Abstract
Quantum sensing has the potential to improve the sensitivity of chemical sensing by exploiting the characteristics of qubits, which are sensitive to the external environment. Modulation of quantum coherence by target analytes can be a useful tool for quantum sensing. Using molecular qubits is expected to provide excellent sensitivity due to the proximity of the sensor to the target analyte. However, many molecular qubits are used at cryogenic temperatures, and how to make molecular qubits respond to specific analytes remains unclear. Here, we propose a material design in which the coherence time changes in response to a variety of analytes at room temperature. We used the photoexcited triplet, which can be initialized at room temperature, as qubits and introduce them to a metal–organic framework that can flexibly change its pore structure in response to guest adsorption. By changing the local molecular density around the triplet qubits by adsorption of a specific analyte, the mobility of the triplet qubit can be changed, and the coherence time can be made responsive.
Subject terms: Metal-organic frameworks, Excited states
Molecular qubits have great potential as quantum sensors due to their ability to be in close proximity to target analytes, but design guidelines on how to make them respond to specific analytes remain unclear. Here authors show that hybrids of triplet qubits and flexible MOFs can change the coherence time of electron spins of photo-excited triplet state upon adsorption of guest.
Introduction
In the second quantum revolution, quantum information science (QIS) comprises the application of quantum mechanics to fields such as quantum computing1,2, quantum communication3,4, and quantum sensing5–10. Quantum bits (qubits) are its building blocks, and manipulating these superposition states creates new properties that cannot be achieved by classical bits11,12. Among the quantum technologies, quantum sensing will have an impact on a wide range of fields, including chemistry9 and biology10. An important parameter for quantum sensing is the time over which the coherence of spin correlations is preserved13. Since the sensitivity of quantum sensing depends on coherence time, a long coherence time is highly desirable5. Defect centers, such as the nitrogen-vacancy (NV) centers in diamond, have long coherence times of more than microseconds at room temperature, enabling sensing of a variety of small molecules and biomolecules8,10. However, defect centers are difficult to place in a specific position, and it is also difficult to selectively address different centers14.
Unlike defect-based systems, molecular qubits14–31 are uniform in structure and can be precisely controlled. Furthermore, their small size allows them to closely approach the target analyte. For electron spin-based systems, the coherence time can be determined as the spin-spin relaxation time (T2). Paramagnetic metal complexes15–18, radical pairs22, and metal complex-radical conjugates24–26 are typical examples of molecular qubits. Although some radical is known to show long T2 over several microseconds32, it is still not easy to achieve microsecond-scale T2 at room temperature for many molecular qubits. Furthermore, it remains unclear how to make molecular qubits responsive and selective to specific analytes.
One strategy to gain responsivity to analytes is employing porous materials such as metal-organic frameworks (MOFs) or covalent–organic frameworks (COFs). There are a few examples of quantum sensing using MOFs containing paramagnetic species33,34. In these reports, the hyperfine interaction between electron spins and nuclear spins was used to sense analytes, but this method left a major challenge in that it could not distinguish compounds composed of the same nuclear species, such as benzene and toluene, for example.
Another way to sense analytes is by using the modulation of the coherence time, known as relaxometry. In general, T2 depends on several factors, such as the motion of the qubit itself and surrounding spins11,35. The motion of the qubit changes the orientation of the molecule relative to the external magnetic field, and since the g factor is usually anisotropic, the interaction with the magnetic field is perturbed, causing relaxation. Other sources of relaxation are the surrounding electron and nuclear spins. In particular, nuclear motion in the surrounding molecules causes fluctuation in the local magnetic field, leading to relaxation. Relaxometry can be a powerful method to detect target analytes based on these mechanisms. However, since qubits exposed in the nanopores of MOFs and COFs can undergo significant relaxation from the motion of the qubit itself, it is necessary to design the environment around the qubit appropriately to have a sufficiently long T2.
