Abstract
Background
In S. cerevisiae, the mitotic exit network (MEN) proteins, including the Polo-like protein kinase Cdc5 and the protein phosphatase Cdc14, are required for exit from mitosis. In pre-anaphase cells, Cdc14 is sequestered to the nucleolus by Net1 as a part of the RENT complex. When cells are primed to exit mitosis, the RENT complex is disassembled and Cdc14 is released from the nucleolus.
Results
Here, we show that Cdc5 is necessary to free nucleolar Cdc14 in late mitosis, that elevated Cdc5 activity provokes ectopic release of Cdc14 in pre-anaphase cells, and that the phosphorylation state of Net1 is regulated by Cdc5 during anaphase. Furthermore, recombinant Cdc5 and Xenopus Polo-like kinase can disassemble the RENT complex in vitro by phosphorylating Net1 and thereby reducing its affinity for Cdc14. Surprisingly, although RENT complexes containing Net1 mutants (Net1(7m) and Net1(19m') lacking sites phosphorylated by Cdc5 in vitro are refractory to disassembly by Polo-like kinases in vitro, net1(7m) and net1(19m') cells grow normally and exhibit only minor defects in releasing Cdc14 during anaphase. However, net1(19m') cells exhibit a synergistic growth defect when combined with mutations in CDC5 or DBF2 (another MEN gene).
Conclusions
We propose that although Cdc5 potentially disassembles RENT by directly phosphorylating Net1, Cdc5 mediates exit from mitosis primarily by phosphorylating other targets. Our study suggests that Cdc5/Polo is unusually promiscuous and highlights the need to validate Cdc5/Polo in vitro phosphorylation sites by direct in vivo mapping experiments.
Background
In the budding yeast Saccharomyces cerevisiae, the mitotic exit network (MEN) plays a pivotal role in driving cells from mitosis to G1. Identified components of the MEN include Tem1 (a GTP-binding protein), Lte1 (a putative guanine nucleotide releasing factor), Nud1 (a spindle-pole-body protein), Cdc15 (a protein kinase), Dbf2/Mob1 (a protein kinase complex), Cdc5 (a Polo-like protein kinase), and Cdc14 (a protein phosphatase). When cells harboring conditional-lethal temperature-sensitive (ts) mutations in any of these proteins are shifted to the restrictive temperature, they uniformly arrest in late anaphase as large-budded cells with segregated chromosomes, fully elongated microtubule spindles, and in all tested cases, elevated Cdc28/Clb2 protein kinase activity (Cdc28 is the major cyclin-dependent kinase Cdk1 and Clb2 the major mitosis-specific cyclin of budding yeast) (reviewed in [1], [2]).
The protein phosphatase Cdc14 is likely to be the most downstream element in the MEN because it can act directly on cell cycle regulators to promote exit from mitosis. By counteracting Cdk1-mediated phosphorylations, Cdc14 activates both Hct1/Cdh1, which targets Clb2 for degradation [3-5], and Swi5, which turns on the transcription of Cdk1 inhibitor Sic1 [6,3]. Degradation of Clb2 and accumulation of Sic1 act in concert to eliminate mitotic Cdk activity, which is required for exit from mitosis [7]. Thus, elucidating how Cdc14 is regulated holds the key to understanding how cells exit mitosis. In pre-anaphase cells, Cdc14 is confined to the nucleolus as a subunit of the RENT complex (which also contains Net1/Cfi1 and Sir2), and its phosphatase activity is inhibited by Net1 [8-11]. Damaged DNA and mis-oriented mitotic spindles delay exit from mitosis (reviewed in [12]), but after errors have been rectified or bypassed, Lte1, Nud1, and possibly other unidentified factors [13] promote Tem1 activation. Tem1 recruits Cdc15 to the spindle pole body, which in turn recruits and phosphorylates the Dbf2/Mob1 protein kinase complex, thereby switching it on [2,14-17]. The Tem1-signaling pathway eventually triggers disassembly of the RENT complex, enabling the release of Cdc14 (and a portion of Sir2) from Net1. Active Cdc14 diffuses into the nucleus and cytoplasm to extinguish mitotic Cdk activity and to promote exit from mitosis.
The relative position of the Polo-like kinase Cdc5 in the MEN is obscure. Cdc5 is required for full activation of the Dbf2-Mob1 protein kinase complex [15-17], and has been proposed to impinge on the MEN both upstream and downstream of Tem1 [15-18]. During exit from mitosis, Cdc5 is required for Clb2 destruction, and similar to over-expression of Cdc14 [3], induction of Cdc5 triggers precocious Clb2 degradation in pre-anaphase cells [19,20]. In addition to its role in exit from mitosis, Cdc5 has other functions during mitosis in budding yeast. Depletion of Cdc5 or Polo-like kinase in cdc5▲ cells arrests a significant fraction of the cells in early anaphase with short mitotic spindles [20,21], and phosphorylation of the cohesin subunit Scc1 by Cdc5 enhances the efficiency of Scc1 cleavage by the separase Esp1 during the metaphase to anaphase transition [22].
Although many proteins are known to be essential for exit from mitosis, the factor that sets Cdc14 free from the nucleolus still remains elusive. In this study, we assess the roles of Cdc5 in disassembly of the RENT complex and exit from mitosis.
Results
Release of Cdc14 from the nucleolus in late anaphase requires Cdc5
In S. cerevisiae, disassembly of the RENT complex and the subsequent release of Cdc14 from the nucleolus drive cells to exit mitosis [8,9]. This event is thought to require two independent signals: an unknown mitosis-specific signal and a signal from the mitotic exit network (MEN) proteins including Tem1, Cdc15, and Dbf2 [8,9]. The Polo-like kinase Cdc5 also belongs to the MEN, thus we tested whether it too is required for release of Cdc14 from the nucleolus by using indirect immunofluorescence to compare Cdc14 localization patterns in wild type and cdc5-1 cells. Wild type cells with short spindles were in pre-anaphase and showed focal nucleolar Cdc14 staining, and those with elongated mitotic spindles were in late anaphase and showed diffused Cdc14 staining (Figure 1, Rows 1 and 4; [8,9]). In contrast, cdc5-1 cells uniformly exhibited focal Cdc14 staining whether they were grown exponentially at permissive temperature (25°C) or arrested in late mitosis at nonpermissive temperature (33°C) (Figure 1, Rows 2 and 5). Furthermore, in all (including late anaphase) cdc5-1 cells, Cdc14 co-localized with A190, a nucleolar protein (Figure 1, Rows 3 and 6), suggesting that when CDC5 is compromised, most or all Cdc14 fails to vacate the nucleolus in late anaphase.
Figure 1.
Release of Cdc14 from the nucleolus requires Cdc5. CDC14-HA3 cells in wild type (+) (RJD 1191) or cdc5-1 (RJD 1217) background were grown at 25°C, and a portion of the cultures were further shifted to 33°C for three hours to arrest cdc5-1 in late anaphase. Cells were subjected to indirect immunofluorescence with HA.11 to visualize Cdc14-HA3 (Column 1) and either anti-tubulin or anti-A190 antibodies to visualize the mitotic spindles and the nucleoli, respectively (Column 2). The positions of nuclei, as indicated by DAPI staining, are shown in Column 3.
