Abstract
The Omicron subvariants of SARS-CoV-2, especially for BA.2.86 and JN.1, have rapidly spread across multiple countries, posing a significant threat in the ongoing COVID-19 pandemic. Distinguished by 34 additional mutations on the Spike (S) protein compared to its BA.2 predecessor, the implications of BA.2.86 and its evolved descendant, JN.1 with additional L455S mutation in receptor-binding domains (RBDs), are of paramount concern. In this work, we systematically examine the neutralization susceptibilities of SARS-CoV-2 Omicron subvariants and reveal the enhanced antibody evasion of BA.2.86 and JN.1. We also determine the cryo-EM structures of the trimeric S proteins from BA.2.86 and JN.1 in complex with the host receptor ACE2, respectively. The mutations within the RBDs of BA.2.86 and JN.1 induce a remodeling of the interaction network between the RBD and ACE2. The L455S mutation of JN.1 further induces a notable shift of the RBD–ACE2 interface, suggesting the notably reduced binding affinity of JN.1 than BA.2.86. An analysis of the broadly neutralizing antibodies possessing core neutralizing epitopes reveals the antibody evasion mechanism underlying the evolution of Omicron BA.2.86 subvariant. In general, we construct a landscape of evolution in virus-receptor of the circulating Omicron subvariants.
Subject terms: Structural biology, Antimicrobial responses, SARS-CoV-2
Here, the authors evaluate the neutralization susceptibilities of BA.2.86 and JN.1 and determine the structural basis for binding to the ACE2 receptor and monoclonal neutralizing antibodies, revealing their evolution and antibody evasion mechanism.
Introduction
SARS-CoV-2, the causative agent of the ongoing COVID-19 pandemic, continues to evolve, posing a persistent health threat1–3. Following the global co-prevalence of XBB.1, BQ.1, BA.2.3.20, and CH.1.1 variants, characterized by notable antibody evasion, the epidemic and evolution of SARS-CoV-2 entered the era of XBB.1-related subvariants, such as XBB.1.5, XBB.1.9, and XBB.1.16 and then the EG.5.1 and HK.3 variants4,5. The EG.5.1 is derived from XBB.1.9, with several additional mutations. The HK.3 further harbors a L455F mutation based on EG.5.1 (Fig. 1a). A new subvariant of BA.2, BA.2.86, was first discovered in Israel and uploaded to the GISAID database, which was increasing in the prevalence and subsequently reported in many other countries (Fig. 1b)6–8. BA.2.86 variant has attracted strong attention due to harboring a large number of amino acid mutations on the spike (S) protein, which is really different from all previous variants. As shown in Fig. 1a and Fig. S1, the S protein of BA.2.86 contained 58, 34, and 36 mutations comparing with the wild-type (WT), BA.2, and XBB.1.5, respectively9–11. Furthermore, the 483del mutation represents the first deletion identified in the receptor-binding domain (RBD) of the SARS-CoV-2 S protein. Even more worryingly, the JN.1 subvariant, carrying an additional L455S mutation on the RBD compared with the BA.2.86, has become a more dominant circulating strain worldwide12,13.
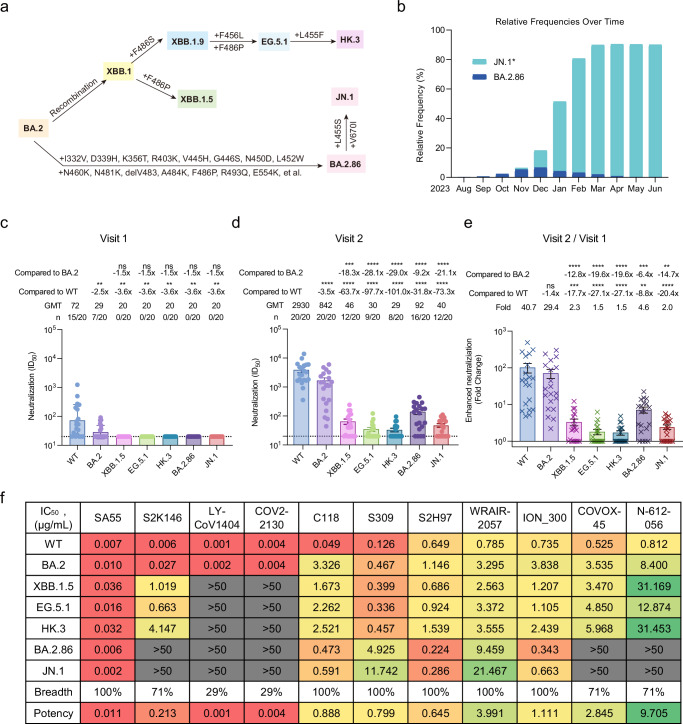
Fig. 1. Overall mutations in the spike, prevalence, and antibody evasion of SARS-CoV-2 BA.2.86 variant.
a The schematic diagram of several SARS-CoV-2 BA.2-related subvariants evolution. Some additional mutations in the spike acquired by XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1 were displayed. b The relative frequencies of BA.2.86 and JN.1* over time. The data to produce the chart were collected from the GISAID database and updated on 30 June 2024. Of which, JN.1* combine the JN.1 and its main sublineages including JN.1.1, JN.1.4, JN.1.4.5, JN.1.7, JN.1.11, JN.1.16, JN.1.16.1, KP.1.1, KP.2, KP.2.3, KP.3, KP.3.1.1, KP.3.2, and KP.3.3 variants. The neutralization of 20 BA.4 or BA.5 breakthrough infected human plasma samples collected in the early stage of breakthrough infection (c: Visit 1) and in another follow-up (d: Visit 2, an interval of 7–15 days) against WT, BA.2, XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1, respectively. The 50% inhibitory dilution (ID50) values are means of two independent experiments. Data are presented as geometric mean values ± standard deviation (SD). The number, GMT, fold change, and significance of difference are labeled on the top. “-” represents decreased value. e Fold change in the enhanced neutralization of WT, BA.2, XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1 by the BA.4 or BA.5 breakthrough infection. The fold change was obtained through the calculation of ID50 in Visit 2 divided by ID50 in Visit 1. Data are presented as geometric mean values ± SD. The statistical significance in (c–e) was performed using two-tailed Kruskal–Wallis test with paired Wilcoxon’s multiple-comparison test. ns: P > 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001. f The neutralization of mAbs against WT, BA.2, XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1 pseudoviruses. The 50% inhibitory concentration (IC50) values are means of two independent experiments. The neutralization potency is marked in the different color. Red: high, yellow: moderate, green: weak, gray: non-neutralization (IC50 > 50 μg/mL). The neutralization breadth is defined as the percentage of pseudoviruses neutralized by each mAb. The neutralization potency is calculated by the geometric mean of neutralizing values <50 μg/mL. Source data are provided as a Source Data file.
Notably, BA.2.86 and JN.1 subvariants still employ ACE2 as the host receptor8,12. During the infection process, the binding of RBD to the peptidase domain (PD) of ACE2 initiates conformational changes in the trimeric S protein. These changes expose the fusion peptide, facilitating membrane fusion with host cells14–20. However, the impact of these additional mutations on the binding pattern between the BA.2.86/JN.1 S protein with ACE2 remains uncharacterized.
Monoclonal antibody (mAb) plays a crucial role in blocking the entry of SARS-CoV-2 into host cells. Several mAbs, such as SA55 and S309 (sotrovimab), have displayed the potent efficacy in neutralizing multiple Omicron subvariants7,21–23. S309 targets a proteoglycan epitope on Asn343 in the front of RBD without competing with ACE2, showing moderate but broad neutralizing activities against Alpha, Beta, Gamma, Delta, and Omicron variants of SARS-CoV-21,7,8,22,24,25. By contrast, SA55 binds to the back of the up RBD and blocks the ACE2, exhibiting high neutralizing potency against SARS-CoV-2 variants including XBB.1.5 and BA.2.86, as well as other sarbecoviruses7,25–27. Given the different binding epitopes of S309 and SA55, the investigation of the cocktail usage of both mAbs is crucial to fight against the SARS-CoV-2 Omicron subvariants.
In this work, we comprehensively evaluate the neutralization susceptibilities of a series of Omicron subvariants to the plasma polyclonal antibodies (pAbs) from BA.4 or BA.5 breakthrough infected donors and several available broadly neutralizing mAbs, indicating the enhanced antibody evasion of BA.2.86 and JN.1. We also characterize the ACE2-bound S protein structures of BA.2.86 and JN.1, and reveal their antibody escape mechanism. These results could pinpoint the specific mutations on the S protein of BA.2.86 and JN.1 accountable for their conformational changes and comprehend the mechanisms underlying their modified properties for antibody evasion.