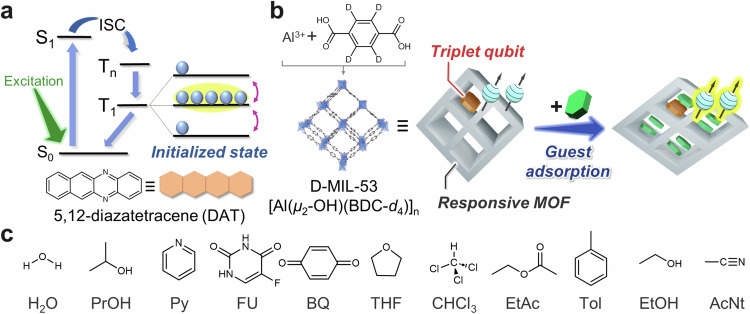
The photoexcited triplet state of organic dyes is an excellent candidate for qubits. Photoexcited triplet states have the advantage that anisotropic spin-orbit coupling allows temperature-independent sublevel selective intersystem crossing (ISC) from photoexcited singlet states, creating highly polarized electronic states, even at room temperature36–39 (Fig. 1a). This is a major difference from the spins of metal complexes and radicals in thermal equilibrium33,34, and triplets can offer higher spin polarization than those systems at room temperature and low magnetic fields. This means that more spins are available as qubits in the triplet state, which is an advantage in multi-qubit systems. Furthermore, it has long been known that the triplet state of pentacene doped in dense p-terphenyl crystals exhibits a long T2 on the microsecond scale even at room temperature37. However, except for a few pioneering examples27–30, photoexcited triplets are largely unexplored in the context of qubits and have never been used for quantum sensing because they are buried in dense matrix crystals and cannot approach the target analyte. As with other paramagnetic qubits, the response of triplet qubits to target analytes based on hyperfine spectroscopy and relaxometry is expected34. A characteristic of triplets is that they have an anisotropic and large (~ GHz) zero-field splitting (ZFS) interaction so that the molecular motion of the qubit causes a pronounced relaxation based on orientation change with respect to the magnetic field40. Therefore, when target analytes are incorporated into the nanopores and the environment around the qubit changes, it is expected that the changes can be sensitively captured through relaxometry.
Fig. 1. Strategy for quantum sensing using triplet qubits and a flexible MOF.
a Initialization of the triplet qubit. Spin polarization between two of the three triplet sublevels generated by photoexcitation and following spin-selective intersystem crossing (ISC) can be used as qubits. b Response of triplet qubits to the guest-induced structural change of a flexible MOF. DAT and D-MIL-53 are employed as triplet qubits and the flexible MOF, respectively. c Guest analyte molecules used in this research. H2O, 2-propanol (PrOH), pyridine (Py), 5-fluorouracil (FU), p-benzoquinone (BQ), THF, chloroform (CHCl3), ethyl acetate (EtAc), toluene (h-Tol), ethanol (EtOH), and acetonitrile (AcNt) are introduced to D-MIL-53⊃DAT.
Here, we propose a material design toward quantum sensing in which triplet qubits are supported in nanoporous metal-organic frameworks (MOFs) to make the analyte molecules accessible and the coherence time T2 of the triplet qubits is changed by flexibly changing the MOF structure through adsorption of the guest analyte. Our method can show different T2 responses for a wider variety of organic compounds. In the present system, MOF has molecular recognition ability by interacting with guest analytes, and triplet qubit is a reporter that outputs it as the change in the coherence time T2. In other words, qubit and analyte do not interact directly but are indirectly influencing each other via MOF. By utilizing the change in molecular motion of qubits due to the introduction of guest molecules, we have succeeded in harnessing the molecular motion, which is normally a source of harsh relaxation at room temperature, to change the quantum coherence.
We selected MIL-53 as a flexible MOF that exhibits guest-responsive structural change, termed breathing behavior41–43. Some MOFs can flexibly change their crystal structure by introduction and removal of guest molecules44–46. The N-substituted tetracene, 5,12-diazatetracene (DAT)36, was employed as the triplet qubit. Compared with conventional pentacene, DAT has the advantages of good solubility and superior air stability. Recently, we reported the successful introduction of DAT into MIL-53 nanopores and polarization transfer from the electron spins of the DAT triplet to the 19F nuclear spins of the guest fluorouracil47.
We introduced a variety of additional guest analyte molecules into DAT-accommodated MIL-53 and evaluated T2 of the DAT triplet using pulsed electron paramagnetic resonance (EPR) (Fig. 1b). Analyte molecules have nuclear spins, such as that on protons, which usually work as noise to shorten T2. Interestingly, the T2 of DAT in MIL-53 shows various T2 depending on the type of guest analyte, with the longest T2 being longer than 1 μs at room temperature. This can be mainly attributed to the local molecular density around the DAT and has led to a new concept of quantum sensing by controlling the dynamics of qubits.
