Abstract
Scientific research has suggested that maize spread from Mexico and arrived in lowland South America in a state of partial domestication. However, archaeological samples with primitive morphological characteristics that corroborate this finding have not been recorded in the region thus far. Unexpectedly, many samples were identified in the Peruaçu Valley with characteristics never previously observed in South America. These archaeological samples with primitive characteristics, which are the focus of this work, represent the furthest records from the center of origin of the species and the longest duration of the maintenance of such characteristics (between 1010 and 570 years before present). The findings of this study, including archaeological samples, native races, and samples of teosinte, attest to a long history of maize diversification in lowland South America.
Archaeological evidence shows primitive characteristics and a long history of floury maize races diversification in South America.
INTRODUCTION
Maize (Zea mays spp. mays L.) is a cereal of great importance for global food security (1), and hypotheses about its domestication and diversification have been the subject of discussion since the 19th century (2–15). The lowland South America encompass regions with elevations less than 1500 m and exhibit extensive genetic diversity of maize (10, 16–22). Recent research has shown that the process of maize domestication is more complex than previously understood (23, 24), suggesting that maize was dispersed from the Rio Balsas Valley, Mexico, after beginning its domestication from populations of parviglumis, about ~9000 years before present (BP), and arrived in Southwestern Amazonia (Fig. 1) in a state of partial domestication (8). However, there is a shortage of macrobotanical remains in general in the earliest time periods of maize cultivation. Archaeological records, mainly in the forms of phytoliths, pollen, and starch, demonstrated the presence of maize for at least 6800 years BP (25) in different locations of lowland South America (25–35). Archaeological samples of cobs, ears, and grains are scarce in the region. However, a large number of samples were found in the Peruaçu Valley, state of Minas Gerais (MG), Brazil (Fig. 1), with characteristics never previously observed in South America; these samples are the focus of the present study. We considered the hypothesis that maize populations that arrived in lowland South America still had primitive characteristics and later diversified, giving rise to different native races. We determined that primitive traits were preserved and spread to locations far from the secondary center of maize diversification in southwestern Amazonia, where they were maintained for long periods of time, lasting at least until very close to the arrival of the first European settlers.
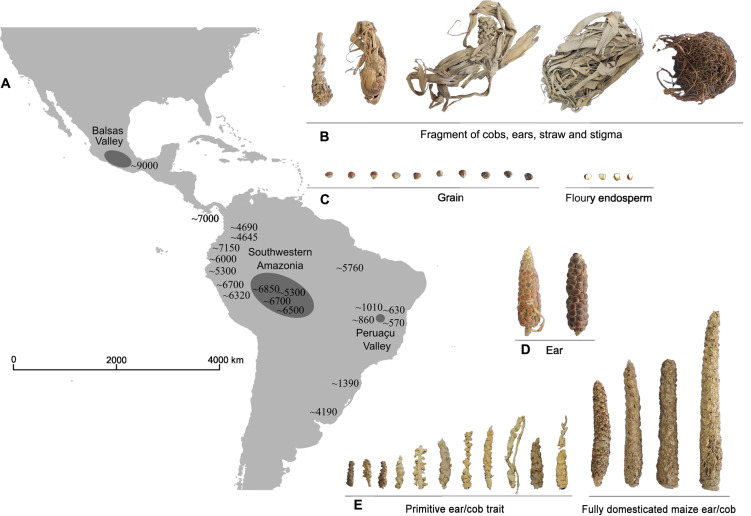
Fig. 1. Geographical distribution of archaeological records of maize throughout the Americas.
(A) Geographical location of Rio Balsas Valley, Southwestern Amazonia, Peruaçu Valley and archaeological records of maize (in years BP) (table S5) across the Americas. (B) Fragments of cobs, ears, straw, and stigma from archaeological samples of maize from Peruaçu Valley, Minas Gerais (MG). (C) Grains and floury endosperm in archaeological maize samples from Peruaçu Valley, MG. (D) Maize ears characterized for the classification of races in archaeological samples from Peruaçu Valley, MG. (E) Samples with primitive traits and typical ears/cobs of fully domesticated maize from archaeological samples from Peruaçu Valley, MG. The photos of the ears are presented on the same scale for each item [(A), (B), (C), (D) and (E)]. Photo credits: Flaviane Malaquias Costa, Universidade de São Paulo.
The species Z. mays comprises four subspecies: Z. mays L. ssp. mays, which is referred to as maize, Z. mays L. ssp. parviglumis, Z. mays L. ssp. mexicana, and Z. mays L. ssp. huehuetenangensis (14, 15). The wild relatives of maize can be called teosinte. Other hypotheses also suggest that a second dispersal event of maize hybridized with mexicana arrived in South America after ~6000 years BP and present evidence that this hybridization may have conferred agronomic advantages to maize, such as increased ear size and the number of grains per row (13). Samples of maize cobs, cob fragments, straw, and maize stigma were identified in the Peruaçu Valley (Fig. 1). The Peruaçu Valley is home to many caves, within which archaeological remains and human bones have been found, which indicate the presence of humans for at least 10,000 years BP (36–38), with crops appearing in the area around 2000 years BP (3, 8, 39).
To explore the study’s hypothesis, we performed an assay that involved the morphological characterization of archaeological samples of maize and modern samples of teosinte and the molecular characterization of accessions representing native races of maize. The following archaeological samples were characterized using qualitative and quantitative descriptors: (i) 282 fragments of ears/cobs (Fig. 1, tables S1 and S2, and data S1), (ii) 12 grains (Fig. 1 and data S2), and (iii) two ears (Fig. 1 and data S3). In addition, we characterized 22 modern teosinte samples (data S4) preserved in the Peabody Museum of Archeology and Ethnology, Harvard University (Peabody number: 2001.1.396) to obtain comparative parameters for the archaeological data. The ear/cob shape was estimated based on the archaeological samples of ear/cob fragments and samples of teosinte, considering the criteria presented in table S3. In addition, the length (cm), base diameter (mm), row number, and number of grains/row of these same samples were characterized. The race classification of the two archaeological ears was performed based on the characterization of 14 morphological descriptors of the crop, considered key to the classification of maize races (table S4) (18, 20, 40). Maize races are defined as groups of related populations with sufficient morphological characteristics in common to allow their recognition as a group (41). These groups occupy a defined geographical region (40) and are associated with specific uses (18, 20, 22).