Results
The antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariant
In a previous study, we established a cohort of BA.4 or BA.5 breakthrough infections and evaluated the antibody escape abilities of BA.2.75, BA.2.3.20, BQ.1.1, XBB.1, and CH.1.128. In this study, we further constructed SARS-CoV-2 pseudoviruses of XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1, and measured their antibody evasion abilities in a head-to-head comparison with the WT and BA.2 using plasma samples from BA.4 or BA.5 breakthrough infected donors (Fig. S2). These plasma samples were obtained in the early stage (Visit 1, 0–5 days post positive PCR test) and late stage of breakthrough infection (Visit 2, 7–15 days after Visit 1) from 20 BA.4 or BA.5-infected individuals who had received at least two doses of SARS-CoV-2 vaccines based on the WT virus or WT spike protein28. In Visit 1, some plasma still retained neutralization against the WT and BA.2 pseudoviruses. However, all plasma lost their neutralizing activities against XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1 (Fig. 1c). In Visit 2, after the BA.4 or BA.5 breakthrough infection, the neutralization of plasma against all of tested variants were enhanced on some extent (Fig. 1d). XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1 showed more striking antibody evasion than BA.2. For the most concerned BA.2.86 and JN.1, these plasma samples against BA.2.86 displayed a 31.8-fold and 9.2-fold reduction in the geometric mean titers (GMTs), respectively, comparing with that against WT and BA.2. JN.1 presented a 73.3-fold and 21.1-fold reduction, respectively, and showed a more significant antibody escape ability than BA.2.86. Due to the immune imprinting (or so-called antigenic sin) of vaccinated WT vaccines29,30, the enhanced neutralization against WT virus (40.7-fold) was still the largest, followed by BA.2 (29.4-fold), BA.2.86 (4.6-fold), XBB.1.5 (2.3-fold), JN.1 (2.0-fold), EG.5.1 (1.5-fold), and HK.3 (1.5-fold) (Fig. 1e). These results suggested that EG.5.1 and HK.3 showed similar and the most striking antibody evasion in these tested SARS-CoV-2 variants. By contrast, BA.2.86, despite having more mutations in the spike, had an obviously weaker antibody escape ability than XBB.1.5, EG.5.1, and HK.3 in this cohort of BA.4 or BA.5 breakthrough infections. At the same time, we found that an additional L455S mutation on the RBD of JN.1 could further enhance the antibody escape ability than BA.2.86.
Previously, we found that only a few of RBS-like (S2K146, LY-CoV1404, COV2-2130, and C118), S309-site (S309), and most of S2H97-like (S2H97, WRAIR-2057, ION_300, COVOX-45, and N-612-056) mAbs maintained certain broad-spectrum neutralizing activities against SARS-CoV-2 variants31. In this study, we further evaluated the neutralizing activities of these broad mAbs against the concerned variants. SA55, another RBS-like mAb, was also included in this tested antibody panel, showing broad and potent neutralizing activities against SARS-CoV-2 variants23,29. Consistent with previous results31,32, all tested mAbs effectively neutralized WT and BA.2 (Fig. 1f and Fig. S3). LY-CoV1404 and COV2-2130 totally lost their neutralizing activities against XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1. In addition, BA.2.86 and JN.1 also escaped from the neutralization of S2K146, COVOX-45, and N-612-056. Among these tested mAbs, SA55 exhibited the most broad-spectrum and potent neutralizing activities (100%, 0.011 μg/mL) against all tested variants, followed by S2H97 (100%, 0.645 μg/mL), S309 (100%, 0.799 μg/mL), C118 (100%, 0.888 μg/mL), etc. These results suggested that BA.2.86 and JN.1 further escaped some available broadly neutralizing mAbs, narrowing potential antibody drug candidates.
Structural determination of the complex formation between BA.2.86 Spike proteins and ACE2
To investigate the biochemical characteristics of BA.2.86 subvariant, we characterized the binding affinity between the RBD and ACE2-PD. The monomeric ACE2-PD binds to the RBD of BA.2 and BA.2.86 with KD values of 16.0 ± 2.5 nM and 7.10 ± 0.66 nM, respectively—approximately 1–3 times higher than that of WT-RBD (25 ± 4.0 nM) (Fig. S4). The trend in the results is consistent with previous findings7,16,33. Notably, the binding affinity of JN.1 (29.0 ± 3.5 nM), with an additional RBD mutation of L455S than BA.2.86, is a bit lower than that of the WT (Fig. S4), which is also consistent with previous findings on the L455F mutation and deep mutational scanning results34,35.
To delve deeper into the interaction details, we then employed single-particle cryo-EM to elucidate the structures of the trimeric BA.2.86 S protein in complex with ACE2-PD. BA.2.86-S was incubated with PD at a molar ratio of about 1:1.2 for 1 h, and the mixture was underwent size-exclusion chromatography to remove excessive PD (Fig. S5). Further details regarding cryo-sample preparation, data acquisition, data processing, and structural determination can be found in the “Methods” section and Figs. S6–S10 and Table S1. The three-dimensional (3D) structure of BA.2.86-S bound with ACE2-PD was revealed at an overall resolution of 3.0 Å (Fig. 2a, b and Fig. S7). A focused refinement was applied to enhance the resolution at the interface between ACE2 and RBD, resulting in a final local resolution of 3.0 Å, supporting reliable modeling and analysis of the interface (Fig. S9).
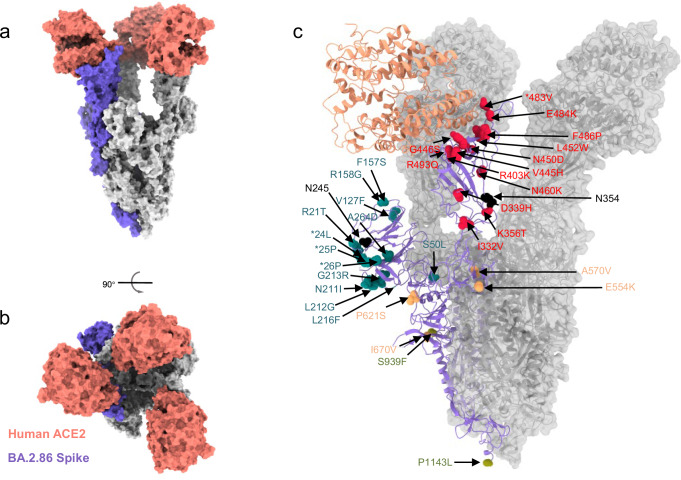
Fig. 2. Structural and biological analysis of Omicron BA.2.86 subvariant.
a, b Surface presentation of domain-colored cryo-EM structures of extracellular domain of S protein (S-ECD) from Omicron BA.2.86, respectively, in complex with the PD of human ACE2, which show top and side view. Three RBDs are extended upward. ACE2 colored salmon. c Location of mutations detected in BA.2.86 spike, relative to its ancestral BA.2 (PDB: 7XO7). The red, cyan, orange, and laurel green mutations are in RBD, NTD, SD, and S2, respectively. The additional N-glycosylation sites colored in black. *deletion.
The trimeric structure of the S protein provides a comprehensive mapping of BA.2.86 mutations, primarily distributed on the protein’s surface (Fig. 2c). In our depiction of mutations within the spike glycoprotein of SARS-CoV-2 Omicron subvariants (BA.2/XBB.1.5), shown in Fig. 2c, BA.2.86 displays 34 additional mutations compared to the BA.2 variant. These include 13 mutations in the N-terminal domain (NTD, shown in cyan), 14 in the RBD (shown in red), 2 in subdomain 1 (SD1, shown in orange), 3 in subdomain 2 (SD2, shown in orange), and 2 in the S2 region (shown in laurel green). Notably, the structure of BA.2.86 reveals two additional N-glycosylation sites located in NTD (Asn245, shown in black) and RBD (Asn354, shown in black).
In the 3D EM reconstruction structure, three RBDs are in the “up” conformation, each bound to ACE2-PD. However, during classification, another subset with 50,714 particles was processed to an overall resolution of 3.8 Å. In this subset, one RBD tilts outside and binds with ACE2 because a nearby ACE2 accommodates a “partial-up” RBD when docked with the structure of BA.2.86 and ACE2 (Fig. S7g, h). Unfortunately, the preferred orientation precluded detailed model building of this conformation. This marks the first reported instance of ACE2 binding to a “partially up” RBD. The binding of the spike protein with ACE2 in this “partially up” conformation may represent an intermediate state between the up and down orientations of the RBD. This intermediate state retains the ability to recognize the receptor.