Results and discussion
DAT was introduced into partially deuterated MIL-53 [Al(µ2-OH)(BDC-d4)]n (denoted as D-MIL-53⊃DAT; BDC-d4 = perdeuterated terephthalate) by simply soaking D-MIL-53 in a dichloromethane solution of DAT, according to our previous report47. Since the MOF was washed several times with dichloromethane during sample preparation, excess DAT was removed and it is unlikely that DAT remains on the MOF crystal surface. As the concentration of the DAT solution is increased, the structure of the obtained MOF changes gradually (Supplementary Fig. 1), supporting the accommodation of DAT in the MOF nanopores. The loading amount of DAT was estimated to be 0.87 wt% by UV–Vis absorbance after digesting D-MIL-53 (Supplementary Fig. 2), similar to our previous report47. The following 11 guests were additionally introduced to D-MIL-53⊃DAT: H2O, 2-propanol (PrOH), pyridine (Py), 5-fluorouracil (FU), p-benzoquinone (BQ), THF, chloroform (CHCl3), ethyl acetate (EtAc), toluene (h-Tol), ethanol (EtOH), and acetonitrile (AcNt). FU and BQ were introduced by sublimation, and other guests were introduced by exposing D-MIL-53 to each vapor. Guest molecules were selected to cover as wide a range of volumes as possible based on the previous reports41–43. In addition, for liquid guests, we selected those with boiling points high enough not to be evacuated during sample preparation and not too high to be introduced by vapor diffusion. For solid guests, we chose those that could be introduced by sublimation.
Guest-dependent coherence time (T2)
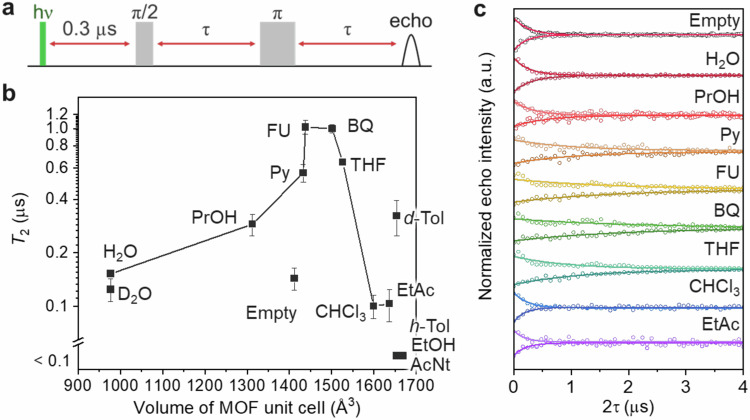
The coherence times (T2) of the photoexcited triplets of DAT in D-MIL-53 were measured by pulsed EPR at room temperature with a spin echo sequence varying the pulse intervals (Fig. 2a). Measurements were taken at the magnetic field of the high-field and low-field peaks of the EPR spectra shown later, respectively, and both decay curves are shown (Fig. 2c). The decay of echo intensity was fitted with a single exponential function and T2 was obtained as the decay constant. While DAT shows electron spin echo envelope modulation (ESEEM) due to hyperfine coupling with 1H nuclei and 14N nuclei at a higher magnetic field, it does not affect the estimation of T2. The T2 values obtained at the higher and lower magnetic fields are similar (Supplementary Table 1). A relatively short T2 of around 0.1 μs is observed for D-MIL-53⊃DAT without any guest analyte (termed empty). No echo signals are detected when h-Tol, EtOH, or AcNt are introduced as guests, probably due to the shortening of T2 (< 0.1 μs). This is not strange, since proton nuclear spins usually work as noise and reduce coherence time13. Meanwhile, interestingly, the introduction of some guest molecules such as Py, FU, BQ, and THF clearly elongated T2 of the DAT triplets, despite the increase in proton and other nuclear spins that could cause spin relaxation. The T2 of DAT in the presence of FU and BQ are over 1 μs, which are notably long as molecular qubits at room temperature48. To confirm the measurements were run in the spin dilute regime, the T2 was measured with different laser powers (Supplementary Fig. 3). As expected from the low concentration of DAT in the sample (0.87 wt%), the change of T2 was small and intermolecular interactions between electron spins of DAT triplets could be ignored.
Fig. 2. Guest-dependent T2 of DAT triplets in D-MIL-53.
a Spin echo sequence used for T2 measurement. The intervals of microwave pulses and echo detection were varied. b Plot of T2 and the fit error obtained by the single-exponential fitting of the spin echo decay curves against the unit cell volume of MIL-53. Only the T2 obtained from the low field peaks where the effect of ESEEM was small is plotted. For the empty, FU, and THF samples, the means and the standard errors of three measurements are shown. Errors of the H2O and THF samples were small and buried in the symbol. c Spin echo decay curves after pulsed photoexcitation at 532 nm for empty (D-MIL-53⊃DAT) and guest-filled (D-MIL-53⊃[DAT + guest]) samples for each guest shown in Fig. 1 at room temperature. The decay curves of each sample at the magnetic field corresponding to the higher and lower EPR peaks (Fig. 3a) are shown at the top and bottom, respectively. Single-exponential fitting curves for each sample are also shown. Echo signals were not observed when the guest was h-Tol, EtOH, or AcNt.