RESULTS
The characterization of the 282 ear/cob fragments and of the teosinte samples showed evident diversity (tables S1 and S2 and Fig. 1), within which we highlight the amplitude of the minimum, average, and maximum number of rows for archaeological samples 4, 12, and 18, respectively, and the number of grains/row for samples 4, 13, and 40. For the teosinte samples, the minimum, average, and maximum number of rows were 2, 4, and 8, respectively, and the numbers of grains/row were 6, 12, and 27. As for the shape of the archeological ear/cob, for the whole fragments, the conical shape was predominant (71%), and for teosinte, the cylindrical shape predominated (55%) (table S2). A “primitive” trait was considered to be cobs with a row number less than eight because 95% of the teosinte samples characterized had a row number of less than eight and because thus far, no maize races with this feature have been found in lowland South America (16–18, 20, 22, 42, 43), which indicates the absence of this trait in modern maize races in the region. On the basis of this parameter, 14 archaeological samples (5%) were identified with this “primitive” characteristic (four and six rows) (table S6). The modern maize races from lowland South America, characterized from 254 landraces, presented from eight to 26 rows per ear, with an average of 12 rows (43).
Among the teosinte samples, there were two patterns in which 14 samples were classified as subgroup 1 and eight samples as subgroup 2 (Fig. 2). Archaeological samples with primitive characteristics from Peruaçu Valley, MG, have morphological characteristics that are more similar to teosinte samples with subgroup 2 (Fig. 2); these samples could be understood as proto-maize (8). This study presents an important record of archaeological samples of maize with primitive characteristics in lowland South America and, by dating the samples, indicates that such primitive characteristics persisted long after the introduction of maize in this area. These results agree with those of other studies (8, 9), which suggest that the alleles of maize domestication were not completely fixed when the species arrived in the region.
Fig. 2. Morphological descriptors of the teosinte samples preserved in the Peabody Museum of Archeology and Ethnology, Harvard University.
Ears of teosinte classified as subgroups 1 (A) and 2 (B) and their predominant characteristics. (C) Number of rows and ear shapes of teosinte samples and archaeological samples of ears/cobs with primitive characteristics from Peruaçu Valley, MG. The photos of the ears are presented on the same scale for each item [(A) and (B)]. Photo credits: Flaviane Malaquias Costa, Universidade de São Paulo.
The archaeological grains characterized in this study had a floury endosperm, with variation in grain shape and pericarp color (table S7 and Fig. 1). Approximately 300 maize races have been described for the American continent (44). Of these, 40% have a floury endosperm. The classification of maize races in Brazil (17) identified 15 races and 19 subraces of maize, among which four “indigenous races,” namely, Entrelaçado, Caingang, Avati Moroti, and Lenha, have floury endosperm. To identify the relationship between the accessions and classify the maize races present in the archaeological samples based on the morphological data (table S4), we performed principal coordinate analysis (PCoA) (Fig. 3) and cluster analysis using a unrooted dendrogram (Fig. 3), which specifies the relationships between the accessions without defining an evolutionary path (45, 46). The PCoA explained 71.14% of the total variation in the data using the first two principal coordinates. The contribution of the variables in the PCs analysis is shown in fig. S1. The Entrelaçado maize race was the most closely related to the archaeological samples. In the analysis, when K = 2, the archaeological samples and all representatives of the Entrelaçado race were found in the same group, and when K = 3 and K = 4, the archaeological samples and Entrelaçado were separated into two groups. When K = 4, the Avati Moroti race was separated from the Caingang and Lenha races, which were more closely related to each other in the other group.
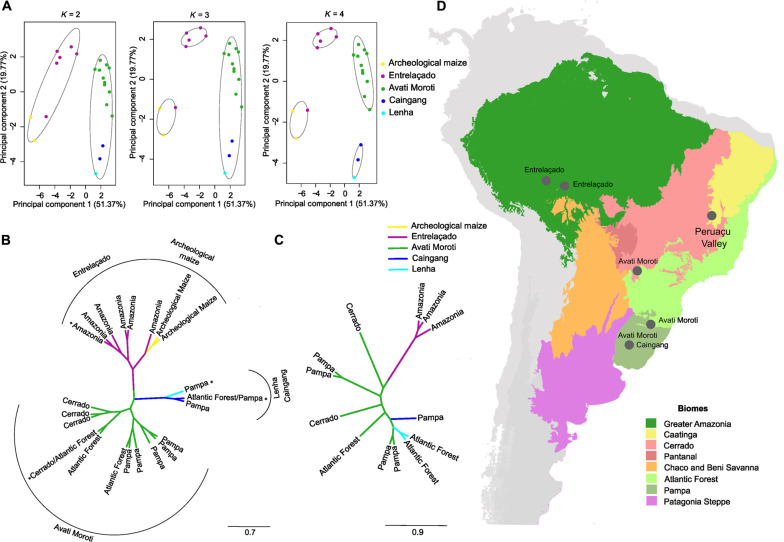
Fig. 3. Genetic structure of the floury maize races based on morphological and genomic data in the main lowland ecoregions of South America.
(A) Principal coordinate analysis (PCoA) was performed based on key descriptors for the classification of maize races from 22 maize accessions, with 20 accessions representing the floury maize races of lowland South America and two archaeological samples of maize from Peruaçu Valley, MG. The graphs generated considered the group numbers of K = 2, K = 3, and K = 4. (B) Ward cluster analysis of the 22 maize accessions based on the Gower index, performed using morphometric descriptors of the maize races. *Races of floury maize from Brazil described in the literature for lowland South America (16). (C) Cluster analysis of 13 accessions representing the floury maize races from lowland South America performed by the neighbor-joining method based on 5313 SNP markers. (D) Main lowland ecoregions of South America (62, 63) and location of the floury maize races. Photo credits: Flaviane Malaquias Costa, Universidade de São Paulo.