The remodeled interaction between BA.2.86 RBD and ACE2-PD
Structural analysis has identified 10 residues on BA.2.86 RBD at the interface with ACE2 (Fig. 3), including the side chains of Asn417, Tyr449, Tyr453, Tyr489, Gln493, Arg498, Thr500, Tyr501 and the main chains of Ala475 and Gly502. The binding pattern is predominantly through hydrogen bonds or salt bridges. Notably, although there are occasional mutations in amino acid residues within the RBD region, accompanied by substitutions in amino acid interaction networks, these changes are still insufficient to alter the overall conformation. The overall interface pattern remains largely unchanged, indicating a conserved binding mode.
Fig. 3. Interactions between SARS-CoV-2 BA.2.86-RBD and ACE2.
a Cryo-EM density of the BA.2.86-RBD–ACE2 interface. The “up” RBD is shown in medium purple. The ACE2 is shown in salmon. This is an overall cryo-EM density of the RBD–ACE2 region. b The cryo-EM density map between the RBD and ACE2 interface. Residues are shown in sticks, with the corresponding cryo-EM density represented in mesh. c Overall structural model of Omicron RBD–ACE2-bound region. The regions enclosed by the red, green, and blue solid lines are illustrated in detail in (d). d Detailed analysis of the interface between RBD and ACE2. Polar interactions are indicated by yellow dotted lines.
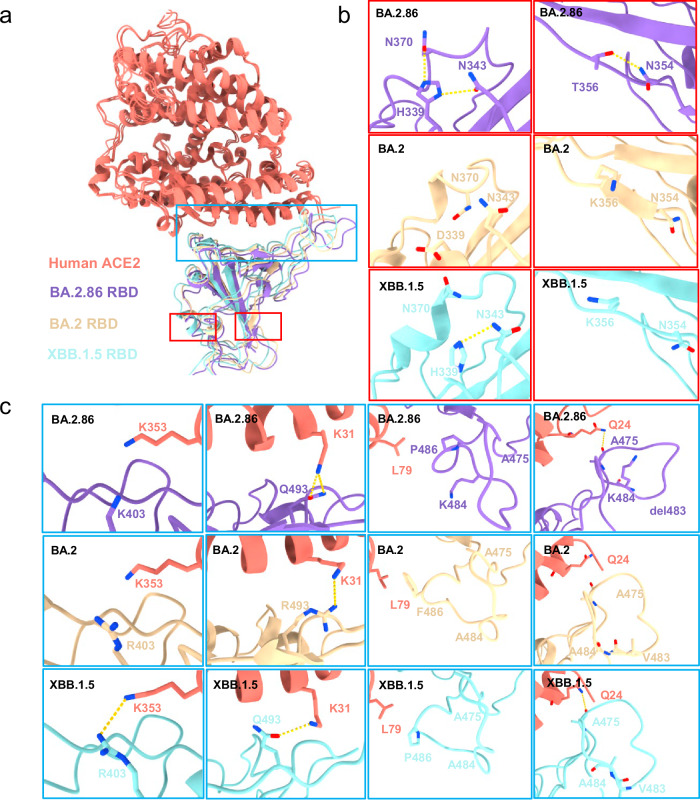
We further compared the interaction between the RBD and ACE2 across different subvariants (Fig. 4a–c). First, it is observed that at the tips of BA.2.86, His339 introduces new interactions with Asn370 and Asn343, whereas in BA.2, it is Asp339, with no interactions (Fig. 4b). This new interaction also triggers the formation of hydrogen bonds between Thr356 and Asn354 of RBD and the main chain of Ala475 with Gln24 of ACE2. Besides, the R403K mutation of BA.2.86 results in the loss of interaction with Glu37 of ACE2, similar to XBB.1.5, while retaining the interactions of Asn417 and Gln493 with Asp30 and Lys31 of ACE2, respectively (Figs. 3d and 4c). The A484K changes the surrounding hydrophobic environment (Ala475, Val483, and Phe/Pro486), which in turn affects the interaction between Pro486 and Leu79 of ACE2. The mutation of Lys484 and the loss of Val483 make the tips significantly shift (T473-P490). Collectively, these changes remodel the interaction network between BA.2.86 and ACE2.
Fig. 4. Interface comparison between BA.2.86-RBD, BA.2-RBD, and XBB.1.5 with ACE2.
a Structural alignment of the BA.2.86-RBD (this paper), BA.2-RBD (PDB:7XO9), and XBB.1.5-RBD (PDB:8WRL). The regions enclosed by the red and blue dashed lines are illustrated in detail in (b) to (c), respectively. BA.2.86-RBD and ACE2 in our cryo-EM structure are colored medium purple and salmon, respectively; BA.2-RBD and XBB.1.5-RBD are colored light yellow and light blue, respectively. b, c Variation of the mutation residues between BA.2.86-RBD (labeled in medium purple), BA.2-RBD (labeled in light yellow), and XBB.1.5-RBD (labeled in light blue).
The neutralizing mechanism of combined SA55 and S309
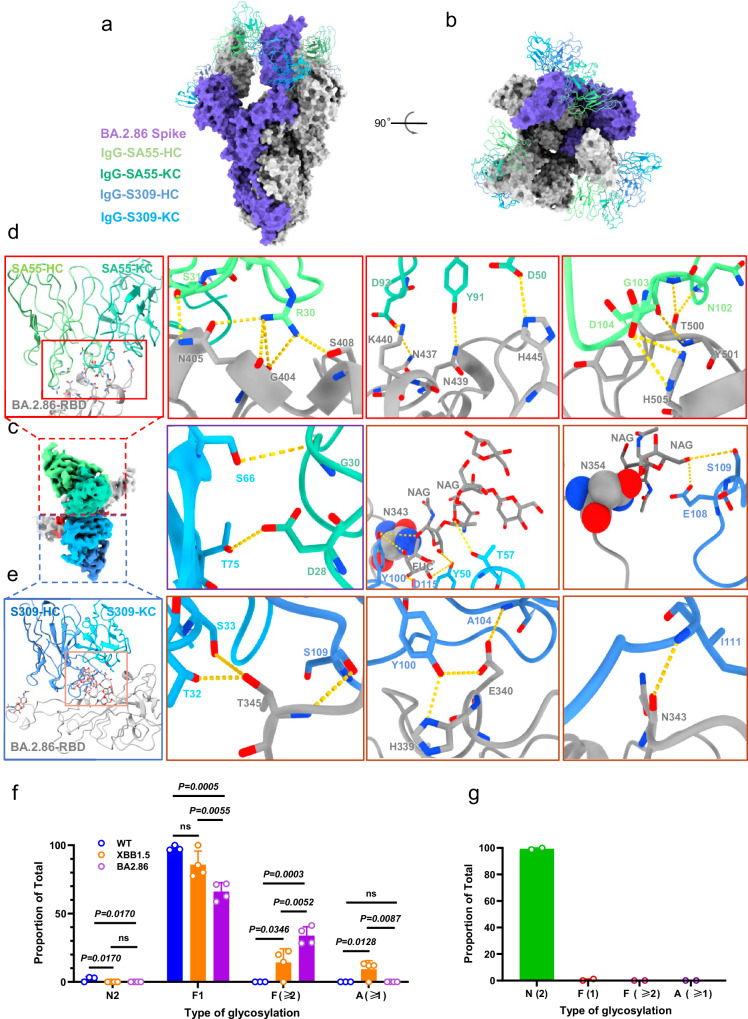
Considering the distinct epitopes of SA55 and S309, we further conducted a detailed analysis of the structure of the BA.2.86 S protein in complex with the SA55 and S309 (Fig. 5a, b, and Figs. S11 and S12). The binding epitopes of SA55 and S309 remain almost consistent with previously reported structures36,37. Specifically, the heavy chain (HC) of SA55 interacts with Gly404, Asn405, Ser408, Thr500, Tyr501, and His505 on BA.2.86 RBD, while the Kappa light chain (KC) interacts with Asn437, Asn439, Lys440, and His445 (Fig. 5d). Conversely, the KC of S309 interacts with Thr345, and the HC interacts with Thr345, His339, Glu340, and the glycosylation moiety of Asn343 on BA.2.86 (Fig. 5e). Notably, we observed additional interactions between the glycan of Asn354 and multiple sites on S309, potentially contributing to its high potency and activity (Fig. 5c). Interestingly, we found two hydrogen bonding interactions between SA55 and S309, involving Ser66 and Thr75 of S309-KC, Ser30 and Asp28 of SA55-KC, respectively (Fig. 5c). This finding suggests potential value as a cocktail therapy targeting the subvariants of SARS-CoV-2.