The T2 value was plotted against the cell volume of pristine MIL-53 (Fig. 2b). With increasing cell volume, T2 shows a non-monotonic trend, becoming longer and then shorter. Interestingly, triplet qubits and flexible MOF hybrids exhibit different coherence times for various guest molecules at room temperature.
Possible mechanism of guest-dependent T2
To obtain information about the interaction of DAT with guest molecules in the ground and excited triplet states, we obtained UV–Vis absorption and time-resolved EPR spectra, respectively. For D-MIL-53⊃[DAT + H2O] (MIL-53 immediately absorbs water and forms a hydrated structure), DAT shows absorption peaks at 450, 480, and 515 nm derived from π–π* transitions. This is consistent with the absorption peaks of DAT molecularly dispersed in p-terphenyl crystal and is clearly different from the red-shifted peaks of DAT in its aggregated solid state, as we previously reported36 (Supplementary Fig. 4). With the exception of BQ, where the analyte guest itself shows absorption, no significant change in the absorption spectrum is observed when various guest molecules are introduced (Supplementary Fig. 5). A plot of the T2 versus the dielectric constants of guests also showed no correlation (Supplementary Fig. 6). This indicates that the interaction between DAT and the guest molecules is small in the ground state and that DAT is not aggregated by the introduction of guests.
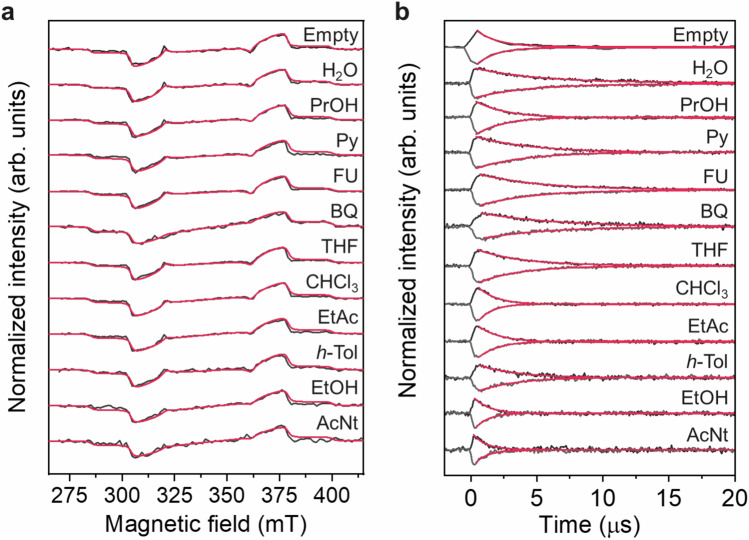
Figure 3a shows the time-resolved EPR spectra of the DAT triplet in D-MIL-53 in the absence and presence of guest molecules. All the samples show similar EPR spectra typical for a polarized DAT triplet, and it is consistent with the echo-detected field-swept spectra obtained from the pulsed EPR measurements (Supplementary Fig. 7). There is no significant motional narrowing as in the EPR spectrum of the triplet state in solution49, indicating that the incorporation of DAT into the MOF nanopores effectively suppresses its rotation. The EPR spectra were simulated using the EasySpin toolbox50 in MATLAB (red lines in Fig. 3a and Supplementary Table 1). The slight changes in the relative zero-field populations (Px: Py: Pz) were observed probably due to the change in vibronic spin-orbit coupling51, but the T2 values were unrelated to the relative populations (Supplementary Fig. 8). There are also no large differences in zero-field splitting parameters (D and E) among the samples, which reflect the electron structure of DAT, indicating the absence of strong electronic interactions between the DAT triplet and guest analytes. In addition, the DAT structure would be kept after guest introductions.
Fig. 3. Time-resolved EPR measurements of DAT in D-MIL-53.
a Time-resolved EPR spectra at 0.1–0.5 µs for empty (D-MIL-53⊃DAT) and guest-filled (D-MIL-53⊃[DAT + guest]) samples for each guest shown in Fig. 1 at room temperature (black). The simulated EPR spectra are shown as red lines. b Decays of the EPR peaks at higher and lower magnetic fields after pulsed photoexcitation at 532 nm for empty (D-MIL-53⊃DAT) and guest-filled (D-MIL-53⊃[DAT + guest]) samples at room temperature. Decay curves were fitted by a single exponential function, and the fitting results are shown as red curves.
The spin-lattice relaxation time (T1) was estimated from the decay of the EPR signal after light irradiation (Fig. 3b). Even under continuous microwave irradiation which makes the decay faster, the T1 value for each sample is more than an order of magnitude longer than the corresponding T2 value, indicating that T2 is not dominated by T1 (Supplementary Fig. 9 and Supplementary Table 1). The guest dependence of T1 shows a similar trend to that of T2, with some exceptions (such as H2O). This result suggests that additional relaxation mechanisms exist for spin-spin relaxation.