The cluster analysis showed the formation of three groups, with a consistent group composed of landraces of the Entrelaçado race from Southwestern Amazonia and from the literature (16, 17), which was grouped with archaeological samples (found in the Cerrado/Caatinga biome, which is at least 2000 km away). The ROF1C variety (a landrace conserved by a ribeirinho, traditional farmer that lives along riversides, from Rondônia State, in Southwestern Amazonia) was closer to the archaeological samples, and the other Entrelaçado accessions were closer to each other. This group of Amazonian maize was isolated from other races of floury maize. The cluster analysis showed that the archaeological samples were more closely related to the Entrelaçado race, which continues to be conserved in Southwestern Amazonia by traditional farmers and indigenous peoples (18). In the group formed by the Avati Moroti race, all samples from the Cerrado ecoregion, one sample from the Atlantic Forest, and the Avati Moroti race in the literature were more closely related to each other, forming a subgroup; similarly, the samples from the Pampa ecoregion and one sample from the Atlantic Forest formed another subgroup. Another group was formed by the Caingang and Lenha races from the Pampa and Atlantic Forest ecoregions.
The genomic characterization of 13 accessions representing the floury maize races from lowland South America was performed using 5013 single-nucleotide polymorphism (SNP) markers to analyze the genetic relationship between the races. Cluster analysis based on the neighbor-joining method and Nei’s genetic distances was performed (47). The results obtained for the genomic data (Fig. 3) corroborated the results of the cluster analysis that included morphological data, where a stronger genetic relationship was observed between the Entrelaçado race and the Avati Moroti race, and a weaker relationship was observed with the Caingang and Lenha races. In addition, the Amazonian Entrelaçado race was more isolated from the other races, and the Avati Moroti was a larger and more widely distributed group, occurring in the Cerrado, Atlantic Forest, and Pampa ecoregions.
Another cluster analysis of the morphological data, including the teosinte data, was performed with the aim of exploring the relationship between the samples. This analysis was rooted in a two-row teosinte sample. The similarity relationships presented by the analysis suggest that the groups formed by the archaeological samples and the Entrelaçado race are the most related to the teosintes subgroup 2. In addition, in the sequences of the branches, the Avati Moroti, Caingang, and Lenha appear (Fig. 4). The relationships observed in the analysis suggest the hypothesis that the number of rows and grain size (length) increased throughout the diversification process of these races, but we highlight that future genetic analyses must be carried out to investigate and confirm these hypotheses. The revolutionary transition of the two-row teosinte to the maize with the highest number of rows during domestication of the species markedly improved crop yields (48–50). Also, loci associated with increased ear size and number of kernels per row were detected when analyzing evidence of the hybridization of maize with mexicana in the highlands of Mexico around 4000 years after the beginning of domestication and its subsequent dispersal throughout the Americas (13). We consider that this analytical approach is not a phylogenetic analysis and has a restricted number of variables. However, the analysis allowed us to explore the relationship between archaeological samples and native races of floury maize from lowland South America.
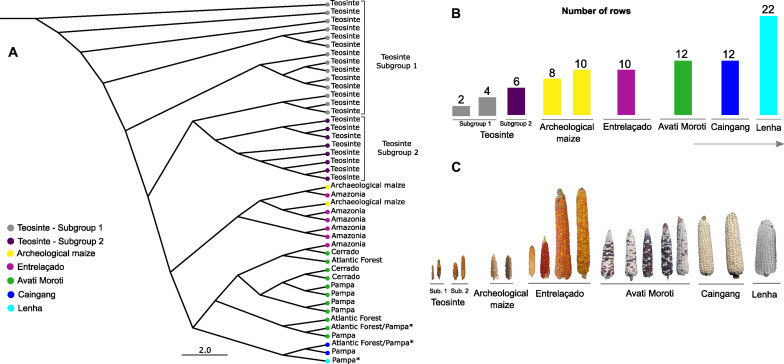
Fig. 4. Similarity relationships between modern teosintes, archaeological samples from the Peruaçu Valley, and floury maize races from lowland South America.
(A) Ward cluster analysis of 44 accessions (22 teosinte, two archaeological samples of maize from Peruaçu Valley, MG, and 20 accessions representing the floury maize races of lowland South America), based on the Gower index (57) of morphometric characteristics of maize races. *Races of floury maize from Brazil described in the scientific literature for lowland South America (16). (B) Number of teosinte rows, archaeological samples of maize, and races of floury maize from lowland South America. (C) Photographic record of teosinte ears (subgroups 1 and 2), archaeological samples of maize and representatives of the floury maize races of lowland South America. The photos of the ears are presented at the same scale. Photo credits: Flaviane Malaquias Costa, Universidade de São Paulo. The photo of the Lenha race was presented by Brieger et al. (17), Universidade de São Paulo.
DISCUSSION
The identification of samples with primitive morphological characteristics in the region of Peruaçu Valley, MG, raises the following scientific questions: (i) How can the presence of samples with primitive morphological characteristics millennia after the beginning of domestication in Mexico be explained (2, 4, 5, 12)? (ii) How can the presence of samples with primitive morphological characteristics millennia after the diversification in Southwestern Amazonia (8, 19) be explained? (iii) How can the presence of samples with primitive morphological characteristics thousands of kilometers away from these two geographic points (~7150 and ~ 2300 km, respectively) and located in different ecoregions be explained? (iv) Would these “more domesticated” and “more primitive” maize types have arrived and evolved together in the Peruaçu Valley, or were they part of distinct introductions and migration routes? The present study points to important evidence of the presence of primitive maize much later than the time of domestication and in a very distant location, which reinforces the need to deepen our understanding of this topic. In particular, the results provide information and suggest a future study, a potential application of archaeogenomics (3, 8, 9, 23, 24), including genome analysis of the 282 archaeological samples of maize, which cover a temporal range of 500 to 1010 years BP (3, 8, 9, 39), to answer these questions.
