Abstract
Mutations in the HCFC1 transcriptional co-factor protein are the cause of cblX syndrome and X-linked intellectual disability (XLID). cblX is the more severe disorder associated with intractable epilepsy, abnormal cobalamin metabolism, facial dysmorphia, cortical gyral malformations, and intellectual disability. In vitro, murine Hcfc1 regulates neural precursor (NPCs) proliferation and number, which has been validated in zebrafish. However, conditional deletion of mouse Hcfc1 in Nkx2.1 + cells increased cell death, reduced Gfap expression, and reduced numbers of GABAergic neurons. Thus, the role of this gene in brain development is not completely understood. Recently, knock-in of both a cblX (HCFC1) and cblX-like (THAP11) allele were created in mice. Knock-in of the cblx-like allele was associated with increased expression of proteins required for ribosome biogenesis. However, the brain phenotypes were not comprehensively studied due to sub-viability. Therefore, a mechanism underlying increased ribosome biogenesis was not described. We used a missense, a nonsense, and two conditional zebrafish alleles to further elucidate this mechanism during brain development. We observed contrasting phenotypes at the level of Akt/mTor activation, the number of radial glial cells, and the expression of two downstream target genes of HCFC1, asxl1 and ywhab. Despite these divergent phenotypes, each allele studied demonstrates with a high degree of face validity when compared to the phenotypes reported in the literature. Collectively, these data suggest that individual mutations in the HCFC1 protein result in differential mTOR activity which may be associated with contrasting cellular phenotypes.
Keywords: HCFC1, Neural development, Akt/mTor, Asxl1, 14-3-3 βα
1. Introduction
Mutations in the human HCFC1 transcriptional co-factor protein cause cblX syndrome (MIM 309541) (Gérard et al., 2015, 1; Yu et al., 2013) and X-linked intellectual disability (XLID) (Huang et al., 2012; Jolly et al., 2015; Koufaris et al., 2016, 1; Piton et al., 2011; Piton et al., 2013). cblX is a multiple congenital anomaly syndrome characterized by abnormal cobalamin metabolism, neurodevelopmental defects, cortical gyral malformations, intractable epilepsy, craniofacial dysmorphic features, and movement disorders. However, XLID is milder and associated only with intellectual disability. The clinical phenotypes of the patients are heterogeneous and varied in severity. Multiple systems and approaches have been undertaken to understand the function of human HCFC1 during development. In vitro over expression assays first revealed decreased neural precursor cell (NPC) proliferation with abnormal differentiation and a bias towards the astrocyte lineage in mouse neuro-spheres (Huang et al., 2012). Later we demonstrated that mutation or knockdown of the zebrafish paralogs of HCFC1 caused increased proliferation of NPCs, but these changes were associated with increased expression of radial glial (RGC) and neuronal markers (Castro et al., 2020; Quintana et al., 2017). Global deletion of Hcfc1 in mice was embryonic lethal (Minocha et al., 2016a), but tissue specific deletion in Nkx2.1 + progenitors caused increased death in the cells of the sub-ventricular zone and mantle, which was associated with a decrease in GABAergic interneurons and glia (Minocha and Herr, 2019). Thus, while model system specific, previous studies collectively demonstrate a function for human HCFC1 in brain development and neural precursors.
Recently, a patient derived cblX mutation (p.A115V) was knocked into the mouse Hcfc1 loci to produce the first in vivo cblX model, but the brain phenotypes were not comprehensively studied. However, these studies suggested a mechanism by which mutations in mouse Hcfc1 cause abnormal development. Concurrently, Chern and colleagues uncovered increased expression of proteins essential for ribosome biogenesis in a cblX-like disorder (Chern et al., 2022) caused by mutations in the mouse THAP11 (p.F80L) protein, a protein that interacts with HCFC1 in human cells and in the mouse (Mazars et al., 2010). These data are supported by previous in vitro studies which have shown that human and mouse HCFC1 regulate genes essential for metabolism and stem cell maintenance (Dejosez et al., 2008; Dejosez et al., 2010). What remains uncharacterized is the mechanism by which in vivo mutations in the HCFC1 protein promote increased ribosome biogenesis as well as the effects of this biogenesis on brain development.
The mechanistic target of rapamycin (mTOR) pathway is a well-known regulator of protein translation and cell growth, particularly as it relates to stem cell regulation (Gabut et al., 2020). Interestingly, the mTOR pathway is a common pathway dysregulated in neurodevelopmental disorders (Parenti et al., 2020). Zebrafish harbor one mTOR ortholog, which is expressed in the brain throughout development (Fleming and Rubinsztein, 2011). mTOR is regulated at many levels, but one positive regulator of this pathway is PI3K/AKT. Activation of PI3K leads to AKT activation, which in turn drives activation of mTOR signaling and promotes ribosomal biogenesis and protein synthesis. We have previously demonstrated that inhibition of PI3K can restore the NPC deficits present in a zebrafish with a nonsense mutation in hcfc1a (co60 allele) (Castro et al., 2020). These data, combined with recent literature, indirectly link regulation of PI3K/AKT signaling with phenotypes that occur in cblX syndrome. PI3K/AKT signaling is conserved in zebrafish, so we characterized this pathway in two germline mutant alleles of the zebrafish hcfc1a gene. In addition, we previously identified a driver of AKT signaling, the zebrafish Asxl1 polycomb group protein, which was up-regulated in the co60 allele, one germline mutant of zebrafish hcfc1a (Castro et al., 2020). These data implicate Asxl1, as an HCFC1 downstream target gene, and a putative mediator of Akt/mTor in zebrafish. Therefore, we hypothesized that mutations in human HCFC1 drive ribosomal biogenesis and disrupt brain development through regulation of AKT/mTOR.
Here we identified divergent activity of the zebrafish Akt/mTor signaling pathway in two independent zebrafish hcfc1a germline mutant alleles. Contrasting Akt/mTor activity was correlated with divergent RGC phenotypes. In addition, we identified disparate expression of two downstream effectors of the human HCFC1 protein in each allele: Asxl1 and 14-3-3βα, both of which are known to regulate PI3K/AKT/mTOR activity, and brain development in mouse and human cells (An et al., 2019, 1; Cornell and Toyo-oka, 2017; Gómez-Suárez et al., 2016; Youn et al., 2017). Further, restoration experiments revealed that zebrafish gfap expression is mTor dependent. Thus, our analysis of distinct zebrafish hcfc1a mutant alleles raises the possibility that unique patient variants can have different effects on human HCFC1 protein dosage/function and may lead to individual cellular and molecular phenotypes.
2. Materials and methods
2.1. Experimental model and subject details.
For the experiments described, embryos were produced from natural spawning’s of the following lines: hcfc1aco64/co64, hcfc1aco60/+, Tg(sox2:2A:EGFP), Tg(gfap:EGFP), Tg(hsp701:HCFC1), Tg(hsp701:HCFC1c.344C>T), AB, or Tupfel long fin. Embryos were maintained in E3 media as described by the protocols on the Zebrafish Information Network (ZFIN) at 28° with a 14/10 light:dark cycle. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas El Paso, protocol number 811869-5. Methods for euthanasia and anesthesia were performed according to guidelines from the 2020 American Veterinary Medical Association guidebook.
The hcfc1aco64/co64 allele was created as previously described (Castro et al., 2020) with an identical guide RNA and with equivalent CRISPR/Cas9 editing technology as described. The hcfc1aco64/co64 was created at the same time as the afore published hcfc1aco60/+ allele except that the insertion/deletion introduced produced a second allele with the sequences described. The offspring of the hcfc1aco64/co64 allele were generated from a single founder (F0), which was outcrossed with 3 independent wildtype fish to generate a minimum of 3 families of F1 carriers. Each family consisted of approximately 20 total fish with equal numbers of males and females. To generate subsequent generations, we outcrossed a minimum of 3-F1 individuals with wildtype to obtain a minimum of 3 families of F2 carriers. We subsequently outcrossed F2 carriers (minimum of 3) with wildtype (AB) fish to produce an F3 generation of approximately 3 total families with equal numbers of males and females. Sanger sequencing confirmed mutation of the allele and experiments were initiated in the F3 generation. The Tg(hsp701:HCFC1c.344C<T) was created using Gateway cloning technology as previously described (Castro et al., 2020; Kwan et al., 2007). Vectors utilized for the HCFC1 open reading frame were previously described (Quintana et al., 2014; Quintana et al., 2017). The experiments described herein were performed in the F2 generation, which was produced from a single founder (F0). The positive F0 carrier was outcrossed with wildtype (AB) to produce 2 families of F1 individuals and a minimum of 3 carriers of the F1 generation were outcrossed to produce 3 families of F2 carriers that were utilized for the experiments described. All shared resources will be shared via ZFIN.
2.2. Genotyping of indicated zebrafish lines
Genotyping was performed with DNA from excised larval tissue or fin clips (adults). Tissue was lysed in 50 millimolar (mM) sodium hydroxide (Fisher Scientific) for 15 min at 95° Celsius and pH adjusted with 1 M Tris-HCl. Primers pairs were developed for each allele that specifically bind to and amplify the mutated allele and will not amplify the wildtype allele (Castro et al., 2020). For the hcfc1aco60/+ the primers specific to the mutated allele are FWD: CCAGTTCGCCTTTTTGTTGT and REV: ACGGGTGGTATGAACCACTGGC. PCR annealing was performed at 64°. For the hcfc1aco64/co64 allele, the mutant allele was amplified by standard PCR at an annealing temperature of 64° with FWD: CCAGTTCGCCTTTTTGTTGT and REV: CTGGAGGGATGTTCATAACGG. For identification of the wildtype allele, the following primers were utilized: FWD: CCAGTTCGCCTTTTTGTTGT and REV: TCCCCACGAACGGCTGGTAT.
Genotyping of the Tg(hsp701:HCFC1) allele was performed with the following primers: FWD: TGAAACAATTGCACCATAAATTG present in the hsp701 promoter and REV: CGTCACACACGAAGCCATAG in the HCFC1 open reading frame. PCR for HCFC1 genotyping was performed at 60° annealing temperature with standard extension times. The two heat shock alleles were genotyped with overlapping primer pairs. Genotyping and validation of this allele was previously described (Castro et al., 2020).
2.3. Protein isolation and western blotting
Protein was isolated from the excised brains of larvae at 5 DPF. Brains were homogenized using a pestle in 1X Radio-Immuno-Precipitation-Assay (RIPA) (Fisher Scientific) or 1X Cell Lysis Buffer (Cell signaling). Protease inhibitors were added at a final 1X concentration (Fisher Scientific). Supernatant was collected after a 10-minute centrifugation at 10,000XG in a cold centrifuge. Protein concentration was quantified using Precision Red (Cytoskeleton) according to manufacturer’s instructions. Western blots were performed using standard techniques using the Novex Tris-Glycine-SDS system (Fisher Scientific). Protein loading was quantified using Ponceau S (Fisher Scientific) and membranes were blocked with 5 % ECL Prime Blocking Agent (Fisher Scientific). Primary antibody concentrations are as follows: pAkt (Thr308) (Cell Signaling Cat. # D25E6) (anti-rabbit) 1:3000, Akt (pan) (Cell Signaling Cat. # C67E7) (anti-rabbit) 1:3000, 14-3-3-beta/alpha (Cell Signaling Cat # 9636S) (anti-rabbit) 1:3000, S6 Ribosomal (5G10) (Cell Signaling Cat # 2217S) (anti-rabbit) 1:3000, P-S6 Ribosomal Protein (S235/236) (Cell Signaling Cat # 4858S) (anti-rabbit) 1:3000, β-actin (Sigma/Millipore Cat # A5441 clone AC-15) (anti-mouse) 1:5000, N-terminal HCFC1 (Sigma/Millipore Cat # QC10816) (anti-rabbit) 1:500, C-terminal HCFC1 (Bethyl labs Cat # 50-156-0286 purchased through Fisher Scientific) (anti-rabbit) 1:100. Secondary antibodies were utilized at a concentration of 1:20,000 and chemiluminescence was performed with ECL Primer Western Blotting detection reagent (Fisher Scientific). Imaging was performed using an iBright chemiblot system.
