Abstract
Strigolactones (SLs), carotenoid-derived plant hormones, govern the growth and development of both monocotyledonous and dicotyledonous plants. DWARF27 (D27), a plastid-targeted protein located at the initiation site of the core pathway in SL synthesis, plays a crucial role in regulating plant tillering (branching). In rice (Oryza sativa) and wheat (Triticum aestivum), OsD27 and TaD27-B proteins modulate the number of plant tillers by participating in SL biosynthesis. Similarly, AtD27 in Arabidopsis thaliana is required for SL production and has a significant impact on phenotypic changes related to branching. At the same time, TaD27 in wheat has been confirmed as a functional orthologue of AtD27 in Arabidopsis, and both Psathyrostachys juncea and wheat belong to the Triticeae, so we speculate that PjD27 gene may also have the same function as AtD27 in Arabidopsis. In this study, we initially screened the PjD27 gene significantly associated with tillering regulation through transcriptome data analysis and subsequently validated its expression levels using qRT-PCR analysis. Furthermore, we conducted phylogenetic analysis using amino acid sequences from 41 species, including P. juncea, to identify closely related species of P. juncea. Here, we analyze the conservation of D27 protein among P. juncea, rice, wheat, and Arabidopsis and provide preliminary evidence suggesting that PjD27 protein is an orthologue of D27 protein in Arabidopsis. Through reverse genetics, we demonstrate the crucial role of PjD27 in regulating plant branching, establishing it as a functional orthologue of D27 in Arabidopsis. Furthermore, following transient expression in tobacco (Nicotiana tabacum), we demonstrate that the subcellular location of the PjD27 protein is consistent with the cellular location of TaD27-B in wheat. Quantitative analysis of SLs shows that PjD27 is a key gene regulating tillering (branching) by participating in SL biosynthesis. By elucidating the function of the PjD27 gene, our findings provide valuable genetic resources for new germplasm creation and improving grain yield in P. juncea.
Keywords: DWARF27, Psathyrostachys juncea, Arabidopsis thaliana, functional orthologue, tillers/branches number, strigolactones
Introduction
Tillering is one of the major components affecting plant yield. Generally, agronomists consider tiller number and tiller angle as the most important tillering traits in crop plants (Ishikawa et al. 2005; Dong et al. 2023; Wang, Tu et al. 2023). Fertilization controls plant growth parameters such as tiller number, leaf size, and panicle number. However, several genes are known to influence tillering. For example, the key gene regulating the number of tillers/branches in wheat (TN1) is highly conserved across wheat varieties. The TN1 gene was cloned from the wheat tn1 mutants and is known to impact the yield of wheat by promoting the growth of wheat tiller buds (Dong et al. 2023). Similarly, the transcriptional regulation of fertilizer-responsive gene Os1900 in rice (Oryza sativa L.) can mediate the increase of tiller number and grain yield (Cui et al. 2023). Tiller angle is also important and can improve photosynthetic efficiency and increase field density by affecting the plant architecture to increase grain yield per unit area (Zhao et al. 2023). Genes play a role in tiller angle, and the Tiller angle control 1 (TAC1) gene is a key gene known to regulate the dynamic expression of tiller angle during plant growth stages in rice (Wang, Tu et al. 2023).
Strigolactones (SLs) are plant hormones that regulate plant growth and development (Proust et al. 2011). SLs inhibit shoot branching in plants of many different species. SLs inhibit auxin synthesis in plant lateral branches and then indirectly act on apical dominance (Agusti et al. 2011). For example, Arabidopsis thaliana treated with SLs produced fewer branches, an effect attributed to inhibition of auxin synthesis (Brewer et al. 2009).
SLs have likely evolved to mitigate undue plant growth when nutrients are limited. Early work reviewing rice, observed that the level of SL production is inversely related to phosphorus availability, inhibiting the growth of rice tiller buds when nutrients are limited (Umehara et al. 2010). RMS3 and RMS4 in pea (Pisum sativum) shoots inhibit the outgrowth of pea buds by participating in the biosynthesis and feedback signal of SLs (Dun et al. 2009). Similarly, D17, D3, and D27 are key genes in the biosynthesis or signaling pathway of SLs in rice, and their defective mutants have an increased number of tillers due to the lack of this SL-mediated axial synthesis inhibition (Cui et al. 2023).
DWARF27 (D27) is a plastid-targeted protein required for the biosynthesis of SLs, which converts all-trans-β-carotene into 9-cis-β-carotene during the biosynthetic pathway of SLs, and further forms the precursor of SLs known as carlactone through the catalysis of Carotenoid Cleavage Dioxygenase 7 (CCD7) and CCD8. Finally, SLs are formed through other reaction paths, thereby inhibiting the number of plant tillers/branches (Brewer et al. 2009; Alder et al. 2012; Waters et al. 2012; Harrison et al. 2015). Some studies have revealed the regulatory function of D27 in plant tillering (branching). Rice d27 mutants were associated with high tillering and severe dwarfism (Lin et al. 2009; Wang and Li 2011). In the case of calcium deficiency, the level of SLs in rice will decrease accordingly, thereby stimulating the outgrowth of tiller buds (Shindo et al. 2018). The Arabidopsis d27-1 mutant had a phenotype of significantly increasing the number of axillary buds. When the exogenous SL analog GR24 was applied, the shoot branching was significantly inhibited in Arabidopsis mutant (Waters et al. 2012). The TaD27 genes which are orthologues of rice D27 in SLs biosynthesis were identified in wheat. TaD27-RNAi plants had an increased number of branches or tillers, and TaD27-B-overexpressing (TaD27-B-OE) plants had fewer tillers (Zhao et al. 2020).
Natural grassland in high-latitude cold steppe regions faced degradation tendency due to overgrazing and climate change (Li, Peng et al. 2022). Divers of P. juncea varieties with good tolerance and high biomass are urgently needed (Fikadu et al. 2022; Main et al. 2023). Psathyrostachys juncea (Fisch.) Nevski, as a high-quality cool season forage grass with good tolerance, has a high breeding value in forage yield improvement (Liu et al. 2006). The study of its tillering-related genes contributes to the production of high-yield and grazing-tolerant P. juncea varieties. In addition, the increase in P. juncea biomass yield contributes to soil surface coverage and ecological restoration (Li et al. 2023), and provides potential genetic resources for improving the tillering traits of other perennial grasses (Li, Yun et al. 2022).
Wheat is one of the most significant cereal crops, which is utilized as a staple food for human consumption and contributes about 20% of proteins worldwide (Shiferaw et al. 2013; Yeni and Teoman 2020). P. juncea with abundant superior germplasm of stress-bearing and disease resistance, is a wheat-related wild, which provides an excellent gene pool of wheat (Wang et al. 2016; Hu et al. 2018). However, the yield traits of P. juncea still need to be improved (Li, Yun et al. 2022). Therefore, we used P. juncea, a perennial bunch grass, as the research material to analyze the function of tillering-related genes, aims to improve the yield of P. juncea.
We screened candidate genes involved in the SL synthesis pathway that are highly correlated with tillering through transcriptome data analysis, and the candidate gene PjD27 was selected as the main focus of our research (Li, Yun et al. 2022). The subcellular location of PjD27 protein was analyzed by transient transfection of tobacco (Nicotiana tabacum), and the 35S:pCambia1300-PjD27-cYFP vector (Supplementary Fig. 1, a and b) was constructed to infect Arabidopsis by Agrobacterium infection. We observed that infection of A. thaliana (Col-0) with the associated gene led to a significant reduction in the number of branches, though the Arabidopsis d27 mutant (At1g03055; SALK_071443.2) restored the number of branches associated with the wild type, consistent with the effect of AtD27 on the phenotype of Arabidopsis (Waters et al. 2012). Our results indicate that PjD27 is the main gene regulating tillering behavior in P. juncea, providing valuable information for alleviating the lack of germplasm resources and improving the yield of P. juncea.