Overexpression of Cdc5 evicts Cdc14 from the nucleolus
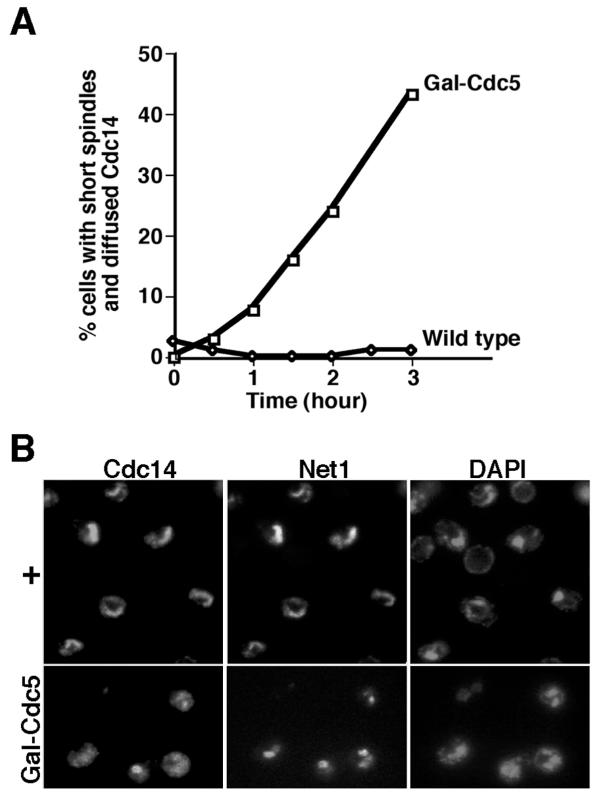
To identify candidate proteins that directly disrupt the RENT complex, we examined whether overexpression of the MEN proteins could release Cdc14 from the nucleolus. We first tested Cdc5, because Cdc5 is required for Cdc14 release (Figure 1), and because overexpression of Cdc5 triggers ectopic degradation of Clb2 [19,20], which normally depends upon active Cdc14 [3]. These experiments employed a GAL-CDC5(DB▲) strain, in which expression of Cdc5(DB▲), a stabilized version of Cdc5 lacking the destruction box, was driven by the inducible GAL promoter [20]. Cells were arrested in G1 by α-factor, supplemented with galactose to induce the GAL promoter, and released into galactose-containing medium. Cdc14 was focal in almost all cells with short mitotic spindles, but became diffused in a significant fraction of these cells after Cdc5(DB▲) was induced (Figure 2A). A similar result was obtained if the total number of cells with diffused Cdc14 was plotted instead (data not shown). To address whether overexpression of Cdc5 could also force Cdc14 out of the nucleolus in cells arrested at a cell cycle stage where Cdc14 is nucleolar, we arrested cells in metaphase using the microtubule polymerization inhibitor nocodazole. After the expression of Cdc5(DB▲) was induced in nocodazole-arrested cells, Cdc14 diffused beyond the domain of Net1 in ~49% of cells (Figure 2B). In contrast, GAL-driven overexpression of Dbf2/(GST-Mob1) protein kinase complex did not dislodge Cdc14 from the nucleolus (data not shown). Taken together, these data suggest that overexpression of Cdc5 – but not the putative downstream-most MEN component Dbf2/Mob1 – activates premature release of Cdc14 from the nucleolus.
Figure 2.
Overexpression of Cdc5(DB▲) triggers Cdc14 release. (A) Overexpression of Cdc5(DB▲) promotes release of Cdc14 from the nucleolus even in cells with short spindles. CDC14-MYC9 cells with (WY333) or without (WY201) integrated GAL-CDC5(DB▲) were grown in YP + 2% raffinose (GAL promoter uninduced), arrested in G1 by α-factor, supplemented with galactose to 2% for 0.5 hour (GAL promoter induced), and released into YP + 2% galactose at time = 0. At indicated time points after release, samples were fixed, and subjected to indirect immunofluorescence with anti-tubulin and 9E10 antibodies to visualize the mitotic spindles and Cdc14-Myc9, respectively. The percentages of cells with short spindles and diffused Cdc14 were calculated. (B) Overexpression of Cdc5(DB▲) triggers Cdc14 release in cells arrested in mitosis. NET1-MYC9 cells with (WY331) or without (WY53) integrated GAL-CDC5(DB▲) were grown to exponential in YP + 2% raffinose, arrested in mitosis by nocodazole (10 μg/ml final), and supplemented with galactose to 2% for four hours. Samples were subjected to indirect immunofluorescence with anti-Cdc14 and 9E10 (to detect Net1-Myc9).
Cdc5 influences the phosphorylation state of Net1 in vivo
Because the overexpression studies implicated Cdc5 as a promising candidate for participating directly in disassembly of the RENT complex, we evaluated further whether the phosphorylation state of Net1 was affected by Cdc5. Net1-Myc9 from a cdc14-1 culture at 37°C (nonpermissive temperature) was hyperphosphorylated and migrated slower in SDS-PAGE [8] than that from identically treated wild-type or cdc5-1 cultures (Figure 3, compare Lane 4 with Lanes 1 and 2). Furthermore, this hyperphosphorylation of Net1 in cdc14-1 cells was significantly reduced upon simultaneous inactivation of CDC5 (Figure 3, compare Lanes 3 and 4), suggesting that Cdc5 is at least partially responsible for the phosphorylation state of Net1 in late mitosis.
Figure 3.
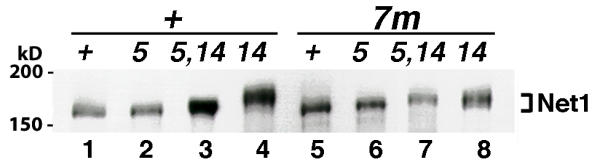
Phosphorylation of Net1 is modulated by Cdc5. NET1-myc9 (+, Lanes 1-4) in the wild-type ("+", RJD1349), cdc5-1 ("5", RJD1417), cdc14-1 ("14", RJD1408) and cdc5-1 cdc14-1 ("5, 14", WY334) backgrounds or net1(7m: S30, S31, S48, S60, S64, S242, S335 -> A)-myc9 (7m, Lanes 5-8) cells in corresponding backgrounds ("+", WY347; "5", WY363; "14", WY351; "5, 14", WY378) were grown in YP + 2% glucose + 1 M sorbitol at 25°C (cdc5-1 cdc14-1 cells seemed to grow more consistently in sorbitol-containing media), and shifted to 37°C for 3 hours when > 90% of cdc5, cdc14, and cdc5 cdc14 cells were arrested at the large-bud stage. Cell extracts were prepared by SDS-boiling method [8], fractionated on a 7.5% SDS-polyacrylamide gel, and Net1-Myc9 was detected by immunoblotting with 9E10 antibodies. The reduced mobility of Net1 isolated from cdc14-1 compared to wild type cells is due to phosphorylation [8].
