Abstract
Neurotransmitters are crucial for the proper functioning of neural systems, with dopamine playing a pivotal role in cognition, emotions, and motor control. Dysregulated dopamine levels are linked to various disorders, underscoring the need for accurate detection in research and diagnostics. Single-stranded DNA (ssDNA) aptamers are promising bioreceptors for dopamine detection due to their selectivity, improved stability, and synthesis feasibility. However, discrepancies in dopamine specificity have presented challenges. Here, we surface-functionalized a nano-plasmonic biosensing platform with a dopamine-specific ssDNA aptamer for selective detection. The biosensor, featuring narrowband hybrid plasmonic resonances, achieves high specificity through functionalization with aptamers and passivation processes. Sensitivity and selectivity for dopamine detection are demonstrated across a wide range of concentrations, including in diverse biological samples like protein solutions, cerebrospinal fluid, and whole blood. These results highlight the potential of plasmonic “aptasensors” for developing rapid and accurate diagnostic tools for disease monitoring, medical diagnostics, and targeted therapies.
An aptamer-based microfluidic integrated nanoplasmonic platform rapidly, selectively, and directly detects dopamine from blood.
INTRODUCTION
Neurotransmitters regulate neural function and well-being in animals, requiring a balanced interplay of neurological hormones for proper bodily function. Among these, dopamine (1) stands out as a critical neuromodulator, playing a pivotal role in regulating cognition (2), emotions such as happiness or pleasure (3, 4), and motor skills (5, 6). Dysregulation of dopamine concentrations in humans is associated with a range of neurodegenerative disorders such as Parkinson’s (7) and Alzheimer’s disease (8), neurodevelopmental conditions like attention-deficit hyperactivity disorder (ADHD) (9) and Tourette syndrome (10), and psychological complications such as bipolar and schizophrenia (11, 12). In addition, abnormal dopamine levels can serve as a diagnostic indicator for specific types of cancers (13–17). Hence, accurate and reliable detection of dopamine concentrations is critically important for the development of pharmaceutical drug research and medical therapies (18).
Conventional techniques use antibody-based enzyme-linked immunosorbent assay (ELISA) (19, 20) or high-performance liquid chromatography (HPLC) for dopamine isolation, coupled with detection methodologies including fluorescence spectrometry (21), colorimetric analysis (22), mass spectrometry (23), or electrochemical reactivity (24–27). These methods, however, suffer from complexities in assay preparation, feasibility, long response time, and selectivity, making them unsuitable for point-of-care applications. Moreover, detecting dopamine directly from unprocessed whole blood with high accuracy and specificity poses considerable challenges, primarily due to its low concentration and the presence of interfering molecules (28). Standard detection methods often use indirect approaches targeting major dopamine metabolites like homovanillic acid (HVA) (29) or use catecholamine tests that measure combined levels of dopamine, epinephrine, and norepinephrine (30, 31) but lack specificity and require complex sample preparation.
In recent decades, electrochemical methods, such as cyclic voltammetry, have emerged as promising tools for the direct, label-free detection of dopamine owing to its simplicity, low cost, and rapid response (32). These methods involve modifying electrodes with various sensing enhancers, offering a viable alternative for detection (33–36). However, because dopamine and several other structurally related catecholamine neurotransmitters have similar redox potentials (37), achieving selectivity remains a major challenge, particularly in complex matrices such as blood or cerebrospinal fluid in the brain. Moreover, an electroactive environment can generate unwanted oxidation products, leading to biofouling of the electrodes, ultimately resulting in poor performance and sensor instability (38). Optical sensors are a strong candidate in the sensitive and label-free detection of such small molecules, with many such studies been shown using photonic, plasmonic, or optoelectronic response for quantitative analyses (39–42). These systems, however, although versatile, still suffer from issues such as selectivity, reproducibility, and reliability.
Aptamers, artificial bioreceptors, have emerged as excellent candidates for the specific detection of several neurotransmitters (43, 44), including dopamine. These are usually short, single-stranded RNA or DNA (ssRNA or ssDNA)–based oligomer proteins, having a particular nucleotide sequence that can selectively bind to a specific target molecule with high affinity (45, 46). Aptamers can target a wide variety of ligands (47), ranging from simple ions and small molecules, such as neurotransmitters, to large macromolecules like proteins, peptides, viruses, and even whole cells (48). Aptamers are typically selected in vitro from an RNA or DNA pool via SELEX (systematic evolution of ligands by exponential enrichment) procedure (49, 50). Because of ease of synthesis, good shelf life, and high specificity, they have found applicability as receptors in biosensors for clinical diagnosis and therapy. Recent studies have shown several dopamine-specific aptamers being developed (51), with the first one being an RNA-based aptamer (52). However, because of limited stability and difficulty in synthesis of RNA (53), a DNA homolog (57 base pairs) was subsequently developed to enhance specificity and affinity for dopamine (54). More recently, a shorter DNA aptamer (44 base pairs), obtained through direct selection (55), has been reported to exhibit even higher affinity and selectivity (56). Although, both DNA aptamers have shown potential for dopamine detection across various studies (57–60), recent contradictory results (61) regarding their specificity have complicated efforts toward a comprehensive understanding.
In this study, we assess the performance of an all-optical, surface-functionalized plasmonic biosensing platform for the detection of low concentrations of neurotransmitter dopamine directly from diverse biological samples, including protein solutions, artificial cerebrospinal fluid (aCSF), and unprocessed whole blood. The proposed sensor exhibits highly sensitive narrowband hybrid plasmonic resonances that is tunable across the visible to near-infrared (NIR) spectral range, making it extremely responsive to local alterations as shown in our earlier works (48, 62, 63). The sensor surface is functionalized with dopamine-specific aptamers, followed by a passivation procedure to mitigate unwanted charge-induced biofouling and nonspecific bindings. Here, we compare the efficacy of two distinct dopamine-specific ssDNA aptamers, named 57-mer and 44-mer based on their respective base-pair lengths, to determine their suitability for detecting dopamine concentrations in general phosphate-buffered saline (PBS) solutions. Our results reveal superior binding affinity and sensitivity of the 44-mer at high concentrations compared to the 57-mer counterpart. We experimentally demonstrate a broad detection range spanning several orders of magnitude, with a sub-nanomolar detection limit in standard 1× PBS solution, bovine serum albumin (BSA) solution, and aCSF. All these cases demonstrate a strong concentration-dependent signal correlation with minimal interference. Additional tests exhibit good selectivity against dopamine-related catecholamines and metabolites. Last, integrating the optical biosensing platform with a flow-based microfluidic channel setup allows real-time monitoring of dopamine levels directly from unprocessed whole blood based on a layout that we reported earlier (63), achieving a detection limit in the range of 1 nM. The versatility of the proposed integrated platform holds promise for simultaneous real-time detection of several neurotransmitters with excellent selectivity. We envision that such aptamer-integrated optical biosensors will serve as a robust platform for label-free, noninvasive, and real-time detection of neurotransmitters with exceptional specificity and sensitivity, with minimal interference, revolutionizing biomedical/clinical diagnostics and monitoring.