The archaeological ear samples analyzed in our study, the origin of which is located in the Cerrado, were all grouped together with the current samples collected in Amazonia. This result demonstrates that time, geographical distance, and change in ecoregion during the diffusion of maize in the past did not separate the archaeological samples of maize found today in the Amazon region throughout evolutionary history. Archaeological samples from Peruaçu Valley, with a morphology closer to that of domesticated maize, have already been genetically analyzed in another study, and a relationship was found with maize samples from Southwestern Amazonia in South America (8). These results corroborate the hypothesis that maize dispersed in lowland South America (19) and reinforce the finding that floury maize was dispersed from Amazonia to the Cerrado, Atlantic Forest, and Pampa, subsequently diversifying into different races.
This study allowed us to identify archaeological samples with primitive characteristics in a region of lowland South America; these samples have been found notably distant from the center of origin of the species and showed a substantial recorded duration of such primitive characteristics (between 1010 and 570 years BP), demonstrating the diversity and importance of such archaeological samples from Peruaçu Valley, MG. These results refute the hypotheses of single domestication for maize (2, 4, 6) and corroborate the hypotheses of stratified domestication for maize (8, 9, 13). The study also made it possible to analyze the relationships between archaeological samples and native races of floury maize from lowland South America. The region comprises exclusive gene pools that evolved over millennia, which leads us to the conclusion that conservation measures for the species on this continent should be a priority. The archaeological samples, dated to between 570 and 1010 years BP, were found far from the center of origin in Central America and from the secondary center of diversification in Southwestern Amazonia in a distinct ecoregion and have maintained morphologically primitive characteristics. These findings draw our attention and make us wonder what environmental and human/cultural components led to the maintenance of these primitive characteristics over such a long distance and for such a long time in the history of this species and its relationship with humans.
MATERIALS AND METHODS
Materials
Details of the archaeological samples used in this study are given in data S1 to S3. Modern teosinte (Peabody number: 2001.1.396) is conserved at the Peabody Museum of Archeology and Ethnology, Harvard University. Details of the teosinte are provided in data S4. Modern maize landrace accessions, collected in traditional agricultural contexts, were sampled in the collection of the “Workshops for Maize Races Conservation” of the University of São Paulo, Brazil, and the Universidad da Republica, Uruguay. Details of the landraces are provided in data S5 to S8.
The term “landraces” is used in this study to refer to local varieties traditionally managed and reproduced by family farmers (10, 18–20). The term “maize races” refers to groups of related populations with sufficient morphological characteristics in common to allow their recognition as a group (41), which occupy a defined geographical region (40) and are associated with particular social and cultural contexts (18–22).
Methods
Dating the archaeological maize samples
Archaeological samples of maize were collected at the archaeological rock shelter sites Lapa do Boquete and Lapa da Hora, located in the Peruaçu Valley, state of MG, Brazil (fig. S2). In these sites, there are traces of continuous human occupation over the last 10,000 years BP, with crop species appearing from around 2000 and remaining until 570 years BP, together with the presence of many human cultural artifacts and traditions, including rock paintings representing crops, which are rare around the world (fig. S3) (3, 8, 39). Samples of maize, beans, and cassava were found inside a kind of basket, deliberately buried in pits, inside those caves. Crop remains had been preserved by desiccation. Each of these baskets has a different age along the period from 2000 to 500 years BP (8, 39).
Of the 282 samples of ear/cob fragments characterized in this study, at least 186 have been dated (data S1). Radiocarbon dating was carried out using two approaches: indirect dating (3) and direct dating (8). Indirect dating was performed due to the large number of samples found in the same basket by dating samples of charcoal and Guariroba coconuts (Syagrus oleracea), which were stored in the same place as the maize samples. The charcoal and Guariroba coconut samples chosen for dating were selected because they were very abundant in the baskets, and as the dating method is destructive, the maize samples were preserved. The indirect dating of the samples was performed at the Center for Nuclear Energy in Agriculture at the University of São Paulo, using the radiocarbon dating methodology by liquid scintillation spectrometry with benzene (3, 51). The ages determined ranged from 1010 to 570 years BP. Direct dating was carried out by Kistler et al. (8) using accelerator mass spectrometry radiocarbon dating (52) of two archaeological maize samples at Beta Analytic to validate the indirect dating and confirm the previously identified dates. Calibration of the Beta Analytic dates was performed in OxCal (51) using the SHCal13 calibration curve (53).
Morphological characterization of archaeological samples of maize and modern teosinte
The archaeological samples were characterized at the Brazilian Company for Agricultural Research, Genetic Resources and Biotechnology (Embrapa Cenargen), in Brasília, Brazil. There were 282 samples of ear/cob fragments (Fig. 1 and data S1), 12 grain samples (Fig. 1 and data S2), and two ear samples (Fig. 1 and data S3). In addition, 22 modern samples recorded as teosinte (Peabody number: 2001.1.396, Data S4) from the Peabody Museum of Archeology and Ethnology, Harvard University, in Cambridge, Massachusetts, United States, were characterized. The probable origin of these samples is Central America. The passport data for these samples have restricted information (how many plants they represent, where they were collected, and what maize was nearby). Photographic records of archaeological samples of maize and modern teosinte were obtained. The ear/cob shape was estimated for the archaeological ear/cob fragments and 22 samples of modern teosinte, considering the criteria presented in table S3. In addition, the length (cm), base diameter (mm), apical diameter (mm), row number, and number of grains/row were characterized for the fragments. A “primitive” trait was considered to be cobs with a row number less than eight because 95% of the teosinte samples characterized had a row number of less than eight and because thus far, no maize races with this feature have been found in lowland South America (16–18, 20, 22, 42, 43), which indicates the absence of this trait in modern maize races in the region.