Our analysis of Cdc5's role in release of Cdc14 from the RENT complex revealed that Cdc5 is required for disassembly of RENT in late anaphase, that Cdc5 can provoke premature disassembly of RENT in pre-anaphase cells, and that Cdc5 controls (at least in part) the phosphorylation state of Net1 during anaphase. Moreover, the arrest of cdc5-1, cdc5-2, cdc5-4, and cdc5-7 temperature-sensitive mutants in late mitosis was potently suppressed by the TAB6-1 allele (Park et al., submitted), which encodes a form of Cdc14 that binds weakly to Net1 and is released from Net1 during mitosis even in the absence of MEN function [23]. Taken together, these data suggested the hypothesis that cdc5 mutants arrest in late anaphase because inadequate Cdc5-dependent phosphorylation of Net1 results in failure to release active Cdc14 from the RENT complex.
Polo-like kinases can disrupt the RENT complex in vitro
Both Net1 and Cdc14 are extensively phosphorylated by Cdc5 in vitro (Figure 4A). To test if Cdc5 could dislodge Cdc14 from Net1, we devised a simplified in vitro release assay using GST-Cdc14 and His6-T7-Net1N expressed and purified from bacteria. Net1N includes the N-terminal 341 amino acids of Net1 and is sufficient to bind and inhibit Cdc14 [11]. When GST-Cdc14/His6-T7-Net1N complex anchored to anti-T7 beads was treated with ATP plus increasing amounts of recombinant Cdc5 purified from insect cells, both Cdc14 and Net1N were phosphorylated as indicated by their increased apparent molecular weight. Furthermore, Cdc14 was released from bead-bound Net1N to the supernatant (Figure 4A), and free Cdc14 was active as a protein phosphatase (data not shown), suggesting that Cdc5 is sufficient to disengage active Cdc14 from Net1.
Figure 4.
Polo-like kinases can phosphorylate and disrupt the RENT complex in vitro. (A) Cdc5 can disrupt recombinant Net1N/Cdc14 complex. GST-Cdc14 was incubated with anti-T7 beads loaded with His6-T7-Net1N (assuming 100% binding efficiency, each reaction contained ~10 pmol of Cdc14 and ~80 pmol of Net1N), where Net1N consists of the N-terminal 341 amino acids of Net1. The beads were divided into equal portions, and treated with indicated amounts of Cdc5 (~10 pmol/μl). Proteins released into the supernatant (sup) or bound to the beads (bead) were fractionated by SDS-PAGE and immunoblotted with anti-T7 and anti-GST to detect His6-T7-Net1N and GST-Cdc14, respectively. (B) The Polo-like kinase Plx1 can also disrupt Net1N/Cdc14 complex. GST-T7-Cdc14 was captured on anti-GST resins, and incubated with His6-T7-Net1N. The resins were divided into two equal portions, and treated with either active (+) or inactive (m) Plx1. Proteins released from or bound to beads were fractionated by SDS-PAGE and immunoblotted with anti-T7 antibodies to detect both GST-T7-Cdc14 and His6-T7-Net1N. Note that phosphorylation by active Plx1 causes both GST-T7-Cdc14 and 1 + 156 - T7 - Metlin to migrate shower in SDS-Page. (C) Plx1 can disassemble immunoprecipitated RENT complex. RENT complex was retrieved from myc9-NET1 CDC14-HA3 cell lysate on a resin coated with 9E10 antibodies. The resin was divided into two equal portions, and treated with active (+) or inactive (m) Plx1. Proteins released into the supernatant or bound to the beads were separated by SDS-PAGE and immunoblotted with 9E10, 12CA5, and anti-Sir2 antibodies to detect Net1, Cdc14, and Sir2 proteins, respectively.
The essential functions of Cdc5 can be performed by mammalian Polo-like kinase Plk1 [21], which led us to test if Cdc5 can also be substituted by other Polo-like kinases in the release assay. His6-T7-Net1N/GST-T7-Cdc14 complex was assembled on anti-GST beads, and treated with either active or N172A kinase-inactive Plx1 (Polo-like kinase from Xenopus; [24]) in the presence of ATP. Active but not inactive Plx1 phosphorylated both Net1N and Cdc14, and completely dislodged Net1N from bead-bound Cdc14 (Figure 4B). Thus, we conclude that like Cdc5, the protein kinase activity of Plx1 can disrupt the interaction between Net1N and Cdc14. Because recombinant Plx1 but not Cdc5 was consistently expressed at high levels in insect cells, and because release assays using Plx1 and Cdc5 yielded similar results, we used Plx1 in most of our release assays.
The Net1N/Cdc14 complex is only a crude approximation of RENT, because it does not contain all components of RENT (such as Sir2), lacks modifications which normally occur in yeast cells, and contains Net1N instead of Net1. Thus, we tested whether Polo-like kinases could disrupt a physiological RENT complex. Myc9-Net1 and the associated RENT subunits were immunopurified from extracts of myc9-NET1 cells on 9E10 beads and treated with Plx1 plus ATP. Active but not inactive Plx1 released a significant portion of Cdc14 and Sir2 from bead-bound Net1 to the supernatant fraction (Figure 4C). In a similar experiment, Cdc5 also disrupted the RENT complex (data not shown). Taken together, these data suggest that Polo-like kinases are sufficient to release Cdc14 and Sir2 from Net1 in vitro.
Polo-like kinase abolishes Cdc14-binding activity of Net1 in vitro
Polo-like kinases phosphorylated and disrupted Net1N/Cdc14 complex, raising the question whether disassembly of the complex in vitro required phosphorylation of Net1N or Cdc14 or both. To address this question, we tested the binding affinity of combinations of Plx1-phosphorylated or unphosphorylated Net1N and Cdc14. Unmodified Net1N bound equally well to beads harboring phosphorylated or unphosphorylated Cdc14 (Figure 5, Lanes 5 and 8, respectively). To the contrary, phosphorylated Net1N remained in the supernatant (Figure 5, Lanes 2 and 3), and failed to bind to Cdc14 beads (Figure 5, Lanes 6 and 7). Thus, we surmise that Polo-like kinase disassembles RENT in vitro by eliminating the Cdc14-affinity of Net1.
Figure 5.