RESULTS
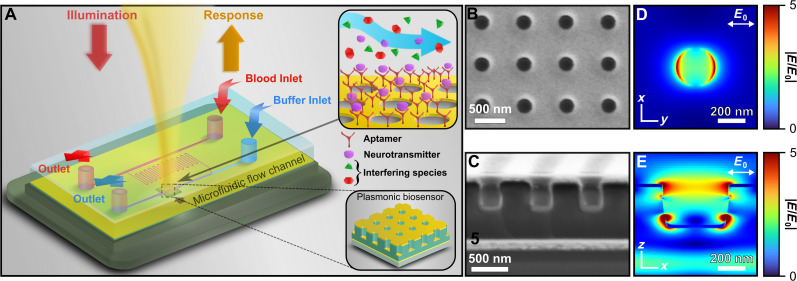
The proposed biosensing platform consists of a surface-functionalized, highly sensitive plasmonic sensor integrated with a polydimethylsiloxane (PDMS)–based microfluidic chip for detection in physiological fluids as shown in Fig. 1A. The plasmonic sensor consists of an array of three-dimensional (3D) hole-disk arrangement of optically thin metal that is asymmetrically coupled to an underlying resonant cavity with a reflector underneath as shown in our previous works (48, 62–65). Figure 1 (B and C) presents scanning electron microscopy (SEM) images of the nanostructured surface (top) and vertical cross section (bottom) of a fabricated plasmonic biosensor, respectively, providing detailed insights into its morphology and structure. The nanostructured surface is fabricated using a large-area parallel nanoimprinting method that enables fabrication of robust and reliable sensors, exhibiting multiple hybrid plasmonic resonances in the visible-NIR spectral region (64, 65). This is due to the strong coupling between the top localized surface plasmon (LSP) mode and the photonic cavity mode, resulting in strong, narrowband hybrid resonances that are highly sensitive to local changes in effective refractive index. Previous works have shown the reliable replication of narrowband plasmonic responses, which can be enhanced through automated imprinting methods (48, 63). The cavity-coupled plasmonic biosensor exhibits multifold enhancements in the local electric field intensity over the nanostructured surface at the spectral hybrid plasmonic resonances. The finite-difference time-domain (FDTD)–simulated electric near-field magnitude profiles at different perspectives at one such LSP resonance (LSPR; wavelength, 842 nm) are shown in Fig. 1 (D, top, and E, cross section). The electric field mode in Fig. 1D (xy plane) appears dipolar due to the polarization of the incident excitation being linear in the xy plane (64). The cavity is composed of an ultraviolet (UV)–cured negative epoxy resist SU-8 that offers structural rigidity. To mitigate epoxy swelling during biosensing applications, which can induce unpredictable and irreversible changes in spectral information (66), a conformal and uniform coating of 20-nm aluminum oxide (Al2O3) layer is deposited using atomic layer deposition (ALD) after nanoimprinting, which prevents undesired fluid diffusion into the epoxy. In addition, the plasmonic surface is made of optically thin gold (30 nm) coated via electron beam (ebeam) deposition, which ensures inertness and thus exhibits negligible oxidation and reactivity in almost all environmental conditions (48).
Fig. 1. Description of plasmonic biosensor platform.
(A) Schematic of integrated plasmonic biosensor platform. Top inset shows specificity of surface-functionalized DNA aptamer in presence of other interfering species, where the aptamer binds selectively to dopamine. Bottom inset shows a cartoon representation of the plasmonic biosensor. (B and C) Scanning electron microscopy (SEM) image of the biosensor showing surface (B) and cross-sectional (C) view. (D and E) Finite-difference time-domain (FDTD)–predicted local electric near-field enhancement at λ = 842 nm, 2 nm above the surface (D) and cross section (bottom).
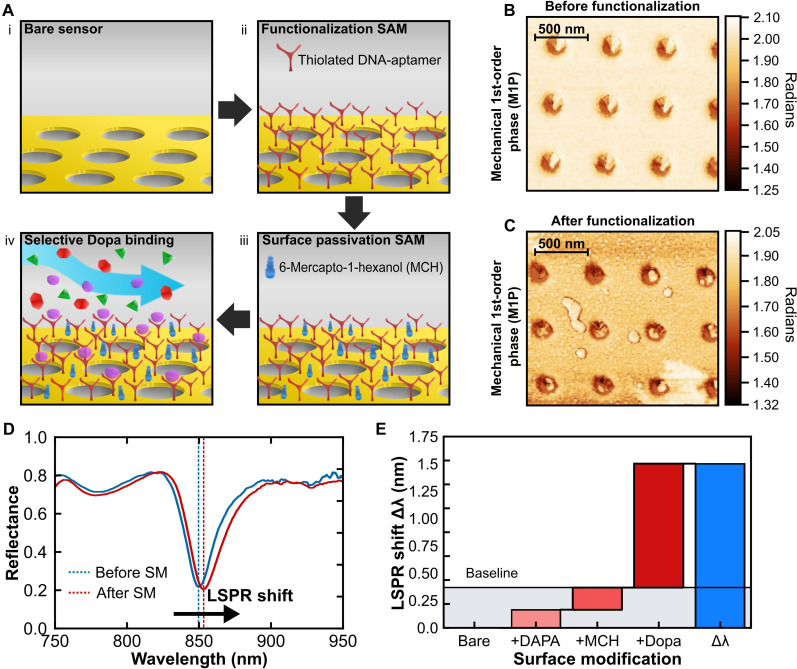
In the first study, we determined the functionalizing assay (67) appropriate for low-concentration dopamine detection. For this, two custom-modified thiol-terminated (─S═S─) ssDNA-based aptamers (Integrated DNA Technologies Inc.) of different base-pair lengths, one having 57 base pairs and the other having 44 base pairs, are chosen. The two custom ssDNA aptamers, named according to their base-pair lengths as “57-mer” and “44-mer” have similar storage conditions, activation, and surface functionalization protocols. Figure 2A shows a schematic of the overall strategy for surface modification and detection. The pristine sensors are initially functionalized with the thiol-activated aptamers that covalently bond to the gold, creating a dopamine-specific selective monolayer. Even so, detecting biomarkers in unfiltered or minimally purified bio-samples (blood, plasma matrix, or cerebrospinal fluid) can be challenging due to electrostatic accumulation of proteins on the negatively charged gold surface, leading to unwanted biofilm adsorption. This can hinder the actual positive signal acquired because of the desired biomarker-selective binding. To mitigate this issue, a surface passivation technique based on self-assembled monolayer of thiol-terminated 6-mercapto-1-hexanol (6-MCH) is subsequently coated on the surface. This approach is a common strategy for passivating gold-based platforms (67), helping to alleviate the problem of protein biofouling. Following the surface modification, the system’s far-field reflectance response undergoes redshifts (Fig. 2C) slightly, which is taken as the baseline for our target analyte detection. After the sensor’s functionalization with thiolated ssDNA aptamers and surface passivation with MCH chemistry, the activated system is ready for subsequent measurements and tests pertaining to aptamer selection and dopamine detection via optical spectroscopy and analysis (see Materials and Methods for details). Figure 2B presents the biosensor surface topography imaged using atomic force microscopy (AFM). The surface profile here is represented by the first-order mechanical phase amplitude (M1P) of the AFM tip, which carries information regarding the “softness” of the surface. Because of minimal thickness of the surface variations following aptameric functionalization (a few nanometers), they are not discernible in the z-variation profile (68), but a distinct change in surface morphology can be observed in the phase profile (Fig. 2, B and C). The detailed interpretation of M1P has been explained in Materials and Methods.