2.4. RNA isolation and quantitative real time PCR
RNA was isolated from embryos at the indicated time points using Trizol (Fisher Scientific) according to manufacturer’s protocol. Reverse transcription was performed using Verso cDNA synthesis (Fisher Scientific) and total RNA was normalized across all samples. For each biological replicate (2-5/experiment) PCR was performed in technical triplicates using an Applied Biosystem’s StepOne Plus machine with Applied Biosystem’s software. Analysis was performed using 2ΔΔct. Sybr green (Fisher Scientific) based primer pairs for each gene analyzed are as follows: hcfc1a fwd: ACAGGGCCTAACACAGGTTG, hcfc1a rev: TCCTGTGACTGTGCCAAGAG, hcfc1b fwd: GGATGGGTCCCTCTGGTTAT, hcfc1b rev: CGGTAACCATCTCGTCCACT, mmachc fwd: CACACTGTCTGCCTGCATCT, mmachc rev: CGGATTGTGGATGTCTGATG, asxl1 fwd: CCAGAGCTGGAAAGAACGTC, asxl1 rev: ACATCTCCAGCTTCGCTCAT, rpl13a fwd: TCCCAGCTGCTCTCAAGATT, rpl13a rev: TTCTTGGAATAGCGCAGCTT.
2.5. Proteomics sample preparation
Samples were isolated from the whole brain as described above and proteins were precipitated from lysis buffer using trichloroacetic acid (TCA) as follows: 50 microliters (μl) of 100 % TCA (Sigma/Millipore – cat# T6399-5G) was added to 200 μL of protein sample at a final concentration of 20 %, then incubated at 4 °C for 10 min. The mixture was centrifuged at 14,000 relative centrifugal force (rcf) for 5 min. The pellet was resuspended in 200 μL of 100 % liquid chromatography-mass spectrometry (LC/MS) grade acetone (Fisher Scientific) and centrifuged at 14,000 rcf for 5 min and supernatant was discarded. This process was repeated two more times to ensure removal of residual detergents. After the third wash, the pellet was heated on a heating block at 95 °C for 2 min to ensure acetone evaporation. Samples were stored at −80 °C and resuspended in 8 M Urea prior enzymatic digestion.
Enzymatic tryptic digestion of proteins was obtained by using iST Sample preparation kit from PreOmics (catalog no. iST 96x 00027) according to manufacturer’s instructions. Briefly, 10 μL of LYSE buffer was added per 1 microgram (μg) of protein from larvae brains and placed in a heating block (80 °C for 20 min, mixing every 5 min) for protein denaturing, alkylation, and reducing disulfide bonds within it. Droplets were subject to brief centrifugation (300 rcf; 10 sec, room temperature). Samples were sonicated using 10-cycles; 30 sec on/off. After sonication, 50 μL of DIGEST buffer was added, gently vortexed, and placed in a heating block at 37 °C for 90 min. Samples were gently vortexed every 10-minutes during the 90-minute incubation period. One hundred microliters of STOP solution was added and thoroughly mixed by pipetting up and down 10-times. Samples were transferred to cartridges using 1.5 ml (mL) microcentrifuge tubes with collar adapters and centrifuged for 1-minute at 3,800 rcf. Loaded peptides were washed once with 200 μL of Wash-1 and Wash-2 solutions, with 1-minute 3,800 rcf centrifugation in between each wash step. Peptides were eluted in a fresh 1.5 mL microcentrifuge tube using 100 μL of ELUTE solution for 1-minute at 3,800 rcf each. Two elution rounds were performed. Eluted peptides were dried completely in a speed vacuum centrifugation at 45 °C, 100 mTorr, for 90 min (Savant; Thermo Fisher Scientific). Dried peptides were stored at −80 °C until ready for LC-MS/MS data acquisition.
2.6. Liquid chromatography–tandem mass spectrometry (LC/MS)
Digested peptide mixtures were resuspended in LC/MS grade 4 % acetonitrile (ACN) (Fisher Scientific), 0.1 % formic acid (FA), at a concentration of 1 μg/μL. One microliter of resuspended samples was loaded onto an Acclaim PepMap rapid separation LC column (75 μm × 50 cm nanoViper, PN 164942, Thermo Scientific), which had been previously equilibrated with 4 % solvent B (100 % acetonitrile, 0.1 % formic acid), and 96 % solvent A (100 % H2O, 0.1 % formic acid). The multi-step gradient started with peptides being loaded with a flow rate of 0.5 μL/min for 15 min using a Dionex Ultimate 3000 RSLCnano (Thermo Scientific). The flow rate was decreased to 0.3 μL/min over 15 min, and the solvent B increased to 20 % at 115 min, then increase to 32 % at 135 min, and finally increased to 95 % solvent B over 1 min. To elute any additional remaining peptides, 95 % solvent B was held for 4 min at a flow rate of 0.4 μL/min. The flow rate was increased to 0.5 μL/min over 1 min. The column was re-equilibrated at 4 % solvent B for 39 min and maintained a flow rate of 0.5 μL/min for a total of 180 min total runtime for each technical duplicate. The column was maintained at 55 °C throughout the entire data acquisition. One blank injection was performed between biological samples using a 60-minute two sawtooth gradient at 4–95 % solvent B and re-equilibrated at 4 % solvent B for the next sample injection.
Eluted peptide information was acquired with a Q-Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific) in positive mode, set to data dependent MS2. Full MS ions were collected at resolution of 70,000, AGC target of 3e6, with a scan range of 375 to 1500 m/z. Ions were fragmented with NCE set to 27 and collected at a resolution of 17,500 with AGC target at 1e5, 2 m/z isolation window, maximum IT set to 60 ms, loop count 10, TopN 10. Charged exclusion ions were set to unassigned, 1, 6 – 8, >8.
2.7. Bioinformatics data analysis
After LC/MS analysis, Proteome Discover (PD) 2.5.0.400 (Fisher Scientific) was utilized to identify the proteins from each peptide mixture. The database for Danio rerio was downloaded from UniProtKB; https://www.uniprot.org/ on 21 October 2021 with a database 61,623 sequences. A contaminant dataset was run in parallel composed of trypsin autolysis fragments, keratins, standards found in CRAPome repository and in-house contaminants. PD analysis parameters are as follows: false-discovery rate (FDR) of 1 %, HCD MS/MS, fully tryptic peptides only, up to 2 missed cleavages, parent-ion mass of 10 ppm (monoisotopic); fragment mass tolerance of 0.6 Da (in Sequest) and 0.02 Da (in PD 2.1.1.21) (monoisotopic). Two-high confidence peptides per protein were applied for identifications. PD dataset was processed through Scaffold Q + S 5.0.1. Scaffold (Proteome Software, Inc., Portland, OR 97219, USA) was used to probabilistically validate protein identifications derived from MS/MS sequencing results using the X! Tandem and Protein Prophet computer algorithms. Data was transferred to Scaffold LFQ (Proteome Software, Portland, Oregon, USA) which was used to validate and statistically compare protein identifications derived from MS/MS search results. A protein threshold of 95 %, peptide threshold of 95 %, and a minimum number of 2 peptides were used for protein validation. Normalized weighted spectral counts were used when comparing the samples. To ascertain p-values, Fisher’s Exact was run with a control FDR level q * 0.05 with standard Benjamini-Hochberg correction.
2.8. Flow cytometry
To identify mutants, we adapted a protocol whereby tissue was excised from caudal most tip of the tail from larvae at 2–3 DPF (Kosuta et al., 2018). Tissue was collected on a filter paper and genotyped as described in the sections above. Equal numbers of mutants and wildtype larvae were euthanized according to standard procedures, then heads were excised from tails to create a whole brain homogenate. Dissociation of heads was followed exactly according to previous protocol (Bresciani et al., 2018). Briefly, tissue was dissociated using Trypsin-EDTA (0.25 %) (Fisher Scientific) with collagenase (100 mg/mL) (Fisher Scientific), applying heating at 30° Celsius and disturbing tissue by pipetting up and down. The reaction was stopped by adding lamb serum (Fisher Scientific) and cells were harvested through centrifugation (5 min at 700g at room temperature). The cells were resuspended in 1X phosphate buffered saline (Fisher Scientific) and filtered through 40-70uM filter before performing flow cytometry. Analysis was performed with Kaluza software at a fixed rate of 10,000 cell events for a maximum of 3 min.
2.9. Immunohistochemistry
Larvae were fixed at the indicated time points in 4 % paraformaldehyde (Electron Microscopy Sciences) for minimum of 1 h at room temperature. Genotyping was performed as described (Kosuta et al., 2018). Brain tissue was embedded in 1.5 % agarose (Fisher Scientific) produced in 5 % sucrose (Fisher Scientific). Embedded blocks were incubated overnight in 30 % sucrose (Fisher Scientific) and then frozen with dry ice before cryosectioning (20–30 μM). Validated transgenic reporter animals were utilized for visualization of Sox2 + and Gfap + cells (ZDB-TGCONSTRCT-070410-2). For DNA content staining, Hoechst (2ug/ml) (Fisher Scientific) dye was utilized. Hoechst stain was performed according to manufacturer’s protocol available as part of the EdU Click-It technology kit (Fisher Scientific). All slides were cover slipped using Vectashield (Vector Laboratories) and imaged on a Zeiss LSM 700 at 20 × −63 × magnification.
2.10. Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Quintana et al., 2011) except that larvae were disassociated into a single cell homogenate prior to cell fixation with formaldehyde. Briefly, equal numbers of non-heat shocked (NHS) and heat-shocked (HS) larvae in either line, Tg(hsp701:HCFC1) or Tg(hsp701:HCFC1 c.344C>T), were euthanized according to standard procedures, then heads were excised from tails to create a whole brain homogenate. Tissue was dissociated as described above for flow cytometry. Shearing was performed in a 200 μL volume of 50 mM Tris pH 8.0 (Millipore/Sigma) by adding 40 units of micrococcal nuclease (Fisher Scientific) for 10 min at 37 °C. Ethylenediaminetetra-acetic acid (EDTA-Fisher Scientific) (10 mM) was used to stop the reaction and nuclei were lysed in 1 % sodium dodecyl sulfate (SDS-Fisher Scientific). ChIP was performed with anti-HCFC1 antibodies (Millipore Sigma AV38600) or anti-IgG antibodies (Santa Cruz Biotechnology, sc-2025). The collected precipitates were washed and de-crosslinked and the DNA was purified using standard protocols (Quintana et al., 2011). qPCR was used to determine enrichment using primers designed to the 1 kilobase region upstream of each start site/gene of interest. Primers to gapdh putative promoter were used as a negative control and each set of enrichment was normalized to gapdh PCR or input as described (Quintana et al., 2011). Primers used for SYBR green (Fisher Scientific) based PCR are as follows: gapdh fwd: GCCTGATTTGGTTGTGTCCT, gapdh rev: CCAATTGGGAAATGCTTGAG, asxl1 fwd: GGCTGTAGGAGCGACTGAAG, asxl1 rev: TAAACACACACAGGGCGAAG.
2.11. Heat shock and drug treatment assays
Heat shock was performed as previously described (Castro et al., 2020; Hudish et al., 2013). Briefly, heat shock was initiated at 24 h post fertilization (HPF) and performed twice daily every 8–12 hours until 5 DPF. Heat shock was performed for a duration of 30–45 min by incubating dishes at 38°. For rapamycin treatment (Selleck Chemicals), the drug was dissolved in 100 % DMSO (Fisher Scientific) and embryos were treated at 24 HPF and 72 HPF with a 0.8 uM concentration for a period of 24 h. Concentration was determined empirically using a gradient coupled with gene expression of ccne1. The concentration utilized reduced ccne1 expression, without any effect on larval viability. Media was removed and no treatment was applied at 48 and 96 HPF. Total treatment time during the 5-day period is 48 h. Total time with treatment off is 72 h.