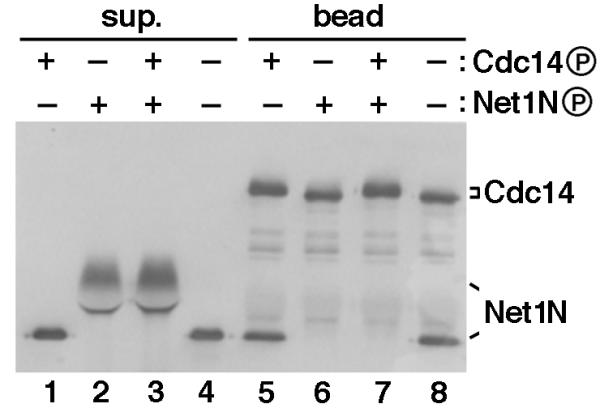
Plx1 abolishes the Cdc14-binding activity of Net1N. Combinations of Plx1-phosphorylated or unphosphorylated Net1N and Cdc14 were tested for their abilities to form protein complexes. All four reactions consisted of identical components and permutations of four steps (1. addition of GST-Cdc14 beads; 2. incubation with Plx1 for 45 min at room temperature; 3. depletion of ATP at room temperature for 20 min; and 4. addition of Net1N). The ATP depletion step ensured that only the component(s) present with Plx1 prior to ATP depletion would be phosphorylated. Thus, Lanes 1 and 5 resulted from 1 -> 2 -> 3 -> 4; Lanes 2 and 6 resulted from 4 -> 2 -> 3 -> 1; Lanes 3 and 7 resulted from 1 -> 4 -> 2 -> 3; and Lanes 4 and 8 resulted from 1 -> 4 -> 3 -> 2. Whether Cdc14 or Net1 was phosphorylated by Plx1 in the presence of ATP (+) or unphosphorylated because of depletion of ATP (-), was indicated above each lane. All reactions were terminated by a final binding reaction at 4°C for 1 hr, and proteins in the supernatant (sup.) and bead fractions were separated by SDS-PAGE and immunoblotted with anti-T7 antibodies to detect both GST-T7-Cdc14 and His6-T7-Net1N.
RENT complexes comprised of Net1 phospho-site mutants are resistant to disassembly by Polo-like kinase, but exhibit only minor phenotypes in vivo
If Cdc5 disassembled the RENT complex through phosphorylating Net1, then a mutant Net1 lacking Cdc5 phosphorylation sites should bind Cdc14 even in the presence of active Cdc5/Polo-like kinase. To test this hypothesis, we prepared Net1N phosphorylated by Cdc5 in vitro, and mapped 12 definitive and more than 20 potential phosphorylation sites using mass spectrometry, which is described in detail elsewhere [25].
We constructed an extensive set of phospho-site mutants of Net1N, which were tested for their ability to remain bound to Cdc14 beads in the presence of Plx1 [25]. By successively combining individual mutations in Cdc5 phosphorylation sites, we created the phosphosite mutants Net1N(7m) (with S30, S31, S48, S60, S64, S242, and S335 all changed to A) and Net1N(19m') (with T16, S30, S31, S48, S60, S64, T194, T195, T196, T197, S223, S224, S228, S231, S242, S295, S301, T302, S335 all changed to A). Net1N(7m) contained mutations required to strongly inhibit Plx1-dependent disruption of Net1N-Cdc14 complexes, whereas Net1N(19m') contained additional mutations without compromising the affinity of Net1N to Cdc14 [25]. Both of these mutant proteins, when expressed in E. coli and assembled with recombinant Cdc14, were found to be severely defective in releasing Cdc14 upon treatment with Plx1 (data not shown).
To evaluate the effects of the Net1(7m) and Net1(19m') mutants in vivo, DNA fragments encoding either the wild type or the mutant NET1N alleles were subcloned into a TRP1-marked integrating plasmid to generate full-length NET1-myc9 [894L1], net1(7m)-myc9 [894L2], and net1(19m')-myc9 [894L3] plasmids, respectively, which were then integrated into the TRP1 locus of net1▲/+ diploid yeast cells. After sporulation, net1▲ NET1-myc9, net1▲ net1(7m)-myc9, and net1▲ net1(19m')-myc9 strains were obtained and analyzed.
First, we compared in vivo phosphorylation states of Net1 and Net1(7m). Net1 and Net1(7m) from asynchronously growing cells migrated identically in an SDS-polyacrylamide gel (Figure 3, Lanes 1 and 5). Net1(7m) from cdc14-1 cells arrested at 37°C appeared to migrate slightly faster than the corresponding Net1 (Figure 3, compare Lanes 8 and 4), and the reduced mobility of Net1(7m) in cdc14 cells was no longer dependent upon CDC5 (compare lanes 7 and 8). Net1(19m') behaved identically to Net1(7m) in this assay (data not shown). We conclude that there is diminished Cdc5-dependent phosphorylation of Net1(7m) in cdc14 cells arrested in anaphase.
Next, we asked if the mutant RENT complex is more resistant toward Plx1. RENT complexes from net1▲ NET1-myc9 and net1▲ net1(7m)-myc9 cell lysates were captured on 9E10 beads and treated with increasing amounts of recombinant Plx1 plus ATP. Whereas ~40% of Cdc14 was released from wild-type RENT complex in the presence of 0.5 μl of Plx1, less than 10% of Cdc14 was released from mutant RENT complex when treated with 15 μl of Plx1 (Figure 6A). Thus, the mutant RENT is at least 30-fold more resistant to Plx1 than wild-type RENT. RENT complexes comprising Net1(19m') yielded nearly identical results (data not shown).
Figure 6.