Materials and methods
Plant materials and treatment conditions
Columbia wild-type A. thaliana (Col-0) and tobacco materials were kindly provided by the Shenzhen Institute of Advanced Technology (SIAT), Chinese Academy of Sciences. The Arabidopsis d27 mutant (At1g03055, SALK_071443.2) was acquired from AraShare Science. Primers LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′), d27-Left Primer (5′-ATCTTCGATCTCTGGAGGCTC-3′), and d27-Right Primer (5′-GGATACGGCAACTAGGGTTTC-3′) were designed by T-DNA Primer Design (http://signal.salk.edu/tdnaprimers.2.html). Both Arabidopsis and tobacco were cultivated in a 1:1 mixture of peat soil and vermiculite. Arabidopsis was grown under controlled conditions with a constant temperature of 23°C, a light cycle of 16 h, followed by an 8-h dark period. Tobacco plants were cultured at 25°C with a light cycle of 12 h, alternating with a dark period of equal duration.
RNA sequencing
Thirty dense tillering (DT) plants and another thirty sparse tillering (ST) plants were selected as materials in P. juncea, with 3 biological replicates for DT and ST plants, respectively. These plants were planted in the forage germplasm nursery of Inner Mongolia Agricultural University in Hohhot, Inner Mongolia, China. Plants were grown in the forage nursery for a period of 2 months before they were destructively sampled. The tiller nodes were meticulously cleaned and subsequently frozen using liquid nitrogen. Total RNA was extracted using a total RNA extraction kit (DP432, TransGen, Beijing, China), and cDNA was synthesized from the total RNA using PrimeScript RT reagent Kit (AW311-02, All-Gold, Beijing, China). The resulting cDNA library was then sequenced by Biomarker Technologies (Beijing, China). The sequencing data were screened to analyze gene expression levels, which were calculated using Fragments per Kilobase of transcript per Million mapped reads (FPKM). In addition, the false discovery rate was used as a key indicator for differentially expressed genes (DEGs) screening to calculate the significance of differences in gene expression levels. Subsequently, the gene expression heatmap was generated using the online platform EHBIO (http://www.ehbio.com/ImageGP/index.php/Home/Index/PHeatmap.html). Pathway analysis of DEGs was conducted based on the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (Supplementary Fig. 3). For detailed test conditions, review our previous publication (Li, Yun et al. 2022).
Sequence and phylogenetic analysis of D27
The coding sequence (CDS) of the PjD27 gene was successfully cloned and subsequently subjected to sequencing analysis by Ruibiotech Company, located in Guangzhou. Homologous sequence alignment was performed using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) in the NCBI database, and the amino acid sequences of D27 proteins derived from 41 species were used for phylogenetic analysis. A neighbor-joining (NJ) phylogenetic tree was constructed using Interactive Tree of Life (iTOL) online platform (https://itol.embl.de/) and Molecular Evolutionary Genetics Analysis 11.0 software (Zhu et al. 2022) with 1,000 bootstrap test replicates. Conserved domains within D27 proteins from P. juncea, wheat, rice, and Arabidopsis were analyzed using the Structure tool at NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), while the amino acid sequences were compared utilizing DNAMAN software.
Generation of 35S:PjD27 Arabidopsis plants
Total RNA was extracted from the tiller nodes of P. juncea individuals using an RNA extraction kit (DP432, TransGen, Beijing, China). Subsequently, cDNA was generated from 1 µg of total RNA by PrimeScript RT reagent Kit (AW311-02, All-Gold, Beijing, China). The full-length CDS of the PjD27 gene was first amplified using D27-F (5′-ATGGCCTTGATGCCCGTGG-3′) and D27-R (5′-GTTCTGAATTTTGGGACATATTGCAGC-3′) primers designed in SnapGene 6.0.2, with 2 × Phanta Max Master Mix enzyme (Vazyme, Nanjing, China) and cDNA as a template. Then, it was ligated to the Blunt vector using a cloning kit (CB501-01, TransGen, Beijing, China), and positive clones were confirmed by sequencing. Approximately 0.5 µL of the cloning products was then reamplified using pCambia-1300-cYFP Recombinant primers 1300-D27-F 5′-CCGAATTCGGAGTCGACACTAGTATGGCCTTGATGCCCGTGG-3′ and 1300-D27-R 5′-TCCACCTCCGACCGGTGCACTAGTGTTCTGAATTTTGGGACATATTGCAGC-3′ (the underlined parts are the adapter primers of the pCambia-1300-cYFP vector). The full-length CDS was then transferred to pCambia-1300-cYFP (an infection vector provided by SIAT) via recombination, generating the final 35S:PjD27 vector. The Col-0 and Arabidopsis d27 mutant were then transformed by the floral dip method. Transgenic seedlings were grown in 16 h light for 11 days on one-half-strength Murashige and Skoog medium containing hygromycin B (50 mg.ml−1) and termentin (100 mg.ml−1).
Western blotting analysis
The leaves of the Arabidopsis d27 mutant, Col-0, and positive plants were pulverized into a fine powder using liquid nitrogen and subsequently transferred to a centrifuge tube containing an equal volume of 2 × SDS loading buffer. After spotting, electrophoresis was performed at a voltage of 120 V for a duration of 1 h. Subsequently, the gel was inverted and positioned beneath the PVDF membrane. It was then covered with filter paper and a sponge before being transferred to the membrane using an electric field strength of 100 V for a period of 60 min at a temperature of 4°C. After incubation of the PVDF membrane with the primary antibody GFP and subsequent treatment with the secondary antibody [HRP-labeled goat anti-mouse lgG (H + L)], dropwise addition of a mixture containing an equal amount of BeyoECL Moon A solution and B solution was performed, followed by imaging using a western blot (WB) exposure instrument (Ren et al. 2023). The aforementioned reagents were supplied by SIAT.
RT-PCR and RT-qPCR analysis
The roots, tillering nodes, and leaves from 32-day-old Arabidopsis d27 mutants, Col-0, transgenic lines expressing PjD27 under the control of the CaMV 35S promoter with both Col-0 background [35S:PjD27 (Col-0)] and Atd27 mutant background [35S:PjD27 (d27)], were used as experimental samples for subsequent analysis using RT-qPCR and RT-PCR techniques. Moreover, the freeze–dried tillering nodes from DT and ST were also included as other samples for subsequent RT-qPCR experiments.
In all cases, total RNA was extracted from the aforementioned materials using a total RNA extraction kit (DP432, TransGen, Beijing, China). cDNA was synthesized from the total RNA using PrimeScript RT reagent Kit (AW311-02, All-Gold, Beijing, China). Various template cDNAs were diluted into 4 concentration gradients (1, 1/2, 1/5, 1/10), and RT-qPCR was performed using Taq Pro Universal SYBR qPCR Master Mix enzyme (Vazyme, Nanjing, China) with a volume of 5 µL. Cycle conditions were as follows: 95°C for 2 min; then 40 cycles of 95°C for 5 s, 60°C for 30 s; 95°C for 15 s; followed by melt-curve analysis. Primer pairs for quantitative RT-PCR were designed using NCBI (https://blast.ncbi.nlm.nih.gov), except for ACTIN (At5g09810) (Waters et al. 2012) and ACTIN-Pj (Li, Yun et al. 2022). All quantitative RT-PCR primers are listed in Supplementary Table 2. PjD27 gene expression in Arabidopsis was quantified based on the 2−ΔΔCt method. RT-PCR primer pairs used for identifying 35S:PjD27 (Col-0) and 35S:PjD27 (d27)-positive plants were designed using NCBI. The primers were RT-D27-F 5′-AGAAGTTCGCCGGAAGGAAGGAAG-3′ and RT-D27-R 5′-TGGGACATATTGCAGCGGAG-3′, resulting in a total product size of 579 bp. The ACTIN (AT3G18780) was provided by SIAT.
Subcellular location analysis of PjD27 protein
The recombinant fusion protein 35S:PjD27, along with the negative control 35S:cYFP, was propagated through Agrobacterium GV3101 and subsequently injected into the dorsal surface of 1-month-old tobacco leaves. YFP (Yellow Fluorescent Protein) fluorescence was captured using a Nikon (A1R) confocal microscope after 2 days of tobacco growth (Bian et al. 2023). The subcellular location of the PjD27 protein was analyzed and documented by comparing it with the autofluorescence emitted by chloroplasts.