3. Results
3.1. Akt/mTor signaling is increased in the co60 allele
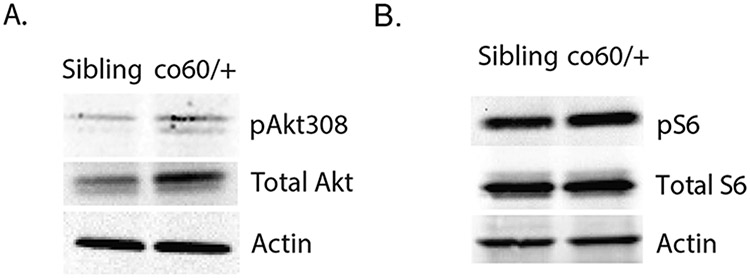
We have previously established that nonsense mutation of zebrafish hcfc1a (co60 allele) causes an increase in asxl1 expression, which is associated with increased proliferation of NPCs (Castro et al., 2020). In mice, ASXL1 promotes AKT phosphorylation (Youn et al., 2017) and we established that inhibition of zebrafish PI3K, an upstream regulator of AKT, is sufficient to restore NPC numbers to normal levels in the co60 allele. Thus, we hypothesized that nonsense mutation of hcfc1a leads to increased activation of zebrafish Akt kinase. We performed western blot analysis using anti-phosho-Akt threonine 308 (pAkt308) and total Akt antibodies. We detected increased expression of total Akt, which was associated with hyperphosphorylation of pAkt308 (Fig. 1A). Since total Akt was routinely higher in the co60 allele, we used β-actin as an additional loading control to validate increased expression of total Akt protein (Fig. 1A). We observed equal β-actin expression, which correlated with Ponceau S staining of the membrane. We did not analyze phosphorylation of Serine 473, as we could not identify an antibody that would cross react with zebrafish Akt at this location.
Fig. 1. Akt/mTor signaling is increased in the hcfc1aco60/+ allele.
A-B. Western blot analysis was performed with anti-phospho Akt (thr308) antibodies (pAkt308), total Akt, anti-phospho p70S6kinase (pS6), total S6, or β-actin using brain homogenates from sibling wildtype or carriers of the hcfc1aco60/+ (co60/+) allele. N = 4/group/biological replicate and performed in 3 independent biological replicates.
AKT kinase is an upstream regulator of the mTOR pathway in mice and humans, which subsequently promotes growth, proliferation, translation, and ribosomal protein synthesis. Chern and colleagues published that cblX is associated with increased expression of proteins required for ribosome biogenesis (Chern et al., 2022). Therefore, we performed western blot analysis to detect phosphorylation of S6 kinase (pS6), which is phosphorylated by active mTOR signaling in humans and mice. We observed increased pS6 kinase in carriers of the co60 allele with equivalent levels of total S6 protein (Fig. 1B), β-actin was used to validate protein loading in addition to total S6. Ponceau S was used to confirm protein loading and concentration. In addition, we did not detect differences in total S6 in any biological replicate. These data demonstrate that nonsense mutation of hcfc1a is associated with increased zebrafish Akt/mTor signaling.
3.2. Missense mutation of the N-terminal kelch domain does not change the expression of hcfc1a
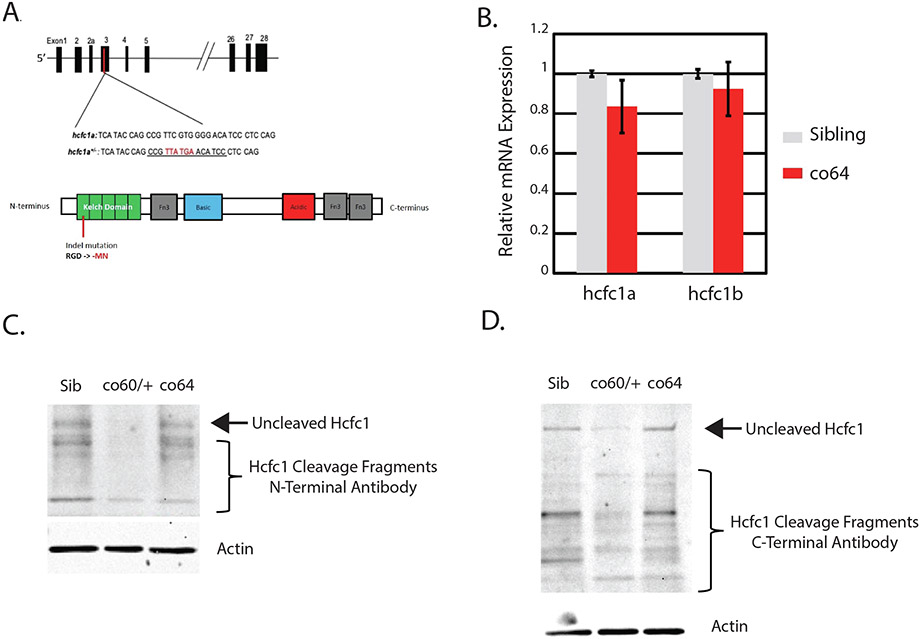
Nonsense mutation of zebrafish hcfc1a results in increased Akt/mTor (Fig. 1). However, nonsense mutation of hcfc1a (co60 allele) is hap-loinsufficient in zebrafish and reduces hcfc1a expression by 50 %, which does not accurately reflect the types of mutations present in cblX syndrome. cblX syndrome is caused by missense mutations that do not reduce overall protein expression (Yu et al., 2013). We generated an additional hcfc1a allele (hcfc1aco64/co64 or co64) which results in an insertion deletion that causes an in-frame deletion of arginine at amino acid position 75 with two missense mutations p.G76M and p.D77N within the kelch protein interaction domain (Fig. 2A). The co64 allele is homozygous viable into adulthood (the only model to date) and homozygous offspring obey Mendelian inheritance patterns. As shown in Fig. 2B, we did not detect any significant difference in the expression of hcfc1a or hcfc1b, a second zebrafish ortholog of HCFC1 in the co64 allele. Since hcfc1b expression is unchanged, we do not anticipate any compensatory mechanisms present in the co64 allele, but we cannot fully rule out the possibility at this time.
Fig. 2. Missense and nonsense alleles of the hcfc1a gene differentially affect Hcfc1 expression.
A. Schematic diagram for the location of the hcfc1aco64/co64 homozygous viable allele. CRISPR/Cas9 created and insertion/deletion in the genomic DNA that causes an in-frame deletion of arginine and a 2-amino acid base pair change (GD > MN) in the kelch domain. B. Quantitative PCR (qPCR) was performed with total RNA isolated from whole brain homogenates from wildtype siblings or homozygous carriers of the hcfc1aco64/co64 allele (co64). The expression of hcfc1a and the hcfc1b paralog were analyzed. No significant difference in expression was observed, consistent with the effects of missense mutations in cblX syndrome. A total of N = 23 wildtype sibling and N = 25 homozygous brains were used to measure hcfc1a expression. The expression of hcfc1b was analyzed with a total number of N = 18 wildtype siblings and N = 20 homozygous carriers. Numbers were obtained over the course of a minimum of 3 biological replicates. C-D. Western blot analysis was performed with N-terminal (C) or C-terminal (D) specific antibodies to the HCFC1 protein. Total protein was detected in brain homogenates from wildtype siblings, heterozygous carriers of the hcfc1aco60/+ (co60/+), and homozygous carriers of the hcfc1aco64/co64 (co64) allele. β-actin was used as a loading control. N = 4/group/biological replicate and performed in 2 independent biological replicates were utilized for western blot analysis.
3.3. The co60 and co64 alleles have unique effects on Hcfc1a protein expression
The two zebrafish orthologs, hcfc1a and hcfc1b, are ubiquitously expressed throughout the developing embryo (Quintana et al., 2014). Human HCFC1 is synthesized as a large preprotein that is cleaved into N and C-terminal fragments (Kapuria et al., 2016). We used two independent antibodies which detect conserved epitopes in the N or C-terminal domains of the human HCFC1 protein. We compared the expression of Hcfc1a using both N and C-terminal specific antibodies in the co60 and co64 alleles using western blot. The co60 allele (nonsense) showed decreased Hcfc1 expression utilizing both N- and C-terminal specific antibodies (Fig. 2C&D). In contrast, the co64 allele (missense) did not markedly reduce overall protein expression (Fig. 2C&D). The conserved N- and C terminal epitopes analyzed are present in both Hcfc1a and Hcfc1b. Therefore, the expression of the bands in Fig. 2C&D indicates levels of both Hcfc1a and Hcfc1b. These data provide evidence that the co64 allele does not reduce overall protein expression, but the co60 allele affects the expression level of Hcfc1a.
3.4. The co64 allele is associated with decreased Akt/mTor signaling.
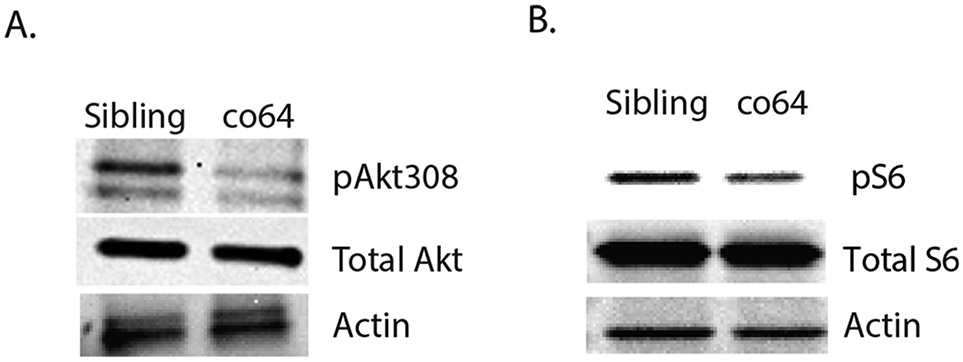
We observed increased Akt/mTor in the co60 allele, but since the co64 and co60 alleles differentially affect protein expression, we hypothesized that dosage changes in Hcfc1a expression (co60) would affect brain development and Akt/mTor signaling differently than missense mutations in the kelch domain. We used western blot analysis to determine the level of Akt/mTor signaling in the co64 allele. We observed hypophosphorylation of Akt at threonine 308 with equivalent levels of total Akt (Fig. 3A). We used total Akt and β-actin as loading controls and did not detect changes in the expression of total Akt or β-actin in any of the biological replicates performed. This contrasts with the co60 allele, which demonstrated consistently with increased total Akt. Decreased Akt phosphorylation was associated with decreased pS6 in the co64 allele (Fig. 3B). Total S6 protein and β-actin were unchanged across multiple biological replicates. These data reveal differential regulation of Akt/mTor in the co60 and co64 alleles, which we confirmed have unique effects on Hcfc1a protein expression (Fig. 2).
Fig. 3. Differential regulation of Akt/mTor in the hcfc1aco64 allele.
A-B. Western blot analysis was performed with anti-phospho Akt (thr308) (pAkt308), total Akt, anti-phospho-p70S6kinase (pS6), total S6, or β-actin antibodies using brain homogenates from sibling wildtype or carriers of the hcfc1aco64/co64 (co64) allele. N = 4/group/biological replicate and performed in 3 independent biological replicates were utilized for western blot analysis.