For the characterization of the two archaeological samples of ears, 14 morphological descriptors of the crop that are considered key for the classification of maize races were considered (table S4) (20, 22, 40). To preserve the archaeological samples, nondestructive descriptors were considered, and in the case of destructive descriptors, only one representative grain was used for characterization.
Statistical analyses aimed at classifying the races and analyzing similarity relationships between floury maize from lowland South America
Descriptive statistics were used to characterize the archaeological samples of maize ear/cob fragments and modern teosinte. To classify the maize races of the archaeological samples, the qualitative variables were obtained based on the absolute frequencies of each variation or category within a trait, and the highest frequency value (mode) was adopted to characterize the variety for that variable. For the quantitative variables, arithmetic means were obtained. The ape package (54) for R software (55) was used for PCoA, and the smallest convex polygon containing all the points of each group was estimated to explore the similarity relationships between the populations and the identified groups. A principal component analysis was performed using the factoextra package (56) in the R software to present the contribution of the variables in the analysis (fig. S1). Cluster analysis was performed using the Ward agglomerative method, and the genetic distance was estimated using the Gower genetic similarity index (57), which combines qualitative and quantitative variables. The analysis was performed using R software (55), the vegan (58), and cluster (59) packages.
PCoA and cluster analysis were performed based on 14 key descriptors for the classification of maize races, including data from 22 maize accessions (data S5). The maize accessions included in the analysis included 16 landraces of floury maize characterized by (43), four floury maize races described for Brazil (Caingang, Entrelaçado, Lenha, and Moroti), whose data are available in the literature (17, 18, 42), and two samples of archaeological maize ears characterized in this study. Considering that an unrooted dendrogram specifies the relationships between the accessions and not their evolutionary origins (45, 46), cluster analysis without roots was performed to validate the presence of clusters and classify the maize races cultivated in the past. In addition, cluster analysis was performed with data from samples of modern teosinte, which rooted the analysis (considering the ancestry of the samples) based on 10 morphological descriptors (data S6) to discuss the similarity relationships between modern teosintes, archaeological samples from the Peruaçu Valley, and floury maize races from lowland South America. The Harvard samples were used in this study with the aim of obtaining a comparative reference parameter for the morphological characteristics analyzed in the archaeological samples from the Peruaçu Valley, since they have characteristics that differ significantly from completely domesticated maize. Cluster analysis was rooted in a two-row Harvard teosinte sample, considered the most ancestral trait.
DNA extraction from plant material
Molecular characterization was performed on 13 accessions representing lowland South American floury maize races (data S7 and S8). DNA extraction from the landraces was performed by forming a bulk of 30 individuals for each accession (60). Fresh young leaf fragments measuring 1 to 1.5 cm2 were collected in the third week after planting. After collection, the leaves were freeze-dried and sent to the Genetic Analysis Service for Agriculture (SAGA) laboratory of the Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT), located in Mexico, where DNA extraction was performed. The samples were extracted using the modified CTAB method (61).
Markers of SNPs identified by the DartSeq technique
The molecular characterization of maize landraces to obtain SNP markers (data S8) was performed using the DarTseq genotyping technique developed by the Diversity Arrays Technology (DarT) company (https://diversityarrays.com/technology-and-resources/dartseq/dartseq-data-types/). The samples were processed, evaluated, and analyzed in the SAGA laboratory based on CIMMYT. The first and most important step of this technique is the fragmentation of the genomic complex, with the objective of developing genomic libraries that represent the evaluated germplasm. A combination of two enzymes was used in the digestion process, one with frequent cutting (Pst I) and the other with rare cutting (Nsp I). Subsequently, primers were added to amplify the fragments created, and adapters (barcodes) were ligated for sample identification. The genomic library was sequenced on the Illumina HiSeq 2500 sequencer. The sequences of each sample were aligned with a library of sequences representative of a variety of previously processed maize genomes. This process was performed by DarTsoft14 software, developed by DarT. Thus, the analytical pipeline (DarTsoft14) identified SNP markers completely independently of any reference genome. This method makes DarTseq markers robust and practically free of any verification bias faced by many other genotyping and SNP generation technologies, which is extremely important when evaluating the genetic diversity of a population.
Statistical analysis of molecular data
The molecular marker SNPs were obtained and filtered based on the following criteria: call rate ≥ 0.95 (proportion of samples for which the corresponding marker information did not correspond to missing data), RepAvg (proportion of pairs of replicated technical assays for which the marker score is consistent) ≥ 0.85, and AvgPIC (mean polymorphism information content of reference alleles and SNP) > 0. To analyze the genetic relationship between races, cluster analysis was performed with the neighbor-joining method based on the Nei genetic distances (47) using the ape package (54) in the R program (55). The dendrogram was constructed and edited in FigTree v. 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Acknowledgments
We thank A. Prous, M. Jacqueline Rodet, and the Federal University of Minas Gerais’ Natural History Museum team for the archaeological maize collection in Peruaçu Valley, MG; C. R. Clement for all contributions to our evolutionary studies of maize in lowland South America; and the Peabody Museum of Archeology and Ethnology, Harvard University, for the teosinte collection (Peabody number: 2001.1.396). We thank especially all local and indigenous farmers and also the Collaborative Research Network of the Interdisciplinary Group of Agrobiodiversity Study (InterABio) for the support provided in the maize race collections.
Funding: This work was supported by the São Paulo Research Foundation (FAPESP; process 2015/26837-0, Brazil), Brazilian National Council for Scientific and Technological Development (CNPq; process 421045/2016-7, Brazil), and Sectoral Commission of Scientific Research (CSIC; process id2016/400, Uruguay). CNPq awarded scholarships and research fellowships to F.M.C. (142371/2016-5 and postdoctoral 162792/2020-4), N.C.d.A.S., E.A.V., and M.I.Z. We thank Fundação de Estudos Agrários Luiz de Queiroz (FEALQ), and Universidad de la República for the financial support of this publication.