Quantification of SL levels
The roots and tiller nodes of Col-0, Atd27 mutants, and homozygous overexpression lines in different backgrounds were hydroponically cultivated using a Hoagland nutrient solution with a 1/8 dilution ratio for ∼3 weeks until reaching the tillering stage, with each sample being replicated 3 times. After cultivation, tissues were collected from the field, rinsed with deionized water, dried, and subsequently pulverized in liquid nitrogen. Approximately 0.5 g of the test sample was weighed into a test tube, followed by the addition of 5 mL acetonitrile solution for overnight extraction at 4°C. Subsequently, samples were centrifuged at a speed of 12,000 r min−1 for 5 min, after which the resulting supernatant was collected and put to the side. A 5-fold volume of acetonitrile solution was added to the precipitate to elucidate 2 further extractions. Subsequently, all the above supernatants were recombined. Following the addition of 200 mg C18 filler, vigorous shaking for 30 s ensued, followed by centrifugation at 10,000 r min−1 for 5 min to obtain the supernatant. After concentration, the supernatant was redissolved in 300 µL methanol and filtered through a 0.22 µm organic phase filter membrane. Subsequently, the sample was subjected to detection using ultra-performance liquid chromatography coupled with tandem mass spectrometry [UPLC–MS/MS (QTRAP6500, AB)]. The reagents utilized in the test included 5-Deoxystrigol [5DS (Anpel, CAS: 151716-18-6)], GR24 (Yuan Ye, Shanghai, CAS: 76974-79-3), strigol (Anpel, CAS: 1820-11-2), acetonitrile and methanol: HPLC (Merck Company), and CNW C18 QuEChers filler (Anpel Company, Shanghai). The chromatographic column employed was Agilent Poroshell120 SB-C18 (2.1 × 150 mm, 2.7µm) with a column temperature of 30°C. Mobile phase A consisted of a mixture of water containing 0.1% formic acid, while mobile phase B comprised acetonitrile; the flow was set at 0.3 mL min−1 [for detailed test conditions, according to the method described previously (Besserer et al. 2006; Jamil et al. 2010; Proust et al. 2011)].
Statistical analysis
The comparison between a treatment and a control was conducted using t-tests (and nonparametric tests). For comparisons between multiple treatments and a control, one-way 2-tailed ANOVAs were performed as described in the figure legends. All data analysis was conducted using GraphPad Prism 9.5 software.
Results
PjD27 downregulated the tiller number of P. juncea
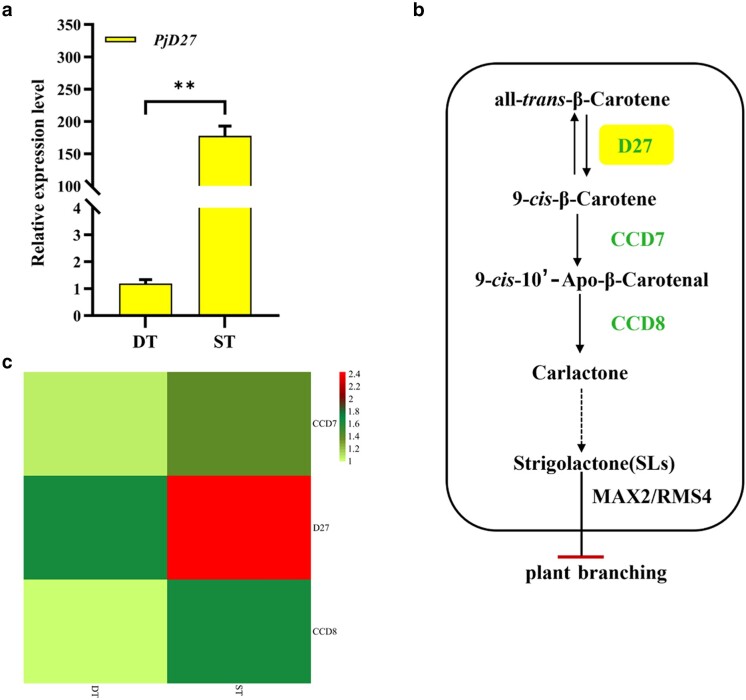
In order to verify the expression level of PjD27 gene in sparse tiller and DT individuals of P. juncea, we used qRT-PCR using the cDNA of P. juncea at the tillering stage as a template. The results showed that the expression level of PjD27 in ST was significantly higher than that in DT (Fig. 1a), indicating that PjD27 downregulated the number of tillers in P. juncea. This result is consistent with the findings from transcriptome analysis. Simultaneously, we observed that CCD7 (BMK_Unigene_196444) and CCD8 (BMK_Unigene_152703), which are located downstream of D27 (BMK_Unigene_142326), exhibited a downregulatory effect on the tiller number of P. juncea (Fig. 1c). In order to further insight into the effect of PjD27 on tillering regulation, we performed a pathway analysis of PjD27 based on the research results of KEGG database (Li, Yun et al. 2022) and others (Brewer et al. 2009; Dun et al. 2009). It is speculated that PjD27 was involved in the biosynthesis of SLs, thereby inhibiting plant branching (Fig. 1b).
Fig. 1.
Regulation of PjD27 on tiller number of P. juncea. a) Expression of PjD27 quantified by qRT-PCR analysis. DT represents dense tiller and ST represents sparse tiller in P. juncea. Asterisks denote significant differences between the ST and DT [**P < 0.01, t-tests (and nonparametric tests)]. b) D27 is involved in the synthesis of SL and regulates the growth of plant branches. The solid arrow indicates the reaction direction of each step, and the virtual arrow indicates the reaction direction of omitting some steps, and the horizontal bar indicates the inhibition. c) Expression of DEGs related to the biosynthesis of SLs.
Evolutionary relationship analysis of D27 genes
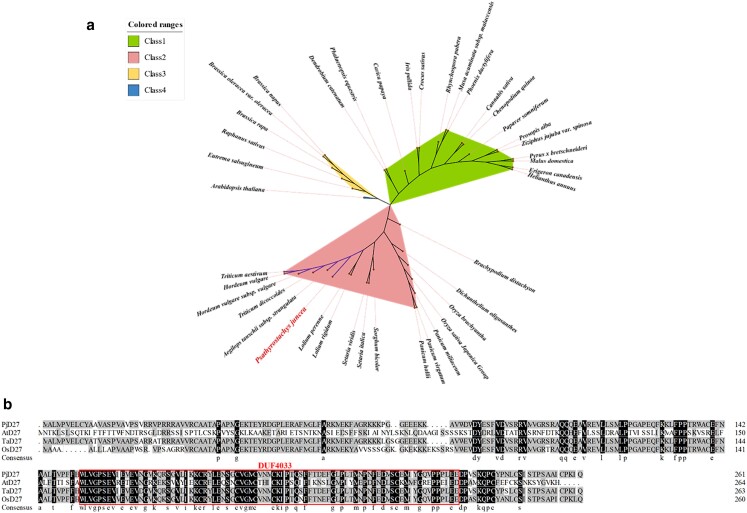
To study the evolutionary relationships among the D27 genes in different species, 41 D27 genes were analyzed to construct an unrooted phylogenetic tree. The D27 genes were classified into 4 groups (Fig. 2a; Supplementary Table 1), which contained 17 (class 1), 18 (class 2), 6 (class 3), and 1 (class 4) members. Our phylogenetic tree suggested that D27 genes evolved to different degrees in the evolutionary process. The D27 genes of P. juncea, wheat, and other Triticeae crops were divided into the same subclass, indicating that they are highly homologous. As further evidence of their shared ancestry, we point out that the PjD27 protein in P. juncea had the same highly conserved domain, named DUF4033, as D27 proteins in Arabidopsis, wheat and rice, suggesting that they were orthologues of PjD27 protein (Fig. 2b). We present this comparison as theoretical evidence for the functional role PjD27 likely plays in P. juncea.