To gain insight into the local molecular density around DAT in the MOF nanopores, thermogravimetric analysis (TGA) was performed (Supplementary Fig. 10). The MOF cell volume minus the framework volume was approximated as the guest accessible pore volume, and the volume fraction of the pore volume occupied by the guest was estimated. Interestingly, the larger the volume fraction occupied by the guest, the longer the T2 (Fig. 4). h-Tol does not fit this trend, and it is possible that the rotation of the methyl group is responsible for shortening T213. When perdeuterated toluene (d-Tol) is introduced as a guest, T2 for the DAT triplet is extended to ~ 0.3 μs (Supplementary Fig. 11), which matches the trend for the other guest molecules. This suggests that, in the absence of a particularly relaxing substituent such as a methyl group, the higher the volume fraction occupied by guest (or local molecular density), the more the motility of DAT is suppressed and the longer T2 is. We also plotted T2 against other MOF parameters, such as guest-accessible pore volume and accommodated guest volume. The former shows a similar trend to MOF volume (Supplementary Fig. 12) and is considered not to determine the coherence time. The latter show weak positive correlations between the two parameters (Supplementary Fig. 13). This might be because guest molecules with larger volumes can fill larger space and suppress the motional relaxation of DAT. As for the terephthalate ligand of MIL-53, there is no change in T2 for the DAT triplet in the deuterated and protonated forms. There is little change in the T2 of the DAT triplet when H2O and D2O are used as guest molecules. Moreover, the increase of numbers of 1H or 19F nucleus due to the guest introduction should normally shorten the relaxation time, but in many of our systems, T2 was rather lengthened (Supplementary Fig. 14). Therefore, the effect of the change in the nuclear spin bath should not be a primary regulator of T2.
Fig. 4. Dependence of T2 on volume fraction occupied by the guest.
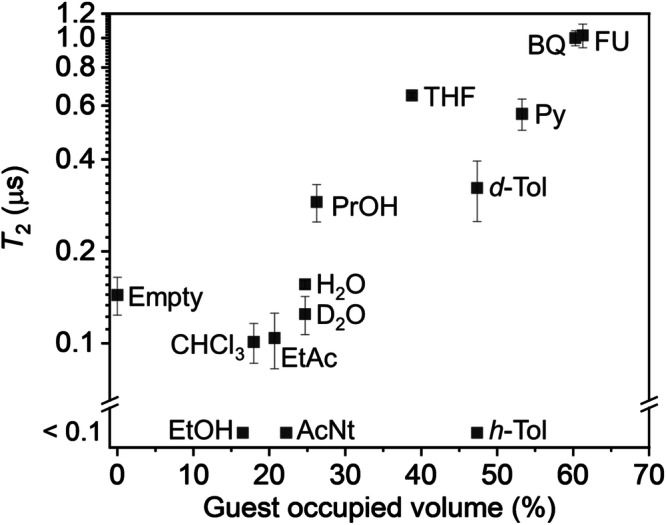
T2 and volume fraction occupied by the guest are plotted for empty (D-MIL-53⊃DAT) and guest-filled (D-MIL-53⊃[DAT + guest]) samples. The means and the standard errors of three measurements are shown for the empty, FU, and THF samples, and the fit errors obtained by the single-exponential fitting of the spin echo decay curves are shown for other samples. For the H2O and THF samples, errors were small and buried in the symbols.
Further analysis using experimental techniques such as solid-state nuclear magnetic resonance (NMR) and terahertz spectroscopy may reveal the detailed dynamics of DAT, but it is difficult to get such insights because the amount of DAT incorporated in the MOF is only 0.87 wt% and there are much larger amounts of the host ligands and other guest molecules. Indeed, we synthesized deuterated DAT (95 % D) and introduced it into MIL-53 composed of protonated (non-deuterated) terephthalate, but we could not detect any 2H NMR signal even after integrating 1000 scans with 15 kHz magic angle spinning at 9.4 T.