Author contributions: Study conceptualization and design: F.M.C., F.d.O.F., and M.I.Z. Sample acquisition: F.d.O.F., F.M.C., N.C.d.A.S., R.V., and E.A.V. Morphological data collection: F.M.C., F.d.O.F., N.C.d.A.S., and R.V. Statistical analysis of morphological data and classification of maize races: F.M.C., N.C.d.A.S., and R.V. Genomic data collection: F.M.C., N.C.d.A.S., R.V., and E.A.V. Genomic data analysis: F.M.C., R.V., N.C.d.A.S., E.A.V., and M.I.Z. Archaeology background and interpretation: F.d.O.F., F.M.C., R.V., N.C.d.A.S. Interpretation and integration of results: F.M.C., F.d.O.F., R.V., N.C.d.A.S., E.A.V., and M.I.Z. Manuscript drafting: F.M.C. and F.d.O.F., with input from R.V., N.C.d.A.S., E.A.V., and M.I.Z. All authors reviewed and contributed to the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S3
Tables S1 to S7
Legends for data S1 to S8
References
Other Supplementary Material for this : manuscript includes the following:
Data S1 to S8
REFERENCES AND NOTES
- 1.FAO, FAOSTAT statistics database. (Food and Agriculture Organization of the United Nations, Rome, 2020).
- 2.Matsuoka Y., Vigouroux Y., Goodman M. M., Sanchez G. J., Buckler E., Doebley J., A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freitas F. O., Bendel G., Allaby R. G., Brown T. A., DNA from primitive maize landraces and archaeological remains: Implications for the domestication of maize and its expansion into South America. J. Archaeol. Sci. 30, 901–908 (2003). [Google Scholar]
- 4.Vigouroux Y., Glaubitz J. C., Matsuoka Y., Goodman M. M., Sánchez G. J., Doebley J., Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am. J. Bot. 95, 1240–1253 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Piperno D. R., Ranere A. J., Holst I., Iriarte J., Dickau R., Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. U.S.A. 106, 5019–5024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heerwaarden J., Doebley J., Briggs W. H., Glaubitz J. C., Goodman M. M., de Jesus Sanchez Gonzalez J., Ross-Ibarra J., Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. U.S.A. 108, 1088–1092 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perales H., Golicher D., Mapping the diversity of maize races in Mexico. PLOS ONE 9, 1–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kistler L., Maezumi S. Y., Gregorio de Souza J., Przelomska N. A. S., Malaquias Costa F., Smith O., Loiselle H., Ramos-Madrigal J., Wales N., Ribeiro E. R., Morrison R. R., Grimaldo C., Prous A. P., Arriaza B., Gilbert M. T., de Oliveira Freitas F., Allaby R. G., Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 362, 1309–1313 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Kistler L., Thakar H. B., VanDerwarker A. M., Domic A., Bergström A., George R. J., Harper T. K., Allaby R. G., Hirth K., Kennett D. J., Archaeological Central American maize genomes suggest ancient gene flow from South America. Proc. Natl. Acad. Sci. U.S.A. 117, 33124–33129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa F. M., Silva N. C. A., Ogliari J. B., Maize diversity in southern Brazil: Indication of a microcenter of Zea mays L. Genet. Resour. Crop Evol. 64, 681–700 (2017). [Google Scholar]
- 11.E. L. Sturtevant, Varieties of Corn (USDA Bulletin, Washington, D.C., 1989). [Google Scholar]
- 12.T. Á. Kato, C. Mapes, J. A. Serratos, R. A. Bye, Origen y diversificacion del maiz: Una revision analítica (Universidad Autónoma de México, Ciudad de Mexico, 2009). [Google Scholar]
- 13.Yang N., Wang Y., Liu X., Jin M., Vallebueno-Estrada M., Calfee E., Chen L., Dilkes B. P., Gui S., Fan X., Harper T. K., Kennett D. J., Li W., Lu Y., Ding J., Chen Z., Luo J., Mambakkam S., Menon M., Snodgrass S., Veller C., Wu S., Wu S., Zhuo L., Xiao Y., Yang X., Stitzer M. C., Runcie D., Yan J., Ross-Ibarra J., Two teosintes made modern maize. Science 382, (2023). [DOI] [PubMed] [Google Scholar]
- 14.Doebley J., Molecular evidence and the evolution of maize. Econ. Bot. 44, 6–27 (1990). [Google Scholar]
- 15.Doebley J. F., Iltis H. H., Taxonomy of zea (Gramineae). I. A subgeneric classification with key to taxa. Am. J. Bot. 67, 982–993 (1980). [Google Scholar]
- 16.E. Paterniani, M. M. Goodman, Races of Maize in Brazil and Adjacent Areas (CIMMYT, Mexico City, 1977). [Google Scholar]
- 17.F. G. Brieger, J. T. A. Gurgel, E. Paterniani, A. Blumenchein, M. R. Alleoni, Races of maize in Brazil and other eastern South American Countries. National Academy of Sciences, National Research Council, Washington DC, 1958.