Fig. 2.
Sequence and phylogenetic analysis of D27. a) Phylogenetic tree of D27 genes from different species. The phylogenetic tree was built using the NJ method with 1,000 bootstrap replications. Arabic numerals (1–4) represent each gene cluster, which are labeled with different colors. P. juncea has been marked in red (a). Purple lines represent a subclass of P. juncea and its highly homologous species. b) Multiple alignments of D27proteins in different species. The red rectangle denotes the conserved domain of DUF4033.Pj, P. juncea, At, A. thaliana, Ta, Triticum aestivum, Os, Oryza sativa (here specifically denotes Oryza sativa Japonica Group).
PjD27 is necessary for regulating the number of plant branches
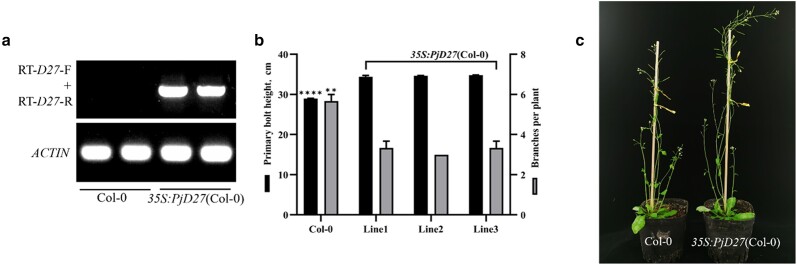
Previous studies have shown that the Arabidopsis d27 mutant plants had a significantly increased branching phenotype (Waters et al. 2012). To verify the biological function of the PjD27, we first introduced the 35S:PjD27 vector into Col-0 plants. RT-PCR analysis showed that introduction of the 35S:PjD27 vector led to the development of complete PjD27 transcripts in Arabidopsis (Fig. 3a). Through the phenotypic analysis of 3 independent PjD27-OE transgenic Arabidopsis lines (Supplementary Fig. 2a) and Col-0, it was found that the increased expression of PjD27 caused an obvious decrease in branches and increase in primary bolt height in the PjD27-OE lines compared with the wild type (Fig. 3, b and c). We took this as evidence that PjD27 plays an important role in regulating the number of plant branches.
Fig. 3.
Overexpression of PjD27 gene downregulates the number of branches in Arabidopsis. a) RT-PCR analysis of PjD27 transcripts in seedlings homozygous. Primers were designed in the CDS of PjD27 gene. Col-0 serves as a control for RT. b) Branch numbers (dark gray bars) and plant height (black bars) in 42-day-old Col-0 and 3 independent PjD27-OE transgenic Arabidopsis lines. Data are means ± SE (n = 30 plants). Asterisks denote significant differences between treated and untreated samples within the same genotype (**P < 0.01, ****P < 0.0001, one-way ANOVA). c) Axillary branching in the representative plants described in a and b.
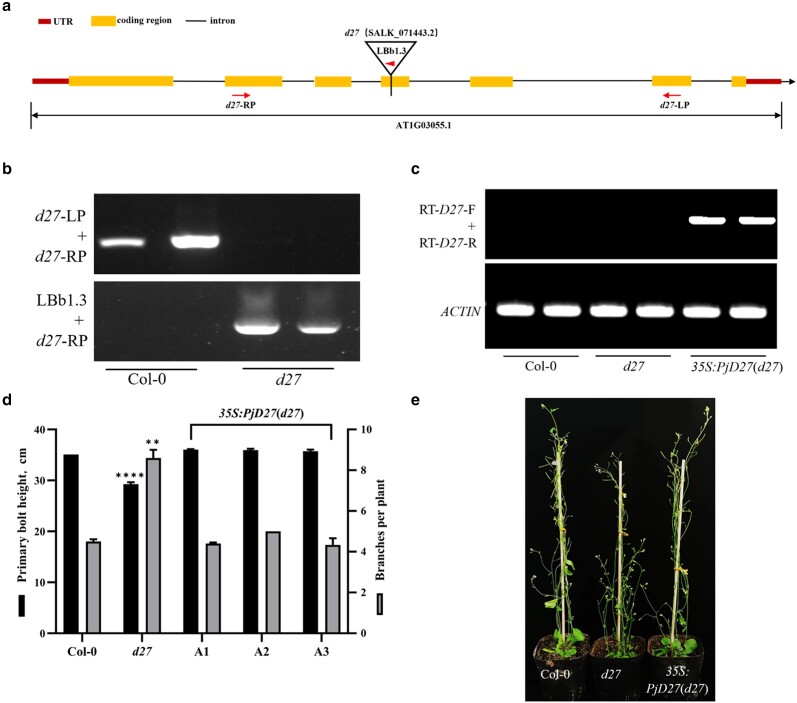
To further understand the function of PjD27 in plant branches, we also introduced the 35S:PjD27 vector into homozygous Arabidopsis d27 mutant plants (Fig. 4, a and b), and we complemented the mutant with a cDNA encoding the PjD27 protein, but designed the cDNA so that the PjD27 protein would only be expressed under the control of the cauliflower mosaic virus 35S promoter. Three independent transgenic complementary lines (35S:PjD27-A1, 35S:PjD27-A2, and 35S:PjD27-A3) were selected for phenotypic analysis (Supplementary Fig. 2b). The result showed that there were no phenotypic differences in plant branches and plant height between the 3 independent transgenic complementary lines and Col-0, and an increased branching and severe dwarf phenotype in Arabidopsis d27 mutant plants was complemented by 35S:PjD27 (Fig. 4, d and e). At the same time, RT-PCR analysis also showed Arabidopsis d27 mutant plants had accumulated PjD27 transcripts (Fig. 4c). The biological role of the PjD27 gene in the regulation of branch number and plant height in Arabidopsis was consistent with the role that the OsD27 gene plays in rice (Lin et al. 2009). We took this experimental result as further evidence that PjD27 likely plays a similar role in regulating the branch number and forage yield of P. juncea.
Fig. 4.
The expression of PjD27 in Atd27 plants completely restored the phenotype back to that of the wild type. a) The primary transcript structure of the Arabidopsis d27. Inverted triangle indicates the position of T-DNA insertion in Atd27, and arrows represent the approximate positions and orientations of PCR primers. UTR, untranslated region. b) RT-PCR analysis of Atd27 transcripts in seedlings homozygous for T-DNA insertion allele. Primers were selected to span insertion point and an upstream region of the transcript. c) RT-PCR verification of 35S:PjD27(d27) plants. The primer pair was designed based on the coding region sequence of PjD27. d) Branch numbers (dark gray bars) and plant height (black bars) in 42-day-old Col-0, Atd27 mutant plants and 3 independent PjD27-OE transgenic Arabidopsis complementary lines. Data are means ± SE (n = 30 plants). d) Asterisks denote significant differences that Atd27 mutant plants compared with Col-0 and complementary lines (**P < 0.01, ****P < 0.0001, one-way ANOVA). e) Phenotypes of representative plants described in c and d.