Instead, we performed a molecular dynamics (MD) simulation to get insights about the DAT dynamics in MIL-53. We selected three representative systems, empty (MIL-53⊃DAT), MIL-53⊃[DAT + H2O], and MIL-53⊃[DAT + BQ], which showed different guest density and either longer or shorter T2. The simulation was conducted at 300 K for 0.2 μs. We attempted to get insight into the rotational and translational motions of DAT based on the distances between the aluminum atom and the four carbon atoms (Supplementary Figs. 15, 16, and Supplementary Table 4). In the empty sample, the distances of all directions changed drastically, indicating the presence of both rotational and translational motions. In the case of MIL-53⊃[DAT + H2O], the displacement in the z (pore) direction was observed, while only fluctuated movements in the x and y directions were observed, which means translational motion in the pore direction is dominant. Importantly, judging from the standard deviation shown in Supplementary Table 4, much fewer displacements were observed for MIL-53⊃[DAT + BQ], which has the longest T2 reported in this paper, implying that DAT mobility is significantly suppressed in this system. These results support that the DAT motions can contribute to spin relaxation processes. Since μ-OH groups bridging aluminum metals were not deuterated, the interaction between the proton and DAT remained and could be modulated by the translational motion of DAT. Moreover, the DAT triplet has anisotropic zero-field splitting parameters, D and E, which can also be altered by the change of molecular orientation by rotational motions known as tumbling. Therefore, the fluctuations of interactions with surrounding proton spins of the OH groups and anisotropic parameters based on the molecular motion of DAT would cause spin-spin relaxation (See Supplementary Discussion for more information)35. Note that the T2 of D-MIL-53⊃[DAT + H2O] was comparable to the empty sample, while the result of MD simulation implies D-MIL-53⊃[DAT + H2O] has less motion than the empty sample. The possible reason is the difference in the dynamics of the Al-OH groups. The root mean square fluctuation (RMSF) value of the OH groups, which indicates how much the OH is dynamic, was found to be larger in MIL-53⊃[DAT + H2O] (Supplementary Fig. 17). This intense motion may bring additional relaxation sources to the D-MIL-53⊃[DAT + H2O] system.
Although the mobility changes in DAT can affect the linewidths of the EPR spectra, almost no change was observed for our system40. Even for the empty sample, which is considered to have the highest mobility of DAT in the present systems, the spectral shape is completely anisotropic and solid-like. This is consistent with the MD simulation result (Supplementary Fig. 16) that the timescale of the rotation is around tens of nanoseconds. Thus, even under an empty situation, the rotational motions of DAT are sufficiently slower than the EPR timescale (~ 100 ps) and might bring no notable changes.
To support the idea that DAT dynamics can be a main factor determining T2, we measured T2 at lower temperatures in the empty, guest-free state (D-MIL-53⊃DAT) (Supplementary Fig. 18). The T2 of the DAT triplet, which is 0.1 μs at room temperature, is longer at 100 and 50 K at 1.0 and 2.8 μs, respectively. Conversely, in p-terphenyl, which is a solid and dense crystal, the DAT triplet shows a long T2 of 2.4 μs, even at room temperature. These results confirm that the dynamics of DAT is important for spin–spin relaxation process.
This study demonstrates the response of quantum coherence to diverse neutral molecules by hybridizing triplet qubits in flexible MOFs. The photoexcited triplet, which is initializable at room temperature and exhibits a long coherence time, was used as qubits, and the MOF, which can flexibly change its structure upon adsorption of various analyte molecules, was used to make the qubits responsive. This proof-of-concept result will stimulate the development of many hybrid quantum sensors using various combinations of MOFs and qubits in the future. For example, if a MOF that strongly interacts with guests having a certain functional group, and a chromophore that can strongly interact with the MOF are used as a qubit, when a molecule that can interact more strongly with the MOF than the qubit is adsorbed as a guest, the qubits may become more mobile and a change in T2 will be observed. Since there are many examples of MOFs selectively adsorbing specific guests or showing abrupt adsorption at a certain pressure, numerous quantum sensors that read the type and concentration of molecules could be realized. This concept can be generalized to create dozens to hundreds of hybrid sensors by combining dozens of MOFs and qubits, and by recognizing the response of each analyte to these sensors as a pattern, it will be possible to selectively sense specific analytes. The current discovery is expected to lead to the creation of a new concept of quantum sensing platform, which could be called a quantum nose. The control of MOF orientation is expected to enable more advanced quantum sensing by sophisticated qubit manipulation. Our system uses photoexcited triplets as qubits, which is highly compatible with optically detected magnetic resonance (ODMR). The implementation of ODMR would lead to highly sensitive sensing and, ultimately, to single molecule quantum sensing27,52–54. The triplet-MOF complex will provide an ultra-sensitive and highly selective quantum sensing platform that will have an impact in a wide range of fields such as analytical chemistry, life sciences, and medicine.