- 18.Costa F. M., de Silva N. C., Vidal R., Clement C. R., Alves R. P., Bianchini P. C., Haverroth M., de Freitas F., Veasey E. A., Entrelaçado, a rare maize race conserved in Southwestern Amazonia. Genet. Resour. Crop Evol. 68, 51–58 (2021). [Google Scholar]
- 19.Costa F. M., Silva N. C. D. A., Vidal R., Clement C. R., De Oliveira Freitas F., Alves-Pereira A., Petroli C. D., Zucchi M. I., Veasey E. A., Maize dispersal patterns associated with different types of endosperm and migration of indigenous groups in lowland South America. Ann. Bot. 129, 737–751 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa F. M., Silva N. C. D. A., Vidal R., Clement C. R., De Oliveira Freitas F., Alves-Pereira A., Petroli C. D., Zucchi M. I., Veasey E. A., A new methodological approach to detect microcenters and regions of maize genetic diversity in different areas of lowland South America. Econ. Bot. 77, 345–371 (2023). [Google Scholar]
- 21.De Almeida Silva N. C., Vidal R., Bernardi Ogliari J., Costich D. E., Chen J., Relationships among American popcorn and their links with landraces conserved in a microcenter of diversity. Genet. Resour. Crop Evol. 67, 1733–1753 (2020). [Google Scholar]
- 22.de Almeida Silva N. C., Vidal R., Ogliari J. B., New popcorn races in a diversity microcenter of Zea mays L. in the Far West of Santa Catarina, Southern Brazil. Genet. Resour. Crop Evol. 64, 1191–1204 (2017). [Google Scholar]
- 23.Vallebueno-Estrada M., Rodríguez-Arévalo I., Rougon-Cardoso A., González J. M., Cook A. G., Montiel R., Vielle-Calzada J. P., The earliest maize from San Marcos Tehuacán is a partial domesticate with genomic evidence of inbreeding. Proc. Natl. Acad. Sci. U.S.A. 113, 14151–14156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Madrigal J., Smith B. D., Moreno-Mayar J. V., Gopalakrishnan S., Ross-Ibarra J., Gilbert M. T. P., Wales N., Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr. Biol. 26, 3195–3201 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lombardo U., Iriarte J., Hilbert L., Ruiz-Pérez J., Capriles J. M., Veit H., Early Holocene crop cultivation and landscape modification in Amazonia. Nature 581, 190–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brugger S. O., Gobet E., van Leeuwen J. F. N., Ledru M. P., Colombaroli D., van der Knaap W. O., Lombardo U., Escobar-Torrez K., Finsinger W., Rodrigues L., Giesche A., Zarate M., Veit H., Tinner W., Long-term man-environment interactions in the Bolivian Amazon: 8000 years of vegetation dynamics. Quat. Sci. Rev. 132, 114–128 (2016). [Google Scholar]
- 27.Bush M. B., Piperno D. R., Colinvaux P. A., A 6,000 year history of Amazonian maize cultivation. Nature 340, 303–305 (1989). [Google Scholar]
- 28.Bush M. B., Miller M. C., De Oliveira P. E., Colinvaux P. A., Two histories of environmental change and human disturbance in eastern lowland Amazonia. Holocene 10, 543–553 (2000). [Google Scholar]
- 29.Bush M. B., Correa-Metrio A., McMichael C. H., Sully S., Shadik C. R., Valencia B. G., Guilderson T., Steinitz-Kannan M., Overpeck J. T., A 6900-year history of landscape modification by humans in lowland Amazonia. Quat. Sci. Rev. 141, 52–64 (2016). [Google Scholar]
- 30.Piperno D. R., The origins of plant cultivation and domestication in the New World Tropics. Curr. Anthropol. 52, S453–S470 (2011). [Google Scholar]
- 31.Hilbert L., Neves E. G., Pugliese F., Whitney B. S., Shock M., Veasey E., Zimpel C. A., Iriarte J., Evidence for mid-Holocene rice domestication in the Americas. Nat. Ecol. Evol. 1, 1693–1698 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Watling J., Castro M. T., Simon M. F., Rodrigues F. O., Brilhante de Medeiros M., De Oliveira P. E., Neves E. G., Phytoliths from native plants and surface soils from the Upper Madeira river, SW Amazonia, and their potential for paleoecological reconstruction. Quat. Int. 550, 85–110 (2020). [Google Scholar]
- 33.Piperno D. R., Aboriginal agriculture and land usage in the Amazon Basin, Ecuador. J. Archaeol. Sci. 17, 665–677 (1990). [Google Scholar]
- 34.Iriarte J., Holst I., Marozzi O., Listopad C., Alonso E., Rinderknecht A., Montaña J., Evidence for cultivar adoption and emerging complexity during the mid-Holocene in the La Plata basin. Nature 432, 614–617 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Wesolowski V., de Souza S. M. F. M., Reinhard K. J., Ceccantini G., Evaluating microfossil content of dental calculus from Brazilian sambaquis. J. Archaeol. Sci. 37, 1326–1338 (2010). [Google Scholar]
- 36.Prous A., As Muitas Arqueologias das Minas Gerais. Rev. Espinhaço 2, 36–54 (2013). [Google Scholar]
- 37.Prous A., Fouilles de l’Abri du Boquete, Minas Gerais, Brésil. J. Soc. Am. 77, 77–109 (1991). [Google Scholar]
- 38.Fogaça E., A Tradição Itaparica e as indústrias líticas pré-cerâmicas da Lapa do Boquete. Rev. Mus. Arqueol. Etnol. 5, 145–158 (1995). [Google Scholar]
- 39.Freitas F. O., Rodet M. J., O que ocorreu nos últimos 2000 anos no vale do Peruaçu? Uma análise multidisciplinar para abordar os padrões culturais e suas mudanças entre as populações humanas daquela região. Rev. Mus. Arqueol. Etnol. 20, 109–126 (2010). [Google Scholar]
- 40.Bird R. M. K., Goodman M. M., The races of maize v: Grouping maize races on the basis of ear morphology. Econ. Bot. 31, 471–481 (1977). [Google Scholar]
- 41.Anderson E., Cutler H., Races of maize: Their recognition and classification. Ann. Mo. Bot. Gard. 29, 69–89 (1942). [Google Scholar]
- 42.F. De María, G. Fernández, G. Zoppolo, Caracterización agronómica y clasificación racial de las muestras de maíz coleccionadas bajo el proyecto IPGRI. Reunión técnica de la Facultad de Agronomía, Universidad de la República, Uruguay (1979).