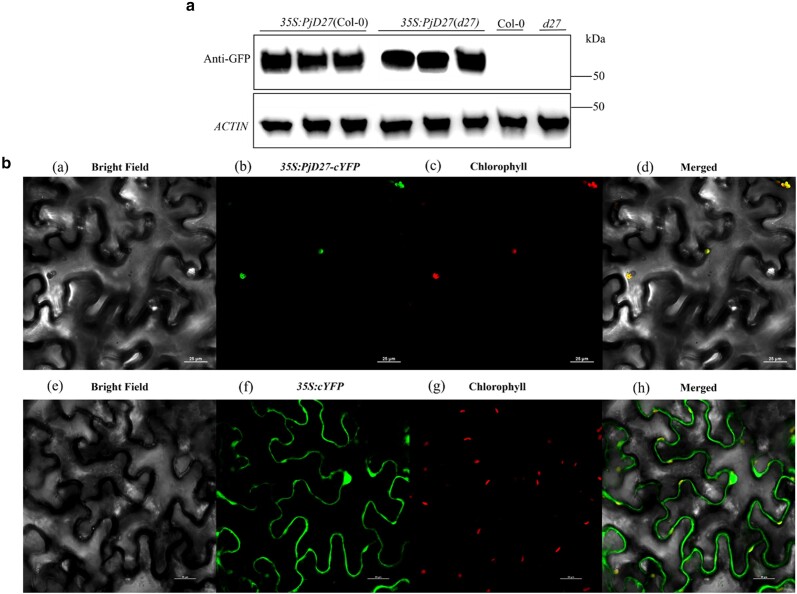
Western blotting and subcellular location of PjD27 protein
We observed that PjD27 was expressed in all the 35S:PjD27 lines, while the PjD27 protein was not expressed in Col-0 and Atd27 mutant plants. To further explore the subcellular location of the PjD27, we generated a chimeric construct encoding the full-length PjD27 protein fused with YFP. Following transient expression in tobacco, imaging of fluorescence showed that the YFP fluorescence fully overlapped with the red fluorescence associated with chloroplasts (Fig. 5b). The result showed that the PjD27-cYFP fusion protein was predominantly localized in the chloroplast, which was consistent with the cellular location of OsD27 in rice (Lin et al. 2009) and TaD27-B in wheat (Zhao et al. 2020).
Fig. 5.
Overexpression characteristics of PjD27 protein. a) Western blotting verification of PjD27. The six protein bands are 6 independent transgenic lines. b) Subcellular localization analysis of PjD27-cYFP fusion protein. b, PjD27-cYFP fusion protein fluorescence, f, 35S: p1300-cYFP fluorescence; c and g, chloroplast autofluorescence; d and h, the merged images denote colocalization of the 2 fluorescent signals; a and e, the cell outline of tobacco in bright field micrographs of the same field of view. The scale size in the picture is 25µm.
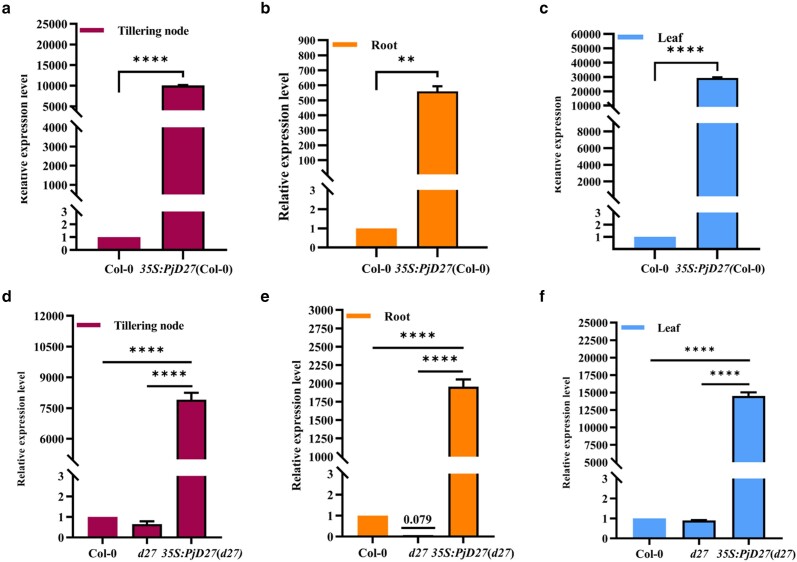
The expression level of PjD27 in Arabidopsis is significantly increased
The primers for qRT-PCR were designed based on the coding region sequence of PjD27. qRT-PCR was performed using cDNAs from roots, tillering nodes, and leaves of Col-0, Arabidopsis d27 mutant plants, and PjD27-OE transgenic Arabidopsis lines to analyze the expression level of the PjD27 gene in each set of plants. We observed that the PjD27 transcript level was significantly increased in tillering nodes (Fig. 6a), roots (Fig. 6b), and leaves (Fig. 6c) of 32-day-old PjD27-OE transgenic Arabidopsis lines compared with the wild type. The expression level of the PjD27 gene in leaves and tillering nodes was orders of magnitude (103 to 104) times greater than expressed in roots. When the cDNA encoding the PjD27 protein complemented Arabidopsis d27 mutant plants, the expression level of the PjD27 gene was also markedly increased relative to the Col-0 and Arabidopsis d27 mutant plants (Fig. 6, d–f).
Fig. 6.
Expression level of PjD27 in different tissues of Arabidopsis. a–c) The analysis of relative expression level of PjD27 in different tissues of 35S:PjD27(Col-0) plants compared with Col-0. d–f) The analysis of relative expression level of PjD27 in different tissues of 35S:PjD27(d27) plants compared with Col-0 and Arabidopsis d27 mutant plants. Transcripts were normalized to Actin gene (At5g09810). The expression level of PjD27 in Col-0 was normalized to 1, which was scaled to the value in the remaining samples. All the templates were the cDNAs of roots, tiller nodes, and leaves of 32-day-old Arabidopsis. The data represent the results of quantitative analysis obtained from 3 biological replicates and 3 technical replicates for each sample. Asterisks denote significant differences between Col-0 and PjD27-OE transgenic Arabidopsis lines, as well as between complementary lines compared to Col-0 and Atd27 mutant plants (**P < 0.01, ****P < 0.0001, one-way ANOVA).
D27 protein feedback to SL
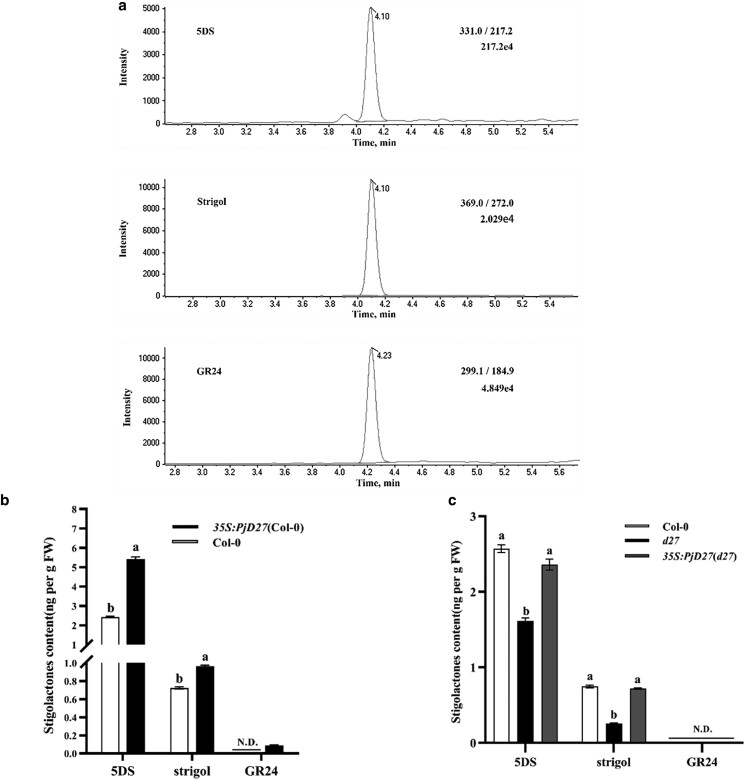
To explore the potential role of PjD27 in SL biosynthesis, we analyzed SLs in tillering nodes and root tissues of 35S:PjD27 plants by UPLC–MS/MS. We used 3 common standards—2 SLs (5DS and strigol), and the synthetic compound GR24 (frequently used as an analog to conventional SLs), as internal standards for quantification to investigate the correlation between PjD27 expression and different types of SLs in Arabidopsis. We selected respectively m/z 331, 369, and 299.1 for 5DS, strigol, and GR24, as parent ions on quadruple mass spectrometry and detected respectively m/z 217.2, 272, and 184.9 for 5DS, strigol and GR24, as fragment ions on time-of-flight mass spectrometry after collision-induced dissociation for quantification (Fig. 7a). To determine whether SLs participate in the branching inhibitor pathway in Arabidopsis, we examined the effect of 5DS, strigol and GR24 on the branching phenotype of Col-0, 35S:PjD27 (Col-0), Atd27 mutants and 35S:PjD27 (d27).