3.5. NPC number is increased in cblX-like syndromes.
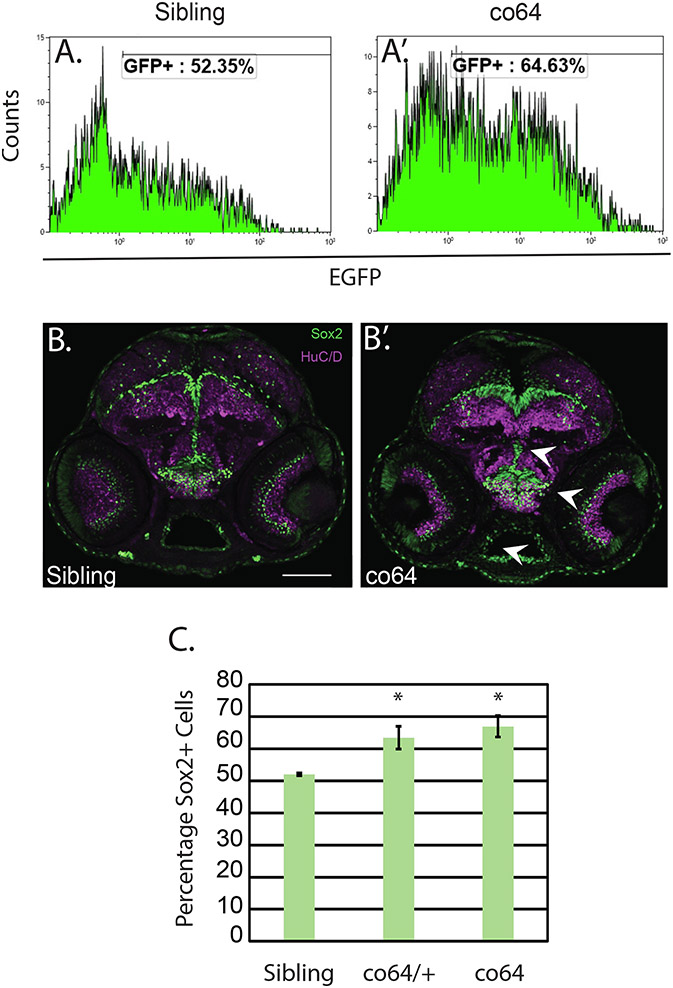
We have previously demonstrated that the co60 allele causes increased numbers of Sox2 + NPCs (Castro et al., 2020). We sought to determine the number of NPCs in the co64 allele given the differential regulation of Akt/mTor. To quantify the number of NPCs, we first crossed the co64 allele into the Tg(sox2:2A:EGFP) reporter. We then produced a single cell homogenate and performed flow cytometry to detect EGFP positive cells. We detected an increase in the number of GFP + cells by flow cytometry at 6 days post fertilization (DPF) (Fig. 4A&A’) and validated this increase using immunohistochemistry at 2 DPF (Fig. 4B&B’, arrows indicate regions of increased cell number). We performed flow cytometry on three independent occasions and found a statistically significant (p < 0.05) increase in NPCs in heterozygous and homozygous carriers of the co64 allele (Fig. 4C). These data suggest that both the co60 and co64 allele have increased NPCs. The presence of overlapping phenotypes provides face validity for the effectiveness of each allele as a putative model system to inform as to the function of human HCFC1 in NPC development.
Fig. 4. Neural precursors (NPCs) are increased in the co64 allele.
A&A’. Flow cytometry analysis was used to detect the number of Sox2 + cells in the hcfc1aco64/co64 allele (co64) and wildtype siblings at 6 days post fertilization (DPF). Histograms represent a single biological replicate with N = 5/group. B&B’. Immunohistochemistry at 2 days post fertilization (DPF) was performed to detect Sox2+ (green) and Huc/D + cells (magenta) in sibling wildtype and the co64 allele. N = 6/group obtained from 2 biological replicates. C. Quantification of percentages obtained by flow cytometry from 3 biological replicates (N = 5/replicate and performed in 3 biological replicates, one is shown in A&A’). The percentage of Sox2 + cells was quantified by flow cytometry and the average of 3 replicates is plotted. *p < 0.05.
3.6. The co60 and co64 alleles differentially disrupt RGC number.
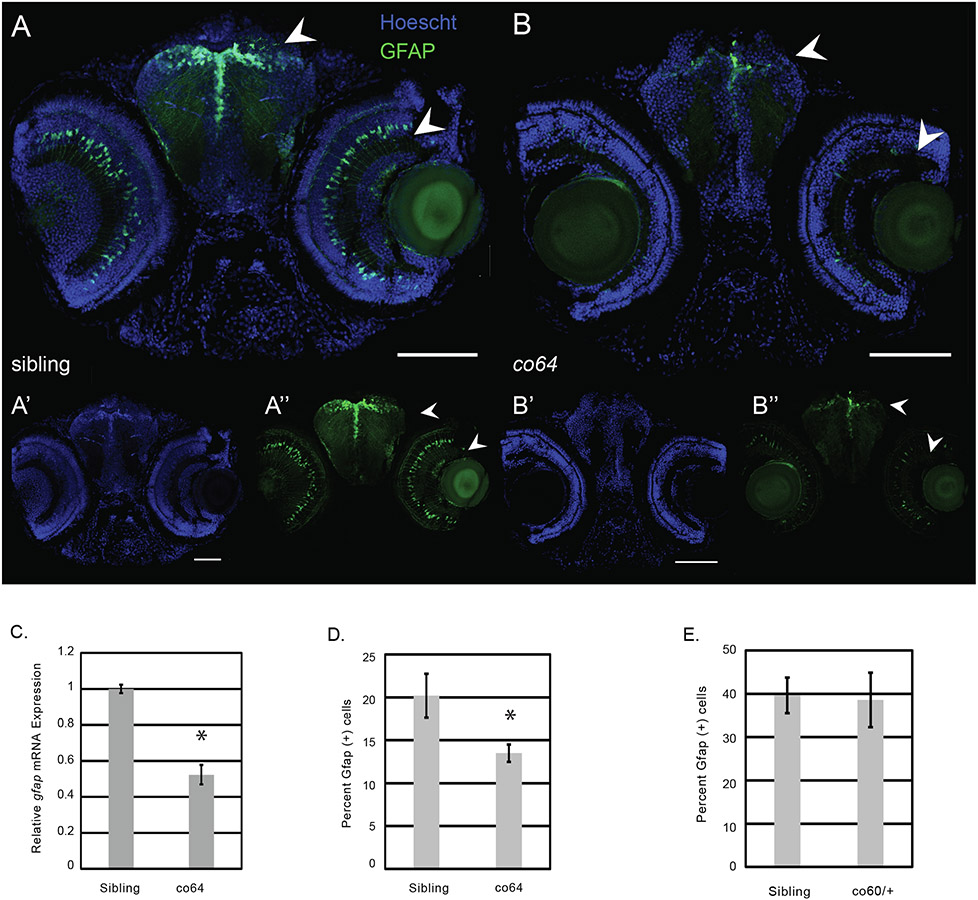
We have previously shown that the co60 allele increases the expression of zebrafish gfap and elavl3, markers of RGCs and neurons, respectively (Castro et al., 2020). Therefore, we measured the expression of both genes in the co64 allele. We observed a statistically significant decrease in zebrafish gfap expression at 2 DPF (Fig. 5C), which we validated by immunohistochemistry at 6 DPF (Fig. 5A&B). We next crossed the co64 allele into the Tg(gfap:EGFP) reporter and performed flow cytometry to quantify the total number of RGCs. We observed a statistically significant decrease in the total number of RGCs (Fig. 5D). Decreased RGC number in the co64 allele contrasted with the number of RGCs in the co60 allele, as we detected equivalent numbers of RGCs using flow cytometry in the co60 allele (Fig. 5E). Interestingly, we validated an increase in the expression of zebrafish gfap in the co60 allele (Fig. 6F, yellow bars, p = 0.06). This display of increasedmRNA with no change in cell number can be owed to similar number of RGCs producing increased levels of zebrafish gfap transcript and protein in the co60 allele (Castro et al., 2020). These data demonstrate that 1) the co64 allele reduces the number of RGCs, 2) the co60 allele increases gfap expression, but has no effect on total RGC number, and 3) the different hcfc1a alleles studied have unique effects on RGC development.
Fig. 5. Radial glial cells (RGCs) are reduced in number and expression in the co64 allele.
A-B. A’-B’, & A”-B”. Immunohistochemistry was used to detect the Gfap + cells in wildtype sibling and homozygous carriers of the hcfc1aco64/co64 allele (co64) at 6 days post fertilization (DPF). The co64 allele was crossed with the Tg(gfap:EGFP) reporter. Hoechst DNA content stain (A’-B’) was used as a control. N = 4 sibling and N = 7 co64. Arrows indicate areas of decreased expression. C. Quantification of gfap expression in sibling wildtype and homozygous carriers of the co64 allele. Total RNA was isolated from whole brain homogenates and gfap expression was quantified by qPCR from 3 biological replicates (N = 3/replicate) (*p < 0.05). D. The number of Gfap + cells was quantified at 5 DPF using flow cytometry in the co64 allele and wildtype sibling. Analysis was performed in biological triplicate with a total N = 14 and the average percentage of positive cells was plotted (p < 0.05). E. The number of Gfap + cells was quantified by flow cytometry in sibling wildtype and the hcfc1aco60/+ allele. Two biological replicates were performed with total N = 6. The average percentage of cells in each biological replicate is plotted with error bars representing standard error of the mean between biological replicates. No significant difference was found.
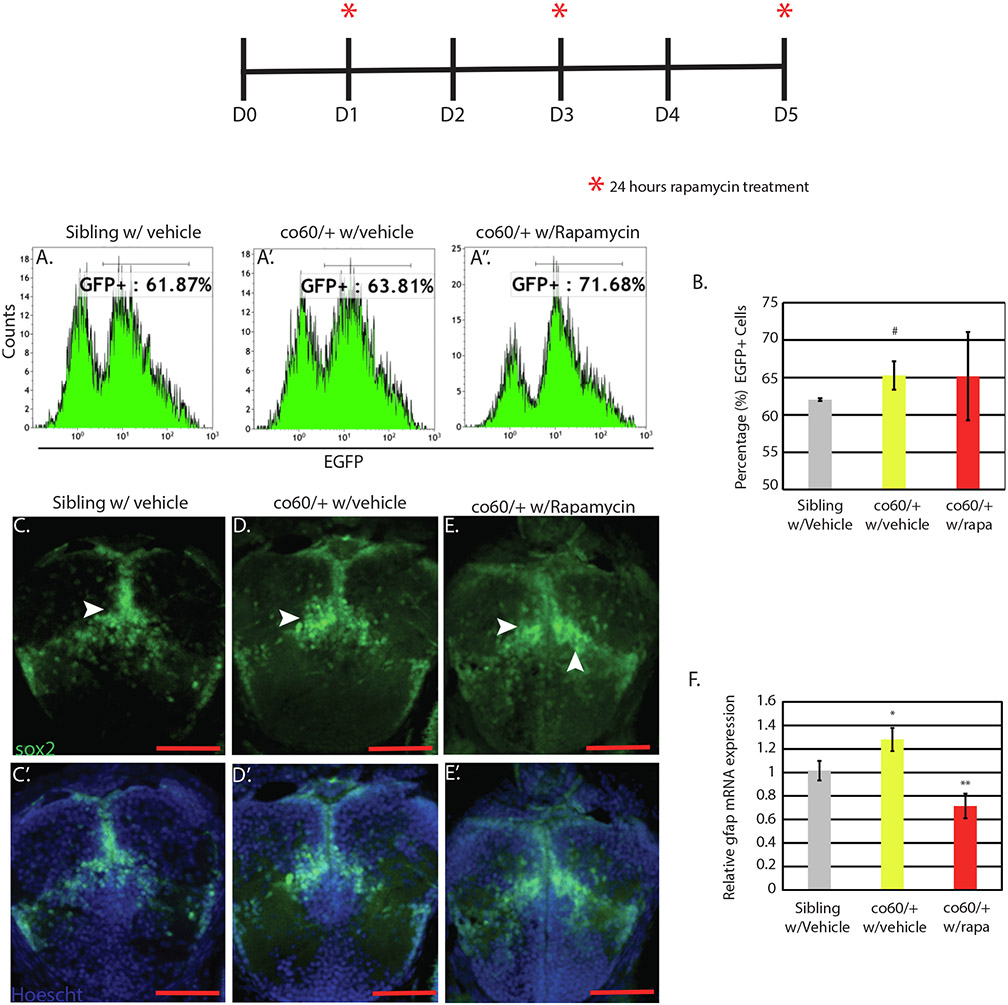
Fig. 6. gfap expression but not Sox2 cell number is mTor dependent.
