Abstract
Mallards (Anas platyrhynchos) and other dabbling ducks in the genus Anas are an important component of the wild bird reservoir for avian influenza (AI) virus; these viruses are maintained in migratory duck populations through a fecal-oral transmission route. We provide a detailed characterization of intestinal viral shedding in Mallards infected with a wild bird-origin low pathogenic (LP) AI virus. Five of eight, 1-mo-old Mallards inoculated with a high dose of an H3N8 LP AI virus became infected as determined by reisolation and seroconversion. Infected birds excreted high concentrations of virus for up to 14 days postinoculation (DPI) without exhibiting overt clinical signs of disease. The pattern of viral shedding was relatively consistent between individual birds, with peak shedding on 2–3 DPI and a progressive decline over the remainder of infection. Detection of viral shedding varied depending on sample type (excrement sample or cloacal swab) and diagnostic test (virus isolation or real-time quantitative reverse transcription polymerase chain reaction). Our data provide detailed insights into the intestinal excretion of an H3N8 LP AI virus in Mallards and the performance of diagnostic assays commonly used in wild bird surveillance. Such information is valuable for estimating potential risks for spillover of LP AI viruses from Mallards to domestic animals, developing accurate transmission models for Mallard populations and facilitating the interpretation and comparison of surveillance results from different studies.
Keywords: Avian influenza virus, cloacal swab, excrement, feces, low pathogenic, Mallard, RT-PCR, virus isolation
INTRODUCTION
Experimental infections of Mallards (Anas platyrhynchos) and related domestic duck breeds (e.g., Pekin ducks) with avian influenza (AI) viruses have clearly demonstrated preferential excretion of AI virus in feces (Slemons and Easterday, 1978; Webster et al., 1978; Kida et al., 1980; Spackman et al., 2009; Costa et al., 2011). This is associated with virus replication in enterocytes lining the distal small and large intestines or in the epithelium of the bursa of Fabricius in young ducks (Slemons and Easterday, 1978; Webster et al., 1978; Kida et al., 1980; Daoust et al., 2011). The duration of fecal shedding varies between low pathogenic (LP) AI virus isolates but routinely exceeds 1 wk (Slemons and Easterday, 1978; Webster et al., 1978; Kida et al., 1980; Spackman et al., 2009; Costa et al., 2011) and can be prolonged, with detectable virus reported up to 28 days postinoculation (DPI; Hinshaw et al., 1980). Similarly, the quantity of virus excreted in the feces varies between LP AI isolates but is typically high with peak infectious titers exceeding 105 median embryo infectious doses (EID50)/ mL of swab media or gram of excrement (Webster et al., 1978; Kida et al., 1980). Mallards also excrete wild birdrigin LP AI viruses via the respiratory tract during early infection; however, this generally occurs at a lower titer or for a shorter duration than in feces (Webster et al., 1978; Kida et al., 1980; Spackman et al., 2009; Costa et al., 2011).
Although there have been experimental studies focusing on wild bird-origin LP AI virus infection in ducks, there is a dearth of experimental data on quantity of viral shedding over the duration of infection (based on titration in embryonating chicken eggs), the relationship between infectious viral titers and viral RNA load in cloacal swabs and excrement samples over time, or whether there are short-term variations in shedding patterns within a given 24-hr period. As recently discussed (Henaux and Samuel, 2011), such information is critical for the parameterization of transmission models, especially related to environmental or fecal-oral transmission of these viruses (Rohani et al., 2009), and to evaluate and interpret field survey results that can be based on different sample types (swab samples collected directly from the bird or environmental feces) or diagnostic tests (virus isolation or real-time quantitative reverse transcription [qRT-] PCR) commonly used in wild bird surveillance efforts (Brown and Stallknecht, 2008).
Our goals were to 1) provide a detailed characterization of viral shedding over 14 days, as measured with virus titration and qRT-PCR, in excrement of Mallards infected with a wild bird-origin LP AI virus and 2) compare the performance of common sampling approaches (cloacal swab vs. excrement samples) and diagnostic tests (virus isolation vs. qRT-PCR) currently used in wild bird surveillance programs over the course of viral infection.