Fig. 2. Aptameric surface functionalization for selective detection.
(A) Representation of the neurotransmitter detection scheme: (i) The sensors are batch-prepared and cleaned, (ii) the sensor surface is functionalized with a thiol-based dopamine specific DNA aptamer to create a monolayer, (iii) the surface is further passivated with 6-mercapto-1-hexanol (6-MCH) to reduce further biofouling from interfering species, and (iv) a buffer matrix containing different neurotransmitters and biomolecules is introduced for selective dopamine (Dopa) binding to the sensor surface. (B and C) Atomic force microscopy (AFM) image of first-order mechanical phase (M1P) showing the surface morphology before (B) and after (C) the binding events. (D) Experimental spectra of one of the plasmonic biosensor surface before (blue curve) and after (red curve) the surface modification events. (E) Bridge plot showing the spectral change for corresponding surface modification events denoted in the x-coordinate. DAPA, thiolated ssDNA aptamer.
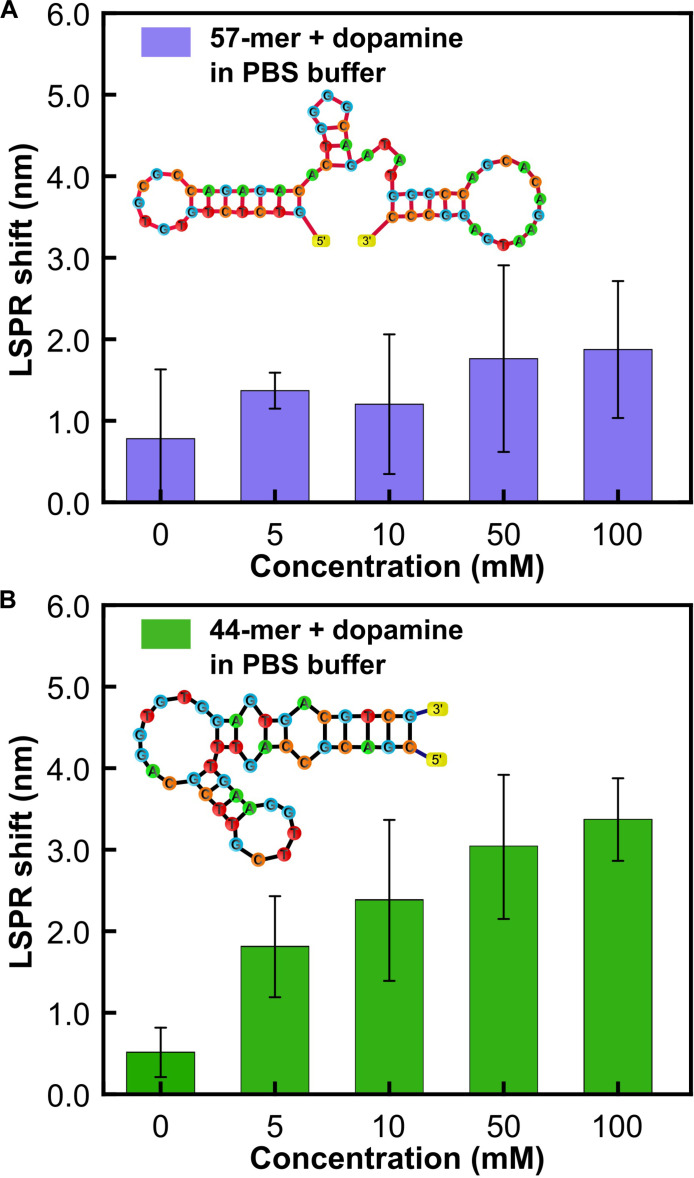
The primary investigation aims to evaluate the effectiveness of two aptamers in detecting elevated levels of dopamine. For this purpose, several concentrations of dopamine, ranging from 5 to 100 mM, in PBS (1×) was prepared. PBS maintains a consistent pH, ensuring biomolecular stability, and provides controlled ionic strength, mimicking physiological conditions, which is crucial for preserving the proper conformation and function of biomolecules. In addition, PBS minimizes nonspecific interactions and biofouling, enhancing sensor specificity and reliability. Subsequently, these concentrations were incubated separately on two distinct biosensors: one functionalized with a 57-mer aptamer and the other with a 44-mer aptamer. After a 20-min incubation period, the biosensor surface is rinsed with molecular-grade water and air-dried at room temperature for 5 min. The thiolated end of the aptamer covalently attaches to the gold surface, while the active end selectively binds to the diluted dopamine, inducing a conformational change that generates local variations in the near field of the biosensing surface. The high sensitivity of the plasmonic sensor’s optical near field to these fluctuations results in a corresponding far-field response, manifesting as a resonance redshift in the optical spectra. Figure 3 shows the corresponding LSPR shifts obtained for the dopamine concentrations in the abovementioned range. The 57-mer (Fig. 3A) shows a higher baseline at control compared to the 44-mer (Fig. 3B), possibly due to being a longer chain oligomer. Furthermore, at higher concentrations, the 57-mer reaches saturation, whereas a linear trend can be seen for the 44-mer. This can be explained by the fact that the LSPR shift being detected is sensitive to near-field changes and that the 57-mer being a long chain does not sufficiently alter the plasmonic near field after dopamine binding beyond a certain concentration. Therefore, for the subsequent dopamine sensing measurements, we chose the 44-mer aptamer for the sensor functionalization.
Fig. 3. Comparison of detection threshold for two different dopamine specific ssDNA aptamers.
Spectral characterization of the sensor responses for different concentrations of dopamine in PBS buffer for surface functionalization with (A) 57–base pair DNA aptamer and (B) 44–base pair DNA aptamer.