Top: Schematic representing the onset of 0.8 micromolar(uM) rapamycin or vehicle dimethyl sulfoxide (DMSO) treatment. Each asterisk represents the onset of a 24- hour period of treatment. Days (D) without an asterisk are those where media did not contain 0.8 uM rapamycin or vehicle control. A-A”. Flow cytometry of the hcfc1aco60/+(co60/+) and wildtype sibling was performed at 5 days post fertilization (DPF). The number of Sox2 + cells were quantified with the Tg(sox2:2A:EGFP) transgene. Flow cytometry shown here is a single representative replicate, but the assay was performed on 4 independent occasions with N = 14/group. B. Graphical representation of the average percentage of Sox2 + cells obtained by flow cytometry across 4 biological replicates. # p = 0.014959. The number of Sox2 + cells was not restored by treatment with 0.8uM rapamycin. C-E. Immunohistochemistry validation of A-A” analyzing Sox2 + cells using the Tg(sox2:2A:EGFP) reporter. Hoechst stain was performed as a control in C’-E’. F. Quantitative PCR (qPCR) was used to determine the expression of gfap in vehicle treated sibling wildtype, the co60/+ allele treated with vehicle control (w/vehicle) or the co60/+ allele treated with rapamycin (w/rapa). The level of gfap expression is increased in the co60/+ allele (*p < 0.06) and restored by treatment with rapamycin (**p < 0.05).
3.7. NPC number is mTor independent in the co60 allele.
Both the co60 and co64 alleles cause an increase in the number of NPCs but demonstrate divergent regulation of Akt/mTor. Based on these data, we surmised that NPC number was Akt/mTor independent. To test this, we treated co60 embryos/larvae with rapamycin (Fig. 6 Schematic) and measured the number of Sox2 + NPCs in whole brain homogenates using flow cytometry. We observed a statistically significant increase in the number of NPCs by flow cytometry in the co60 allele (Fig. 6A-A”&B). These data are consistent with previous studies, which find a higher percentage increase of NPCs in a region specific manner across the forebrain, midbrain, and hindbrain (Castro et al., 2020). Treatment of rapamycin did not reduce the number of NPCs at 5 DPF (Fig. 6A-A”&B) and exacerbated the phenotype leading to an even higher number of total EGFP Sox2 + cells in specific replicates. We validated these results with immunohistochemistry (Fig. 6C-E & C’-E’).
3.8. Gfap expression is partially regulated by mTor in the co60 allele.
The co60 and co64 alleles demonstrated with differential expression of gfap as documented in Fig. 5C and (Castro et al., 2020). In the co60 allele, increased gfap expression is not due to increased numbers of cells (Fig. 5E). We validated the increased expression of gfap expression in the co60 allele in Fig. 6F (yellow bars, p = 0.06). Since gfap and mTor activity were differentially regulated in the co60 and co64 alleles, we sought to determine if gfap expression was mTor dependent. Wildtype and co60 carriers were treated with rapamycin as shown in Fig. 6 schematic and the expression of gfap was analyzed by qPCR analysis. Treatment with rapamycin caused a statistically significant decrease in the expression of gfap in mutant animals (co60) (Fig. 6F, red bars). These data suggest that gfap expression is partially regulated by mTor signaling in the co60 allele.
3.9. Asxl1 expression correlates with Akt/mTor activity in the co60 and co64 alleles.
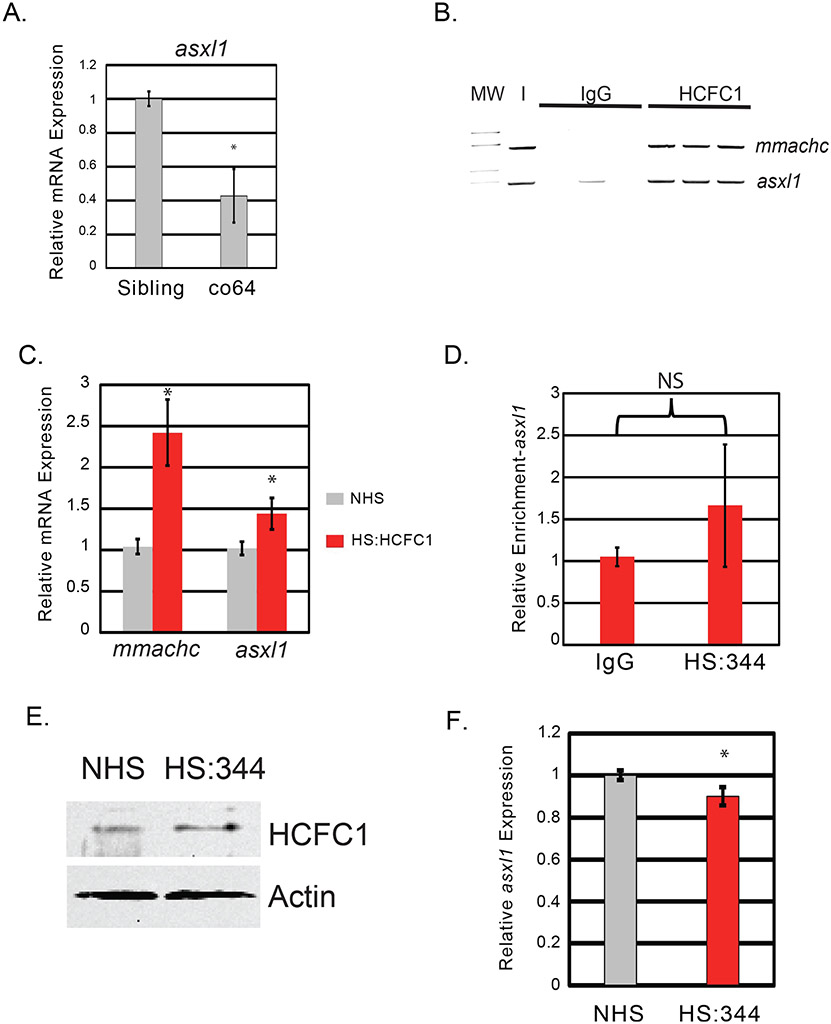
We have previously established that the co60 allele caused increased expression of zebrafish asxl1, a mediator of AKT signaling in mice (Castro et al., 2020; Youn et al., 2017). Thus, we hypothesized that differential expression of asxl1 was mediating contrasting Akt/mTor signatures in each allele. We measured the expression of asxl1 in the co64 and observed it to be significantly decreased relative to sibling wildtype (Fig. 7A). Human HCFC1 protein has been shown to bind to the human ASXL1 promoter (Dehaene et al., 2020) and therefore, we hypothesized that the expression of zebrafish asxl1 was reduced in the co64 allele due to abnormal binding of mutated Hcfc1a to the zebrafish asxl1 promoter. To test this hypothesis, we utilized the Tg(hsp701:HCFC1) transgene (Castro et al., 2020). After performing a heat shock protocol (Castro et al., 2020; Hudish et al., 2013), we performed chromatin immunoprecipitation (ChIP) and used PCR to detect binding of wildtype human HCFC1 protein to the zebrafish mmachc and asxl1 promoters. Human and mouse MMACHC is a known downstream target gene of HCFC1 and used here as a positive control (Dejosez et al., 2010). Semi-quantitative PCR was performed in technical triplicate and demonstrated amplification of the zebrafish mmachc and asxl1 promoter regions (Fig. 7B). We did not detect amplification after 40 cycles in the IgG antibody control lanes, except in one sample designed to amplify the asxl1 promoter region. However, this amplification was far less than the enrichment detected using HCFC1 specific antibodies. These results indicate putative binding to the zebrafish mmachc and asxl1 promoters by human HCFC1. Positive binding was associated with increased mRNA expression of zebrafish mmachc and asxl1 following heat shock protocol (Fig. 7C).
Fig. 7. cblX mutations differentially regulate asxl1 expression.
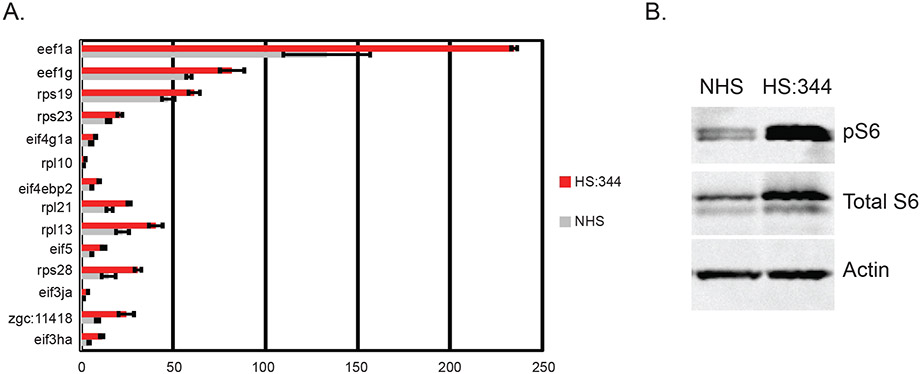
A. Quantitative PCR (qPCR) was performed with total RNA isolated from whole brain homogenates of wildtype siblings or homozygous carriers of the hcfc1aco64/co64 allele (co64) to measure the expression level of asxl1. A total N = 15 animals were used in two biological replicates. B. Chromatin Immunoprecipitation (ChIP) with semi-quantitative PCR was used to detect binding to the endogenous mmachc (+control) or asxl1 promoters. ChIP was performed using the Tg(hsp701:HCFC1). Binding was detected with anti-HCFC1 antibodies (HCFC1) relative to the IgG control (IgG). Lanes represent triplicate PCR from one biological replicate. Additional replicates were performed and validated using qPCR. Approximately 10 % of the lysate (I) was utilized as an input control. N = 20 heads per group were isolated and used for analysis. Assay was repeated on 3 independent occasions for a total N = 60. C. qPCR was used to measure the expression of mmachc and asxl1 in no heat shock (NHS) and heat shocked (HS:HCFC1) fish carrying the Tg(hsp701:HCFC1) transgene. Error bars represent standard error of the mean from biological replicates. Each biological replicate was performed with a pool of embryos from which total RNA was isolated. *p < 0.05. N = 20 heads per group across all replicates. D. ChIP was performed with anti-HCFC1 antibodies (HS:344) or IgG control (IgG) following a heat shock protocol with Tg(hsp701:HCFC1 c.344C>T) larvae. N = 20 heads per group were isolated and used for analysis for each biological replicate (3 replicates were performed). E. Western blot was performed with anti-HCFC1 antibodies or β-actin in no heat shock (NHS) or heat shocked Tg(hsp701:HCFC1 c.344C>T) larvae (HS:344). N = 4/group/biological replicate and performed in 2 biological replicates. F. qPCR analysis was utilized to determine the expression of asxl1 in carriers of the Tg(hsp701:HCFC1 c.344C>T) allele. *p < 0.05. N = 20 heads per group were isolated and used for analysis. Error bars represent standard error of the mean between biological replicates as indicated in materials and methods.