MATERIALS AND METHODS
Animals
Ten 1-day-old Mallards were purchased from a commercial waterfowl breeder (McMurray Hatchery, Webster City, Iowa, USA) and raised under indoor confinement until they were 4 wk old at the College of Veterinary Medicine, The University of Georgia (UGA), Athens, Georgia, USA. At 4 wk, the ducks were transferred to a biosafety level-2Ag+ facility at UGA, where the experimental trial was conducted. General animal care was provided and experimental sampling was performed according to an animal care and use protocol approved by the Institutional Animal Care and Use Committee at UGA.
Virus
The LP AI virus A/Mallard/Minnesota/199106/1999 (H3N8) was used in the trial. This strain was selected because H3N8 viruses represent one of the most common subtypes reported from wild ducks (Wilcox et al., 2011), and previous experimental trials indicated that this strain replicates efficiently in Mallards (Costa et al., 2010, 2011). Viral stock was propagated by second passage in 9- to 11-day-old specific pathogen-free (SPF) embryonating chicken eggs (Swayne et al., 2008). The infectious titer of the stock was determined in SPF chicken eggs, as described below, and calculated using the method described by Reed and Muench (1938). The viral inoculum was prepared by diluting infective amnioallantoic fluid in sterile brain-heart-infusion (BHI) media to yield a final titer of 106 EID50/0.1 mL. The back titer of the inoculum, determined immediately after Mallard-inoculation, was 106.5 EID50/0.1 mL. The sham inoculum consisted of sterile BHI media.
Experimental design
All 10 Mallards were individually housed in biocontainment isolation units ventilated under negative pressure with high-efficiency particulate air-(HEPA-) filtered air. After a 3-day acclimation, eight virus-exposed and two negative control Mallards were intranasally (IN; via the choanal cleft) inoculated with 0.1 mL of BHI medium containing 106.5 EID50 of the H3N8 LP AI virus or with 0.1 mL of BHI medium, respectively. After inoculation, the ducks were monitored twice daily at 9:00 AM and 4:00 PM for behavioral changes or any clinical signs of disease. Less than 1% blood volume based on body weight was collected from the right jugular vein and placed into serum separator tubes (Becton, Dickinson and Company, Franklin Lakes, New Jersey, USA) prior to inoculation and at 7 and 14 DPI. Fresh excrement samples, consisting of varying amounts of feces, urine, and urates, were collected from each duck prior to inoculation (0 DPI PM) on 1 to 9 DPI (AM and PM), 10 DPI (AM), 12 DPI (AM and PM), and 14 DPI (AM). Excrement samples were collected by placing the duck in a cardboard dog carrier (C Specialties Inc., Indianapolis, Indiana, USA) lined with a sheet of Benchkote™ with the laminated surface facing up. A separate dog carrier was dedicated to each individual bird, and Benchkote sheets were changed after each sampling time point. After 15–30 sec, the duck was removed from the carrier, and excrement was placed into preweighed 4 mL cryogenic vials (Corning Inc., Corning, New York, USA) containing 3.0 mL of BHI medium with supplemental antimicrobial drugs (100 μg/mL gentamicin, 100 units/mL penicillin, and 5 μg/mL amphotericin B). The weight of the cryogenic vial was measured again immediately after the sample was added to determine the quantity (grams) of excrement. Cloacal swabs were collected from each duck prior to inoculation (0 DPI PM only) on 1 to 10, 12, and 14 DPI (AM only). Cloacal swabs were collected immediately after the ducks were removed from the carriers used to collect the excrement samples and were stored in 2 mL of the viral transport medium described above. The trial was terminated on 14 DPI at which time birds were euthanized by CO2 inhalation.
Virus isolation and titration
Virus isolation and titration were performed on all excrement samples and cloacal swabs using 9- to 11-day-old SPF embryonating chicken eggs (Swayne et al., 2008). Briefly, both sample types were thawed on the day of testing, vortexed, and maintained at room temperature for 30 min to allow for antimicrobial activity of the transport media. Samples were centrifuged at 1,610 × G for 15 min. For virus isolation, 0.25 mL of supernatant was inoculated into the allantoic sac of four eggs. For virus titrations, tenfold serial dilutions were made in sterile BHI media, and 0.1 mL of diluted sample was inoculated into five eggs per dilution. After inoculation, eggs used for virus isolation or titration were incubated at 37 C for 5 days. Amnioallantoic fluid was harvested and tested for hemagglutination activity (Swayne et al., 2008), and viral titers were calculated as described above.