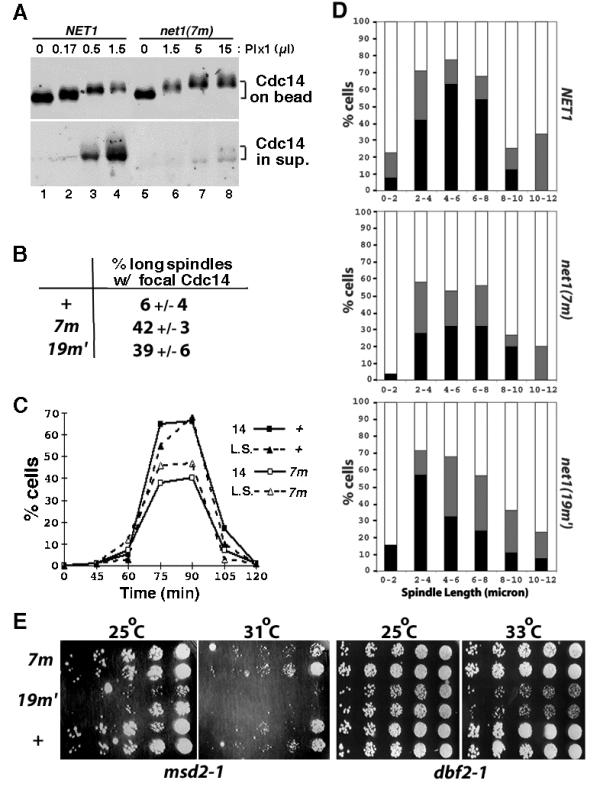
RENT complexes containing phospho-site mutant Net1(7m) is refractory to disassembly by Polo-like kinase in vitro, but exhibit minor phenotype in vivo. (A) RENT complexes from isogenic NET1-myc9 (WY374) and net1(7m)-myc9 (WY347) cells were immunoprecipitated on 9E10 beads. Beads were divided into equal portions, and treated with indicated amounts of Plx1. Cdc14 retained on Net1-bound beads or released into the supernatant was fractionated by SDS-PAGE and detected by immunoblotting with anti-Cdc14 antibodies. (B) Asynchronous NET1-myc9 (+), net1(7m)-myc9 (7m), and net1(19m')-myc9 (19m') cultures were subjected to indirect immunofluorescence using anti-Cdc14 and anti-tubulin antibodies. Cells with long mitotic spindles (~10% of total) were examined further to calculate the percentage that displayed focal Cdc14 staining. (C) NET1-myc9 (+) and net1(7m)-myc9 (7m) cells were arrested in G1 with α factor, and released into YP + 2% glucose media (T = 0) at 25°C. Samples withdrawn and fixed at the indicated time points were double-labeled with anti-Cdc14 and anti-tubulin antibodies. The percentage of cells with long mitotic spindles (L.S.) and the percentage of cells with delocalized Cdc14 (14) were calculated and plotted independently. (D) Cells harboring the dbf2-1 mutation and either a wild-type or mutant net1 allele were grown in YPD at 25°C, arrested in G1 with α-factor (12 μg/ml), released into YPD prewarmed to 37°C, and incubated at 37°C thereafter. Cells collected at 70–110 minutes after α factor release were double-labeled with anti-Cdc14 and anti-tubulin antibodies. Spindle length was measured and localization of Cdc14 was determined to be in one of the three categories: 1. full release (black box): complete release of Cdc14 from the nucleolus into the nucleus; 2. partial release (gray box): Cdc14 was nuclear in one of the DAPI masses and nucleolar in the other DAPI mass in the same cell, or Cdc14 was nuclear but with stronger nucleolar staining; and 3. no release (clear box): Cdc14 was strictly nucleolar. In each of the three panels, more than 300 cells were counted. (E) Starting with 3000 cells, three-fold serial dilutions of NET1-myc9 (+), net1(7m)-myc9 (7m), and net1(19m')-myc9 (19m') cells in msd2-1 or dbf2-1 background were spotted on a YPD plate from right to left, and incubated at the indicated temperature for 2 days before the picture was taken. Two independent isolates for each strain were used.
Despite the striking defects of the phosphosite mutant proteins in vitro, neither net1(7m)-myc9 nor net1(19m')-myc9 cells expressed a dramatic phenotype. We observed that ~40% of net1(7m)-myc9 and net1(19m')-myc9 cells with long mitotic spindles showed focal (i.e. nucleolar) Cdc14 localization, compared to ~6% of wild type cells (Figure 6B). However, this result indicates that the majority (~ 60%) of net1 phosphosite-mutant cells released Cdc14 from the nucleolus during anaphase with normal kinetics. Indeed, the kinetics of cell cycle progression (monitored by either elongation of the mitotic spindle or delocalization of Cdc14 from the nucleolus) in the wild-type and the net1(7m) mutant strains were almost identical (Figure 6C).
While this manuscript was under consideration, Cdc5 was shown to be involved in promoting 'Cdc14 early anaphase release' (FEAR, [26]). FEAR is a transient phenomenon that depends on Cdc5, but does not depend upon other components of the MEN pathway. It was reported that in early anaphase, Cdc14 is transiently released from the nucleolus by the action of the FEAR pathway, but quickly returns to the nucleolus unless the MEN is activated to promote sustained release of Cdc14 and subsequent exit from mitosis. To evaluate whether the net1(7m) and net1(19m') phosphosite mutants have an impact on FEAR, we analyzed Cdc14 localization in synchronous populations of dbf2-1, net1(7m) dbf2-1, and net1(19m') dbf2-1 cells following release from an α-factor block in G1 phase at 25°C to α-factor-free medium prewarmed to 37°C (nonpermissive temperature for dbf2-1). As shown in Figure 6D, the percentage of cells that completely released Cdc14 during early anaphase (spindle length 0–8 μm) was reduced by ~50% in net1(7m) and ~25% in net1(19m') cells compared with wild-type cells.
To test for the possibility that the proteins encoded by net1(7m) and net1(19m') might be hypomorphic with respect to their resistance toward phosphorylation by Cdc5 (i.e., not all Cdc5 phosphorylation sites on Net1 were mapped and mutated), we evaluated whether the net1 phosphosite-mutants exhibited a synthetic growth phenotype when combined with msd2-1 (a mutant allele of CDC5). This experiment was based on the idea that a reduction in phosphorylation sites on Net1 coupled with a reduction in Cdc5 activity might synergize to prevent efficient release of Cdc14 during anaphase, resulting in failure to exit mitosis. The net1(19m') but not net1(7m) allele showed a synthetic growth defect when combined with msd2-1 (Figure 6E). The net1(19m') allele also showed a synthetic growth defect when combined with dbf2-1. These observations imply that some of the residues mutated in net1(19m') represent in vivo phosphorylation sites that influence the MEN-dependent release of Cdc14 during anaphase.
Discussion
The 'RENT control' hypothesis posits that disassembly of the RENT complex and subsequent release of Cdc14 from the nucleolus drive cells from mitosis to interphase. This study analyzes how the Polo-like kinase Cdc5 participates in this key step of the mitotic exit program.
Cdc5 participates in disassembly of the RENT complex during anaphase
CDC5 was originally discovered as a gene essential for exit from mitosis [27]. We found that Cdc5 was required to release Cdc14 from the nucleolus: cdc5-1 cells in late anaphase displayed focal nucleolar Cdc14 staining, whether grown at the permissive or restrictive temperature. By contrast, Cdc14 was released from the nucleolus in the great majority of wild type cells by late anaphase. Surprisingly, cdc5-1 cells grown at 25°C could exit mitosis despite persistent nucleolar localization of Cdc14, which may have been due to release of a small fraction of Cdc14 that was nevertheless sufficient to drive exit from mitosis in these cells. Alternatively, it is possible that complete release of Cdc14 from the nucleolus in cdc5-1 cells was so brief that it escaped our detection. A ramification of this observation is that indirect immunofluorescence assays based on Cdc14 localization may not correctly predict whether a cell can exit mitosis.
Not only was Cdc5 required for Cdc14 release, but a stabilized mutant lacking the destruction box (Cdc5(DB▲)) triggered inappropriate Cdc14 release in pre-anaphase cells when overexpressed from the GAL1,10 promoter. Ectopic release of Cdc14 might contribute to the proliferation block and Clb2 degradation caused by induction of GAL-CDC5(DB▲) [19,20]. Overexpression of Cdc5(DB▲) also altered nucleolar structure in nocodazole-arrested cells (Figure 2B), the reason for which is not understood. In contrast, GAL-mediated overexpression of Tem1 (R. Li, personal communication), Cdc15 [28], or Dbf2/GST-Mob1 (RA, unpublished data) had little or no detectable effect on either exit from mitosis or cell proliferation, suggesting that the Tem1/Cdc15/Dbf2/Mob1 branch of the MEN plays a more restricted role in promoting the exit from mitosis.