We next produced a heat shock transgene that would express the human cblX (c.344C > T) variant (Tg(hsp701:HCFC1c.344C>T)) located in the kelch domain and previously described (Yu et al., 2013). Interestingly, this patient variant is the same variant produced in mice by Chern and colleagues. We performed ChIP using anti-IgG or anti-HCFC1 antibodies but did not detect binding of HCFC1 to the zebrafish asxl1 promoter by qPCR (Fig. 7D). Loss of enrichment was not the result of an inability of the antibody to bind the C.344C > T mutant variant, as western blot was used to validate the specificity of the HCFC1 antibody to the cblX variant protein (Fig. 7E). Importantly, endogenous zebrafish Hcfc1 was detected in the no heat shock control as the antibody cross reacts with zebrafish Hcfc1 protein. Consistent with a loss of binding, we did not detect up-regulation of asxl1 mRNA after heat shock protocol (Fig. 7F). Collectively, these data suggest that human HCFC1 binds to and activates the zebrafish asxl1 promoter and that this function may be disrupted in cblX.
3.10. NPC/RGC number are independent of murine Asxl1 over-expression.
Our data uncovered contrasting activity of Akt/mTor and differential gfap expression in the co60 and co64 alleles. These phenotypes correlate with unique signatures of asxl1 expression, whereby high Akt/mTor activity is associated with increased asxl1 expression. Furthermore, cblX mutations, including the c.344C > T variant, disrupted binding and activation to the zebrafish asxl1 promoter. Therefore, we hypothesized that the mTor dependent phenotypes in the co60 allele were Asxl1 dependent. To test this, we injected mRNA encoding mouse Asxl1 into single cell embryos harboring either the Sox2 or Gfap EGFP reporters. We then monitored the number of total NPCs and RGCs by flow cytometry. As shown in Fig. 8A, we did not detect any significant change in the number of Sox2 + or Gfap + cells after over expression of Asxl1. We did not anticipate changes in the number of Sox2 + cells after Asxl1 injection because this is an overlapping phenotype present in the co60 and co64 alleles. Furthermore, we were not surprised that the number of Gfap + cells was normal after injection because the co60 allele disrupts the expression of gfap and not cell number (Fig. 5E and 6F). However, we did not detect increased expression of gfap or sox2 relative to non-injected control (Fig. 8B). Importantly, injection of Asxl1 encoding mRNA induced blood cell phenotypes consistent with a previously described function for this gene in hematopoiesis (Gjini et al., 2019; Uni and Kurokawa, 2018) (Supplementary Fig. 1). The presence of blood cell phenotypes indicates the functional translation of Asxl1 mRNA after injection.
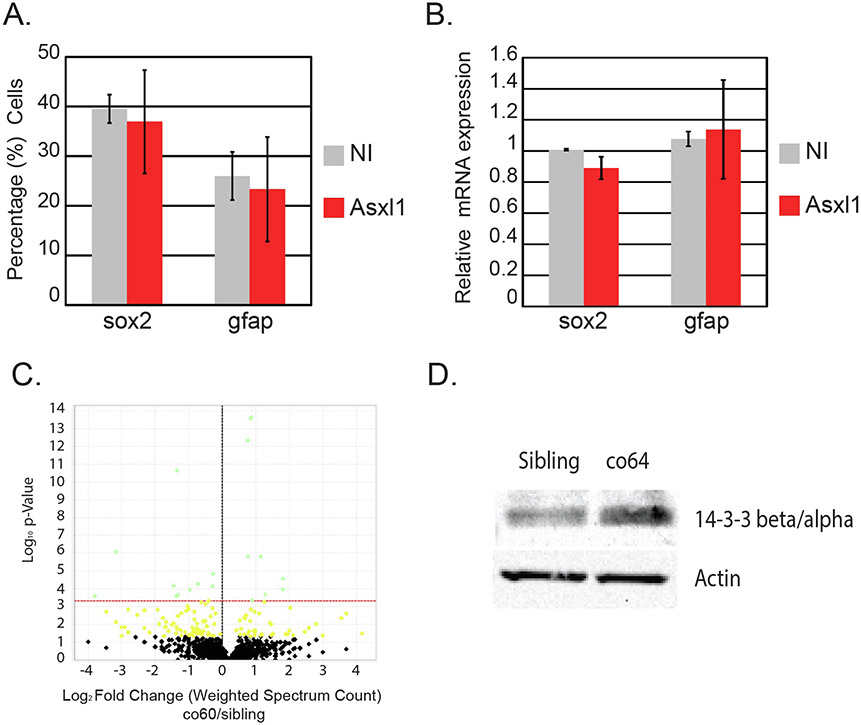
Fig. 8. Forced expression of Asxl1 does not drive NPC/RGC development but proteomics reveals differential expression of 14-3-3 βα in each cblX allele.
A. The number of Gfap + or Sox2 + cells were quantified by flow cytometry in non-injected (NI) Tg(gfap:EGFP), or Tg(sox2:2A:EGFP) injected with mRNA encoding the murine Asxl1 gene (Asxl1). Analysis was performed in two biological replicates. Error bars represent the standard error of the mean between biological replicates. N = 14–20/group across 2 biological replicates. B. The relative expression of gfap or sox2 was quantified in non-injected (NI) or wildtype embryos injected with mRNA encoding the murine Asxl1 gene (Asxl1). N = 12/group across 3 biological replicates. Error bars represent standard error of the mean from biological replicates. C. Mass spectrometry was performed to detect abnormal protein expression from N = 13 brain homogenates obtained from sibling wildtype or carriers of the hcfc1aco60/+. Volcano plot describes the proteins that were significantly different between groups. D. Western blot analysis was performed to determine the expression of 14-3-3 βα in total brain homogenates of the co64 allele or sibling control, β-actin was used as a loading control. N = 4/group/biological replicate. A total of 2 biological replicates were performed.
3.11. Proteome analysis reveals abnormal expression of ribosomal proteins and 14-3-3 βα in the co60 allele.
Over-expression of mouse Asxl1 did not alter RGC expression or number and therefore, we used proteomics analysis to identify novel mediators of Akt/mTor in the co60 allele. Our analysis identified a total of 2172 proteins that were differentially expressed with 159 of those proteins statistically different in the co60 allele. We identified many proteins belonging to biological processes such as hematopoiesis, erythrocyte development, the ribosome, and/or structural constituents of the ribosome. After secondary Benjamini-Hochberg correction, only 20 proteins were significantly different in the co60 allele relative to sibling control. Zebrafish Asxl1 protein was not detected as abnormal. Literature analysis of the 20 significant proteins demonstrated increased expression of ribosomal proteins: Rpl36a, Rpl38a, Rps7, Rpl18, Rpl9, Rpl30, Rpl18a, and Rpl3 (Supplementary Data File 1). Our identification of increased expression of proteins required for ribosome biogenesis is consistent with results obtained from a patient derived cblX-like knock-in allele (human THAP11 gene), which adds additional face validity to our model system (Chern et al., 2022). Additional literature characterization of the remaining proteins revealed a statistically significant decrease in the expression of 14-3-3 βα (Fig. 8C). 14-3-3 proteins are a family of signaling proteins that bind to phosphorylated serine and threonine residues. They are known to regulate brain development (Cornell and Toyo-oka, 2017) and have been shown to reduce AKT phosphorylation at threonine 308 in human cell lines (Gómez-Suárez et al., 2016). Based on the function of 14-3-3 proteins, we surmised that the co60 and co64 alleles would have opposite expression profiles as it relates to 14-3-3 βα. We used western blotting to determine the expression of 14-3-3 βα in the co64 allele and observed increased expression relative to sibling wildtype (Fig. 8D), which contrasts with reduced expression in the co60 allele (Fig. 8C). We next used the Harmonizome and The Encyclopedia of DNA Elements (ENCODE) databases to determine if human HCFC1 binds to and regulates the expression of 14-3-3- β/α. 14-3-3- β/α is encoded by the YWHAB gene and we validated by database that human HCFC1 binds to the YWHAB promoter (ENCODE Project Consortium, 2012; ENCODE Project Consortium, 2012; Luo et al., 2020; Rouillard et al., 2016). Thus, 14-3-3- β/α is an HCFC1 target gene and we detected abnormal expression in the co60 and co64 alleles indicating conservation of this regulatory mechanism.
3.12. Phenotypes in the co60 allele overlap with phenotypes in the c.344C > T cblX variant
We observed two independent mTor signatures in the co60 and co64 alleles. One of which is associated with increased mTor and increased expression of proteins required for ribosome biogenesis. Thus, we questioned which of our alleles, and which mTor signature most closely resembles the molecular mechanisms underlying cblX syndrome. To begin to address this, we performed proteomics analysis using the Tg(hsp701:HCFC1c.344C>T) at 5 DPF. We observed a statistically significant up regulation of proteins essential for ribosome biogenesis and elongation initiation factors which are required for protein translation (Fig. 9A and Supplemental File 2). These data support and align with previous literature and therefore, provided additional face validity of heat shock HCFC1 transgenes as systems to help understand cblX syndrome. We plotted the average spectral count from biological replicates for each of those proteins whose function is linked to ribosome biogenesis and translation. Each protein was increased in expression after induction of the c.344C > T patient variant (Fig. 9A and Supplemental File 2). We next hypothesized that an increase in these proteins was a consequence of increased mTor signaling in the Tg(hsp701:HCFC1c.344C>T). We compared pS6 phosphorylation in heat shocked induced and non-heat shock control samples. We observed increased pS6 kinase and increased total S6, with equivalent levels of β-actin as a loading control (Fig. 9B). We used β-actin as a control because we observed an increase in total S6 in multiple biological replicates. These data suggest that cblX mutations induce defects in the synthesis of proteins important for ribosomal biogenesis, consistent with previous literature.
Fig. 9. Proteomics demonstrates abnormal expression of ribosomal proteins and activation of Akt/mTor in the Tg(hsp701:HCFC1c.344C>T).
A. Graph depicting average spectral counts of genes encoding proteins associated with ribosomal biogenesis and translation initiation. Proteomics was performed in biological triplicates and average spectral count in non-heat shock (NHS) and heat shocked (HS:344) animals carrying the Tg(hsp701:HCFC1c.344C>T) allele were averaged and plotted. Proteins shown were statistically up regulated in the c.344C > T allele. Error bars represent standard deviation between spectral counts obtained from biological and technical replicates. A total of N = 45 fish across 2 biological replicates were used for NHS and HS (HS:344) and N = 46 fish across 2 biological replicates were used for NHS and HS Tg(hsp701:HCFC1) for analysis. B. Western blot was performed on whole brain homogenates with phospho-S6 kinase, total S6, or β-actin antibodies in no heat shock (NHS) or heat shocked animals carrying the Tg(hsp701:HCFC1c.344C>T) allele. Analysis was performed at 5 days post fertilization for both data in A and B. For western blot, a total N = 4/group/biological replicate was utilized in 2 biological replicates.
4. Discussion
Mutations in the human HCFC1 gene cause cblX syndrome and XLID (Gérard et al., 2015, 1; Huang et al., 2012; Jolly et al., 2015, 1; Koufaris et al., 2016; Scalais et al., 2017, 1; Yu et al., 2013). Various model systems have been developed and suggest that human HCFC1 is critical for the function of NPCs (Castro et al., 2020; Chern et al., 2022; Huang et al., 2012; Jolly et al., 2015; Minocha and Herr, 2019; Minocha et al., 2016a; Minocha et al., 2016b; Quintana et al., 2017). Most recently, Chern and colleagues characterized the cellular and molecular phenotypes associated with a cblX patient knock-in allele (Chern et al., 2022). These analyses focused on craniofacial development and demonstrated only limited characterization of brain development, likely due to sub-viability. Despite limitations associated with viability, analysis of a related disorder caused by mutations in the human THAP11 protein (p.F80L), which interacts with HCFC1, revealed increased expression of proteins associated with ribosome biogenesis. These studies complement existing work in the zebrafish in which Castro and colleagues found brain phenotypes to be mmachc independent (Castro et al., 2020).