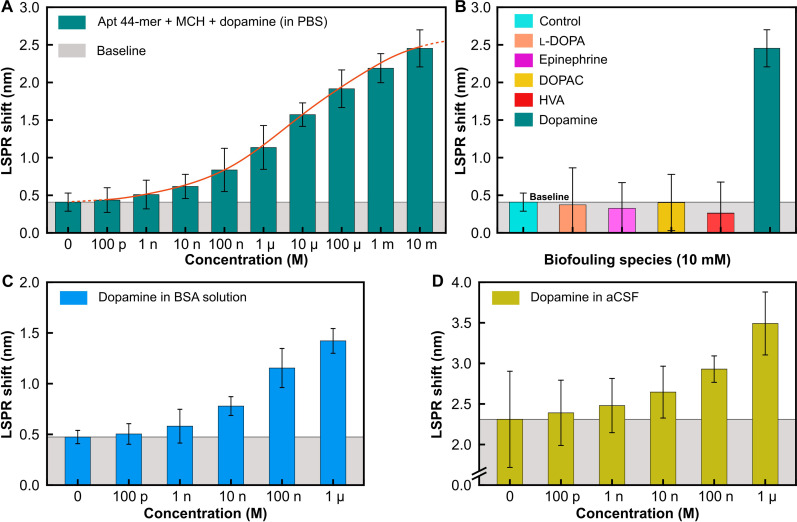
To assess surface coverage for efficient detection of low dopamine concentrations, we initially tested two concentrations of aptamers, 1 and 10 μM, shown in fig. S3. For low dopamine concentration (1 nM) in buffered solution, the device with 1 μM aptameric coverage falls within the baseline resolution. The subsequent measurements have therefore been performed with an aptamer concentration of 10 μM. Following this, we prepare several different concentrations of dopamine in PBS solution and incubate them onto functionalized sensor surfaces for 20 min. The incubated sensors are thoroughly rinsed with molecular-grade water and air-dried, followed by subsequent optical spectra measurements. Figure 4A shows the LSPR shift for various concentrations of dopamine in PBS up to a low concentration of 1 nM. We observe a good sigmoidal trend (like a cumulative distribution curve) in the measured LSPR shift with increasing dopamine concentration, shown here up to 10 mM, indicating the wide dynamic concentration range of the measurement setup. In addition, dopamine presents several structurally related precursors and metabolites that could potentially interfere with accurate detection and hinder the reliability of the assay. Subsequently, we conducted measurements to check for the specificity of the proposed detection platform in presence of such interfering neurotransmitter species closely related to dopamine. Figure 4B shows the LSPR shift obtained on incubation of the biosensor with several potential interfering neurotransmitter molecules like l-3,4-dihydroxyphenylalanine (l-DOPA), epinephrine, 3,4-dihydroxyphenylacetic acid (DOPAC), and HVA, prepared at a concentration of 10 mM in PBS, following a similar assay protocol as the prior experiments. A baseline control measurement is established by incubating unperturbed PBS on the biosensor. The observed shift for the equivalent concentration of dopamine in PBS is also depicted. All measured responses for interfering species are consistently at or below the baseline, in contrast to the response observed for a similar concentration of dopamine, demonstrating exceptional specificity and resistance to biofouling of the functionalized and passivated biosensing platform.
Fig. 4. Detection and biofouling studies in different biological matrices.
(A) LSPR shift versus concentration of dopamine in PBS. (B) Spectral characterization of sensor response in the presence of interfering species [l-3,4-dihydroxyphenylalanine (l-DOPA), epinephrine, and HVA] compared to that of dopamine for the same concentration (10 mM) in PBS buffer. (C) LSPR shift versus concentration of dopamine in BSA-spiked PBS solution and (D) in aCSF (1×). p, pico; n, nano, μ, micro; m, milli.
In real-world applications, a biosensor’s performance is crucial, particularly in dynamic assays containing diverse interfering molecules such as proteins, lipids, blood cells, and neurotransmitters. To evaluate the biosensor’s efficacy in such scenarios, assessments were conducted across two different biological matrices: BSA and aCSF. BSA, a standard protein with chemical similarity to human serum albumin, was used for its established protein concentration benchmark. Lyophilized BSA was dissolved in PBS buffer to create a 0.1% w/v solution, to which varying concentrations of dopamine were added and drop-casted onto the post-functionalized biosensor surface. Similarly, aCSF spiked with dopamine was drop-casted onto the passivated biosensors. After a 20-min incubation at room temperature, the biosensors were gently rinsed, air-dried, and subjected to optical spectrum collection. Figure 4 (C and D) illustrates the LSPR shift for different dopamine concentrations in spiked BSA and aCSF solutions, respectively. The LSPR shift observed for incubation with unspiked BSA was ~0.47 ± 0.07 nm, slightly higher than in the previous case. In contrast, the LSPR shift for aCSF without dopamine was notably higher at 2.31 ± 0.59 nm. Nevertheless, a consistent increasing trend in LSPR shift with dopamine concentration was observed, indicative of the biosensor’s specificity and resistance to surface binding effects. Furthermore, a consistent monotonic trend in the LSPR shift for various dopamine concentrations underscores the biosensor’s superior selectivity, with measured detection limits of about 100 pM for BSA and 90 pM for aCSF.
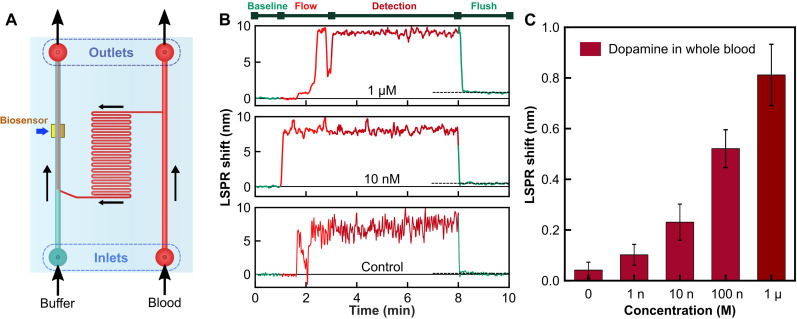
The main hurdle in point-of-care diagnostics and portable biosensor applications lies in effectively detecting biomarkers within unprocessed or minimally processed complex samples like whole blood, where biofouling as well as harsh environmental conditions can disrupt bio-affinity layers, rendering them ineffective for detection. To address this, we use a flow-based system incorporating a functionalized plasmonic biosensor integrated with an on-chip, transparent microfluidic flow channel derived from our earlier works (48, 63). This setup, as schematically depicted in Fig. 5A, features dual inlets and outlets for introducing a buffer solution and the target detection matrix (e.g., whole blood). In our experimental protocol, bovine whole blood (~35% hematocrit) is spiked with dopamine in PBS solution to generate varying dopamine concentrations while maintaining consistent hematocrit levels (refer to Materials and Methods for detailed procedures). Control samples consist of whole blood mixed with an equivalent volume of unspiked PBS.
Fig. 5. Detection of dopamine in unprocessed whole blood.
(A) Microfluidics integrated biosensor schematic representation. (B) Time dynamics of the LSPR shift signal at different stages during measurement. (C) LSPR shift versus concentration of dopamine spiked in bovine whole blood.
To initiate the experiment, PBS buffer is first introduced into the functionalized sensor region via one inlet at a low flow rate until the LSPR signal stabilizes. Subsequently, the PBS channel is sealed, and the blood sample is introduced through the second inlet, covering the sensing region. After a 2-min flow period, the system is allowed to incubate for 5 min. Following this incubation, the PBS solution is reintroduced to remove excess blood components, including blood cells, plasma, and proteins. Figure 5B illustrates the dynamic evolution of the LSPR signal during these flow steps for varying concentrations of dopamine in blood, including the unspiked blood (control). Throughout the blood flow phase, a considerable redshift in the tracked LSPR signal is observed, persisting during the incubation period with occasional perturbations due to dynamic changes in the near-field refractive index resulting from events like the introduction of macromolecular blood cells and temporary loss of spectral tracking. However, flushing with the PBS buffer restores the LSPR tracking signal, albeit with a slight baseline shift attributable to dopamine binding. This change in the baseline LSPR shift after flushing (Fig. 5B) is indicative of surface modification via dopamine capture by the functionalized aptamers. Notably, the control set, devoid of dopamine, exhibits an extremely small baseline shift (~0.042 ± 0.031 nm), indicating the exceptional stability and resistance of the biosensing surface against external contamination. Figure 5C presents the experimentally determined differential baseline LSPR shifts corresponding to different concentrations of spiked dopamine in blood, suggesting a limit of detection within the range of 1 nM. Compared to conventional detection techniques requiring extensive sample preparation, our proposed biosensing platform offers direct and highly specific detection of neurotransmitter dopamine from whole blood within a rapid response time of 5 min, paving the path forward for real-time monitoring of dopamine directly from unprocessed blood.