Extraction of RNA and real-time-PCR
The RNA was extracted from all excrement samples and cloacal swab samples as they were thawed and processed for virus isolation and titration using the QIAamp® Viral RNA Mini Kit (QIAGEN, Valencia, California, USA). The extracted samples were tested for influenza A nucleic acid using qRT-PCR targeting the influenza matrix gene (Spackman and Suarez, 2008). For the statistical analysis described below, any cycle threshold (Ct) value <45 was considered positive by qRT-PCR.
Serology
Blood samples were centrifuged at 1,610 × G for 15 min and serum stored at −20 C until testing for antibodies to the nucleoprotein of influenza A virus with a commercially available blocking enzyme-linked immunosorbent assay (FlockChek AI MultiS-Screen antibody test kit; IDEXX Laboratories, Westbrook, Maine, USA) according to manufacturer’s instructions.
Statistical analyses
The agreement between virus isolation and qRT-PCR for excrement samples and cloacal swabs was estimated based on percentage agreement and kappa statistics (κ). The kappa value was interpreted based on the Landis and Koch classification (1977), where ≤0.2 = slight agreement, 0.2–0.45fair agreement, 0.4–0.6 = moderate agreement, 0.6–0.8 = substantial agreement, and ≥0.85almost perfect agreement. Repeated measures analysis of variance (ANOVA) was used to compare viral shedding over time and between sample types. Degrees of freedom for F-tests of within-subject factors were adjusted using the Green-house-Geisser estimate of epsilon to correct for departures from the sphericity assumption (Stevens, 2002). Post hoc testing was performed using the Bonferroni procedure to limit the type I error rate to 5% over all comparisons. Linear regression with robust standard errors was used to evaluate the relationship between Ct values and infectious titers. Robust standard errors were obtained by specifying the vce (cluster) option in Stata (Stata version 11.1, StataCorp LP, College Station, Texas, USA) to provide accurate variance estimates for the coefficients despite the lack of independence between repeated measurements (Williams, 2000). The percentage of variance explained by the independent variables was estimated by the coefficient of determination (R2) for the regression models. Samples with a negative qRT-PCR result were arbitrarily assigned a Ct value of 45 for statistical analyses. Samples that were positive by virus isolation but did not have a quantifiable infectious titer were assigned a titer of 101 EID50/mL (swabs) or per gram (excrement samples). Receiver operator characteristic (ROC) curves were generated for both excrement samples and cloacal swabs, which graphically plot the true positive rate against the false-positive rate for different qRT-PCR diagnostic cutoffs. The area under the ROC curve was used to evaluate the accuracy of qRT-PCR (sensitivity and specificity) for the prediction of virus isolation testing results for both sample types. All testing assumed a two-sided alternative hypothesis, and P<0.05 was considered statistically significant. Analyses were performed using commercially available statistical software, Stata version 11.1 (StataCorp).
RESULTS
No virus was isolated from the negative control Mallards during the trial, and all serum samples from both ducks were negative for antibodies to AI virus at 0, 7, and 14 DPI. Five of the eight Mallards (63%) inoculated with the H3N8 LP AI virus were infected based on postinoculation seroconversion and viral shedding. None of these infected Mallards exhibited behavioral changes or clinical signs of disease, nor did they have any differences in weight gain relative to the negative controls (data not shown). The five infected Mallards were antibody-negative prior to inoculation (0 DPI), but antibodies directed against AI virus were detected in all ducks at both 7 and 14 DPI. There was no evidence of infection in the remaining three inoculated Mallards based on virus isolation or serology.