The observations that Cdc5 was both necessary and sufficient for Cdc14 release and that Cdc5 influenced phosphorylation state of Net1 in vivo prompted us to examine whether Cdc5 directly catalyzes disassembly of the RENT complex. Indeed, Cdc5 and the Xenopus polo-like kinase Plx1 reduced the binding affinity between recombinant Cdc14 and Net1 by phosphorylating Net1, and dislodged both Cdc14 and Sir2 from the RENT complex in vitro.
Net1 mutants lacking critical Cdc5 phosphorylation sites support normal progression through mitosis
If Cdc5 directly promotes disassembly of RENT in vivo by a mechanism similar to that observed in vitro, then a Net1 mutant that cannot be phosphorylated by Cdc5 should not allow release of Cdc14 from the nucleolus in anaphase cells. To address this prediction, we mapped in vitro Cdc5-phosphorylation sites in the N-terminal part of Net1 which is sufficient to bind and inhibit Cdc14 [11]. The details of this study are described elsewhere [25]. Among the thirty or so mapped sites (with some being more efficiently phosphorylated than others), no strict consensus sequence for Cdc5 phosphorylation could be derived. However, the data from this and other [22,29] studies suggest a clear preference for phenyl or aliphatic side chains (F, Y, L, I, V, M) in the +1 position.
Mutating all mapped phosphorylation sites was ruled out as being uninformative, because a Net1 mutant lacking 22 sites [Net1(22m)] already suffered significantly reduced binding affinity for Cdc14 (data not shown). Thus, we prioritized phospho-sites based on their stoichiometry of phosphorylation and on homology to sites whose phosphorylation was demonstrated to be important in earlier release assays. In total, we tested more than 20 phospho-site mutants of Net1, and identified two – Net1(7m) and Net1(19m') – that still bound Cdc14 normally, but displayed a strong defect in releasing Cdc14 upon treatment with Plx1. Full length net1(7m)-myc9 and net1(19m')-myc9 alleles were constructed and expressed in net1▲ background. Mutant RENT complexes retrieved from these cells were significantly more resistant to the Polo-like kinase Plx1 in release assays (Fig. 6A and data not shown). However, when net1(7m) and net1(19m') cells were evaluated by immunofluorescence, the majority released Cdc14 in late anaphase, and the subset that exhibited a modest delay in Cdc14 mobilization probably released enough Cdc14 to exit mitosis in a timely manner such that essentially no cell cycle progression defect was evident (Fig. 6C). In addition, Cdc14 remained nucleolar in cdc5-1 net1▲ net1(7m)-myc9 cells (data not shown), suggesting that the release event was still regulated properly.
How does Cdc5 promote disassembly of the RENT complex and exit from mitosis?
There are two extreme interpretations of our analysis of the Net1(7m) and Net1 (19m') mutants: First, Cdc5 is not directly involved in disrupting the RENT complex, and second, Cdc5 directly disassembles the RENT complex via phosphorylation of Net1, but because it is not feasible to evaluate all possible 109 (=230) combinations of phospho-site mutants or because the sites phosphorylated by Cdc5 in vivo are different from those in vitro due to the presence of factors such as Sir2, Pol I, and rDNA [10,23], we missed key phosphorylation sites that would have given a more definitive phenotype when mutated. A variation on this theme is that it might never be possible to construct a phosphosite mutant of Net1 that binds Cdc14 but fails to release it during anaphase, if the hydroxyl groups that are phosphorylated normally form critical hydrogen bonds that tether Cdc14 to Net1.
A third hypothesis that stakes middle ground between these polar extremes is that Cdc5 affects RENT stability both directly and indirectly, with the former role being less significant than the latter. Several lines of evidence are consistent with this view. First, net1(7m) and net1(19m') mutants exerted a modest effect on release of Cdc14 from the nucleolus, in that ~40% of cells with long mitotic spindles retained focal localization of Cdc14 and that both net1 phospho-site mutants displayed a modest defect in 'Cdc14 early anaphase release' (FEAR; [26]). Second, net1(19m') showed a synthetic growth defect when combined with dbf2-1 or the msd2-1 allele of CDC5. Third, phosphorylation site mapping of Net1 purified from arrested cdc14-1 cells (WS and SC, unpublished data) revealed that at least one (S231) of the five in vivo phosphorylation sites in the first 341 amino acids was directly phosphorylated by Cdc5 in vitro. Nevertheless, because expression of neither Net1(7m) nor Net1(19m') had a major impact on exit from mitosis, any direct effect of Cdc5 on Net1 appears to be minor, and the requirement for Cdc5 in the release of Cdc14 might arise principally from phosphorylation of other release-promoting proteins.
If Cdc5 plays a minor role in directly disassembling RENT, then how does it promote exit from mitosis? Cdc15, Dbf2, and Mob1 are not properly localized to the spindle pole body in a significant fraction of cdc5-1 cells [30,31,17], and Dbf2/Mob1 protein kinase activity is diminished in cdc5 cells [15-17], suggesting that at least one function of Cdc5 is to generate maximally active Dbf2/Mob1, which can then trigger Cdc14 release. This explanation is unlikely to account fully for Cdc5's role in exit from mitosis, however, because (1) overexpression of Cdc5, but not Dbf2/Mob1, provoked ectopic release of Cdc14 from the nucleolus; and (2) the Dbf2/Mob1 kinase complex is at least partially activated and is correctly localized to the spindle pole body in 50% of cdc5 cells at the nonpermissive temperature [16,17].
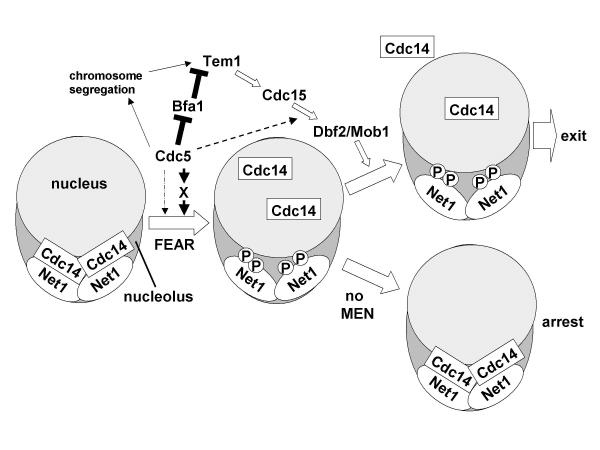
While this manuscript was under consideration, two papers presented new evidence on how Cdc5 contributes to exit from mitosis [29,26]. To summarize our study and these recent findings, we present a model (Figure 7) that is consistent with all of the available data. Upon initiation of chromosome segregation, Cdc5 promotes the transient 'Cdc14 early anaphase release' [26]. Our data suggest that Cdc5 can promote FEAR by directly phosphorylating Net1 as well as other yet-to-be-identified FEAR factors. Cdc5 also helps to activate the MEN by directly phosphorylating and disabling Bfa1 [29], which is thought to inhibit Tem1 by serving as a GTPase-activating protein. Cdc5 presumably impinges on the MEN by other means as well, because Cdc5 is required for maximal Dbf2 protein kinase activity even in the absence of Bfa1's obligate partner Bub2 [18]. Once activated, the MEN in turn sustains the transient Cdc14 release enabled by Cdc5 and other FEAR pathway components. The unique ability of overexpressed Cdc5 to expel Cdc14 from the nucleolus may thus be a consequence of Cdc5's abilities to directly disassemble Net1-Cdc14 complexes, promote FEAR, and activate the MEN. A complete understanding of Cdc5's role in mitotic exit will ultimately require identification of all of its direct substrates in the MEN and FEAR pathways.