We sought to build on these findings by comparing the molecular and cellular phenotypes present in 2 zebrafish hcfc1a germline mutant alleles. We have previously characterized the cellular and molecular phenotypes present in a nonsense allele of hcfc1a (co60). However, cblX is the result of missense mutations and therefore, we created the co64 allele, the first homozygous viable germline mutant in vivo. We demonstrated that the co60 and co64 alleles differentially affect Hcfc1a protein expression. The co60 allele reduces total protein expression whereas the co64 allele does not, producing a protein with a missense mutation in the kelch domain. Multiple different mutations in human HCFC1 have been reported in the literature including those that affect dosage and cause XLID (Huang et al., 2012). The co60 reduces protein expression and the dosage of zebrafish Hcfc1a resulting in cellular and molecular phenotypes that are unique from the co64 allele. Protein analysis confirmed the co64 allele did not affect dosage and genomic sequencing confirmed that the co64 allele resulted in a 2 amino acid modification in the N-terminal kelch domain. It is not surprising that mutations affecting dosage have distinct and overlapping phenotypes when compared with those that alter protein function rather than dosage. Particularly as it is related to human HCFC1, as the protein is proteolytically cleaved and each fragment is known to have unique functions (Julien and Herr, 2003; Julien and Herr, 2004; Luciano and Wilson, 2002; Mangone et al., 2010). In addition, previous studies have established that unique mutations in other genes (GLI3) can cause different diseases (Johnston et al., 2005).
Our comparison of the co60 and co64 alleles revealed contrasting phenotypes at the level of Akt/mTor activation. Nonsense mutation of hcfc1a, which reduced expression of total Hcfc1a protein, resulted in hyperphosphorylation and activation of Akt and mTor. These data strongly support the observations from Chern and colleagues, as activation of mTor promotes translation and ribosome biogenesis, which was elevated in a mouse knock-in allele of Thap11 (cblX-like) (Chern et al., 2022). Thus, the co60 is a valid model system to study the mechanisms by which changes in protein dosage affect brain development and understand the mechanisms driving increased expression of ribosome biogenesis. Mutation of human THAP11 causes a cblX-like syndrome (Quintana et al., 2017), but whether the underlying mechanisms associated with mutation of THAP11 are identical to cblX syndrome has not been elucidated. The expression level of proteins associated with ribosome biogenesis was not directly tested in cblX, but genetic complementation assays with germline mutants that cause ribosomopathies were performed. Here we more directly tested the role of ribosome biogenesis using the Tg(hsp701:HCFC1c.344C>T) transgene. This allele conditionally, but ubiquitously expresses the human patient variant previously modeled by Chern and colleagues (Chern et al., 2022). We observed increased expression of proteins associated with ribosome biogenesis and translation. Therefore, we provide face validity for the Tg(hsp701:HCFC1c.344C>T) as a model of cblX syndrome.
Interestingly, we observed hypophosphorylation of Akt/mTor after missense mutation of hcfc1a. These data contrast with the co60 allele and the Tg(hsp701:HCFC1c.344C>T) allele. We anticipate that the mechanisms present in the co64 allele also differ from the genetic knock-in created by Chern and colleagues (Chern et al., 2022). This is because the co64 deletes an in frame arginine and results in two amino acid changes within the kelch domain. Interestingly, despite differences in Akt/mTor signaling, the co60 and co64 have overlapping effects on the number of NPCs. These overlapping phenotypes provide evidence that Hcfc1a is essential for proper control of the NPC compartment and that the presence of similar phenotypes across different alleles is indicative that these phenotypes are a direct result of mutation in Hcfc1a and not due to any unknown off-target effect. The two zebrafish orthologs, Hcfc1a and Hcfc1b, contain > 75 % sequence identity with human HCFC1 (Quintana et al., 2014). Knockdown of hcfc1a and hcfc1b by morpholino resulted in an increase in the number of NPCs (Quintana et al., 2017), providing additional validity for both the co60 and co64 alleles. While these two genes have overlapping NPC functions, they have divergent craniofacial functions (Quintana et al., 2014). Therefore, future studies will be aimed to characterize the functions of both paralogs in NPC development. For example, we observed differential expression of the zebrafish asxl1 gene in the co60 and co64 alleles. These data warrant additional studies as to the function of asxl1 in both alleles. Our zebrafish studies are supported by in vitro knockdown of Hcfc1, which resulted in an increase in NPCs (Jolly et al., 2015). Given the strong validity of each allele, our data raise the possibility that unique mutations in hcfc1a can lead to overlapping and individual phenotypes. We do not yet understand if individual human disease variants will have differential regulation of mTOR, but such studies are underway and may reveal unique signatures of AKT/mTOR across disease variants. A role for human mTOR signaling in cblX is further supported by a recent review that suggests the phenotypes of multiple neurodevelopmental disorders converge on the mTOR pathway (Parenti et al., 2020).
Previously studies in the co60 allele uncovered increased expression of asxl1, which encodes a chromatin binding protein capable of interacting with Akt in the cytoplasm and promoting Akt phosphorylation (Castro et al., 2020; Youn et al., 2017). Interestingly, asxl1 transcript was increased at the transcriptional level, but not elevated according to proteomics analysis. This can be attributed to unknown post-transcriptional mechanisms or the stringency of bioinformatics performed at the protein level. Additionally, whole brain homogenates were analyzed, which can cause limited sensitivity or ability to detect protein levels above background. We previously used inhibitors of PI3K, an upstream regulator of AKT phosphorylation to indirectly link the NPC phenotypes present in the co60 allele to zebrafish Akt signaling (Castro et al., 2020). Castro and colleagues suggest that PI3K regulates NPC number, therefore we followed up on analyzing the role of AKT/mTOR, a common downstream target of PI3K. However, inhibition of Akt/mTor activity in zebrafish did not restore NPCs. To note, downstream targets of PI3K are vast and therefore, additional studies with which inhibition of other targets are underway. Here we uncovered differential expression of asxl1 in the co60 and co64 alleles. Most interesting was the fact that asxl1 expression was correlated with hyper or hypo activation of Akt/mTor in each allele. We hypothesized that dysregulation of asxl1 in cblX was the mechanism by which Hcfc1a regulates RGC development. However, forced expression of murine Asxl1 did not result in changes to the NPC or RGC populations. These data are inconsistent with morpholino mediated knockdown of asxl1 in the co60 allele, which led to a full restoration of the number of NPCs (Castro et al., 2020). At the present time we cannot explain the mechanism by which morpholino knockdown of asxl1 restored NPC phenotypes. But it is well-known that morpholinos can have off-target effects.
Here we found that heat shock directed expression of wildtype human HCFC1 caused an increase in asxl1 expression, and we detected human HCFC1 bound to the zebrafish asxl1 promoter. These data suggest that asxl1 is a bonafide target of human HCFC1. Since the cblX mutation p.Ala115Val disrupted binding of HCFC1 to this promoter, we investigated whether HCFC1 regulated NPC and RGC phenotypes via modulation of asxl1 expression. However, forced expression of murine asxl1 did not affect the total numbers of NPCs or RGCs and had no effect on gfap expression. Interestingly, we also noted increased asxl1 expression upon over expression of wildtype HCFC1. In our previous study, we also observed decreased sox2 and gfap expression after over expression of wildtype HCFC1 (Castro et al., 2020). Thus, asxl1 expression did not correlate with changes in sox2 and gfap expression, which supports the data we observed after injection of murine asxl1 mRNA. Our collective data suggests that although asxl1 expression is disrupted in the co60 and co64 alleles, it is not likely to be the sole mediator of the Sox2 + NPC or Gfap + RGC phenotypes in cblX or related disorders. Hence, we did not attempt to restore the phenotypes present in the co64 allele with forced asxl1 expression.
Our proteomics analysis of the co60 allele uncovered increased expression of proteins essential for ribosome biogenesis (Supplementary Data File 1). These data are consistent with proteomics analysis using the Tg(hsp701:HCFC1c.344C>T) allele where we identified increased expression of several elongation initiation factor proteins (Supplementary File 2). Given that these two alleles demonstrate phenotypes consistent with the recent knock-in mouse models, we propose that both are valid systems to understand the mechanisms by which mutations in cblX cause disease. We also identified a second putative regulator of zebrafish Akt signaling using proteomics, the 14-3-3- β/α protein. 14-3-3 proteins have been implicated in brain development (Cornell and Toyo-oka, 2017) and some members of the larger family have been shown to increase the number of NPCs (Cornell and Toyo-oka, 2017) and inhibit AKT phosphorylation in cell lines (Gómez-Suárez et al., 2016). We found reduced expression of 14-3-3- β/α in the co60 allele and increased expression in the co64 allele, which correspond with hyperactivated Akt/mTor and reduced Akt/mTor signaling, respectively. We validated through available data in the ENCODE database that human HCFC1 binds to the human YWHAB promoter. Future experiments to characterize the role of 14-3-3- β/α in cblX syndrome are warranted.
Collectively, our data suggest that distinct types of mutations, even in the same domain of Hcfc1a, can have contrasting phenotypes at the level of Akt/mTor, asxl1 expression, and gfap expression. Initially, the presence of contrasting phenotypes may call into question the physiological relevance of the co64 allele because the phenotypes in the co60 allele, such as increased synthesis of proteins required for ribosomal biogenesis, have been validated with recent patient derived models (Chern et al., 2022). However, cell type specific deletion of mouse Hcfc1 in the Nkx + 2.1 + sub-compartment of progenitors is associated with reduced mouse Gfap expression (Minocha and Herr, 2019), data that supports reduced zebrafish gfap expression in the co64 allele. Interestingly, the mouse Thapl1 p.F80L allele demonstrated decreased Gfap expression after knock-in. These data provide additional evidence for a role of HCFC1 and its partner THAP11 in the regulation of Gfap expression. The mouse Thap11 F80L mutation also has increased expression of proteins needed for ribosomal biogenesis. Interestingly, the mTor dependent regulation of gfap expression in the co60 allele combined with decreased mTor and gfap expression in the co64 allele further substantiates the validity for the co64 allele as a model to inform about cblX syndrome. Unfortunately, the co64 could not be directly rescued because over expression of human HCFC1 and heat shock induced expression of human HCFC1 cause independent phenotypes (Castro et al., 2020; Quintana et al., 2017), complicating the interpretation of restoration assays. However, the co60 and co64 alleles have been outcrossed for 15–20 generations each, limiting the potential effects of off-target effects.
5. Conclusions
In conclusion, our study uncovers differential regulation of zebrafish Akt/mTor across different types of germline mutations in the hcfc1a gene. Our data correlate contrasting levels of Akt/mTor to unique cellular defects of the Gfap + RGC cellular compartment. Collectively, this work provides a fundamental mechanism (AKT/mTOR) by which increased synthesis of proteins required for ribosome biogenesis may occur in cblX syndrome. However, our work also suggests that independent mutations with unique effects on overall protein expression/function could cause disease by unique mechanisms that may or may not include AKT/mTOR perturbation. Future studies characterizing the function of unique disease variants in various animal models are underway.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Bruce Appel and Tamim Shaikh for their sharing of model organisms associated with this manuscript. Special thanks to Yahir Davila, Jennifer Davila, Isaiah Perez, and Nayeli Reyes-Nava for their role in genotyping and animal husbandry. Nayeli Reyes-Nava played a significant role in the mentoring and teaching of Valeria Virrueta, an undergraduate student in the laboratory. Her mentorship facilitated the success of experiments performed by Valeria Virrueta. The proteomics analysis was performed in conjunction with the UTEP core facilities, and we graciously thank the directorship of Dr. Igor Almeida and Dr. Renato Aguilera. Additional support was provided by the College of Science and Drs. Robert Kirken and Michael Kenney. Additional feedback and commentary were provided by Dr. Charlotte Vines prior to submission. Thank you to Briana Pinales for figure development and editing. Flow cytometry was performed with the help and guidance of Dr. Armando Varela and Dr. Charles Spencer. All members of the Quintana laboratory from 2019-present aided in animal husbandry and fish care to help facilitate the work described.