Fig. 5. Overall cryo-EM structure of S-ECD (BA.2.86) in complex with S309 and SA55.
a, b Overall structural model of RBD-SA55-S309–bound region. The SA55 and S309 were shown as cartoons and the spike of BA.2.86 was shown with surface map. c Cryo-EM density of the BA.2.86-RBD-S309-SA55 interface. BA.2.86-RBD are colored dark gray. d The regions enclosed by the red solid lines are illustrated in detail in (d). Residues are shown in sticks. The heavy and light chains of SA55 are colored pale green and medium aquamarine, respectively. e The regions enclosed by the pink solid lines are illustrated in detail in (e). The heavy and light chains of S309 are colored cornflower blue and deep sky blue, respectively. The ball indicates the N-glycosylation sites in N343 and N354. The purple solid lines box shows the interaction SA55 with S309. f Comparison of N-linked glycosylation profiles of the site Asn343 derived from S proteins of WT, XBB.1.5, and BA.2.86. Data are presented as mean values ± SD. Each experiment was repeated three times. g N-linked glycosylation profile of the site Asn354 that was identified from S protein of SARS-CoV-2 BA.2.86. N-linked glycans were divided into four basic classifications, including high mannose glycosylation (N2), mono-fucosylation (F1), multi-fucosylation (with ≥2 fucose residues, F ≥ 2), and sialylation (A ≥ 1). H represents hexose, N represents N-acetylglucosamine, F represents fucose, A represents sialic acid. Data are presented as mean values ± SD. Each experiment was repeated four times. In two of these sets, the site was not detected. Source data are provided as a Source Data file.
In comparing the binding of the two antibodies to distinct subvariant RBDs, structural shifts were observed, with BA.2.86 demonstrating the most significant shift, particularly at the tip regions (Fig. 5 and Fig. S13a, b). The G339H mutation in BA.2.86, located at the interface of S309, aligns with G339D in BA.2 (Fig. S13c), but it does not account for the different sensitivities to S309. Similarly, the D405N, R408S, N440K, V445H, N501Y, and Y505H mutations near the epitopes of SA55 in BA.2.86 could have a slight impact on neutralization ability (Fig. S13b).
To dissect the potential roles of N-linked glycans attached to Asn343 and Asn354 in antibody neutralization, we characterized N-linked glycosylation profiles of trimeric S proteins of SARS-CoV-2 WT, XBB.1.5, and BA.2.86 (Fig. 5f, g and Fig. S14). Complex-type glycosylation was predominantly detected at Asn343 across the three S proteins, in line with previous studies38,39. Strikingly, levels of sialylation as well as mono-fucosylation were significantly decreased in BA.2.86 compared to WT and/or XBB.1.5, while multi-fucosylation was significantly increased in BA.2.86 relative to WT and XBB.1.5 (Fig. 5f). The later phenomenon might account for neutralization of S309 to BA.2.86 evidenced by structural analysis, in which core fucose residue at Asn343 interacted with multiple amino acid residues in the antibody (Fig. 5e). The decrease of negatively charged glycans with sialylation at Asn343 of BA.2.86 likely increased the binding affinity of S309 via interactions between the glycan moiety at Asn343 and acidic amino acid residue Asp115 of the antibody. In addition, Asn354, a unique glycosylation site of BA.2.86, was exclusively occupied by less processed high mannose glycans, suggesting less accessability of glycan processing enzymes to this region (Fig. 5g). The interactions between neutral high mannose glycans at Asn354 and acidic amino acid residue Glu108 on S309 likely increase the binding affinity of the antibody to BA.2.86. Of note, another newly acquired glycosylation site Asn245 in BA.2.86 was mainly occupied by high mannose glycans (Fig. S15).
The comparison of neutralizing epitopes among various antibodies
Through model alignment, we identified distinct disparities in the binding region of the RBD among five reported antibodies, namely S309, SA55, S2K146, N-612-056, and COVOX-45 (Fig. 6a, b and Table S2). This further elucidates that the binding affinity and broad-spectrum neutralizing activity of these antibodies may undergo alterations with the variations of SARS-CoV-2.
Fig. 6. The comparison of neutralizing epitopes among various antibodies.
a, b The comparison of overall structural model of RBD-antibody-bound region. S309 (this paper) is colored medium blue. SA55 (this paper) is colored lime green. S2K146 (PDB:7TAT) are colored light yellow. N-612-056 (PDB:7S0B) are colored light cyan. COVOX-45 (PDB:7PRY) is colored orange. WT (PDB:6WPS) are colored plum. XBB.1.5 (PDB:8WRL) are colored wheat. BA.2 (PDB:7XO9) is colored sky blue. BA.2.86 (this paper) is colored medium purple. c, d Compared to WT, XBB.1.5, and BA.2, the specific details of the antigenic epitopes on BA.2.86 for the five antibodies S309, SA55, S2K146, N-612-056, and COVOX-45 are mapped.
From the perspective of antigen–antibody structural biology analysis (Fig. 6c, d), S309 demonstrates exceptionally potent broad-spectrum neutralizing activity. Despite the ongoing evolution of SARS-CoV-2, with N440K and G339H mutations observed in BA.2.86 subvariant compared to the WT strain, and D339H, T346R, and K356T compared to XBB.1.5 or BA.2 strains, the core neutralizing epitope of S309 is not affected.
SA55 and S2K146, by binding to RBD region in competition with ACE2 receptor, exert broad-spectrum neutralizing effects. In comparison to other prevalent strains, BA.2.86 exhibits significant shift in the tip regions, and these structural alterations directly impact their neutralizing activity, with S2K146 demonstrating a more pronounced effect. What is particularly noteworthy is that SA55 neutralizing activity remains largely unimpaired, even in the face of significant alterations compared to the WT strain, such as S375F, D405N, R408S, N440K, V445H, Q498R, N501Y, and Y505H. Moreover, despite the pronounced R403K mutation observed in BA.2.86 relative to XBB.1.5 and BA.2, this change does not impact the core neutralizing epitope of SA55, thus preserving SA55 high neutralizing potency.
Variants with glycosylation modifications play a crucial role in viral immune evasion. We speculate that the glycosylation modification at the neutralizing epitope Asn354 (Fig. 5e) enhances the broad-spectrum neutralizing activity of S309, while concurrently inhibiting the neutralizing capabilities of N-612-056 and COVOX-45. Regarding N-612-056 and COVOX-45, their inability to neutralize the BA.2.86 strain might be attributed to the fact that their binding regions are in close proximity to the tip regions, too. Additionally, from the genetic evolution of the strains, it is evident that their neutralizing activity exhibits pronounced strain specificity (Fig. 6c, d and Fig. S13).
The remodeling of the RBD–ACE2 interface following L455S mutation
The underlying mechanism that the JN.1’s lower binding affinity to ACE2 receptor and enhanced antibody evasion than BA.2.86 remains unclear. To investigate JN.1 variation characteristics, we also employed cryo-EM to elucidate the structures of the trimeric JN.1 S proteins in complex with ACE2. To improve the resolution between RBD–ACE2 interface, we also determined the cryo-EM structure the JN.1 RBD in complex with ACE2-B0AT1 complex40. The 3D structure of JN.1-S bound with ACE2-PD was revealed at an overall resolution of 3.16 Å (Figs. S15, S16 and Table S3), the three RBDs remaining in the “up” conformation, each bound to ACE2-PD (Figs. S15, S16a). Meanwhile, the local resolution at the interface between ACE2 and RBD within JN.1 RBD–ACE2-B0AT1 is 3.03 Å, clearly showing the interaction details between RBD and ACE2 (Fig. 7a, b).