Methods
All reagents were used as received unless otherwise noted. Aluminum chloride hexahydrate (AlCl3• 6H2O) and p-benzoquinone (BQ) were purchased from Sigma-Aldrich. p-Terphenyl was purchased from FUJIFILM Wako Pure Chemical and purified by zone melting. o-Phenylenediamine, potassium dichromate (K2Cr2O7), and ethylenediamine-N,N,N’,N’-tetraacetic acid, tetrasodium tetrahydrate salt (Na4-EDTA) were purchased from FUJIFILM Wako Pure Chemical. 2,3-Dihydroxynaphthalene and 5-fluorouracil (FU) were purchased from TCI. Glacial acetic acid and oxalic acid dihydrate were purchased from Kishida Chemical. Terephthalic acid-d4 (BDC-d4) (ring-d4, 98 atom%) was purchased from Cambridge Isotope Laboratories, Inc. 5,12-Diazatetracene (DAT) and deuterated MIL-53 (D-MIL-53) were prepared following the literature with slight modifications36,47.
General characterization
Solution-state 1H-NMR (400 MHz) spectra were recorded on a JEOL JNM-ECZ400 spectrometer using tetramethylsilane (TMS) as the internal standard. Elemental analysis was carried out using a Yanaco CHN Corder MT-5 at the Elemental Analysis Center, Kyushu University. UV–Vis absorption spectra were obtained on a JASCO V-670 spectrophotometer. Fluorescence spectra were obtained using a JASCO FP-8700 fluorescence spectrometer. Thermogravimetric analysis curves were obtained on a Rigaku Thermo Plus EVO2 under N2.
Sample preparation
The samples were prepared according to a previously reported method with slight modifications36,47.
Synthesis of DAT
A mixture of 2,3-dihydroxynaphthalene (2.00 g, 12.5 mmol) and o-phenylenediamine (1.35 g, 12.5 mmol) was put into a 50 mL flask and heated to 200 °C for 1 h under N2 atmosphere. After filtering and washing the crude product with methanol, 5,12-dihydrodibenzo[b]phenazine was obtained as a yellow solid and used in the next step without further purification.
5,12-Dihydrodibenzo[b]phenazine (2.04 g, 8.69 mmol) was added to 60 mL of glacial acetic acid to produce a suspension. K2Cr2O7 (5.32 g, 18.0 mmol) in 20 mL water was slowly added dropwise, and the mixture was stirred overnight in the dark at room temperature. After neutralization, the organic layer was extracted with CHCl3, dried with anhydrous Na2SO4, and concentrated under reduced pressure. Recrystallization from toluene gave 5,12-diazatetracene (DAT) as a reddish-orange solid (yield: 47%).
1H-NMR (400 MHz, CDCl3, TMS): δ (ppm) = 8.92 (s, 2H), 8.23 (dd, 2H), 8.13 (dd, 2H), 7.81 (dd, 2H), 7.54 (dd, 2H).
Elemental analysis for C16H10N2: calculated (%) H 4.38, C 83.46, N 12.17; found (%) H 4.30, C 83.50, N 12.03.
Synthesis of D-MIL-53
AlCl3• 6H2O (966 mg, 4 mmol), oxalic acid dihydrate (504 mg, 4 mmol), and BDC-d4 were added to 30 mL deionized H2O and put into a 50 mL Teflon-lined autoclave. The mixture was sonicated, heated at 220 °C for 72 h, and cooled to room temperature over 24 h in an oven. After filtering and washing the product with DMF (N,N-dimethylformamide) and methanol, a colorless powder was obtained.
To remove the residual BDC-d4 ligands from D-MIL-53, the obtained powder was added to 15 mL DMF and heated to 120 °C for 16 h in the oven again. The resulting product was filtered and washed with DMF and methanol. These procedures were repeated until the peak from BDC-d4 was no longer observed in thermogravimetric measurements (at least three times). The product was then activated at 150 °C for 3 h.
In ambient air, activated MIL-53 immediately adsorbs water molecules and forms hydrated structures42 (denoted as D-MIL-53⊃ H2O).
Elemental analysis for [C8H1.08D3.92O5Al1]• 1.0(H2O): calculated (%) H 3.06, C 41.75, N 0.00; found (%) H 3.05, C 41.80, N 0.00.
Introduction of DAT into D-MIL-53
D-MIL-53 (50 mg) was put into a 6 mL vial and dried under reduced pressure at room temperature over 3 h. The vial was placed in the dark for 1 h at room temperature after adding 2.5 mL dichloromethane solution of DAT (1 mM). The mixture was centrifuged, washed with fresh dichloromethane (1.0 mL, seven times), and dried under vacuum at 100 °C for 3 h. D-MIL-53⊃DAT was obtained as a yellow powder.
In the ambient air, D-MIL-53⊃DAT quickly absorbs water and forms the hydrated structure (denoted as D-MIL-53⊃[DAT + H2O]).