Figure 7.
Role of Cdc5 in exit from mitosis. Cdc5 may promote Cdc14 early anaphase release (FEAR) in part by phosphorylating Net1 directly (dotted arrow), and by activating an unknown Cdc14 dissociation factor (X). Subsequent activation of the MEN is important to sustain the transient Cdc14 release effected by the FEAR pathway. In the absence of MEN activity, Cdc14 returns to the nucleolus and cells arrest in late anaphase (bottom right). In the presence of MEN activity, Clb/CDK is shut off and cells exit mitosis (top right). The mechanism of action of MEN remains unknown, but activation of MEN correlates with appearance of Cdc14 in the cytoplasm [26], and mutation of known nuclear transport regulators renders mitotic exit independent of CDC15 function [35]. Together, these observations suggest that the MEN may promote exit by biasing the nucleocytoplasmic distribution of Cdc14. In addition to its role in the FEAR pathway, Cdc5 impinges positively on the MEN by several mechanisms, including: (i) Cdc5 promotes Tem1 activation by promoting chromosome segregation [22], which by enabling optimal spindle elongation and penetration into the bud, helps satisfy the spindle positioning checkpoint [12]; (ii) Cdc5 promotes activation of Tem1 GTP-binding protein by inhibiting its negative regulator, the Bfa1/Bub2 GTPase-activating protein [29], and (iii) Cdc5 promotes activation of Dbf2 by an unknown, BUB2-independent mechanism (dashed line) [18].
A word of caution on phosphosite mapping
Cdc5 can phosphorylate up to 30 residues in the amino terminal domain of Net1 in vitro. Remarkably, only five sites in this same segment were confirmed to be phosphorylated in vivo in cdc14 cells, and only one of these corresponded to a site phosphorylated by Cdc5 in vitro. Whereas it is possible that Cdc5 protein kinase activity is qualitatively or quantitatively altered in cells held at the cdc14 block, resulting in reduced phosphorylation of Cdc5 target sites on Net1, our data suggest that Cdc5 is a promiscuous protein kinase. This raises significant concerns about the interpretation of numerous studies in the literature that report effects of Cdc5/Polo on in vitro processes without mapping and genetic confirmation of phosphorylation sites. Moreover, several recent studies have reported direct mapping of sites phosphorylated by Cdc5/Polo in vitro [22,29,32], but in two of these cases the authors relied exclusively on phosphoshift analysis of mutant proteins to confirm that the same sites are phosphorylated in vivo. Given the significant possibility that multiply mutated proteins are not properly phosphorylated in vivo for indirect reasons, or migrate aberrantly upon SDS-PAGE, it may be prudent to perform direct mapping experiments to confirm unambiguously the physiological significance of sites phosphorylated by Cdc5/Polo in vitro.
Conclusions
Although Cdc5 activity (1) is necessary to free nucleolar Cdc14 in late mitosis, (2) can elicit ectopic release of Cdc14 in pre-anaphase cells when overproduced, (3) regulates the phosphorylation state of Net1 during anaphase, and (4) can disassemble native and recombinant RENT complexes in vitro by phosphorylating Net1, a combination of in vivo and in vitro phosphosite mapping and mutagenesis experiments suggest that Cdc5 mediates exit from mitosis primarily by phosphorylating targets other than Net1. The unusual promiscuity of Cdc5/Polo highlights the need to validate Cdc5/Polo in vitro phosphorylation sites by direct in vivo mapping experiments.
Materials and Methods
Protein detection
The SDS-boiling method for yeast extract preparation to visualize Net1 phosphorylation state in vivo and methods for protein detection were described previously [8]. For indirect immunofluorescence, we used 1:1000 rabbit α-A190, 1:2000 rabbit α-tubulin, 1:200 rabbit α-Cdc14, 1:1000 rat YL1/2 (against tubulin), and 1:1000 mouse monoclonal HA.11 and 9E10 antibodies (Babco/CRP). For Western blots, we used 1:2000 9E10, 1:2500 α-Sir2, 1:1000 rabbit α-Cdc14 (crude serum), 1:6000 α-GST, and 1:5000 α-T7 (Novagen).
Recombinant protein expression
GST-T7-Cdc14 (RDB 872) and GST-Cdc14 (RDB 1183) were expressed in E. coli BL21(DE3) pLysS cells, and purified on glutathione-agarose beads (Sigma) as described previously [33]. His6-T7-Net1(1-341) (Net1N, RDB1153) [11] was expressed in E. coli BL21(DE3) pLysS cells, and purified on Ni-NTA agarose beads (Qiagen) as described previously for p28Kix1[34]. Purification of His6-Plx1, His6-Plx1(N172), and His6-HA-Cdc5 from baculovirus-infected insect cells followed an earlier protocol [24]. The concentrations of purified proteins were in the ranges of: Net1N~0.4 mg/ml; GST-Cdc14~0.1–0.2 mg/ml; Plx1~0.3 mg/ml; Plx1(N172)~0.1 mg/ml; and Cdc5~1 mg/ml (although the level of Cdc5 expression varied drastically from one batch to another).