Fig. 7. Interactions and interface comparison between SARS-CoV-2 BA.2.86-RBD and JN.1-RBD with ACE2.
a Cryo-EM density of the JN.1-RBD–ACE2 interface. The JN.1 RBD is shown in cyan. The ACE2 is shown in salmon. This is an overall cryo-EM density of the RBD–ACE2 region. b The cryo-EM structural alignment of JN.1 and BA.2.86 between the RBD and ACE2 interface. Residues are shown in sticks, with the corresponding cryo-EM density represented in mesh. The BA.2.86 RBD is shown in medium purple. c Detailed analysis of 455 site in JN.1 or BA.2.86 RBD–ACE2-bound region. The region illustrates the disappearance of hydrophobic interactions at 455 site. L455S generates the hydrogen bond interaction with Gln493 in intra-RBD. Polar interactions are indicated by yellow dotted lines. d Detailed analysis of the same interacting residues between RBD and ACE2. The region displays the variation in interaction bond lengths between JN.1 and BA.2.86 at the same interacting residue positions. Polar interactions are indicated by yellow dotted lines. e Detailed analysis of the different interacting residues between RBD and ACE2. The region shows the interacting residues altered in JN.1 compared to BA.2.86. Polar interactions are indicated by yellow dotted lines.
We then compared the difference between JN.1 and BA.2.86 in the RBD–ACE2 interface (Fig. 7b–e). Notably, despite the residue 455 of RBD does not directly participate in the interaction with ACE2, the L455S of JN.1 might change the hydrophobic interactions core while it enhances the hydrogen bond interaction with Gln493 in intra-RBD (Fig. 7c). Moreover, L455S moves the RBD upward, and this movement also indirectly causes the tilt of the three α-helix in ACE2. Similarly, the phenomenon is observed in the structure of XBB.1.5-ACE2 complexes belonging to different clades with JN. 1 (Fig. S18). The involved interaction in both JN.1 and BA.2.86, namely the Tyr499, Thr500, Gly502, and Arg498, which alterations in the length of hydrogen bonds may affect their bonding patterns (Fig. 7d). The Tyr449, Ser494, Asn477, Asn487, and Arg498 in JN.1 participate in the interaction with ACE2 relative to the Asn417, Tyr453, Ala475, Tyr489, Gln493, and Tyr501 in BA.2.86, which work together to formulate new combination pattern leading to significant shift (Fig. 7e). In summary, the L455S mutation disrupt the conserved interaction pattern between RBD and ACE2, which significantly alters the interaction landscape. These factors may contribute to the observed increase in antibody evasion and decrease in the binding affinity.
Discussion
In this study, we evaluated the ability of some concerned SARS-CoV-2 subvariants, including XBB.1.5, EG.5.1, HK.3, BA.2.86, and JN.1, to escape from the neutralization of plasma pAbs elicited from BA.4 or BA.5 breakthrough infections and broadly neutralizing mAbs. BA.2.86, characterized by specific spike protein mutations, exhibited significant evasion from pAbs and varying sensitivities to mAbs, with the L455S mutation in JN.1 subvariant exacerbating this characteristic. Notably, certain mAbs, such as SA55, C118, S309, S2H97, WRAIR-2057, and ION_300, remained effective in neutralizing BA.2.86 and JN.1, suggesting their potential utility against concerned SARS-CoV-2 Omicron subvariants. SA55 and S309 recognized two distinct epitopes and exhibited robust broad-spectrum neutralizing activity, even when facing with significant deviations in the RBD region as observed in BA.2.86. This further suggests that a cocktail approach, involving the combination of multiple broad mAbs, should be employed for the prevention and treatment for SARS-CoV-2 variant infection. Moreover, we can leverage the core neutralizing epitopes of elite antibodies with broad-spectrum neutralizing activity to design antibodies with enhanced cross-neutralization capabilities and vaccines with broad preventative functionalities.
Apart from diverse affinities with the cellular receptor ACE2, the dynamic transition between the “up” and “down” states of RBD is crucial for SARS-CoV-2 infectivity. Previous studies on the S protein of SARS-CoV-2 primarily demonstrated either a complete “up” or “down” conformation in RBDs15,16,41. Our investigation revealed a preference for “up” RBDs in Omicron variants, contributing to the increased infectivity of BA.2 and BA.516. It is interesting to note that there exists a kind of “partial-up” RBDs in some Spike alone conformations42,43. However, our study unveiled, for the first time, the presence of a “partial-up” RBD in the BA.2.86 S-ACE2-PD protein, albeit at a low ratio. This suggests that evolved SARS-CoV-2 variants may exhibit a propensity to bind to the partial “up” state of RBDs, possibly indicating an intermediate conformational state or an evolutionary trend. Targeting BA.2.86-like subvariants may involve the use of ACE2-mimicking neutralizing antibodies.
SARS-CoV-2 S protein is a heavily glycosylated protein with 22 potential N-linked glycosylation sites in the WT S protein. In particular, BA.2.86 acquired two additional glycosylation sites, including Asn245 in the NTD region and Asn354 in the RBD region. Both of the two sites were predominantly occupied by high mannose glycans, suggesting less accessibility of neutralizing antibodies to these regions (Fig. 5g and Figs. S14, S15). Previous studies revealed that the glycosylation site Asn343 in the RBD region was associated with binding affinities and neutralization of antibodies, such as S30944. Our mass spectrometry analysis indicated that the level of neutral glycans with multi-fucosylation was significantly increased in BA.2.86 compared to XBB.1.5 and WT, whereas the level of negatively charged glycans with sialylation was significantly decreased in BA.2.86 compared to XBB.1.5 (Fig. 5f). This likely increases binding affinity of S309 to BA.2.86 via the interactions between core fucose residue at Asn343 and acidic amino acid residue Asp115 (Fig. 5e). In addition, the neutral high mannose glycans at Asn354 could boost the binding affinity of S309 to BA.2.86 via interacting with acidic amino acid residue Glu108 (Fig. 5e). Thus, N-linked glycans on SARS-CoV-2 S protein should be taken into account during vaccine development as well as treatment of the infection.
Given the sustained attention toward the emergence of the JN.1 subvariant, its concomitant increase in antibody evasion and decrease in affinity to ACE2 underscore the evolutionary trajectory of SARS-CoV-212,13. This is particularly crucial during the current period marked by significantly reduced detection capabilities. The mutation dynamics within the SARS-CoV-2 RBD have long served as crucial indicators for SARS-CoV-2 surveillance45. The likelihood of mutation at the Leu455 site has been identified through deep mutational scanning and subsequently confirmed in the ongoing evolution of the virus34,35. In our current study, we discovered that the L455S mutation alters the interacting residues at the interface and induces a noticeable shift (Fig. 7 and Fig. S18). This structural alteration reduces the affinity of JN.1 to ACE2 and possibly affects its antibody evasion capability. This is consistent with the previously predicted trend from deep mutational scanning, where L455S is expected to decrease affinity to ACE235. However, it cannot be ruled out that other mutations within BA.2.86, in synergy with L455S, may exacerbate the decline in affinity, suggesting the involvement of epistatic effects.
In summary, our studies provide a molecular understanding of how the ACE2 receptor is engaged by the S proteins of Omicron BA.2.86 and JN.1 subvariants. The analyses presented here offer molecular insights into the higher antibody evasion and interaction pattern of Omicron subvariants, contributing valuable information for therapeutic interventions against SARS-CoV-2 variants.
Methods
Study approval and plasma samples
This study was approved by the Ethics Committee of Shenzhen Third People’s Hospital, China (approval number: 2021-030). All participants had provided written informed consent for sample collection and subsequent analysis. The 20 individuals were infected with BA.4 or BA.5 who had received at least two doses of SARS-CoV-2 vaccines based on the WT virus or WT spike protein28. Of these participants, 6 (30%) were female, and the median age at initial sample collection was 35 (IQR 27–52) years old. The plasma samples were collected in the early stage (Visit 1, 0–5 days post positive PCR test) and late stage of breakthrough infection (Visit 2, 7–15 days after Visit 1), respectively. All plasma samples were stored at −80 °C in the Biobank of Shenzhen Third People’s Hospital and heat-inactivated at 56 °C for 30 min and centrifuged at 12,000 × g for 10 min at 4 °C before use28.
Expression and purification of mAbs targeting RBD
All gene sequences of mAbs targeting RBD were downloaded from the National Center of Biotechnology Information and synthesized and then cloned into the full-length IgG1 expression vectors by Sangon Biotech and GenScript. Paired heavy- and light-chain plasmids were co-transfected into HEK293F cells using polyethylenimines (PEIs) (Polysciences). And then, mAbs were purified from HEK293F cell supernatants after a 5- or 6-day culture using protein-A columns by the manufacturer’s instructions (Senhui Microsphere Technology). The purified mAbs were quantified by NanoDrop Spectrophotometer (Thermo Fisher Scientific)28,31.