DISCUSSION
In recent years, there has been an increasing demand for blood-based neurotransmitter detection methods that not only provide sensitive and selective responses but also are affordable, reliable, and user-friendly. Conventional techniques like HPLC and ELISA, paired with various detection methods, are limited by complexities in sample preparation, response time, and selectivity. In addition, traditional blood detection methods often target major dopamine metabolites like HVA or use catecholamine tests, but they lack specificity. Overall, the lack of sensitivity, specificity, long response times, high complexity, and cost render these methods inadequate for point-of-care applications.
In this context, aptamers, as artificial bioreceptors, present a promising avenue for specific neurotransmitter detection. This work presents an all-optical, aptamer-based plasmonic biosensor showcasing exceptional sensitivity and selectivity in detecting the neurotransmitter dopamine. The biosensor is surface-functionalized with a thiol-modified aptamer with remarkable specificity toward dopamine, while a self-assembled passivation layer based on MCH minimizes biofouling. Optical sensing performance comparison between two distinct thiol-modified ssDNA aptamers reveals the shorter-chained 44-mer as the optimal candidate. In addition, sensing experiments conducted in PBS solutions and biological matrices like BSA-spiked solution and aCSF demonstrate excellent picomolar-level concentration-dependent correlation and minimal cross-interference from similar dopamine-related agents. Furthermore, the biosensor’s ability to detect dopamine directly from whole blood, with a detection limit of 1 nM and a rapid response time of 5 min, exhibits superior selectivity and sensitivity, underscoring its potential for point-of-care diagnostics and portable biosensor applications. Because of the stability of the thiol-gold covalent bond between the modified aptamer and the sensor surface, sensor reusability is difficult, requiring an extensive chemical cleaning protocol, and hence is discarded after every detection measurement. Nonetheless, the ease of fabrication allows mass production of such biosensors in batches, making it robust and reliable. Moreover, the microfluidic integrated biosensing chip is capable of measuring neurotransmitter directly and rapidly from minimally processed/unprocessed complex matrices via simple optical response. This platform holds promise for addressing unmet needs in real-time monitoring of neurological biomarkers and improving patient care through early and accurate detection of neurotransmitter imbalances.
MATERIALS AND METHODS
Biosensor fabrication
Inverse PDMS stamp fabrication
As an initial step, a pristine silicon wafer is cut and cleaned with acetone, isopropyl alcohol, and deionized water, followed by nitrogen blow-dry. A thin layer of electron-resist (PMMA C4, Kayaku Advanced Materials) is spin-coated on the wafer to generate a thickness of about 350 nm. An ebeam lithography system (Raith Nanofabrication GmbH) is used to generate the nanostructured hole-disk pattern onto the coated wafer, followed by post-baking at 180°C and then developed in methyl isobutyl ketone/isopropyl alcohol (1:3) developer for 50 s to generate a master design pattern. This master pattern is then used to create an inverse PDMS-based stamp that would be used for subsequent nanoimprint lithography.
Plasmonic sensor fabrication
The fabrication protocol for the biosensors begins with fused silica glass slides as the base substrate. These glass slides are cut into square shapes and sonicated in an acetone bath for 1 hour, following which they are cleaned with isopropyl alcohol and deionized water, and blow-dried with inert nitrogen gas. The pristine glass slides are then coated with a thin layer of 5-nm titanium (Ti) as an adhesion layer, followed by 100 nm of gold (Au) via ebeam evaporation (AJA International Inc.), to create the reflector base. This is followed by spin coating of a negative-toned photoresist SU-8 2000.5 (Kayaku Advanced Materials) to generate a dielectric cavity (~760 nm). The substrate is then pre-baked at 95°C for 1 min. The nanostructured pattern is thermally nanoimprinted onto the SU-8 using the PDMS inverse stamp fabricated before. This is followed by an exposure under UV light (365 nm) for 5 min to cross-link and cure the patterned photoresist. A 20-nm conformal layer of aluminum oxide (Al2O3) is deposited on the nanopatterned device using ALD (Ultratech Savannah S200). Last, the substrate is coated with a 3-nm Ti and 30-nm Au via ebeam evaporation to generate the 3D separated hole and disk metallic pattern. This concludes the fabrication process of the plasmonic biosensor, which enables the production of many samples in one batch.
Biosensor surface functionalization and passivation protocols
Materials and reagents
The thiol-modified ssDNA aptamers were custom-synthesized and received from Integrated DNA Technologies Inc. The two aptamers, named aptly based on their base-pair lengths as 57-mer and 44-mer, have the following base-pair sequences, respectively: (i) 5′–/ThioMC6-D/–GTC TCT GTG TGC GCC AGA GAC ACT GGG GCA GAT ATG GGC CAG CAC AGA ATG AGG CCC and (ii) 5′–/ThioMC6-D/CGA CGC CAG TTT GAA GGT TCG TTC GCA GGT GTG GAG TGA CGT CG–3′. Both aptamers were centrifuged at 1000 rpm for 5 min, and each of them was divided into 3-μl aliquots of 100 μM concentration, after which they were stored in the freezer at −20°C until further use. Sodium chloride (NaCl; ≥99%), sodium phosphate dibasic (Na2HPO4; ≥99%), potassium phosphate monobasic (KH2PO4; ≥99%), magnesium chloride (MgCl2; >95%), 6-MCH (99%), hydrolyzed polyvinyl alcohol (≥99%), tris(2-carboxyethyl) phosphine hydrochloride (TCEP), and dopamine hydrochloride powder [(HO)2C6H3CH2CH2NH2·HCl] were all purchased from Sigma-Aldrich. Potassium chloride (KCl; ≥99%) was purchased from Thermo Fisher Scientific. Potassium hydroxide (KOH; 10 M) was procured from LabChem (TCP Analytical Group). Molecular-grade water was used for all experiments and was purchased from InterMountain Life Sciences. BSA (lyophilized powder, ≥96%) was purchased from Sigma-Aldrich. aCSF (sterile) solution was purchased from BioChemazone, for which the pH was verified to be ~7.36. Bovine whole blood (in sodium EDTA) was purchased from Lampire Biological Laboratories.
Aptamer and buffer preparation
The PBS solution was freshly prepared by diluting NaCl, KCL, Na2HPO4, and KHPO4 in the standard ratio in molecular-grade water. The pH was adjusted to be about 7.4 ± 0.1. The folding buffer for the aptamers was prepared by adding 2 mM MgCl2 to PBS. The reducing buffer was prepared by mixing TCEP in cold molecular-grade water to achieve an initial stock concentration of 100 mM. The pH is made neutral (~7.0) by adding 10 M KOH. Before aptamer reduction, the reducing buffer is diluted with PBS to prepare 10 mM solutions. The protocol for aptamer activation is similar for both 57-mer and 44-mer and is given as follows: A 3-μl frozen aptamer aliquot is thawed at room temperature, after which it is diluted with the folding buffer to prepare a concentration of 10 μM. This mixture is placed in a water bath set at 95°C for 5 min to induce aptamer folding. The mixture is then removed and allowed to cool down at room temperature for 10 min. It is diluted further with the reducing buffer in a 1:1 ratio and incubated for 15 min to allow thiol-reduction. The final working concentration of the aptamer solution is 10 μM.