Release assays
The substrates for release assays were RENT (isolated from yeast cells) or Cdc14/Net1N (assembled in vitro) complexes anchored on resins through tags on either Net1 (Figures 4A, 4C, and 6A) or Cdc14 (Figures 4B and 5), and were prepared at 4°C. For experiments in Figures 4C and 6A, immunoprecipitation of RENT through Myc9-Net1 was carried out as described earlier [8], except that EDTA (final 2 mM) was included in the lysis buffer. Each release reaction contained RENT from ~10 OD600 units (1 unit equals 1 ml of culture at OD600 = 1.0) of cells bound to 15 μl of 9E10 beads. To assemble Cdc14 beads with Net1N for two assays (Figure 4B, detection method: Western blotting), 2 μl of rabbit-anti-GST antibodies were absorbed to 40 μl of protein A beads (Sigma) in 100 μl of HBS [20 mM HEPES (pH 7.2), 150 mM NaCl] + 2 mM EDTA + 0.1 mg/ml BSA + 1 mM DTT (HEBD) buffer for 1 hr. The beads were washed twice with HBS + 2 mM DTT + 0.2% Triton and once with HBS + 1 mM DTT. The beads were incubated with 250 ng of GST-T7-Cdc14 in 100 μl of HEBD for 1 hr, followed by the same sequence of washes. Finally, the beads were incubated with Net1N (450 ng) for 1 hr. To assemble Net1N beads with Cdc14 for six reactions (Figure 4A, detection method: Coomassie Blue staining), 105 μl of anti-T7 beads (Novagen) were incubated with 24 μg of His6-T7-Net1N in 100 μl of HEBD for 1 hr, washed four times with HBS + 0.2% n-octylglucoside + 2 mM DTT, and incubated with 6 μg of GST-Cdc14 in 100 μl of HEBD for 1 hr. After the final incubation step of the protein complete assembly process, beads were extensively washed, followed by a final wash with Kinase Buffer [5 mM Tris·HCl (pH 8.0), 25 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 1 mM ATP, 10 μg/ml pepstatin + chymostatin + leupeptin (PCL)]. The beads were then distributed into equal aliquots for release assays. Kinase reactions were carried out in 30 μl Kinase Buffer at room temperature for 0.5–1 hr, and the supernatant was transferred to new tubes at the end of assays. The beads were washed once with 70 μl of HEBD, and the wash was pooled with the supernatant, and precipitated with TCA (10% final) on ice for 30 min. After centrifugation in a microfuge for 10 min, the pellet was saved as "sup". Proteins present on the beads or in the sup were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
For experiments in Figure 5, all four reactions consisted of 125 ng GST-T7-Cdc14 adsorbed to 20 μl anti-GST beads, 200 ng His6-T7-Net1N, 1 μl Plx1, 20 μl kinase buffer containing ATP, and the ATP depletion system (see below).
ATP depletion
Kinase reaction was terminated by depleting ATP with 1 μl of hexokinase (1 U/μl) and 2 μl of 10 × G+ I (1 μl 10 mg/ml PCL+ 20 μl 50% glucose + 79 μl H2O) for 20 min at room temperature.
Authors' contributions
WS, RA, and RJD contributed to Figures 1 – Figure 6B, Figures 6C-6E, and Figure 7, respectively. Together, RA and WS did many experiments which, although invisible here, are essential to the development of this project. SLC, MJH, RSA and SAC mapped in vitro Cdc5 phosphorylation sites on Net1N and in vivo phosphorylation sites on Net1. CB and HC mapped the Cdc14-interaction domain of Net1 to the first 341 amino acids, and constructed the corresponding Net1N plasmid.
Acknowledgments
Acknowledgements
We thank R. Verma for advice on phospho-site mapping experiments; R. Verma, B. Hay, and D. Chan for discussions; and W. Seufert and E. Traverso for sharing unpublished data. We thank G. Alexandru, D. Chan, R. Lipford, E. Traverso, and R. Verma for critically reading the manuscript. We also thank J. Charles (Morgan lab), A. Kumagai (Dunphy lab), and M. Shirayama (Nasmyth lab) for the Cdc5 baculoviruses, Plx1 baculoviruses, and GAL-CDC5(DB▲) strain, respectively; M. Nomura and F. Soloman labs provided antibodies against A190 and tubulin, respectively. WS was an HHMI predoctoral fellow. This research was supported by an NIH grant to RJD. WS dedicates this paper to Maste R. Brain, Rubbe R. Stamp, and their invaluable yet dispensable lessons.
Contributor Information
Wenying Shou, Email: shouw@its.caltech.edu.
Ramzi Azzam, Email: azzam@its.caltech.edu.
Susan L Chen, Email: Susa.L.Chen@gsk.com.
Michael J Huddleston, Email: michael_j_huddleston@gsk.com.
Christopher Baskerville, Email: hound@scripps.edu.
Harry Charbonneau, Email: charb@purdue.edu.
Roland S Annan, Email: roland_s_annan@gsk.com.
Steve A Carr, Email: carr@mpi.com.
Raymond J Deshaies, Email: deshaies@its.caltech.edu.
References
- Morgan DO. Regulation of the APC and the exit from mitosis. Nature Cell Biology. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. Embo Journal. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of CDK-dependent phosphorylation. Molecular Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Current Biology. 1999;9:227–236. doi: 10.1016/S0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The Role of Phosphorylation and the Cdc28 Protein-Kinase in Cell-Cycle Regulated Nuclear Import of the Saccharomyces-Cerevisiae Transcription Factor-Swi5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shou WY, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZWS, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou WY, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Traverso EE, Baskerville C, Liu Y, Shou WY, James P, Deshaies RJ, Charbonneau H. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. Journal of Biological Chemistry. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN (vol 11, pg 89, 2001). Trends in Cell Biology. 2001;11:166–166. doi: 10.1016/S0962-8924(01)01958-410.1016/S0962-8924(01)01958-4. [DOI] [PubMed] [Google Scholar]
- Adames NR, Oberle JR, Cooper JA. The surveillance mechanism of the spindle position checkpoint in yeast. Journal of Cell Biology. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Tohe A. The Yeast Tem1 Gene, Which Encodes a Gtp-Binding Protein, Is Involved in Termination of M-Phase. Molecular and Cellular Biology. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Current Biology. 2001;11:784–788. doi: 10.1016/S0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Amon A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Molecular Biology of the Cell. 2001;12:2961–2974. doi: 10.1091/mbc.12.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Jensen S, Frenz LM, Johnson AL, Fesquet D, Johnston LH. The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. Journal of Cell Science. 2001;114:2345–2354. doi: 10.1242/jcs.114.12.2345. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jespersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Current Biology. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The polo-like kinase Cdc5p and the WD-repeat protein Cdc2Op/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. Embo Journal. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SG, Lee KS. A novel function of Saccharomyces cerevisiae Cdc5 in cytokinesis. Journal of Cell Biology. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/S0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Shou WY, Sakamoto KM, Keener J, Morimoto KW, Traverso EE, Azzam R, Hoppe GJ, Feldman RMR, DeModena J, Moazed D, Charbonneau H, Nomura M, Deshaies RJ. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Molecular Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- Chen SL, Huddleston MJ, Shou WY, Deshaies RJ, Annan RS, Carr SA. Mass spectrometry based methods for phosphorylation site mapping of hyperphosphorylated proteins applied to Net1, a regulator of exit from mitosis in yeast. Mol Cell Proteomics. 2002;1:186–196. doi: 10.1074/mcp.M100032-MCP200. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19 and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic-Control of Cell-Division Cycle in Yeast .5. Genetic Analysis of Cdc Mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Molecular Biology of the Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FH, Wang YC, Liu D, Li YM, Qin J, Elledge SJ. Regulation of the Bub2/Bfal GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. Journal of Cell Science. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Molecular and Cellular Biology. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410:215–220. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. Journal of Biological Chemistry. 1997;272:24054–24063. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- Shou WY, Dunphy WG. Cell cycle control by Xenopus p28(Kix1) developmentally regulated inhibitor of cyclin-dependent kinases. Molecular Biology of the Cell. 1996;7:457–469. doi: 10.1091/mbc.7.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou WY, Deshaies RJ. Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae. BMC Genetics. 2002;3:4. doi: 10.1186/1471-2156-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]