A sample of excrement was successfully collected from the five infected Mallards during 111 of the 115 attempts. Weight of excrement samples ranged from 0.10–1.28 g (average: 0.66 g) and did not relate significantly to viral titer (P=0.995). The agreement between virus isolation and qRT-PCR ranged from fair for cloacal swabs (n=65; agreement=69%; κ=0.34) to substantial for excrement samples (n=111; agreement=89%; κ=0.76). Measures of agreement between excrement samples and cloacal swabs ranged from almost perfect for virus isolation (n=63; agreement=90%; κ=0.81) to fair for qRT-PCR (n=63; agreement=67%; κ=0.31). Virus isolation was significantly less sensitive than qRT-PCR for cloacal swabs (McNemar’s χ2; P<0.001); however, there was no significant difference in sensitivities between the two diagnostic approaches for excrement samples (McNemar’s χ2; P=0.388).
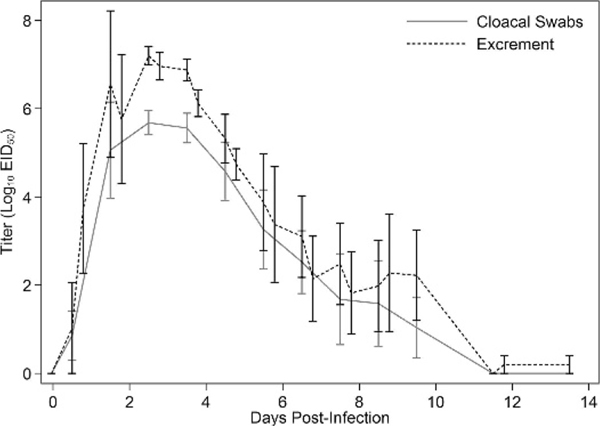
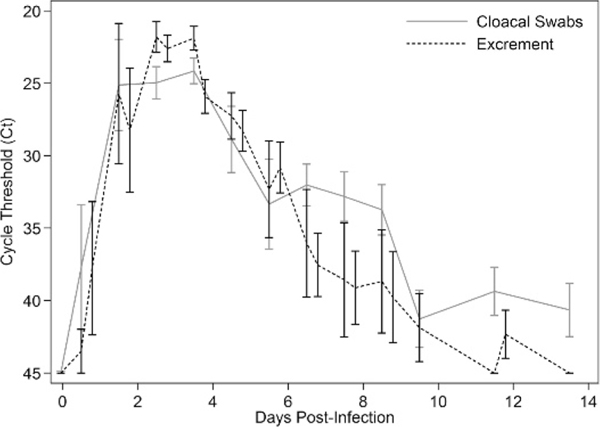
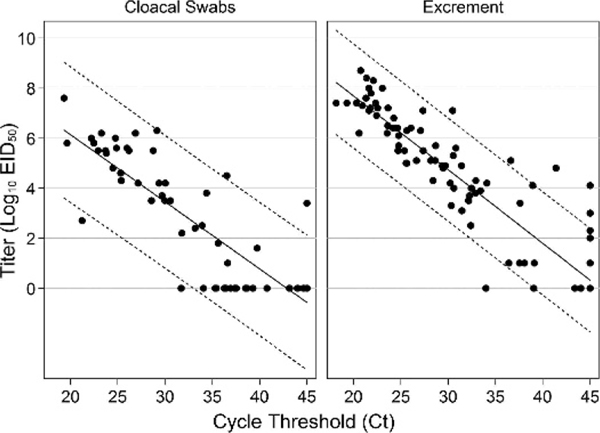
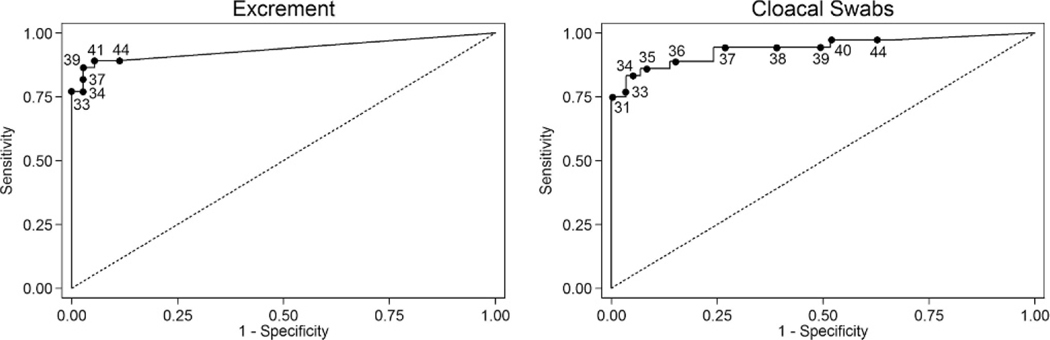
For viral titers, there was a significant effect of both time (P=0.003) and sample type (P=0.010), but there was no significant time x sample type interaction (P=0.124; Fig. 2). For Ct values, there also was a significant effect of both time (P<0.001) and sample type (P=0.017), but there was no significant time x sample type interaction (P=0.153; Fig. 1). Ct values were significant predictors of infectious titers for both sample types, with the univariate model explaining 73% of the variation in infectious titers for cloacal swabs and 88% for excrement samples. For every one unit increase in Ct value, infectious titers decreased by an average of 100.27 EID50/mL of cloacal swab media and 100.30 EID50/g of excrement sample (Fig. 3). Relative to virus isolation, the sensitivity and specificity of qRT-PCR were maximized using a Ct value cutoff of 41.4 for excrement samples and 33.1 for cloacal swabs. Based on the area under the ROC curve, qRT-PCR was an excellent predictor of virus isolation results for both excrement samples (0.933; 95% CI: 0.874, 0.974) and cloacal swabs (0.941; 95% CI: 0.850, 0.983; Fig. 4).
FIGURE 2.
Daily mean (SE) infectious titers for the detection of avian influenza virus by viral titration in excrement samples and cloacal swabs collected from five Mallards (Anas platyrhynchos) after experimental infection with avian influenza virus. Excrement samples were collected twice (9:00 AM and 4:00 PM) on most days, while cloacal swabs were collected once per day (9:00 AM). Viral titers are reported in log10 EID50/mL (cloacal swabs) or per gram (excrement samples).