Fig. 7.
SL analysis in Arabidopsis. a) UPLC–MS/MS chromatograms for internal standard of 5DS, Strigol and GR24. b) UPLC–MS/MS analysis of SLs levels in wild-type (Col-0) and 35S:PjD27(Col-0) plants (mean + s.d., n = 3). FW, fresh weight. c) UPLC–MS/MS analysis of SLs levels in wild-type (Col-0), Atd27 mutants, and 35S:PjD27(d27) plants (mean + s.d., n = 3). FW, fresh weight. Different letters indicate significant differences (P < 0.05, one-way ANOVA). N.D. denotes that the SL content was either not detected or below the detection limit.
We observed that the levels of 5DS in 35S:PjD27 (Col-0) plants were significantly elevated compared with the wild type. Furthermore, quantitative analysis of strigol and GR24 levels also revealed a higher production of SLs in 35S:PjD27 (Col-0) plants compared to Col-0 (Fig. 7b). Upon complementation of AtD27 mutants with a cDNA encoding the PjD27 protein, we analyzed the levels of SLs in 35S:PjD27 (d27) plants, Atd27 mutants, and Col-0. Remarkably, SL levels in 35S:PjD27 (d27) plants were restored to those observed in Col-0, both exhibiting significantly higher levels compared to Atd27 mutants (Fig. 7c). Among the SLs observed in plants, 5DS content was highest, suggesting a predominant involvement of PjD27 in regulating plant branching through 5DS biosynthesis in Arabidopsis. However, the presence of GR24 was either undetected or below the detection limit. These results indicate that the overexpression of the PjD27 gene leads to an increase in overall SL levels in Arabidopsis.
Discussion
Previous research has employed forward genetics to identify tiller-related genes in different phenotypes of P. juncea (Li, Yun et al. 2022). This study uses a revere genetics approach to the candidate gene PjD27 to infer the role it plays in tillering in P. juncea. Prior to that, to further elucidate the regulatory role of PjD27 in the shoot branching of P. juncea, we conducted an analysis to determine the expression level of PjD27 in both dense and sparse tiller samples during the vigorous tillering stage. The finding was consistent with the result obtained from transcriptome analysis using the FPKM method, indicating that PjD27 downregulated the tiller number of P. juncea (Fig. 1, a and c). Through a conserved domain analysis, we identified a highly conserved domain-containing protein, known as DUF4033, within the PjD27 protein. This domain-containing protein is also present in D27 proteins found in wheat, rice, and Arabidopsis, suggesting that they were orthologues of the PjD27 protein (Fig. 2b).
We felt this was sufficient evidence that PjD27 would be related to tillering in P. juncea. Phenotypic analysis showed the number of branches and plant height of PjD27-OE transgenic Arabidopsis lines were markedly changed (Fig. 3). In addition, the expression of PjD27 in Arabidopsis d27 mutant plants almost completely restored the phenotype back to that of the Col-0 (Fig. 4). The 35S:PjD27 (Col-0) plants exhibited a reduced branching phenotype similar to that of TaD27-B-OE plants in wheat, when compared with wild-type plants. The complementation function of 35S:PjD27 (d27) was consistent with that of TaD27-B in Atd27 plants. We found this to be evidence that PjD27 is a functional ortholog of d27 in Arabidopsis (Zhao et al. 2020). D27 is well established as playing a pivotal role in regulating the number of plant branches and possesses a conserved function in suppressing branch formation in both monocotyledonous and dicotyledonous plants (Lin et al. 2009).
The expression of the PjD27 protein in Arabidopsis materials, including the wild-type and Atd27 mutants, was analyzed using a WB (Fig. 5a). The PjD27 protein was robustly expressed in Arabidopsis and was associated with significant changes in plant height and branch number in the plant's phenotype (Figs. 3 and 4).
The subcellular location of proteins is intricately linked to their biological functions (Martin-Merchan et al. 2023). The OsPAP3 protein of rice is located within the chloroplast nucleoids, exhibiting a close association with the abundance of chloroplasts. The overexpression of the OsPAP3 gene in plants led to an increase in both chloroplast abundance and tiller number, thereby playing a pivotal role in enhancing rice yield (Seo et al. 2024). To investigate the function of the PjD27 protein, we conducted a study on its subcellular location in tobacco through transient transfection (Fig. 5b). We observed that—similar to OsPAP3 in rice—the subcellular location of the PjD27 protein is exclusively confined to the chloroplast, a pivotal organelle crucial for optimal plant growth (Fan et al. 2023). Deciphering the precise subcellular location of the PjD27 protein provides valuable insights into unraveling the underlying mechanism governing tillering in P. juncea.
RT-qPCR can rapidly and accurately validate gene expression results at the transcriptional level (Liu et al. 2022; Yang et al. 2022). The analysis of gene expression can reflect the impact of genes on the physiological state of plants (Asif et al. 2023). The expression level of PjD27 gene exhibited variation across different plant tissues in PjD27-OE transgenic Arabidopsis plants. Specifically, the PjD27 transcript levels in tillering nodes and leaves were significantly higher compared to those in roots, indicating a correlation between tissue-specific expression and gene function (Qi et al. 2023). The PjD27 gene was implicated in the regulation of branch numbers in Arabidopsis. Its specific expression in tillering nodes may be associated with its role in regulating tillering, while the increased expression in leaves could be attributed to the location of the PjD27 protein in chloroplasts.
Numerous studies have consistently demonstrated that D27 primarily participates in the biosynthesis of SLs and exerts a pivotal role in the regulation of plant branching (Lin et al. 2009; Xie et al. 2010; Harrison et al. 2015; Wang, Tu et al. 2023; Wang, Yang et al. 2023). Our findings demonstrate the involvement of PjD27 in SL biosynthesis (Fig. 7), thereby regulating branching in Arabidopsis, analogous to the regulatory roles of pea (P. sativum L.) CCD8 (Gomez-Roldan et al. 2008) and rice d17 (Chen et al. 2023). The CCD8 mutant in peas almost completely restored the phenotype back to that of the wild type through the exogenously applied SL-related inhibitor. Furthermore, we observed that our samples had high levels of 5DS, consistent with phosphorus-deficient d3-1, d10-1, and d17-1 rice mutants, suggesting 5DS plays an inhibitory role in Arabidopsis and P. juncea (Umehara et al. 2008). Several studies have demonstrated the inhibitory effects of naturally occurring SLs, namely strigol and 5DS, as well as the SL analog GR24 and its downstream metabolites, on branching processes (Burger 2022; Yamaguchi et al. 2023), which further substantiates our research findings. The determined content of SLs in our research results was relatively low, potentially due to the need for further optimization of sampling time and extraction methods. Nevertheless, the discovery regarding PjD27's involvement in regulating plant branching through its participation in SL biosynthesis establishes a strong theoretical basis and provides valuable experimental resources for subsequent studies on tillering regulation in P. juncea.
Distant hybridization is an important approach for broadening the genetic base of plant cultivars. In many breeding programs, researchers will transfer genes that improve plant productivity from wild relatives to the plant (Kong et al. 2018). For example, a wheat-Haynaldia villosa derived line carried 2 translocations: 6VS/6AL and 4VS/4DL resisted powdery mildew and stripe mosaic virus, respectively (Chen et al. 1995; Zhang et al. 2005). Barley (Hordeum vulgare) yellow dwarf viruses (BYDVs) result in considerable yield losses in cereals. In order to solve this problem, the genome-wide miRNA expression profiling in P. huashanica was analyzed, which revealed the resistance response of miRNAs to BYDV-GAV infection (Shen and Li 2022). P. juncea is a perennial Triticeae forage that can restrain weeds. Halogeton glomeratus is a particularly difficult weed to manage. However, the planting of P. juncea and forage kochia (Bassia prostrata) can reduce halogeton frequency, thereby providing an opportunity for the survival of cultivated plants (Smith et al. 2016). In addition, P. juncea also has an excellent gene pool and is a potential resource for the genetic improvement of wheat (Dong et al. 2008). For example, the novel D-hordeins encoded by Ns 1.3, Ns 2.6, and Ns 2.9 genes in P. juncea have a very important potential for enhancing the end-use quality of wheat flour. Furthermore, these D-hordeins can also contribute to the understanding of Triticeae prolamins’ evolution (Hu et al. 2018). Therefore, the utilization prospects of P. juncea for wheat improvement are promising.