The generation of SARS-CoV-2 pseudoviruses
The pseudoviruses of WT SARS-CoV-2 and subvariants were generated as previously described28,31. The codon-optimized spike gene of SARS-CoV-2 Wuhan reference strain (WT), BA.2, XBB.1.5, BA.2.86 and JN.1 subvariants lacking C-terminal 19 amino acids (a.a) were synthesized (GenScript) and cloned into pVAX1 vector (Spike-Δc19) between the Hind III and Xho I sites. The expression vectors of EG.5.1 and HK.3 spike protein were constructed by point mutation based on the XBB.1.5 using the Mut Express II Fast Mutagenesis Kit V2 according to the manufacturer’s instructions (Vazyme Biotech). Pseudoviruses are generated by co-transfecting 5 μg Spike-Δc19 and 10 μg env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-E-) into HEK293T cells using EZ transfection reagent according to the manufacturer’s instruction (Life-iLab). After 2 days co-transfection, the culture supernatants were harvested, clarified by centrifugation and stored at −80 °C for subsequent assays. Optimal infectious titer was determined by measuring the luciferase activity in the HEK293T-hACE2 cells, which stably overexpress human ACE2, using Bright-Lite Luciferase reagent (Vazyme Biotech).
Detailed sequence information of spike protein for generation of pseudoviruses and expression of Spike and RBD protein in this study was listed below, respectively.
WT SARS-CoV-2: accession number: NC_045512;
SARS-CoV-2 BA.2: accession number: EPI_ISL_9652748;
SARS-CoV-2 XBB.1.5: accession number: EPI_ISL_16071118;
SARS-CoV-2 EG.5.1: accession number: EPI_ISL_17854292;
SARS-CoV-2 HK.3: accession number: EPI_ISL_17978556;
SARS-CoV-2 BA.2.86: accession number: EPI_ISL_18110065;
SARS-CoV-2 JN.1: accession number: EPI_ISL_18313756.
SARS-CoV-2 pseudovirus-based neutralizing assay
The neutralization activity of plasma and mAbs was measured by the pseudovirus neutralizing assay as previously described28. To measure the neutralizing activity, serial threefold dilution of inactivated-plasma samples from the highest dilution of 1:20 and serial fivefold dilution of mAbs from the highest concentration of 50 μg/mL were prepared, respectively. SARS-CoV-2 and related variant pseudoviruses were mixed with serially diluted plasma or mAbs and incubated at 37 °C for 1 h. The pseudovirus without plasma or mAbs was used as the virus control (VC). The mixture was then added into the HEK293T-hACE2 cells in 96 white cell wells and incubated for 48 h at 37 °C. The culture medium was removed and 100 μL of the Bright-Lite Luciferase reagent (Vazyme Biotech) was added and shaken at room temperature for 3 min. The luciferase activity was measured using the Varioskan LUX multimode microplate reader (Thermo Fisher Scientific). The inhibition of plasma or mAbs was determined by comparing with the VC. The 50% inhibitory dilution (ID50) or inhibitory concentration (IC50) was calculated using GraphPad Prism 9.0 software by log (inhibitor) vs. normalized response—variable slope (four parameters) model. The cut-off value of neutralization was set as 1:20 dilution for plasma and 50 μg/mL for mAbs, respectively.
Protein expression and purification of trimeric S proteins and RBDs
The extracellular domain (ECD) (1–1208 a.a) of S protein of SARS-CoV-2 Omicron variant BA.2.86 and JN.1 were cloned into the pCAG vector (Invitrogen) with six proline substitutions at residues 817, 892, 899, 942, 986, and 987 and a C-terminal T4 fibritin trimerization motif followed by Flag and 10×His tag, respectively. A “GSAS” mutation at residues 682–685 was introduced into ECD to prevent the host furin protease digestion. These constructs were hereafter referred to as WT-S, XBB.1.5-S, and BA.2.86-S.
The RBD (319–541 a.a) of S protein from SARS-CoV-2 WT strain and Omicron variant BA.2, BA.2.86, and JN.1 were cloned into the pCAG vector (Invitrogen) with an N-terminal signal peptide of secreted luciferase and a C-terminal 6×His tag, respectively. These residue numbers mentioned above are that relative to the spike (WT). The PD (19–615 a.a) of human ACE2 was also cloned into the pCAG vector (Invitrogen) with an N-terminal signal peptide of secreted luciferase and a C-terminal Flag tag.
The recombinant protein was overexpressed using the HEK293F mammalian cells at 37 °C under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm). When the cell density reached 2.0 × 106 cells/mL, the plasmid was transiently transfected into the cells. To transfect one liter of cell culture, about 1.5 mg of the plasmid was premixed with 3 mg of PEIs in 50 mL of fresh medium for 15 mins before adding to cell culture. Cells or medium was collected by centrifugation at 4000 × g for 15 min after 60 h transfection.
The secreted ECD and RBD of S protein were purified by Ni-NTA affinity resin (Qiagen). The nickel resin loaded was rinsed with the wash buffer 1 containing 25 mM HEPES (pH 7.5), 500 mM NaCl and washed with wash buffer 2 containing 25 mM HEPES (pH 7.5), 150 mM NaCl and 30 mM imidazole. Protein was eluted by wash buffer 2 plus 270 mM imidazole. Then the Ni-NTA eluent of ECD was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl. The peak fractions were collected and stored at −80 °C. The Ni-NTA eluent of RBD was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl. The factions were collected for structural analysis and measurement of RBD binding to human ACE2-PD by BLI.
The secreted PD was purified by anti-FLAG M2 affinity resin (Sigma Aldrich). After loading two times, the anti-FLAG M2 resin was washed with the wash buffer 3 containing 25 mM HEPES (pH 7.5), 150 mM NaCl. The protein was eluted with the wash buffer 3 plus 0.2 mg/mL flag peptide. The eluent of PD was then concentrated and subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl.
The BA.2.86/JN.1-S was incubated with IgG/PD at a molar ratio of about 1:1.2 for 1 h. To remove excessive IgG/PD, the mixture was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl. The peak fractions containing protein complex were collected for EM analysis.
Protein preparation of JN.1 RBD-SLC6A19-ACE2
The cDNAs for full-length human SLC6A19 (B0AT1, accession number: NM_001003841) and ACE2 (accession number: NM_001371415) were subcloned into pCAG, respectively. An N-terminal FLAG tag was fused to B0AT1, and one Strep tag was fused after the N-terminal signal peptide of ACE2 using a standard two-step PCR. HEK293F cells (Invitrogen) were cultured in SMM 293T-II medium (Sino Biological Inc.) at 37 °C under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm). To co-express B0AT1 and ACE2, the cells were transiently transfected into HEK293F cells with the plasmids and PEIs when the cell density reached ~2.0 × 106/mL. For transfection one liter of cell culture, about 0.75 mg plasmids for B0AT1 and 0.75 mg plasmids for ACE2 were premixed with 3 mg PEIs in 50 mL of fresh medium for 15 min before adding to cell culture. The transfected cells were cultured for 48–60 h before harvesting. For purification of the B0AT1 and ACE2 complex, the cells were collected in a buffer containing 25 mM HEPES, pH 7.0, 150 mM NaCl, and three protease inhibitors, aprotinin (1.3 μg/mL, AMRESCO), pepstatin (0.7 μg/mL, AMRESCO), and leupeptin (5 μg/mL, AMRESCO). The membrane fraction was solubilized at 4 °C for 2 h with 1% (w/v) glycodiosgenin (GDN, Anatrace) and the cell debris was removed by centrifugation at 14,700 × g for 45 min. The supernatant was loaded to anti-FLAG M2 affinity resin (Sigma). After rinsing with a wash buffer containing 25 mM HEPES, pH 7.0, 150 mM NaCl, and 0.02% GDN (w/v), the protein was eluted with wash buffer plus 0.2 mg/mL FLAG peptide. The eluent was further purified by Ni-NTA affinity resin (Qiagen). After eluted with the wash buffer supplemented with 300 mM imidazole, the eluent was then concentrated and incubated with JN.1 RBD at a molar ratio of about 1:2.4 for 30 min. Then the protein mixture was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES, pH 7.0, 150 mM NaCl, and 0.01% GDN. The peak fractions were collected and concentrated for EM analysis.
Measurement of Omicron subvariants and WT strain RBD binding to human ACE2-PD by biolayer interferometry
The binding between PD and RBD of WT and other subvariants were performed using Octet Red96e (ForteBio). The PD of ACE2 was biotinylated using Biotinylation Kit (Genemore, 1828M) and loaded to octet SA biosensor (Sartorius). The association and dissociation of PD-coated biosensors with different concentrations of RBD of SARS-CoV-2 S protein were recorded in binding buffer (25 mM HEPES, 150 mM NaCl pH 7.5, 0.04% Tween-20). Data were analyzed by Octet Data Analysis HT 12.0 software. The reference sample and reference sensor were subtracted, and KD values were analyzed using a 1:1 global fit model. Data were plotted using Prism V8.0 software (GraphPad).