FIGURE 1.
Daily mean (SE) cycle threshold (Ct) values for the detection of avian influenza virus by real-time quantitative reverse transcription-PCR in excrement samples and cloacal swabs collected from five Mallards (Anas platyrhynchos) after experimental infection with avian influenza virus. Excrement samples were collected twice (9:00 AM and 4:00 PM) on most days, while cloacal swabs were collected once per day (9:00 AM).
FIGURE 3.
Scatter plot of infectious titers versus cycle threshold (Ct) values for cloacal swabs and excrement samples collected from five Mallards (Anas platyrhynchos) during a 2-wk period after experimental infection with avian influenza virus. The solid lines are estimated regression lines and dotted lines are 95% prediction intervals. Viral titers are reported in log10 EID50/mL (cloacal swabs) or per gram (excrement samples).
FIGURE 4.
Receiver operating characteristic curve for the prediction of avian influenza virus isolation results by real-time quantitative reverse transcription-PCR in excrement samples (n=111) and cloacal swabs (n=65) from Mallards (Anas platyrhynchos). Annotated points on the curve correspond to different cycle threshold (Ct) value cutoffs for the prediction of virus isolation status.
DISCUSSION
Three of the eight (38%) Mallards inoculated with the H3N8 LP AI virus in this study did not become infected. Most experimental trials in Mallards or related domestic duck breeds using waterfowl-origin LP AI viruses have resulted in near 100% infection. We do not know why the three Mallards did not become infected; however, we suspect this may relate to the fact that we housed birds individually. Few experimental trials in ducks have used individually housed birds, and under group-housing conditions, it is impossible to distinguish infection resulting from the inoculum versus other cohoused birds that are infected and excreting virus, especially as Mallards reportedly can begin shedding virus <24 hr after inoculation (Webster et al., 1978; Kida et al., 1980). This possibility is supported by a previous experimental trial conducted in group-housed, age-matched Mallards using this same H3N8 LPAI virus isolate, which resulted in 100% of the IN inoculated birds becoming infected (Costa et al., 2010).
