature09865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miceli S, Negwer M, van Eijs F, Kalkhoven C, van Lierop I, Homberg J, and Schubert D (2013). High serotonin levels during brain development alter the structural input-output connectivity of neural networks in the rat somatosensory layer IV. Front. Cell. Neurosci. 7, 88. 10.3389/fncel.2013.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Bendito G, and Molnar Z (2003). Thalamocortical development: how are we going to get there? Nat. Rev. Neurosci. 4, 276–289. 10.1038/nrn1075. [DOI] [PubMed] [Google Scholar]
- 50.Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, and Rubenstein JL (1999). Early neocortical regionalization in the absence of thalamic innervation. Science 285, 906–909. 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- 51.Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, and Hevner R (1999). Genetic control of cortical regionalization and connectivity. Cerebr. Cortex 9, 524–532. 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- 52.Akhmetshina D, Nasretdinov A, Zakharov A, Valeeva G, and Khazipov R (2016). The Nature of the Sensory Input to the Neonatal Rat Barrel Cortex. J. Neurosci. 36, 9922–9932. 10.1523/JNEUROSCI.1781-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cossart R, and Garel S (2022). Step by step: cells with multiple functions in cortical circuit assembly. Nat. Rev. Neurosci. 23, 395–410. 10.1038/s41583-022-00585-6. [DOI] [PubMed] [Google Scholar]
- 54.Genescu I, Martinez-Anibal M, Kouskoff V, Chenouard N, Mailhes-Hamon C, Cartonnet H, Lokmane L, Rijli FM, Lopez-Bendito G, Gambino F, and Garesl S (2022). Dynamic Interplay between Thalamic Activity and Cajal-Retzius Cells Regulates the Wiring of Cortical Layer 1. Cell Rep. 39, 110667. 10.1016/j.celrep.2022.110667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, and Pachnis V (2012). Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2, 1351–1362. 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akgul G, Abebe D, Yuan XQ, Auville K, and McBain CJ (2019). The Role of AMPARs in the Maturation and Integration of Caudal Ganglionic Eminence-Derived Interneurons into Developing Hippocampal Microcircuits. Sci. Rep. 9, 5435. 10.1038/s41598-019-41920-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Modol L, Bollmann Y, Tressard T, Baude A, Che A, Duan ZRS, Babij R, De Marco Garcia NV, and Cossart R (2020). Assemblies of Perisomatic GABAergic Neurons in the Developing Barrel Cortex. Neuron 105, 93–105.e4. 10.1016/j.neuron.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duan ZRS, Che A, Chu P, Modol L, Bollmann Y, Babij R, Fetcho RN, Otsuka T, Fuccillo MV, Liston C, et al. (2020). GABAergic Restriction of Network Dynamics Regulates Interneuron Survival in the Developing Cortex. Neuron 105, 75–92.e5. 10.1016/j.neuron.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong FK, Bercsenyi K, Sreenivasan V, Portales A, Fernandez-Otero M, and Marin O (2018). Pyramidal cell regulation of interneuron survival sculpts cortical networks. Nature 557, 668–673. 10.1038/s41586-018-0139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Priya R, Paredes MF, Karayannis T, Yusuf N, Liu X, Jaglin X, Graef I, Alvarez-Buylla A, and Fishell G (2018). Activity Regulates Cell Death within Cortical Interneurons through a Calcineurin-Dependent Mechanism. Cell Rep. 22, 1695–1709. 10.1016/j.celrep.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong FK, Selten M, Roses-Novella C, Sreenivasan V, Pallas-Bazarra N, Serafeimidou-Pouliou E, Hanusz-Godoy A, Oozeer F, Edwards R, and Marin O (2022). Serotonergic regulation of bipolar cell survival in the developing cerebral cortex. Cell Rep. 40, 111037. 10.1016/j.celrep.2022.111037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lieberam I, Agalliu D, Nagasawa T, Ericson J, and Jessell TM (2005). A Cxcl12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron 47, 667–679. 10.1016/j.neuron.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 63.Chalasani SH, Sabelko KA, Sunshine MJ, Littman DR, and Raper JA (2003). A chemokine, SDF-1, reduces the effectiveness of multiple axonal repellents and is required for normal axon pathfinding. J. Neurosci. 23, 1360–1371. 10.1523/JNEUROSCI.23-04-01360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stumm R, and Hollt V (2007). CXC chemokine receptor 4 regulates neuronal migration and axonal pathfinding in the developing nervous system: implications for neuronal regeneration in the adult brain. J. Mol. Endocrinol. 38, 377–382. 10.1677/JME-06-0032. [DOI] [PubMed] [Google Scholar]