Surface functionalization and passivation
Before functionalization, the biosensors are cleaned with ethanol and dried with nitrogen gas, followed by 2 min of oxygen plasma cleaning in a plasma chamber. Thick PDMS films (~2 mm) with 3-mm-diameter holes were prepared for aptamer confinement over the sensor surface. The PDMS surface was made hydrophilic by activation in an oxygen plasma chamber. The PDMS wells were then placed on top of the sensors, and 4 μl of aptamer solution was drop-casted in them. The samples are then sealed and allowed to incubate for 4 hours. After incubation, the sensor surface is washed with molecular-grade water three times to remove excess unbounded aptamers. After the functionalization procedure, 4 μl of 1 mM ethanolic solution of 6-MCH is added to each sensor region for surface passivation. The samples are resealed in a petri dish and stored at 4°C for 1 hour. After passivation, the unbounded 6-MCH was removed by cleaning the surface three times with molecular-grade water. This concludes the functionalization and passivation protocol. The functionalized sensor surface is kept hydrated with a small amount of binding buffer until it is used for the bio-detection experiment.
Experimental characterization and analysis
Sample preparation
Dopamine hydrochloride powder is diluted in PBS solution to make an initial concentration of 100 mM. This is further serially diluted with PBS to prepare subsequent lower order of magnitude concentrations. The precursors and metabolites of dopamine (l-DOPA, epinephrine, DOPAC, and HVA) were all prepared accordingly by dilution in PBS. For dilution in BSA, an initial concentration of 100 μM of BSA in molecular-grade water is prepared. This is further serial diluted with dopamine in PBS solution to achieve the final appropriate concentration. Similarly, dopamine is diluted in aCSF (as purchased) to create an initial concentration of 100 mM, which is further serially diluted in aCSF to generate subsequent lower concentrations. For dopamine solutions in whole blood, 30 μl of 10 μM dopamine in PBS was mixed with 270 μl of whole blood to generate 1 μM concentration of dopamine in blood. The same protocol was followed to prepare all subsequent concentrations of dopamine, including the control solution, for which unspiked PBS was mixed with whole blood.
Optical characterization
A single illumination wavelength intensity–based detection schemes do not provide nano- to picomolar-level detection resolution of small molecules like dopamine. A spectrometer (or plate reader in medical terminology) is a common medical device nowadays present in most diagnostic centers/clinics/hospitals, making spectroscopic detection a viable, accurate, and technologically relevant solution. For the optical measurements, we used a custom-build setup comprising of a grating spectrometer (HR2000+, Ocean Optics) with a 0.035-nm spectral resolution, in reflection mode. The LSPR in the collected reflectance spectra is fitted with a quadratic function around the tracked resonance. The sensor’s LSPR shift response is calculated by subtracting the sensor’s initial LSPR response from the final LSPR response achieved after biofluid drop-casting, incubation, and flushing. The error bars shown on the bar charts (Figs. 3 to 5) are calculated by the SD of multiple spectral LSPR shifts measured. The measurement for each dopamine concentration in the fluid matrix has been performed at least five times. For the dopamine spiked in aCSF-based LSPR shifts, measurements were performed at least eight times for each dopamine concentration before averaging.
Microfluidic integration
For microfluidic measurements, a PDMS-based transparent microfluidic chip, fabricated using a standard soft lithography technique, is laminated onto the functionalized biosensor’s surface. Before lamination, the chip’s surface is treated in oxygen plasma for 4 min to promote adhesion and induce hydrophilicity. The chip is then affixed in place using an acrylic clamp. A customized optical setup integrated with a flow-based system is used. The optical setup consists of the same grating spectrometer in reflection configuration, being used with a LabVIEW (National Instrument)–based spectra logging and resonance tracking software that collects reflectance spectra dynamically every second. The software also controls the integrated flow system (ElveFlow Microfluidics) that redirects the desired fluids through the microfluidic channels over the measurement region. After each measurement, the microfluidic chip was thoroughly cleaned with deionized water and ethanol, followed by 10 min of sonication with acetone, and stored in scotch tapes for further measurements.
AFM measurements
The surface morphology experiments were performed by an AFM setup (NeaSpec GmbH). In addition to the surface topography mapping, first-order mechanical amplitude (M1A) and phase (M1P) maps were simultaneously acquired by the system. The M1P maps are useful in identifying separate sample species showing inhomogeneity in adhesion and stiffness (68). Sub-nanometer thin layer of aptamers on the gold surface can be readily isolated from the M1P maps in contrast to the topography maps, where the height variations are close to the surface roughness.
Simulations
We conducted numerical simulations to determine the reflectance spectra and spatial electromagnetic profile at resonance using an FDTD-based software (Ansys Lumerical FDTD). The simulation geometry consists of multiple stacked layers: an initial 100-nm gold backside reflector and a dielectric cavity having variable thickness denoted as L. The top surface of the cavity consists of a hole, generated as a custom surface to replicate the actual depressed hole as seen in the SEM images. The relief depth and diameter of the hole were both kept fixed at 300 nm. The hole was coated conformally with a 20-nm layer of aluminum oxide (Al2O3). Last, a 30-nm gold-based separated disk and hole is generated on the top. The system period is kept fixed at 580 nm. Antisymmetric boundary conditions were applied along the x-boundaries and symmetric boundary conditions along the y-boundaries. The refractive index of the dielectric cavity was set at 1.59 for the whole range, equivalent to SU-8’s index. The dispersion data for gold and Al2O3 were taken from Palik’s handbook (69). Two profile monitors were used to calculate the electric field intensity profile at resonance: one vertically through the middle of the unit cell and one horizontally 2 nm above the gold surface of the unit cell.
Acknowledgments
Funding: This work at the University of Central Florida was supported by NSF grant no. ECCS-1800845.