Although viral titers were lower in cloacal swabs than in the excrement samples, the former accurately characterized viral shedding trends over the course of infection evaluated (Figs. 1, 2). The concentration and duration of viral shedding in this experiment were consistent with previous studies conducted with LP AI viruses in Mallards as evidenced by the comparability of our data to the shedding curves estimated by Henaux and Samuel (2011) based on existing experimental data on LP AI virus infection in ducks. The shape of the shedding curves for both excrement samples and cloacal swabs and the maximum titers for excrement samples were similar between the two studies; however, we observed less of a discrepancy between curves generated for excrement samples and cloacal swabs and a shorter duration of viral shedding in excrement samples. For the latter, although our study captured the majority of LP AI virus shedding in Mallards, the design precluded any conclusions regarding shedding beyond 14 DPI.
Overall, mean titers exceeding 102 EID50/g in the excrement were only observed during the first 10 DPI, and mean titers exceeding 104 EID50/g were limited to 1–5 DPI. There was variation in viral shedding between birds, however, with titers exceeding 104 EID50/g observed in individual Mallards up to 10 DPI. Although, long-term shedding (28 days) of LP AI viruses have been reported in ducks (Hinshaw et al., 1980), our results with this H3N8 isolate suggest that after 7 DPI, viral shedding decreases as would, presumably, the risk of environmental transmission. However, no studies have evaluated transmission between ducks throughout the period of infection and the significance of this prolonged, but reduced shedding related to efficient transmission currently is not understood.
No obvious diurnal pattern in viral shedding was identified, as has been reported for some coccidian parasites excreted in the feces (Lopez et al., 2007; Dolnik et al., 2011); however, there were multiple complicating factors that affected this analysis, including a small sample size and the fact that after peak viral shedding the continual downward trajectories of the excretion curves favored lower evening titers. While there may be a small diurnal effect in LP AI viral shedding in Mallards, these data suggest it is minimal compared to the overall trends reported herein.
Virus isolation was less sensitive than qRT-PCR for cloacal swabs but not for excrement samples. This was reflected in the lower optimum Ct value cutoff to correspond with isolation results for cloacal swabs (33.9) than excrement samples (41.4). These experimental results for cloacal swabs are consistent with a previous analysis of a large waterfowl surveillance data set (Munster et al., 2009) in which AI virus was only isolated from 40% of RT-PCR-positive swab samples that were stored under ideal conditions (−80 C). Presumably, the discrepancy between cloacal swabs and excrement samples reflects a lower amount of infectious virus particles in cloacal swab samples. This discrepancy also may reflect the presence of fecal inhibitors that can increase Ct values relative to infectious titers in individual samples (Spackman and Suarez, 2008). This effect can be seen in our data where infectious titers are consistently higher for excrement samples versus cloacal swabs throughout the infectious period evaluated (Fig. 1); however, Ct values are generally lower in cloacal swabs especially during the early and later stages of infection (Fig. 2). The variation in discordant results as determined by virus isolation and qRT-PCR also follow this temporal pattern; none of the cloacal swabs that were positive by qRT-PCR and negative by virus isolation were collected between 2 and 6 DPI when the infectious titers were highest. This implies diagnostic comparison, in this case virus isolation versus qRT-PCR, will vary greatly depending on when samples were collected in relation to the stage of infection. This complicates comparisons of these diagnostic tools based on field samples, where this variable is undefined.
Results (Ct values) from qRT-PCR assays for AI virus are often used either as a screening tool to identify samples for further testing with virus isolation or as a stand-alone testing platform. Most laboratories have an established diagnostic cutoff for Ct values that are considered positive, which typically ranges from 35 to 40. Based on these experimental results, interpretation of Ct values varies with sample type; cloacal swab samples with high Ct values (>35) are much less likely positive by virus isolation than excrement samples. Such information is important for interpreting surveillance results, as discrepancies between qRT-PCR and virus isolation should be expected. Alternatively, laboratories that test surveillance samples using virus isolation without qRT-PCR prescreening are likely underestimating the prevalence of AI virus infection in ducks.