- 65.Aberle H (2019). Axon Guidance and Collective Cell Migration by Substrate-Derived Attractants. Front. Mol. Neurosci. 12, 148. 10.3389/fnmol.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atanes P, Hawkes RG, Olaniru OE, Ruz-Maldonado I, Amisten S, and Persaud SJ (2019). CXCL14 Inhibits Insulin Secretion Independently of CXCR4 or CXCR7 Receptor Activation or cAMP Inhibition. Cell. Physiol. Biochem. 52, 879–892. 10.33594/000000061. [DOI] [PubMed] [Google Scholar]
- 67.Yuasa-Kawada J, Kinoshita-Kawada M, Tsuboi Y, and Wu JY (2023). Neuronal guidance genes in health and diseases. Protein Cell 14, 238–261. 10.1093/procel/pwac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allen Institute for Brain Science (2023). Transcriptomics Explorer. https://celltypes.brain-map.org/rnaseq/mouse_ctx-hpf_10x;. [Google Scholar]
- 69.Paredes MF, Li G, Berger O, Baraban SC, and Pleasure SJ (2006). Stromal-derived factor-1 (CXCL12) regulates laminar position of Cajal-Retzius cells in normal and dysplastic brains. J. Neurosci. 26, 9404–9412. 10.1523/JNEUROSCI.2575-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tiveron MC, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, Konig N, and Cremer H (2006). Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J. Neurosci. 26, 13273–13278. 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banisadr G, Bhattacharyya BJ, Belmadani A, Izen SC, Ren D, Tran PB, and Miller RJ (2011). The chemokine BRAK/CXCL14 regulates synaptic transmission in the adult mouse dentate gyrus stem cell niche. J. Neurochem. 119, 1173–1182. 10.1111/j.1471-4159.2011.07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gesuita L, Cavaccini A, Argunsah AO, Favuzzi E, Ibrahim LA, Stachniak TJ, De Gennaro M, Utz S, Greter M, and Karayannis T (2022). Microglia contribute to the postnatal development of cortical somatostatin-positive inhibitory cells and to whisker-evoked cortical activity. Cell Rep. 40, 111209. 10.1016/j.celrep.2022.111209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez-Alcaniz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, Lopez-Bendito G, Stumm R, and Marin O (2011). Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron 69, 77–90. 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Guyon A, and Nahon JL (2007). Multiple actions of the chemokine stromal cell-derived factor-1alpha on neuronal activity. J. Mol. Endocrinol. 38, 365–376. 10.1677/JME-06-0013. [DOI] [PubMed] [Google Scholar]
- 75.Marchionni I, Beaumont M, and Maccaferri G (2012). The chemokine CXCL12 and the HIV-1 envelope protein gp120 regulate spontaneous activity of Cajal-Retzius cells in opposite directions. J. Physiol. 590, 3185–3202. 10.1113/jphysiol.2011.224873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicolai J, Burbassi S, Rubin J, and Meucci O (2010). CXCL12 inhibits expression of the NMDA receptor’s NR2B subunit through a histone deacetylase-dependent pathway contributing to neuronal survival. Cell Death Dis. 1, e33. 10.1038/cddis.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petersen CCH (2019). Sensorimotor processing in the rodent barrel cortex. Nat. Rev. Neurosci. 20, 533–546. 10.1038/s41583-019-0200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Molnar Z, Luhmann HJ, and Kanold PO (2020). Transient cortical circuits match spontaneous and sensory-driven activity during development. Science 370, 308–317. 10.1126/science.abb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minlebaev M, Colonnese M, Tsintsadze T, Sirota A, and Khazipov R (2011). Early gamma oscillations synchronize developing thalamus and cortex. Science 334, 226–229. 10.1126/science.1210574. [DOI] [PubMed] [Google Scholar]