Cryo-EM sample preparation and data acquisition
The ECD-PD and ECD-IgG complex was concentrated to ~3 mg/mL applied to the grids, JN.1 RBD-SLC6A19-ACE2 was concentrated to ~8 mg/mL applied to the grids. Aliquots (3.3 μL) of the protein were placed on glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3). The grids were blotted for 3.0 s and flash-frozen in liquid ethane cooled by liquid nitrogen with FEI Mark IV Vitrobot at 8 °C and 100% humidity at 8 °C and 100% humidity. The prepared grids were transferred to a Titan Krios operating at 300 kV equipped with Gatan K3 detector and GIF Quantum energy filter. Movie stacks were automatically collected using AutoEMation46 and EPU software, with a slit width of 20 eV on the energy filter and a defocus range from −1.2 to −1.6 µm in super-resolution mode at a nominal magnification of 105,000×. Each stack was exposed for 1.14 s with an exposure time of 0.035 s/frame, resulting in a total of 32 frames/stack. The total dose rate was ~50 e−/Å2 for each stack. The stacks were motion corrected with MotionCor247 and binned twofold, resulting in a pixel size of 0.855, 0.83, and 1.1 Å/pixel. Meanwhile, dose weighting was performed48. The defocus values were estimated with Gctf49.
Data processing
The Cryo-EM structure of S protein from BA.2.86/JN.1 and JN.1 RBD-SLC6A19-ACE2 has been solved first50 and identical protocol was applied to the complex of other ECD-PD and ECD-IgG complex. Particles for all samples were automatically picked using cryoSPARC from manually selected micrographs. After 2D classification with cryoSPARC, good particles were selected and subject to multiple cycle of heterogeneous refinement without symmetry using cryoSPARC51. The good particles were selected and subjected to local CTF refinement with C1 symmetry, non-uniform refinement, resulting in the 3D reconstruction for the whole structures.
To further improve the map quality of ACE2–RBD and IgG–RBD interfaces, the particles were expanded and re-extracted at the location of these interfaces. The re-extracted dataset was subject to several cycles of 3D classified and focused refinement, resulting in a 3D reconstruction with better quality for ACE2–RBD and IgG–RBD interface. The resolution was estimated with the gold-standard Fourier shell correlation 0.143 criterion52 with high-resolution noise substitution53. Refer to Figs. S6, S11, S16 and Tables S1, S3 for details of data collection and processing.
Model building and structure refinement
For model building of the complex of ECD-PD, the atomic model of the BA.2.86 S protein in complex with PD of ACE2 (PDB ID: 7Y1Y) was used as templates, which were molecular dynamics flexible fitting (MDFF)54 into the whole cryo-EM map of the complex and the focused-refined cryo-EM map of the RBD-PD sub-complex, respectively.
For model building of the complex of RBD-Fab, the atomic model of the IgG-BA.2 RBD (PDB ID: 7XSW) was sequence-substituted to the BA.2.86, in chainsaw and fitted into focused-refined maps of IgG–RBD interface using MDFF.
For model building of the complex of JN.1 RBD-SLC6A19-ACE2, the atomic model of the WT RBD-SLC6A19-ACE2 (PDB ID: 6M17) was used as templates, which were MDFF11 into the whole cryo-EM map of the complex and the focused-refined cryo-EM map of the RBD–ACE2 sub-complex, respectively.
Each residue was manually checked with the chemical properties taken into consideration during model building. Several segments, whose corresponding densities were invisible, were not modeled. Structural refinement was performed in Phenix55 with secondary structure and geometry restraints to prevent overfitting. To monitor the potential overfitting, the model was refined against one of the two independent half maps from the gold-standard 3D refinement approach. Then, the refined model was tested against the other map. Statistics associated with data collection, 3D reconstruction, and model building were summarized in Tables S1 and S3.
Proteolytic digestion of recombinant S proteins
The recombinant proteins were dissolved in 8 M urea in the buffer consisting of 25 mM HEPES and 500 mM NaCl. Dithiothreitol (5 mM) and iodoacetamide (10 mM) were added into the solution to reduce and alkylate the denatured proteins, respectively. The proteins were then divided into two equal aliquots for the proteolytic digestion. The first aliquot was treated with chymotrypsin (Promega, enzyme/substrate ratio was 1:10), while the second aliquot was digested with trypsin (Sigma, enzyme/substrate ratio was 1:10) followed by chymotrypsin (Promega, enzyme/substrate ratio was 1:10). The resulting peptides were desalted with Sep-Pak C18 reverse-phase filter cartridge (Waters). The N-linked glycopeptide fraction of the recombinant S proteins was then purified with Oasis Max extraction cartridge (30 mg, Waters). The peptides were dried using Speed-Vac (JM Technology Co.).
Mass spectrometry analysis
A 100 μm i.d. capillary packed with 1.9 μm C18 resin (Dr. Maisch GmbH) was attached to an EASY-nLC 1000 system (Thermo Fisher Scientific) and placed in line with the heated capillary of the Fusion Orbitrap Tribrid mass spectrometer (Thermo Fisher Scientific). Approximately 2 μg of the N-linked glycopeptides was loaded onto the column. An 87 min gradient was deployed: 0–2 min 3–6% B, 2–62 min 6–30% B, 62–71 min 30–60% B, 71–72 min 60–90% B, 72–77 min 90% B, 77–78 min 90–50% B, and 78–87 min 50% B. The flow rate was set as 250 nL min−1. The mass spectrometer was operated in a data-dependent mode. The parameters of the mass spectrometer were set as follows: MS1 resolution 60 K, scan range 500–2000, AGC target 5E5, maximum injection time 50 ms, dynamic exclusion 45 s, charge inclusion 2–6, cycle time 3 s; MS2 resolution 15 K, AGC target 1E5, collision energy 34.
Mass spectrometry data processing
Mass spectrometry data of N-linked glycopeptides were searched using pGlyco 3.0. The parameters were set as follows: fragmentation HCD, human protein database downloaded from UniProt (in January 2019) including the sequences of trimeric S proteins analyzed in this study, enzyme chymotrypsin (or a combination of trypsin and chymotrypsin), maximal missed cleavage 6, peptide length 6–40, peptide mass 600–4000, fixed modification carbamidomethylation (C +57.022 Da), variable modifications oxidation methionine (M +15.995 Da) and acetylation on protein N-terminus (+42.011 Da), glycan database default human N-linked glycan database, precursor tolerance 10 ppm, fragment tolerance 20 ppm, glycopeptide FDR 0.01. The software pGlycoQuant was employed to extract the intensities of glycopeptides. The parameters were set as follows: type of identification results pGlyco, threshold FDR 0.01, type of quantification DDA label free. The statistical analyses were performed with GraphPad 9.0 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the Cryo-EM Facility and Supercomputer Center of Westlake University for providing cryo-EM and computation support, respectively. We would like to acknowledge Dr. Hua Li at the core research facilities of the Southern University of Science and Technology for the support on mass spectrometry analysis. This work was funded by the National Science Fund for Distinguished Young Scholars (82025022 to Z.Z.), the National Science Fund for Outstanding Young Scholars (82322040 to B.J.), the National Natural Science Foundation of China (92169204 to Z.Z., 82171752 to B.J., and 22374063 to L.C.), the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034 to B.J.), the Guangdong Science and Technology Plan Project, construction of high-level biosafety laboratories (2021B1212030010 to Z.Z.), the Shenzhen Science and Technology Program (RCYX20231211090342048 to R.Y., RCYX20200714114700046 to B.J., and ZDSYS20210623091810030 to Z.Z.), the Science and Technology Innovation Commission of Shenzhen Municipality (JSGG20220226085550001 to R.Y. and Z.Z., JCYJ20220530163413031 to H.G., and JCYJ20200109144201725 to Z.Z.), the Chinese Academy of Medical Sciences Clinical and Translational Medicine Research Project (2022-I2M-C&T-B-113 to Z.Z.), and the Shenzhen High-level Hospital Construction Fund (23250G1002 to B.J.).