An increasing recent trend among AI experimental infection trials is to estimate infectious titers based on Ct values by generating a standard curve from RNA extracted from dilutions of the challenge virus. However, whether Ct values accurately reflect infectious viral titers over the course of LP AI virus infection in ducks has not been adequately examined in existing experimental data. Based on our results, Ct values for both sample types were generally reliable predictors of infectious titer; however, Ct values were less accurate predictors of infectious titer when using cloacal swabs and during early and late infection when Ct values were high. Collectively, these data suggest that the practice of using Ct values to estimate the quantity of infectious virus in a sample collected during an experimental studies should be used with caution, as infectious titers will likely be overestimated, particularly in cloacal swabs and when lower concentrations of virus are being excreted during infection.
In nature, transmission of LP AI viruses in duck populations occurs through the fecal-oral route, which is thought to be enhanced on aquatic habitats due to the tenacity of LP AI viruses in water (Hinshaw et al., 1979; Stallknecht et al., 1990). Recent models indicate that environmental transmission via contaminated water sources is crucial for the long-term maintenance of AI virus in duck populations (Breban et al., 2009; Rohani et al., 2009). Additionally, fecal excretion of LP AI virus into aquatic environments can serve as an important mechanism for spillover into domestic animals with access to the contaminated habitat or water source (Halvorson et al., 1985). The extent of viral shedding in the feces of an infected duck is a critical factor that drives the dynamics of LP AI virus transmission, both within duck populations and across the duck-domestic animal interface. Our study provides a detailed characterization of intestinal excretion of an H3N8 LP AI virus in Mallards, and as such, will allow for more accurate parameterization of future models focusing on LP AI virus transmission.
Numerous AI experimental challenge studies have been conducted in wild and domestic birds over the last 40 yr. Collectively, these studies have identified multiple variables that can influence susceptibility of the host or some measurable outcome of infection (i.e., viral shedding), including factors relating to virus (strain, dose), host (species, age, exposure history), and study design (route of exposure). These factors often complicate comparisons of data between studies conducted under varying conditions. Additionally, although our shedding data are consistent with predictive curves derived from multiple LP AI challenge studies in Mallards (Henaux and Samuels, 2011), these data cannot be generalized for all wild bird-origin LP AI viruses or all avian hosts.
ACKNOWLEDGMENTS
Funding for this work was provided by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), and the Department of Health and Human Services under contract HHSN266200700007C. The opinions expressed herein are those of the authors and do not necessarily reflect the views of any of the funding agencies.
LITERATURE CITED
- BREBAN R, DRAKE JM, STALLKNECHT DE, ANDROHANI P. 2009. The role of environmental transmission in recurrent avian influenza epidemics, PLoS Computational Biology 5: e1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN JD, AND STALLKNECHT DE. 2008. Wild bird surveillance for avian influenza virus. In Avian influenza virus, Spackman E (ed.). Humana Press, Totowa, New Jersey, pp. 85–97. [Google Scholar]
- COSTA TP, BROWN JD, HOWERTH EW, AND STALLKNECHT DE. 2010. The effect of age on avian influenza viral shedding in Mallards (Anas platyrhynchos). Avian Diseases 54 (1 Suppl.): 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSTA TP, BROWN JD, HOWERTH EW, AND STALLKNECHT DE. 2011. Variation in viral shedding patterns between different wild bird species infected experimentally with low-pathogenicity avian influenza viruses that originated from wild birds. Avian Pathology 40: 119–124. [DOI] [PubMed] [Google Scholar]