- 80.Anton-Bolanos N, Sempere-Ferrandez A, Guillamon-Vivancos T, Martini FJ, Perez-Saiz L, Gezelius H, Filipchuk A, Valdeolmillos M, and Lopez-Bendito G (2019). Prenatal activity from thalamic neurons governs the emergence of functional cortical maps in mice. Science 364, 987–990. 10.1126/science.aav7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mizuno H, Ikezoe K, Nakazawa S, Sato T, Kitamura K, and Iwasato T (2018). Patchwork-Type Spontaneous Activity in Neonatal Barrel Cortex Layer 4 Transmitted via Thalamocortical Projections. Cell Rep. 22, 123–135. 10.1016/j.celrep.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 82.Jiang X, Wang G, Lee AJ, Stornetta RL, and Zhu JJ (2013). The organization of two new cortical interneuronal circuits. Nat. Neurosci. 16, 210–218. 10.1038/nn.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y, and Zhu JJ (2004). Rapid arrival and integration of ascending sensory information in layer 1 nonpyramidal neurons and tuft dendrites of layer 5 pyramidal neurons of the neocortex. J. Neurosci. 24, 1272–1279. 10.1523/JNEUROSCI.4805-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee AJ, Wang G, Jiang X, Johnson SM, Hoang ET, Lante F, Stornetta RL, Beenhakker MP, Shen Y, and Julius Zhu J (2015). Canonical Organization of Layer 1 Neuron-Led Cortical Inhibitory and Disinhibitory Interneuronal Circuits. Cerebr. Cortex 25, 2114–2126. 10.1093/cercor/bhu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chittajallu R, Pelkey KA, and McBain CJ (2013). Neurogliaform cells dynamically regulate somatosensory integration via synapse-specific modulation. Nat. Neurosci. 16, 13–15. 10.1038/nn.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ledderose JMT, Zolnik TA, Toumazou M, Trimbuch T, Rosenmund C, Eickholt BJ, Jaeger D, Larkum ME, and Sachdev RNS (2023). Layer 1 of somatosensory cortex: an important site for input to a tiny cortical compartment. Cerebr. Cortex 33, 11354–11372. 10.1093/cercor/bhad371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Siddoway B, Hou H, and Xia H (2014). Molecular mechanisms of homeostatic synaptic downscaling. Neuropharmacology 78, 38–44. 10.1016/j.neuropharm.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marques-Smith A, Lyngholm D, Kaufmann AK, Stacey JA, Hoerder-Suabedissen A, Becker EB, Wilson MC, Molnar Z, and Butt SJ (2016). A Transient Translaminar GABAergic Interneuron Circuit Connects Thalamocortical Recipient Layers in Neonatal Somatosensory Cortex. Neuron 89, 536–549. 10.1016/j.neuron.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuncdemir SN, Wamsley B, Stam FJ, Osakada F, Goulding M, Callaway EM, Rudy B, and Fishell G (2016). Early Somatostatin Interneuron Connectivity Mediates the Maturation of Deep Layer Cortical Circuits. Neuron 89, 521–535. 10.1016/j.neuron.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takesian AE, Bogart LJ, Lichtman JW, and Hensch TK (2018). Inhibitory circuit gating of auditory critical-period plasticity. Nat. Neurosci. 21, 218–227. 10.1038/s41593-017-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, and Luo L (2015). Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634. 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Preibisch S, Saalfeld S, and Tomancak P (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465. 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ridler TW and Calward S (1978). Picture Thresholding Using an Iterative Selection Method. IEEE Transactions on Systems, Men, And Cybernetics SMC-8, 630–632. [Google Scholar]
- 95.Ackman JB, Burbridge TJ, and Crair MC (2012). Retinal waves coordinate patterned activity throughout the developing visual system. Nature 490, 219–225. 10.1038/nature11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jagadeesh B, Gray CM, and Ferster D (1992). Visually evoked oscillations of membrane potential in cells of cat visual cortex. Science 257, 552–554. 10.1126/science.1636094. [DOI] [PubMed] [Google Scholar]
- 97.Steriade M, Contreras D, Amzica F, and Timofeev I (1996). Synchronization of fast (30–40 Hz) spontaneous oscillations in intrathalamic and thalamocortical networks. J. Neurosci. 16, 2788–2808. 10.1523/JNEUROSCI.16-08-02788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Allene C, Cattani A, Ackman JB, Bonifazi P, Aniksztejn L, Ben-Ari Y, and Cossart R (2008). Sequential generation of two distinct synapse-driven network patterns in developing neocortex. J. Neurosci. 28, 12851–12863. 10.1523/JNEUROSCI.3733-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request