Author contributions: Research design and conceptualization: A.B., S.L., and D.C. Investigation: A.B. and S.L. Methodology: A.B. and S.L. Experiment: A.B., S.L., J.P., and M.S. Funding acquisition: D.C. Data curator: A.B. and S.L. Supervision: A.B. and D.C. Formal analysis: A.B., S.L., and D.C. Software: A.B. Visualization: A.B. Writing—original draft: A.B. and D.C. Writing—reviewing and editing: A.B., P.C.-A., M.K., and D.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.Costa K. M., Schoenbaum G., Dopamine. Curr. Biol. 32, R817–R824 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Nieoullon A., Dopamine and the regulation of cognition and attention. Prog. Neurobiol. 67, 53–83 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Bromberg-Martin E. S., Matsumoto M., Hikosaka O., Dopamine in motivational control: Rewarding, aversive, and alerting. Neuron 68, 815–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise R. A., Jordan C. J., Dopamine, behavior, and addiction. J. Biomed. Sci. 28, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood A. N., New roles for dopamine in motor skill acquisition: Lessons from primates, rodents, and songbirds. J. Neurophysiol. 125, 2361–2374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bova A., Gaidica M., Hurst A., Iwai Y., Hunter J., D. K. Leventha, Precisely-timed dopamine signals establish distinct kinematic representations of skilled movements. eLife 9, 1–141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankovic J., Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 79, 368–376 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Pan X., Kaminga A. C., Wen S. W., Wu X., Acheampong K., Liu A., Dopamine and dopamine receptors in Alzheimer’s disease: A systematic review and network meta-analysis. Front. Aging Neurosci. 10, 175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum K., Chen A. L. C., Braverman E. R., Comings D. E., Chen T. J. H., Arcuri V., Blum S. H., Downs B. W., Waite R. L., Notaro A., Lubar J., Williams L., Prihoda T. J., Palomo T., Oscar-Berman M., Attention-deficit-hyperactivity disorder and reward deficiency syndrome. Neuropsychiatr. Dis. Treat. 4, 893–918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer H. S., Butler I. J., Tune L. E., Seifert W. E., Coyle J. T., Dopaminergic dysfunction in tourette syndrome. Ann. Neurol. 12, 361–366 (1982). [DOI] [PubMed] [Google Scholar]
- 11.Brisch R., Saniotis A., Wolf R., Bielau H., Bernstein H. G., Steiner J., Bogerts B., Braun K., Jankowski Z., Kumaratilake J., Henneberg M., Gos T., Corrigendum: The role of dopamine in schizophrenia from a neurobiological and evolutionary perspective: Old fashioned, but still in vogue. Front. Psych. 5, 110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis K. L., Kahn R. S., Ko G., Davidson M., Dopamine in schizophrenia: A review and reconceptualization. Am. J. Psychiatry 148, 1474–1486 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Grant C. E., Flis A. L., Ryan B. M., Understanding the role of dopamine in cancer: Past, present and future. Carcinogenesis 43, 517–527 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Y., Pan J., Chen Y., Xing W., Li Q., Wang D., Zhou X., Xie J., Miao C., Yuan Y., Zeng W., Chen D., Increased dopamine and its receptor dopamine receptor D1 promote tumor growth in human hepatocellular carcinoma. Cancer Commun. 40, 694–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto S., Yoshida Y., Ozeki Y., Okamoto M., Gotoh K., Masaki T., Nishida H., Shibuya T., Shin T., Daa T., Mimata H., Kimura N., Shibata H., Dopamine-secreting pheochromocytoma and paraganglioma. J. Endocr. Soc. 5, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Horst-Schrivers A., Osinga T. E., Kema I. P., B. F. A. M. VAN DER. Laan, Dullaart R. P. F., Dopamine excess in patients with head and neck paragangliomas. Anticancer Res. 30, 5153–5158 (2010). [PubMed] [Google Scholar]
- 17.Eisenhofer G., Goldstein D. S., Sullivan P., Csako G., Brouwers F. M., Lai E. W., Adams K. T., Pacak K., Biochemical and clinical manifestations of dopamine-producing paragangliomas: Utility of plasma methoxytyramine. J. Clin. Endocrinol. Metab. 90, 2068–2075 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Goldstein D. S., Eisenhofer G., Kopin I. J., Sources and significance of plasma levels of catechols and their metabolites in humans. J. Pharmacol. Exp. Ther. 305, 800–811 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Park H., Paeng I. R., Development of direct competitive enzyme-linked aptamer assay for determination of dopamine in serum. Anal. Chim. Acta 685, 65–73 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Kim J., Jeon M., Paeng K. J., Paeng I. R., Competitive enzyme-linked immunosorbent assay for the determination of catecholamine, dopamine in serum. Anal. Chim. Acta 619, 87–93 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Tian M., Gao W., Zhao J., A simple, rapid, fluorometric assay for dopamine by in situ reaction of boronic acids and cis-diol. J. Anal. Methods Chem. 2019, 6540397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang L., Zhao Z., Ye F., Zhao S., Rapid and sensitive colorimetric detection of dopamine based on the enhanced-oxidase mimicking activity of cerium(IV). New J. Chem. 45, 6780–6786 (2021). [Google Scholar]
- 23.Park J. Y., Myung S. W., Kim I. S., Choi D. K., Kwon S. J., Yoon S. H., Simultaneous measurement of serotonin, dopamine and their metabolites in mouse brain extracts by high-performance liquid chromatography with mass spectrometry following derivatization with ethyl chloroformate. Biol. Pharm. Bull. 36, 252–258 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Cannazza G., Di Stefano A., Mosciatti B., Braghiroli D., Baraldi M., Pinnen F., Sozio P., Benatti C., Parenti C., Detection of levodopa, dopamine and its metabolites in rat striatum dialysates following peripheral administration of L-DOPA prodrugs by mean of HPLC-EC. J. Pharm. Biomed. Anal. 36, 1079–1084 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Rao P. S., Rujikarn N., Luber J. M., Tyras D. H., A specific sensitive HPLC method for determination of plasma dopamine. Chromatographia 28, 307–310 (1989). [Google Scholar]
- 26.Jing F.-C., Chen H., Li C.-L., Rapid determination of dopamine and its metabolites during in vivo cerebral microdialysis by routine high performance liquid chromatography with electrochemical detection. Biomed. Environ. Sci. 20, 317–320 (2007). [PubMed] [Google Scholar]
- 27.Sasa S., Blank C. L. R., Determination of serotonin and dopamine in mouse brain tissue by high performance liquid chromatography with electrochemical detection. Anal. Chem. 49, 354–359 (1977). [DOI] [PubMed] [Google Scholar]
- 28.Stewart A. J., Hendry J., Dennany L., Whole blood electrochemiluminescent detection of dopamine. Anal. Chem. 87, 11847–11853 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Lambert G. W., Eisenhofer G., Cox H. S., Horne M., Kalff V., Kelly M., Jennings G. L., Esler M. D., Direct determination of homovanillic acid release from the human brain, an indicator of central dopaminergic activity. Life Sci. 49, 1061–1072 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Goyal R. N., Bishnoi S., Simultaneous determination of epinephrine and norepinephrine in human blood plasma and urine samples using nanotubes modified edge plane pyrolytic graphite electrode. Talanta 84, 78–83 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Fice D. S., Yeung P. K. F., A simple high-performance liquid chromatography assay for simultaneous determination of plasma norepinephrine, epinephrine, dopamine and 3,4-dihydroxyphenyl acetic acid. J. Pharm. Biomed. Anal. 21, 519–525 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Ning Q., Feng S., Cheng Y., Li T., Cui D., Wang K., Point-of-care biochemical assays using electrochemical technologies: Approaches, applications, and opportunities. Mikrochim. Acta 189, 3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakard S., Pavel I. A., Lakard B., Electrochemical biosensing of dopamine neurotransmitter: A review. Biosensors 11, 179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D. S., Kang E. S., Baek S., Choo S. S., Chung Y. H., Lee D., Min J., Kim T. H., Electrochemical detection of dopamine using periodic cylindrical gold nanoelectrode arrays. Scientific Reports 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsine Z., Mlika R., Jaffrezic-Renault N., Korri-Youssoufi H., Review—Recent Progress in graphene based modified electrodes for electrochemical detection of dopamine. Chemosensors 10, 249 (2022). [Google Scholar]