- DAOUST PY, KIBENGE FS, FOUCHIER RA, VAN DE BILDT MW, VAN RIEL D, AND KUIKEN T. 2011. Replication of low pathogenic avian influenza virus in naturally infected Mallard ducks (Anas platyrhynchos) causes no morphologic lesions. Journal of Wildlife Diseases 47: 401–409. [DOI] [PubMed] [Google Scholar]
- DOLNIK OV, METZGER BJ, AND LOONEN MJ. 2011. Keeping the clock set under midnight sun: Diurnal periodicity and synchrony of avian Isospera parasites cycles in the high Artic. Parasitology 138: 1077–1081. [DOI] [PubMed] [Google Scholar]
- HÉNAUX V, AND SAMUEL MD. 2011. Avian influenza shedding patterns in waterfowl: Implications for surveillance, environmental transmission, and disease spread. Journal of Wildlife Diseases 47: 566–578. [DOI] [PubMed] [Google Scholar]
- HINSHAW VS, WEBSTER RG, AND TURNER B 1979.Water-borne transmission of influenza A viruses? Intervirology 11: 66–68. [DOI] [PubMed] [Google Scholar]
- HINSHAW VS, WEBSTER RG, AND TURNER B, BEAN WJ, WEBSTER RG, AND SRIRAM G 1980. Genetic reassortment of influenza A viruses in the intestinal tract of ducks. Virology 102: 412–419. [DOI] [PubMed] [Google Scholar]
- KIDA H, YANAGAWA R, AND MATSUOKA Y. 1980. Duck influenza lacking evidence of disease signs and immune response. Infection and Immunity 30: 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDIS JR, AND KOCH GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33: 159–174. [PubMed] [Google Scholar]
- LOPEZ G, FIGUEROLA J, AND SORIGUER R. 2007. Time of day, age, and feeding habits influenza coccidian oocyst shedding in wild passerines. International Journal of Parasitology 37: 559–564. [DOI] [PubMed] [Google Scholar]
- MUNSTER VJ, BAAS C, LEXMOND P, BESTEBROER TM, GULDEMEESTER J, BEYER WEP, DE WIT E, SCHUTTEN M, RIMMELZWAAN GF, OSTERHAUS ADME, AND FOUCHIER RAM. 2009. Practical considerations for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. Journal of Clinical Microbiology 47: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REED LJ, AND MUENCH H. 1938. A simple method of estimating fifty-percent endpoints. American Journal of Hygiene 27: 493–497. [Google Scholar]
- ROHANI P, BREBAN R, STALLKNECHT DE, AND DRAKE JM. 2009. Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proceedings of the National Academy of Sciences of the United States of America 106: 10365–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SLEMONS RD, AND EASTERDAY BC. 1978. Virus replication in the digestive tract of ducks exposed by aerosol to type-A influenza. Avian Diseases 22: 367–377. [PubMed] [Google Scholar]
- SPACKMAN E, AND SUAREZ DL. 2008. Type A influenza virus detection and quantitation by real-time RT-PCR. In Avian influenza virus, Spackman E (ed.). Humana Press, Totowa, New Jersey, pp. 19–26. [Google Scholar]
- SPACKMAN E, AND SUAREZ DL, PANTIN-JACKWOOD MJ, SWAYNE DE, AND SUAREZ DL 2009. An evaluation of avian influenza diagnostic methods with domestic duck specimens. Avian Diseases 53: 276–280. [DOI] [PubMed] [Google Scholar]
- STALLKNECHT DE, SHANE SM, KEARNEY MT, AND ZWANK PJ. 1990. Persistence of avian influenza virus in water. Avian Diseases 34: 406–411. [PubMed] [Google Scholar]
- STEVENS JP 2002. Repeated measures analysis. In Applied multivariate statistics for the social sciences, 4th Edition. Lawrence Erlbaum Associates, Mahwah, New Jersey, pp. 500–502. [Google Scholar]
- SWAYNE DE, SENNE DA, AND SUAREZ DL. 2008. Influenza. In A laboratory manual for the isolation and identification of avian pathogens, 5th Edition, Dufour-Zavala L, Swayne DE, Glisson JR, Pearson JE, Reed WM, Jackwood MW, and Woolcock PR (eds.). American Association of Avian Pathologists, Kennett Square, Pennsylvania, pp. 128–134. [Google Scholar]
- WEBSTER RG, YAKHNO M, HINSHAW VS, BEAN WJ, AND MURTI KG. 1978. Intestinal influenza: Replication and characterization of influenza viruses in ducks. Virology 84: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILCOX BR, KNUTSEN GA, BERDEEN J, GOEKJIAN V, POULSON R, GOYAL S, SREEVATSAN S, CARDONA C, BERGHAUS RD, SWAYNE DE, YABSLEY MJ, AND STALLKNECHT DE. 2011. Influenza-A viruses in ducks in Northern Minnesota: Fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One 6: e24010. doi: 10.1371/journal.pone.0024010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS R 2000. A note on robust variance estimation for cluster-correlated data. Biometrics 56: 645–646. [DOI] [PubMed] [Google Scholar]