In summary, PjD27 represents a new genetic tool for exploring shoot branching mechanisms in P. juncea, and it plays an important role in cultivating high-yield P. juncea. Furthermore, functional analysis of the PjD27 gene also contributes to promoting high-quality wheat production.
Conclusion
The present study demonstrates that PjD27 plays critical roles in regulating Arabidopsis branching number by participating in SL biosynthesis. Thus, PjD27 is a functional orthologue of D27 in Arabidopsis. In addition, we also found that the PjD27 protein was mainly localized in the chloroplast, suggesting a potential functional association with tillering similar to that observed in other chloroplast-localized proteins. The functional analysis of the PjD27 gene can provide a theoretical basis for multitillering germplasm creation in P. juncea and hold significant research implications for enhancing grain yield in P. juncea.
Supplementary Material
Acknowledgments
We thank the laboratory staff for kindly supporting our test (the Ministry of Education Key Laboratory of Grassland Resources and Key Laboratory of Forage Cultivation, Processing and High Efficient Utilization of the Ministry of Agriculture, Grassland Resource and Environment College, Inner Mongolia Agricultural University). We would also like to thank Dr Daniel Petticord at the University of Cornell for his assistance with the English language and grammatical editing of the manuscript.
Contributor Information
Xiaomin Ren, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot 010018, China.
Qian Ai, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot 010018, China.
Zhen Li, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot 010018, China.
Qiao Zhao, Shenzhen Key Laboratory of Synthetic Genomics, Shenzhen Institute of Advanced Technology of Chinese Academy of Sciences, Shenzhen, Guangdong 518055, China.
Lan Yun, College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot 010018, China; Key Laboratory of Grassland Resources Ministry of Education, Inner Mongolia Agricultural University, Hohhot 010011, China.
Data availability
Strains and plasmids are available upon request. The Sequence data are available at GenBank; the accession numbers are listed in Supplementary Table 1. The addresses are as follows: https://www.ncbi.nlm.nih.gov/search/all/?term=OR865585, and the amino acid sequence is translated from the online website ORFfinder Home—NCBI (nih.gov). The raw sequencing reads of transcriptome data in this study are available in Sequence Read Archive database (accession number PRJNA789128). The addresses are as follows: (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789128/).
Supplemental material available at G3 online.
Funding
This research was supported by the National Natural Science Foundation of China (NSFC, 32371762) and the Natural Science Foundation of Inner Mongolia (2023ZD07).
Author contributions
XR performed the whole research and drafted the manuscript. ZL and QA helped in experiment and data analysis. LY designed the research, collected research material, and revised the manuscript. QZ helped with experimental design. All authors contributed to the article and approved the submitted version.
Literature cited
- Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, et al. 2011. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci U S A. 108(50):20242–20247. doi: 10.1073/pnas.1111902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science. 335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- Asif S, Jan R, Kim N, Asaf S, Lubna, Khan MA, Kim E-G, Jang Y-H, Bhatta D, Lee I-J, et al. 2023. Halotolerant endophytic bacteria alleviate salinity stress in rice (oryza sativa L.) by modulating ion content, endogenous hormones, the antioxidant system and gene expression. BMC Plant Biol. 23(1):494. doi: 10.1186/s12870-023-04517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pages V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4(7):e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Cui Y, Li J, Guan Y, Tian S, Liu X. 2023. Genome-wide analysis of PIN genes in cultivated peanuts (Arachis hypogaea L.): identification, subcellular localization, evolution, and expression patterns. BMC Genomics. 24(1):629. doi: 10.1186/s12864-023-09723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. 2009. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150(1):482–493. doi: 10.1104/pp.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger M. 2022. Splicing up strigolactone signaling. Plant Cell. 34(9):3160–3161. doi: 10.1093/plcell/koac168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ. 1995. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet. 91(6-7):1125–1128. doi: 10.1007/BF00223930. [DOI] [PubMed] [Google Scholar]
- Chen GE, Wang JY, Votta C, Braguy J, Jamil M, Kirschner GK, Fiorilli V, Berqdar L, Balakrishna A, Blilou I, et al. 2023. Disruption of the rice 4-DEOXYOROBANCHOL HYDROXYLASE unravels specific functions of canonical strigolactones. Proc Natl Acad Sci U S A. 120(42):e2306263120. doi: 10.1073/pnas.2306263120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Nishide N, Mashiguchi K, Kuroha K, Miya M, Sugimoto K, Itoh J-I, Yamaguchi S, Izawa T. 2023. Fertilization controls tiller numbers via transcriptional regulation of a MAX1-like gene in rice cultivation. Nat Commun. 14(1):3191. doi: 10.1038/s41467-023-38670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Zhang L, Zhang Q, Yang Y, Li D, Xie Z, Cui G, Chen Y, Wu L, Li Z, et al. 2023. Author correction: Tiller Number1 encodes an ankyrin repeat protein that controls tillering in bread wheat. Nat Commun. 14(1):3523. doi: 10.1038/s41467-023-39365-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YS, Zhou RH, Li SJXH, Cauderon Y, Wang RRC. 2008. Desirable characteristics in perennial Triticeae collected in China for wheat improvement. Hereditas. 116:175–178. doi: 10.1111/j.1601-5223.1992.tb00819.x. [DOI] [Google Scholar]
- Dun EA, Hanan J, Beveridge CA. 2009. Computational modeling and molecular physiology experiments reveal new insights into shoot branching in pea. Plant Cell. 21(11):3459–3472. doi: 10.1105/tpc.109.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Hou Y, Zheng L, Shi H, Liu Z, Wang Y, Li S, Liu L, Guo M, Yang Z, et al. 2023. Characterization and fine mapping of a yellow leaf gene regulating chlorophyll biosynthesis and chloroplast development in cotton (Gossypium arboreum). Gene. 885:147712. doi: 10.1016/j.gene.2023.147712. [DOI] [PubMed] [Google Scholar]
- Fikadu T, Tesfaye W, Furgasa M, Bekuma W, Legesse F. 2022. Brachiaria grass herbage yield potential and nutritional quality at midland agro ecology of East Hararghe Zone, Ethiopia. Am J Agric For. 10(5):199–203. doi: 10.11648/j.ajaf.20221005.16. [DOI] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, et al. 2008. Strigolactone inhibition of shoot branching. Nature. 455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Newgas SA, Descombes F, Shepherd SA, Thompson AJ, Bugg TDH. 2015. Biochemical characterization and selective inhibition of beta-carotene cis-trans isomerase D27 and carotenoid cleavage dioxygenase CCD8 on the strigolactone biosynthetic pathway. FEBS J. 282(20):3986–4000. doi: 10.1111/febs.13400. [DOI] [PubMed] [Google Scholar]
- Hu XK, Dai SF, Ouellet T, Balcerzak M, Rocheleau H, Khanizadeh S, Pu ZJ, Yan ZH. 2018. Characterization of novel D-hordeins from Psathyrostachys juncea. Biol Plant. 62(2):369–378. doi: 10.1007/s10535-018-0775-6. [DOI] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46(1):79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- Jamil M, Charnikhova T, Verstappen F, Bouwmeester H. 2010. Carotenoid inhibitors reduce strigolactone production and Striga hermonthica infection in rice. Arch Biochem Biophys. 504(1):123–131. doi: 10.1016/j.abb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Kong L, Song X, Xiao J, Sun H, Dai K, Lan C, Singh P, Yuan C, Zhang S, Singh R, et al. 2018. Development and characterization of a complete set of Triticum aestivum–Roegneria ciliaris disomic addition lines. Theor Appl Genet. 131(8):1793–1806. doi: 10.1007/s00122-018-3114-6. [DOI] [PubMed] [Google Scholar]