- 43.N. C. A. Silva, R. Vidal, F. M. Costa, E. A. Veasey, “Clasificación de las razas de maíz de Brasil y Uruguay: Enfoque metodológico y principales resultados” in Maíces de las Tierras Bajas de América del Sur y Conservación de la Agrobiodiversidad en Brasil y Uruguay, N. C. A. Silva, R. Vidal, F. M. Costa, E. A. Veasey, Eds. (Ponta Grossa, Atena, 2020), pp. 87–109. [Google Scholar]
- 44.M. M. Goodman, W. L. Brown, “Races of Corn” in Corn and Corn Improvement, G. F. Sprague, J. W. Dudley, Eds. (American Society of Agronomy, Madison, WI, 1988), pp. 33–79. [Google Scholar]
- 45.M. Ridley, Evolution (Wiley-Blackwell, Oxford, 2003). [Google Scholar]
- 46.N. H. Barton, D. Briggs, J. A. Eisen, D. B. Goldstein, N. H. Patel, Evolution (Cold Sprin, New York, 2007). [Google Scholar]
- 47.Nei M., Genetic distance between populations. Am. Nat. 106, 283–292 (1972). [Google Scholar]
- 48.Doebley J., The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Bommert P., Nagasawa N. S., Jackson D., Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 45, 334–337 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Janzen G. M., Wang L., Hufford M. B., The extent of adaptive wild introgression in crops. New Phytol. 221, 1279–1288 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Pessenda L. C. R., Camargo P. B., Datação radiocarbônica de amostras de interesse arqueológico e geológico por espectrometria de cintilação líquida de baixo nível de radiações de fundo. Quim. Nova 14, 98–103 (1991). [Google Scholar]
- 52.Hall S. A., Early maize pollen from Chaco Canyon, New Mexico, USA. Palynology 34, 125–137 (2010). [Google Scholar]
- 53.Bronk Ramsey C., Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009). [Google Scholar]
- 54.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 55.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
- 56.A. Kassambara, F. Mundt. Factoextra: Extract and visualize the results of multivariate data analyses (2020); https://CRAN.R-project.org/package=factoextra.
- 57.Gower J. C. A., A general coefficient of similarity and some of its properties. Biometrics 27, 857–871 (1971). [Google Scholar]
- 58.J. Oksanen, G. L. Simpson, F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, R. B. O’Hara, P. Solymos, M. H. H. Stevens, E. Szoecs, H. Wagner, M. Barbour, M. Bedward, B. Bolker, D. Borcard, G. Carvalho, M. Chirico, M. De Caceres, S. Durand, H. B. A. Evangelista, R. FitzJohn, M. Friendly, B. Furneaux, G. Hannigan, M. O. Hill, L. Lahti, D. McGlinn, M. H. Ouellette, E. R. Cunha, T. Smith, A. Stier, C. J. F. Ter Braak, J. Weedon, Package “Vegan” Title Community Ecology Package (2022); https://github.com/vegandevs/vegan.
- 59.M. Maechler, P. Rousseeuw, A. Struyf, M. Hubert, K. Hornik, Cluster: Cluster analysis basics and extensions (2015); https://CRAN.R-project.org/package=cluster.
- 60.Bedoya C. A., Dreisigacker S., Hearne S., Franco J., Mir C., Prasanna B. M., Taba S., Charcosset A., Warburton M. L., Genetic diversity and population structure of native maize populations in Latin America and the Caribbean. PLOS ONE 12, 1–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doyle J. J., Doyle J. L., Genomic plant DNA preparation from fresh tissue-CTAB. Phytochem. Bull. 19, 11–15 (1987). [Google Scholar]
- 62.IBGE, Biomes of Brazil (2021); https://ibge.gov.br/en/geosciences/maps/brazil-environmental-information/18341-biomes.html?lang=en-GB.
- 63.Dinerstein E., Olson D., Joshi A., Vynne C., Burgess N. D., Wikramanayake E., Hahn N., Palminteri S., Hedao P., Noss R., Hansen M., Locke H., Ellis E. C., Jones B., Barber C. V., Hayes R., Kormos C., Martin V., Crist E., Sechrest W., Price L., Baillie J. E. M., Weeden D., Suckling K., Davis C., Sizer N., Moore R., Thau D., Birch T., Potapov P., Turubanova S., Tyukavina A., De Souza N., Pintea L., Brito J. C., Llewellyn O. A., Miller A. G., Patzelt A., Ghazanfar S. A., Timberlake J., Klöser H., Shennan-Farpón Y., Kindt R., Lillesø J. P. B., Van Breugel P., Graudal L., Voge M., Al-Shammari K. F., Saleem M., An ecoregion-based approach to protecting half the terrestrial realm. Bioscience 67, 534–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.C. Senigagliesi, C. O. Scoppa, D. A. Freggiaro, A. J. Martínez, A. Clausen, O. Polidoro, M. Ferrer, Catálogo de germoplasma de maíz de Argentina (Instituto Agronomico per L’outremare, Firenze, 1997). [Google Scholar]
- 65.Stothert K. E., The preceramic las vegas culture of coastal Ecuador. Am. Antiq. 50, 613–637 (1985). [Google Scholar]
- 66.Pearsall D. M., Piperno D. R., Antiquity of maize cultivation in Ecuador: Summary and reevaluation of the evidence. Am. Antiq. 55, 324–337 (1990). [Google Scholar]
- 67.Piperno D. R., Ranere A. J., Holst I., Hansell P., Starch grains reveal early root crop horticulture in the Panamanian tropical forest. Nature 407, 894–898 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Grobman A., Bonavia D., Dillehay T. D., Piperno D. R., Iriarte J., Holst I., Preceramic maize from Paredones and Huaca Prieta, Peru. Proc. Natl. Acad. Sci. U.S.A. 109, 1755–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herrera L. F., Cavelier I., Rodríguez C., Mora S., The technical transformation of an agricultural system in the Colombian Amazon. World Archaeol. 24, 98–113 (1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Tables S1 to S7
Legends for data S1 to S8
References
Data S1 to S8