- 36.Rahman M. M., Lee J. J., Electrochemical dopamine sensors based on graphene. J. Electrochem. Sci. Technol. 10, 185–195 (2019). [Google Scholar]
- 37.Suzuki A., Ivandini T. A., Yoshimi K., Fujishima A., Oyama G., Nakazato T., Hattori N., Kitazawa S., Einaga Y., Fabrication, characterization, and application of boron-doped diamond microelectrodes for in vivo dopamine detection. Anal. Chem. 79, 8608–8615 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Wang H. S., Li T. H., Jia W. L., Xu H. Y., Highly selective and sensitive determination of dopamine using a nafion/carbon nanotubes coated poly(3-methylthiophene) modified electrode. Biosens. Bioelectron. 22, 664–669 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Linh V. T. N., Lee M. Y., Mun J., Kim Y., Kim H., Han I. W., Park S. G., Choi S., Kim D. H., Rho J., Jung H. S., 3D plasmonic coral nanoarchitecture paper for label-free human urine sensing and deep learning-assisted cancer screening. Biosens. Bioelectron. 224, 115076 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Linh V. T. N., Kim H., Lee M. Y., Mun J., Kim Y., Jeong B. H., Park S. G., Kim D. H., Rho J., Jung H. S., 3D plasmonic hexaplex paper sensor for label-free human saliva sensing and machine learning-assisted early-stage lung cancer screening. Biosens. Bioelectron. 244, 115779 (2024). [DOI] [PubMed] [Google Scholar]
- 41.Kim W. G., Lee J. M., Yang Y., Kim H., Devaraj V., Kim M., Jeong H., Choi E. J., Yang J., Jang Y., Badloe T., Lee D., Rho J., Kim J. T., Oh J. W., Three-dimensional plasmonic nanocluster-driven light-matter interaction for photoluminescence enhancement and picomolar-level biosensing. Nano Lett. 22, 4702–4711 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Kim I., Kim H., Han S., Kim J., Kim Y., Eom S., Barulin A., Choi I., Rho J., Lee L. P., Metasurfaces-driven hyperspectral imaging via multiplexed plasmonic resonance energy transfer. Adv. Mater. 35, e2300229 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Park J. H., Eom Y. S., Kim T. H., Recent advances in aptamer-based sensors for sensitive detection of neurotransmitters. Biosensors 13, 413 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha K., Mukhopadhyay C. D., Quantitative detection of neurotransmitter using aptamer: From diagnosis to therapeutics. J. Biosci. 45, 44 (2020). [PubMed] [Google Scholar]
- 45.Saraf N., Woods E. R., Peppler M., Seal S., Highly selective aptamer based organic electrochemical biosensor with pico-level detection. Biosens. Bioelectron. 117, 40–46 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Zhou J., Battig M. R., Wang Y., Aptamer-based molecular recognition for biosensor development. Anal. Bioanal. Chem. 398, 2471–2480 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Lai B. S., Juhas M., Recent advances in aptamer discovery and applications. Molecules 24, 941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vázquez-Guardado A., Mehta F., Jimenez B., Biswas A., Ray K., Baksh A., Lee S., Saraf N., Seal S., Chanda D., DNA-modified plasmonic sensor for the direct detection of virus biomarkers from the blood. Nano Lett. 21, 7505–7511 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Tuerk C., Gold L., Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Ellington A. D., Szostak J. W., In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 51.Liu X., Liu J., Biosensors and sensors for dopamine detection. View 2, 20200102 (2021). [Google Scholar]
- 52.Mannironi C., Di Nardo A., Fruscoloni P., Tocchini-Valentini G. P., In Vitro selection of dopamine RNA ligands. Biochemistry 36, 9726–9734 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Farjami E., Campos R., Nielsen J. S., Gothelf K. V., Kjems J., Ferapontova E. E., RNA aptamer-based electrochemical biosensor for selective and label-free analysis of dopamine. Anal. Chem. 85, 121–128 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Walsh R., DeRosa M. C., Retention of function in the DNA homolog of the RNA dopamine aptamer. Biochem. Biophys. Res. Commun. 388, 732–735 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Nakatsuka N., Yang K.-A., Abendroth J. M., Cheung K. M., Xu X., Yang H., Zhao C., Zhu B., Rim Y. S., Yang Y., Weiss P. S., Stojanović M. N., Andrews A. M., Aptamer–field-effect transistors overcome debye length limitations for small-molecule sensing. Science 362, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou Y., Hou J., Liu X., Comparison of two DNA aptamers for dopamine using homogeneous binding assays. Chembiochem 22, 1948–1954 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Zhou J., Wang W., Yu P., Xiong E., Zhang X., Chen J., A simple label-free electrochemical aptasensor for dopamine detection. RSC Adv. 4, 52250–52255 (2014). [Google Scholar]
- 58.Jarczewska M., Sheelam S. R., Ziółkowski R., Górski Ł., A label-free electrochemical DNA aptasensor for the detection of dopamine. J. Electrochem. Soc. 163, B26–B31 (2016). [Google Scholar]
- 59.Jarczewska M., Górski Ł., Malinowska E., Electrochemical aptamer-based biosensors as potential tools for clinical diagnostics. Anal. Methods 8, 3861–3877 (2016). [Google Scholar]
- 60.Abu-Ali H., Ozkaya C., Davis F., Walch N., Nabok A., Electrochemical aptasensor for detection of dopamine. Chemosensors 8, 28 (2020). [Google Scholar]
- 61.Álvarez-Martos I., Ferapontova E. E., A DNA sequence obtained by replacement of the dopamine RNA aptamer bases is not an aptamer. Biochem. Biophys. Res. Commun. 489, 381–385 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Vázquez-Guardado A., Smith A., Wilson W., Ortega J., Perez J. M., Chanda D., Hybrid cavity-coupled plasmonic biosensors for low concentration, label-free and selective biomolecular detection. Opt. Express 24, 25785–25796 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Vázquez-Guardado A., Barkam S., Peppler M., Biswas A., Dennis W., Das S., Seal S., Chanda D., Enzyme-free plasmonic biosensor for direct detection of neurotransmitter dopamine from whole blood. Nano Lett. 19, 449–454 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Vázquez-Guardado A., Safaei A., Modak S., Franklin D., Chanda D., Hybrid coupling mechanism in a system supporting high order diffraction, plasmonic, and cavity resonances. Phys. Rev. Lett. 113, 263902 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Chanda D., Shigeta K., Truong T., Lui E., Mihi A., Schulmerich M., Braun P. V., Bhargava R., Rogers J. A., Coupling of plasmonic and optical cavity modes in quasi-three-dimensional plasmonic crystals. Nat. Commun. 2, 479 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Wouters K., Puers R., Diffusing and swelling in SU-8: Insight in material properties and processing. J. Micromech. Microeng. 20, 095013 (2010). [Google Scholar]
- 67.Oberhaus F. V., Frense D., Beckmann D., Immobilization techniques for aptamers on gold electrodes for the electrochemical detection of proteins: A review. Biosensors 10, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan E. P. S., Lim C. T., Effects of annealing on the structural and mechanical properties of electrospun polymeric nanofibres. Nanotechnology 17, 2649–2654 (2006). [DOI] [PubMed] [Google Scholar]
- 69.E. Palik, Handbook of Optical Constants of Solids (Academic Press, 1998), vol. 3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6