Funding
Partial funding for this project was provided by K01NS099153 to AMQ, R03DE029517 to AMQ, NIMHD Grant No 5U54MD007592 to University of Texas El Paso, NIGMS linked awards RL5GM118969, TL4GM118971, and UL1GM118970 to the University of Texas El Paso. VLC was partially supported by 1F99NS125690-01A1 and the Keelung-Hong Fellowship. We thank the Biomolecule Analysis and Omics Unit (BAOU) at BBRC/UTEP for the full access to the nanoUHPLC-ESI-Q Exactive Plus orbitrap MS system used in this study. AMQ was provided pilot grant funds that partially funded this award as a component of the 5U54MD007592 award. The content is solely the responsibility of the authors and does not represent the official views of the funding agencies.
Abbreviations:
- cblX
methylmalonic acidemia and homocysteinemia cblX type
- XLID
X-linked intellectual disability
- NPCs
neural precursor cells
- RGC
radial glial cells
- mTor
mechanistic target of rapamycin
- PI3K
phosphatidylinositol-3-kinase
- Akt
protein kinase B
- pAkt308
phosphorylated protein kinase B @ amino acid 308
- EGFP
Enhanced green fluorescent protein
- ChIP
chromatin immunoprecipitation
- PCR
polymerase chain reaction
- IgG
immunoglobulin G
- ENCODE
Encyclopedia of DNA Elements
- DPF
days post fertilization
- HPF
hours post fertilization
- ZFIN
Zebrafish Information Network
- CRISPR/Cas9
Clustered Regularly Interspaced Short Palindromic Repeats/Cas9
- DNA
deoxyribonucleic acid
- FWD
forward
- REV
reverse
- RIPA
radio immunoprecipitation assay buffer
- SDS
sodium dodecyl sulfate
- ECL
enhanced chemiluminescence
- qPCR
quantitative polymerase chain reaction
- TCA
trichloroacetic acid
- LC/MS
liquid chromatography-mass spectrometry
- RCF
relative centrifugal force
- ACN
acetonitrile
- FA
formic acid
- PD
proteome discover
- FDR
false discovery rate
- EDTA
ethylenediaminetetraacetic acid
- NHS
no heat shock
- HS
heat shock
Inclusion and diversity
Six of the authors in this manuscript, including the corresponding authors and senior author, self-identifies as an underrepresented ethnic minority in science. All authors included in this manuscript support inclusive, diverse, equitable and responsible conduct of research.
Footnotes
Institutional review board statement
All procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas El Paso, protocol number 811869-5. Methods for euthanasia and anesthesia were performed according to guidelines from the 2020 American Veterinary Medical Association guidebook.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2023.147290. Supplemental File 1 contains an excel spreadsheet of the significant and differentially expressed proteins in the co60 allele generated from proteomics analysis. Supplemental File 2 is an excel spreadsheet with multiple worksheets containing complete analysis of differentially expressed proteins in the Tg(hsp701:HCFC1c.344C>T) allele generated after proteomics. Supplemental Fig. 1 is a word document containing image analysis and figure legend description of larvae injected with murine Asxl1 mRNA.
Data availability
All data associated with this publication will be publicly available through database or contact with the corresponding authors upon publication.
Data will be made available on request.
References
- An S, Park U-H, Moon S, Kang M, Youn H, Hwang J-T, Kim E-J, Um S-J, 2019. Asxl1 ablation in mouse embryonic stem cells impairs neural differentiation without affecting self-renewal. Biochem. Biophys. Res. Commun 508, 907–913. [DOI] [PubMed] [Google Scholar]
- Bresciani E, Broadbridge E, Liu PP, 2018. An efficient dissociation protocol for generation of single cell suspension from zebrafish embryos and larvae. MethodsX 5, 1287–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro VL, Reyes JF, Reyes-Nava NG, Paz D, Quintana AM, 2020. Hcfc1a regulates neural precursor proliferation and asxl1 expression in the developing brain. BMC Neurosci 21, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern T, Achilleos A, Tong X, Hill MC, Saltzman AB, Reineke LC,Chaudhury A, Dasgupta SK, Redhead Y, Watkins D, et al. , 2022. Mutations in Hcfc1 and Ronin result in an inborn error of cobalamin metabolism and ribosomopathy. Nat Commun 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell B, Toyo-oka K, 2017. 14-3-3 Proteins in Brain Development: Neurogenesis, Neuronal Migration and Neuromorphogenesis. Front Mol Neurosci 10, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaene H, Praz V, Lhôte P, Lopes M, Herr W, 2020. THAP11F80L cobalamin disorder-associated mutation reveals normal and pathogenic THAP11 functions in gene expression and cell proliferation. PLoS One 15, e0224646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passed M, Chu L-F, Songyang Z, Thomson JA, Zwaka TP, 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133, 1162–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP, 2010. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24, 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, Rubinsztein DC, 2011. Zebrafish as a model to understand autophagy and its role in neurological disease. Biochim Biophys Acta 1812, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Bourdelais F, Durand S, 2020. Ribosome and Translational Control in Stem Cells. Cells 9, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard M, Morin G, Bourillon A, Colson C, Mathieu S, Rabier D, Billette de Villemeur T, Ogier de Baulny H, Benoist JF, 2015. Multiple congenital anomalies in two boys with mutation in HCFC1 and cobalamin disorder. Eur J Med Genet 58, 148–153. [DOI] [PubMed] [Google Scholar]
- Gjini E, Jing C-B, Nguyen AT, Reyon D, Gans E, Kesarsing M, Peterson J, Pozdnyakova O, Rodig SJ, Mansour MR, et al. , 2019. Disruption of asxl1 results in myeloproliferative neoplasms in zebrafish. Dis Model Mech 12, dmm035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Suárez M, Gutiérrez-Martínez IZ, Hernández-Trejo JA, Hernández-Ruiz M, Suárez-Pérez D, Candelario A, Kamekura R, Medina-Contreras O, Schnoor M, Ortiz-Navarrete V, et al. , 2016. 14-3-3 Proteins regulate Akt Thr308 phosphorylation in intestinal epithelial cells. Cell Death Differ 23, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jolly LA, Willis-Owen S, Gardner A, Kumar R, Douglas E, Shoubridge C, Wieczorek D, Tzschach A, Cohen M, et al. , 2012. A Noncoding, Regulatory Mutation Implicates HCFC1 in Nonsyndromic Intellectual Disability. Am. J. Hum. Genet 91, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudish LI, Blasky AJ, Appel B, 2013. miR-219 regulates neural precursor differentiation by direct inhibition of apical par polarity proteins. Dev. Cell 27, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, et al. , 2005. Molecular and Clinical Analyses of Greig Cephalopolysyndactyly and Pallister-Hall Syndromes: Robust Phenotype Prediction from the Type and Position of GLI3 Mutations. Am J Hum Genet 76, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly LA, Nguyen LS, Domingo D, Sun Y, Barry S, Hancarova M, Plevova P, Vlckova M, Havlovicova M, Kalscheuer VM, et al. , 2015. HCFC1 loss-of-function mutations disrupt neuronal and neural progenitor cells of the developing brain. Hum. Mol. Genet 24, 3335–3347. [DOI] [PubMed] [Google Scholar]
- Julien E, Herr W, 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22, 2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W, 2004. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol. Cell 14, 713–725. [DOI] [PubMed] [Google Scholar]
- Kapuria V, Röhrig UF, Bhuiyan T, Borodkin VS, van Aalten DMF, Zoete V, Herr W, 2016. Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase:UDP-GlcNAc complexes. Genes Dev 30, 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta C, Daniel K, Johnstone DL, Mongeon K, Ban K, LeBlanc S, MacLeod S, Et-Tahiry K, Ekker M, MacKenzie A, et al. , 2018. High-throughput DNA Extraction and Genotyping of 3dpf Zebrafish Larvae by Fin Clipping. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufaris C, Alexandrou A, Tanteles GA, Anastasiadou V, Sismani C, 2016. A novel HCFC1 variant in male siblings with intellectual disability and microcephaly in the absence of cobalamin disorder. Biomed Rep 4, 215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B, 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Luciano RL, Wilson AC, 2002. An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc. Natl. Acad. Sci. U.S.A 99, 13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. , 2020. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res 48, D882–D889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Myers MP, Herr W, 2010. Role of the HCF-1 basic region in sustaining cell proliferation. PLoS ONE 5, e9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne A-C, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, et al. , 2010. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem 285, 13364–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha S, Herr W, 2019. Cortical and Commissural Defects Upon HCF-1 Loss in Nkx2.1-Derived Embryonic Neurons and Glia. Dev Neurobiol 79, 578–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha S, Sung T-L, Villeneuve D, Lammers F, Herr W, 2016a. Compensatory embryonic response to allele-specific inactivation of the murine X-linked gene Hcfc1. Developmental Biology 412, 1–17. [DOI] [PubMed] [Google Scholar]
- Minocha S, Bessonnard S, Sung T-L, Moret C, Constam DB, Herr W, 2016b. Epiblast-specific loss of HCF-1 leads to failure in anterior-posterior axis specification. Dev. Biol 418, 75–88. [DOI] [PubMed] [Google Scholar]
- Parenti I, Rabaneda LG, Schoen H, Novarino G, 2020. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 43, 608–621. [DOI] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafrenière RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, et al. , 2011. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol. Psych 16, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Redin C, Mandel J-L, 2013. XLID-Causing Mutations and Associated Genes Challenged in Light of Data From Large-Scale Human Exome Sequencing. Am. J. Hum. Genet 93, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana AM, Liu F, O’Rourke JP, Ness SA, 2011. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer 11, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana AM, Geiger EA, Achilly N, Rosenblatt DS, Maclean KN, Stabler SP, Artinger KB, Appel B, Shaikh TH, 2014. Hcfc1b, a zebrafish ortholog of HCFC1, regulates craniofacial development by modulating mmachc expression. Dev. Biol 396, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana AM, Yu H-C, Brebner A, Pupavac M, Geiger EA, Watson A, Castro VL, Cheung W, Chen S-H, Watkins D, et al. , 2017. Mutations in THAP11 cause an inborn error of cobalamin metabolism and developmental abnormalities. Human Mol Genet 26, 2838–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG and Ma’ayan A, 2016. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016, baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalais E, Osterheld E, Weitzel C, De Meirleir L, Mataigne F, Martens G, Shaikh TH, Coughlin CR, Yu H-C, Swanson M, et al. , 2017. X-Linked Cobalamin Disorder (HCFC1) Mimicking Nonketotic Hyperglycinemia With Increased Both Cerebrospinal Fluid Glycine and Methylmalonic Acid. Pediatr. Neurol 71, 65–69. [DOI] [PubMed] [Google Scholar]
- Uni M, Kurokawa M, 2018. Role of ASXL1 mutation in impaired hematopoiesis and cellular senescence. Oncotarget 9, 36828–36829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HS, Kim T-Y, Park U-H, Moon S-T, An S-J, Lee Y-K, Hwang J-T, Kim E-J, Um S-J, 2017. Asxl1 deficiency in embryonic fibroblasts leads to cellular senescence via impairment of the AKT-E2F pathway and Ezh2 inactivation. Sci Rep 7, 5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-C, Sloan JL, Scharer G, Brebner A, Quintana AM, Achilly NP, Manoli I, Coughlin CR 2nd, Geiger EA, Schneck U, et al. , 2013. An X-Linked Cobalamin Disorder Caused by Mutations in Transcriptional Coregulator HCFC1. Am. J. Hum. Genet 93, 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this publication will be publicly available through database or contact with the corresponding authors upon publication.
Data will be made available on request.