- Li T, Peng L, Wang H, Zhang Y, Wang Y, Cheng Y, Hou F. 2022. Multi-Cutting improves forage yield and nutritional value and maintains the soil nutrient balance in a rainfed agroecosystem. Front Plant Sci. 13:825117. doi: 10.3389/fpls.2022.825117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang T, Yun L, Ren XM, Wang Y, Shi F. 2023. Association analysis of tiller-related traits with EST-SSR markers in Psathyrostachys juncea. Genes (Basel). 14(10):1970. doi: 10.3390/genes14101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yun L, Ren X, Shi F, Mi F. 2022. Analysis of controlling genes for tiller growth of Psathyrostachys juncea based on transcriptome sequencing technology. BMC Plant Biol. 22(1):456. doi: 10.1186/s12870-022-03837-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X. 2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell. 21(5):1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang C, Bai M, Yan F, Qin H, Wang R, Wan Y, Li G. 2022. Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia. Open Life Sci. 17(1):1155–1164. doi: 10.1515/biol-2022-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WX, Li LH, Zhan ZM. 2006. SSR analysis on the sampling strategy of Psathyrostachys Huashanickeng population. Acta Tritici Sin. 26:16–20. doi: 10.7606/j.issn.1009-1041.2006.02.052. [DOI] [Google Scholar]
- Main O, Jacquemot MP, Griveau Y, Guillaume S, Demonceaux C, Lopez-Marnet P-L, Rey S, Fargier S, Sartre P, Montagnier C, et al. 2023. Precise control of water stress in the field reveals different response thresholds for forage yield and digestibility of maize hybrids. Front Plant Sci. 14:1142462. doi: 10.3389/fpls.2023.1142462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Merchan A, Moro B, Bouet A, Bologna NG. 2023. Domain organization, expression, subcellular localization, and biological roles of ARGONAUTE proteins in Arabidopsis. J Exp Bot. 74(7):2374–2388. doi: 10.1093/jxb/erad030. [DOI] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogué F, Rameau C. 2011. Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development. 138(8):1531–1539. doi: 10.1242/dev.058495. [DOI] [PubMed] [Google Scholar]
- Qi F, Wang F, Xiaoyang C, Wang Z, Lin Y, Peng Z, Zhang J, Wang N, Zhang J. 2023. Gene expression analysis of different organs and identification of AP2 transcription factors in flax (Linum usitatissimum L.). Plants (Basel). 12(18):3260. doi: 10.3390/plants12183260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XM, Yun L, Ai Q, Li Z, Zhao Q, Shi FL. 2023. Subcellular localization, overexpression vector construction and identification of IPT gene in Psathyrostachys juncea. Acta Botanica Boreali-Occidentalia Sinica. 43(9):1441–1449. doi: 10.7606/j.issn.1000-4025.2023.09.1441. [DOI] [Google Scholar]
- Seo DH, Jang J, Park D, Yoon Y, Choi YD, Jang G. 2024. PEP-ASSOCIATED PROTEIN 3 regulates rice tiller formation and grain yield by controlling chloroplast biogenesis. Plant Physiol. 194(2):805–818. doi: 10.1093/plphys/kiad536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Li X. 2022. Genome-wide miRNA expression profiling in Psathyrostachys huashanica reveals insights into the resistance response to barley yellow dwarf virus-GAV infection. Physiol Mol Plant Pathol. 121:101888. doi: 10.1016/j.pmpp.2022.101888. [DOI] [Google Scholar]
- Shiferaw B, Smale M, Braun H, Duveiller E, Reynolds M, Muricho G. 2013. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5(3):291–317. doi: 10.1007/s12571-013-0263-y. [DOI] [Google Scholar]
- Shindo M, Shimomura K, Yamaguchi S, Umehara M. 2018. Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct. 2(4):e50. doi: 10.1002/pld3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Waldron BL, Creech JE, Zobell RA, ZoBell DR. 2016. Forage Kochia and Russian wildrye potential for rehabilitating Gardner's saltbush ecosystems degraded by Halogeton. Rangeland Ecol Manag. 69(5):390–398. doi: 10.1016/j.rama.2016.06.001. [DOI] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. 2010. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51(7):1118–1126. doi: 10.1093/pcp/pcq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. 2011. Branching in rice. Curr Opin Plant Biol. 14(1):94–99. doi: 10.1016/j.pbi.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Wang H, Tu R, Ruan Z, Chen C, Peng Z, Zhou X, Sun L, Hong Y, Chen D, Liu Q, et al. 2023. Photoperiod and gravistimulation-associated tiller angle control 1 modulates dynamic changes in rice plant architecture. Theor Appl Genet. 136(7):160. doi: 10.1007/s00122-023-04404-z. [DOI] [PubMed] [Google Scholar]
- Wang Q, Yang P, Maharajan T, Ramakrishnan M, Asim M, Gui R, Zhou M. 2023. Strigolactone mediates moso bamboo root response to phosphate stress. J Agric Food Chem. 71(20):7921–7936. doi: 10.1021/acs.jafc.2c08925. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao Y, Ray I, Song M. 2016. Transcriptome responses in alfalfa associated with tolerance to intensive animal grazing. Sci Rep. 6(1):19438. doi: 10.1038/srep19438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. 2012. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159(3):1073–1085. doi: 10.1104/pp.112.196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annu Rev Phytopathol. 48(1):93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Seto Y, Kyozuka J. 2023. What's new in strigolactone research?. Plant Cell Physiol. 64(9):933–935. doi: 10.1093/pcp/pcad095. [DOI] [PubMed] [Google Scholar]
- Yang J, Han F, Yang L, Wang J, Jin F, Luo A, Zhao F. 2022. Identification of reference genes for RT-qPCR analysis in Gleditsia microphylla under abiotic stress and hormone treatment. Genes (Basel). 13(7):1227. doi: 10.3390/genes13071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeni O, Teoman Ö. 2020. The agriculture–environment relationship and environment-based agricultural support instruments in Turkey. Eur Rev Med Pharmacol Sci. 30(2):194–218. doi: 10.1017/S1062798720001015. [DOI] [Google Scholar]
- Zhang Q, Li Q, Wang X, Wang H, Lang S, Wang Y, Wang S, Chen P, Liu D. 2005. Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS⋅4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica. 145(3):317–320. doi: 10.1007/s10681-005-1743-8. [DOI] [Google Scholar]
- Zhao L, Wang C, Wang T, Liu J, Qiao Q, Yang Y, Hu P, Zhang L, Zhao S, Chen D, et al. 2023. Identification of the candidate gene controlling tiller angle in common wheat through genome-wide association study and linkage analysis. Crop J. 11(3):870–877. doi: 10.1016/j.cj.2023.01.004. [DOI] [Google Scholar]
- Zhao B, Wu TT, Ma SS, Jiang DJ, Bie XM, Sui N, Zhang XS, Wang F. 2020. TaD27-B gene controls the tiller number in hexaploid wheat. Plant Biotechnol J. 18(2):513–525. doi: 10.1111/pbi.13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Wang Y, Lu S, Yang L, Zhuang M, Zhang Y, Lv H, Fang Z, Hou X. 2022. Genome-wide identification and analysis of cytokinin dehydrogenase/oxidase(CKX) family genes in Brassica oleracea L. Reveals their involvement in response to Plasmodiophora brassicae infections. Hortic Plant J. 8(1):68–80. doi: 10.1016/j.hpj.2021.05.003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The Sequence data are available at GenBank; the accession numbers are listed in Supplementary Table 1. The addresses are as follows: https://www.ncbi.nlm.nih.gov/search/all/?term=OR865585, and the amino acid sequence is translated from the online website ORFfinder Home—NCBI (nih.gov). The raw sequencing reads of transcriptome data in this study are available in Sequence Read Archive database (accession number PRJNA789128). The addresses are as follows: (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA789128/).
Supplemental material available at G3 online.