Author contributions
R.Y., L.C., M.L., B.J., and Z.Z. conceived the project. L.C., M.L., R.Y., B.J., and Z.Z. designed the experiments. H.Y. did the molecular cloning, protein purification, and cryo-EM sample preparation. H.G. did the pseudovirus neutralizing assay. A.W. did the structure determination and model building. H.Y., H.G., A.W., and L.C. performed most experiments and analyzed the data together with assistance from Q.F., J.J., M.W., L.L., X.G., H.W., and R.Z. R.Y., L.C., B.J., and Z.Z. wrote the manuscript. All authors read and approved this version of the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Atomic coordinates and cryo-EM density maps of Omicron BA.2.86 S protein in complex with PD of ACE2 (PDB: 8WYH, whole map: EMD-37928), the interface between BA.2.86 RBD and ACE2 (local map: EMD-37927), BA.2.86 S protein in complex with S309 and SA55 (PDB: 8WYJ, whole map: EMD-37930), the interface between BA.2.86 RBD in complex with S309 and SA55 (local map: EMD-37918), cryo-EM density maps of Omicron JN.1 S protein in complex with PD of ACE2 (whole map: EMD-60028), the interface between JN.1 RBD and ACE2 (PDB: 8ZBQ, local map: EMD-39907), have been deposited to the Protein Data Bank (http://www.rcsb.org) and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/), respectively. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Haonan Yang, Huimin Guo, Aojie Wang, Liwei Cao.
Contributor Information
Liwei Cao, Email: caolw@sustech.edu.cn.
Ming Liao, Email: mliao@scau.edu.cn.
Renhong Yan, Email: yanrh@sustech.edu.cn.
Bin Ju, Email: jubin2013@163.com.
Zheng Zhang, Email: zhangzheng1975@aliyun.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51973-8.
References
- 1.Iketani, S. et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature604, 553–556 (2022). 10.1038/s41586-022-04594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller, J. et al. Substantial neutralization escape by SARS-CoV-2 Omicron variants BQ.1.1 and XBB.1. N. Engl. J. Med.388, 662–664 (2023). 10.1056/NEJMc2214314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yisimayi, A. et al. Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting. Nature625, 148–156 (2024). [DOI] [PMC free article] [PubMed]
- 4.Chen, X. et al. Protective effect of plasma neutralization from prior SARS-CoV-2 Omicron infection against BA.5 subvariant symptomatic reinfection. Lancet Reg. Health West. Pac.33, 100758 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nehlmeier, I. et al. Host cell entry and neutralisation sensitivity of the SARS-CoV-2 XBB.1.16 lineage. Cell. Mol. Immunol.20, 969–971 (2023). 10.1038/s41423-023-01030-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen, M. et al. First cases of SARS-CoV-2 BA.2.86 in Denmark, 2023. Eurosurveillance28, 2300460 (2023). 10.2807/1560-7917.ES.2023.28.36.2300460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang, S. et al. Antigenicity and infectivity characterisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis.23, e457–e459 (2023). 10.1016/S1473-3099(23)00573-X [DOI] [PubMed] [Google Scholar]
- 8.Wang, Q. et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature624, 639–644 (2023). [DOI] [PubMed]
- 9.Lasrado, N. et al. Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86. Vaccine41, 6904–6909 (2023). 10.1016/j.vaccine.2023.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khare, S. et al. GISAID’s role in pandemic response. China CDC Wkly.3, 1049–1051 (2021). 10.46234/ccdcw2021.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meo, S. A., Meo, A. S. & Klonoff, D. C. Omicron new variant BA.2.86 (Pirola): epidemiological, biological, and clinical characteristics - a global data-based analysis. Eur. Rev. Med. Pharmacol. Sci.27, 9470–9476 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Yang, S. et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis.24, e70–e72 (2024). 10.1016/S1473-3099(23)00744-2 [DOI] [PubMed] [Google Scholar]
- 13.Kaku, Y. et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis.24, e82 (2024). 10.1016/S1473-3099(23)00813-7 [DOI] [PubMed] [Google Scholar]
- 14.Yan, R. et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science367, 1444–1448 (2020). 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan, R. et al. Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2. Cell Res.31, 717–719 (2021). 10.1038/s41422-021-00490-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., Shen, Y., Zhang, Y. & Yan, R. Structural basis for the enhanced infectivity and immune evasion of Omicron subvariants. Viruses15, 1398 (2023). 10.3390/v15061398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai, Y. et al. Distinct conformational states of SARS-CoV-2 spike protein. Science369, 1586–1592 (2020). 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, J. et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science372, 525–530 (2021). 10.1126/science.abf2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang, J. et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA117, 11727–11734 (2020). 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol.23, 3–20 (2022). 10.1038/s41580-021-00418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.San Filippo, S. et al. Comparative efficacy of early COVID-19 monoclonal antibody therapies: a retrospective analysis. Open Forum Infect. Dis.9, ofac080 (2022). 10.1093/ofid/ofac080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature583, 290–295 (2020). 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 23.Cao, Y. et al. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep.41, 111845 (2022). 10.1016/j.celrep.2022.111845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaoka, Y. et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Res. Sq.607, 119–127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao, Y. et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe30, 1527–1539.e1525 (2022). 10.1016/j.chom.2022.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao, Y. et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature608, 593–602 (2022). 10.1038/s41586-022-04980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue, C. et al. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis.23, 278–280 (2023). 10.1016/S1473-3099(23)00010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju, B. et al. Striking antibody evasion of SARS-CoV-2 Omicron sub-lineages BQ.1.1, XBB.1 and CH.1.1. Natl Sci. Rev.10, nwad148 (2023). 10.1093/nsr/nwad148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature614, 521–529 (2023). [DOI] [PMC free article] [PubMed]
- 30.Röltgen, K. et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell185, 1025–1040.e1014 (2022). 10.1016/j.cell.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, H. et al. Additional mutations based on Omicron BA.2.75 mediate its further evasion from broadly neutralizing antibodies. iScience26, 106283 (2023). 10.1016/j.isci.2023.106283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasoba, D. et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell185, 2103–2115.e2119 (2022). 10.1016/j.cell.2022.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen, Y. et al. Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2. Cell Discov.8, 123 (2022). 10.1038/s41421-022-00488-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jian, F. et al. Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding. PLoS Pathog.19, e1011868 (2023). 10.1371/journal.ppat.1011868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr, T. N. et al. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog.18, e1010951 (2022). 10.1371/journal.ppat.1010951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magaret, C. A. et al. Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features. Nat. Commun.15, 2175 (2024). 10.1038/s41467-024-46536-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, Z. et al. Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape. Nat. Commun.13, 4958 (2022). 10.1038/s41467-022-32665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Y. N. et al. Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor. Cell Discov.9, 108 (2023). 10.1038/s41421-023-00607-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe, Y. et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science369, 330–333 (2020). [DOI] [PMC free article] [PubMed]
- 40.Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science367, 1444–1448 (2020). 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science375, 864–868 (2022). 10.1126/science.abn8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stalls, V. et al. Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike. Cell Rep.39, 111009 (2022). 10.1016/j.celrep.2022.111009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gobeil, S. M. et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science373, eabi6226 (2021). 10.1126/science.abi6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S. & Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science369, 330–333 (2020). 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell182, 1295–1310 e1220 (2020). 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol.150, 69–80 (2005). 10.1016/j.jsb.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 47.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife4, e06980 (2015). 10.7554/eLife.06980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol.193, 1–12 (2016). 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaning, L. I. et al. Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants. iScience25, 104431 (2022). 10.1016/j.isci.2022.104431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol.333, 721–745 (2003). 10.1016/j.jmb.2003.07.013 [DOI] [PubMed] [Google Scholar]
- 53.Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy135, 24–35 (2013). 10.1016/j.ultramic.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure16, 673–683 (2008). 10.1016/j.str.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr.66, 213–221 (2010). 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and cryo-EM density maps of Omicron BA.2.86 S protein in complex with PD of ACE2 (PDB: 8WYH, whole map: EMD-37928), the interface between BA.2.86 RBD and ACE2 (local map: EMD-37927), BA.2.86 S protein in complex with S309 and SA55 (PDB: 8WYJ, whole map: EMD-37930), the interface between BA.2.86 RBD in complex with S309 and SA55 (local map: EMD-37918), cryo-EM density maps of Omicron JN.1 S protein in complex with PD of ACE2 (whole map: EMD-60028), the interface between JN.1 RBD and ACE2 (PDB: 8ZBQ, local map: EMD-39907), have been deposited to the Protein Data Bank (http://www.rcsb.org) and the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/), respectively. Source data are provided with this paper.