Quantification of the number of DAT molecules in D-MIL-53⊃DAT
The amount of DAT molecules in D-MIL-53 was calculated by UV–Vis absorbance after digesting D-MIL-53⊃DAT in aqueous Na4-EDTA solution. DAT was extracted from 20.0 mg D-MIL-53⊃DAT using 4.0 mL dichloromethane after adding 4.0 mL aqueous Na4-EDTA solution (125 mM). The absorbance at 475 nm of the dichloromethane solution of DAT was 0.055 in a 1 mm cell for 20.0 mg of D-MIL-53⊃[DAT + H2O]. From the calibration curve, the concentration of the solution was calculated to be 0.175 mM. Therefore, the total amount of DAT in the 4.0 mL dichloromethane solution was found to be 0.70 µmol ( = 0.16 mg). Since 7.8 wt% of water was contained in D-MIL-53⊃[DAT + H2O] (from the TGA curve), the weight of D-MIL-53⊃DAT without water was determined to be 18.44 mg (21.74 µmol). Thus, the number of DAT molecules per unit cell of D-MIL-53 was estimated to be 0.70/21.74 = 0.032, which corresponds to 0.87 wt% of DAT in D-MIL-53⊃DAT.
Introduction of guests into D-MIL-53⊃DAT
Guest analytes (except for FU and BQ) were introduced by exposing D-MIL-53⊃DAT to each guest vapor. D-MIL-53⊃DAT was put into a 6 mL vial and dried under reduced pressure at 80 °C for 3 h. The vial was placed in a Schlenk flask containing a small amount of solvent containing activated 3 A molecular sieves and stored in the dark overnight.
The introduction of FU and BQ into the nanochannels of D-MIL-53⊃DAT was performed by the sublimation method55. D-MIL-53⊃DAT was evacuated at room temperature for 3 h. FU (35 mg) or BQ (60 mg) were added to D-MIL-53⊃DAT, and the mixtures were put into a 30 mL Schlenk flask. After 1 h of evacuation, guests were sublimated and absorbed directly into the nanopores of the MOF at 175 °C for FU and 80 °C for BQ under reduced pressure for 1 h. Residual guests were eliminated at 175 °C or 80 °C under vacuum until the powder X-ray diffraction (PXRD) peaks of bulk FU and BQ were no longer observed. D-MIL-53⊃[DAT + FU] and D-MIL-53⊃[DAT + BQ] were obtained as a pale-yellow powder and a yellow powder, respectively.
Sample preparation for EPR measurements
Samples used in the EPR measurements were prepared as follows: D-MIL-53⊃[DAT + guests] were put into 2 mm capillaries, which were plugged with cotton wool to avoid sample scattering. For liquid guests, a small amount of guest was added to the cotton wool to maintain the vapor pressure. The capillaries were degassed under 77 K for more than 30 min with an oil pump and sealed with a flame.
EPR spectroscopy
EPR measurements were performed on a Bruker E680 operated at X-band (~ 9.6 GHz) using 2 mm glass capillaries placed into 4 mm quartz EPR tubes. A Bruker standard dielectric resonator was used as a microwave resonator (ER4118X-MD5W) using TE011 mode. The samples were excited at a wavelength of 532 nm, excitation light intensity of 2–3 mJ/pulse, pulse width of 5–12 ns, and repetition rate of 30 Hz using a Spectra-Physics Quanta-Ray YAG laser. The microwave intensity was ~ 0.06 mW for time-resolved measurements. For pulsed EPR measurements, ~ 2 μW microwave was amplified by a 1 kW amplifier and then attenuated by an attenuator to π/2 pulses with a pulse width of 16 ns. 16 ns and 24 ns microwave pulses were used as π/2 pulse and π pulse, respectively. Temperature was controlled by flowing liquid helium using an Oxford Mercury iTC. The echo decay measurements were conducted using a two-pulse spin echo sequence. The first π/2 pulse was irradiated 0.3 μs after laser excitation, and the pulse interval τ was varied. T2 was obtained by single exponential fitting of decay curves.
Estimation of guest density
The number of guest molecules per unit cell of D-MIL-53 (nguest) was calculated from the following equation (Supplementary Table 2):
| 1 |
where wloss, nDAT, mMOF, mDAT and mguest are weight loss (weight of guest) as a percentage of total weight, the number of DAT molecules per unit cell of D-MIL-53, the molecular weight of a unit cell of D-MIL-53 (equivalent to 4 times chemical formula), the molecular weight of DAT, and the molecular weight of the guest, respectively. The guest accessible pore volume per MOF unit cell (Vpore) of MIL-53⊃water was obtained as 638 Å3 by Platon software. By subtracting this Vpore from the volume of the MOF unit cell of MIL-53⊃H2O (VMOF, 977 Å3)42, the volume of the framework (Vstructure) was estimated as 339 Å3. This Vstructure was assumed to be same for all the D-MIL-53⊃[DAT + guest]. The percentage of guest occupied volume was estimated by the following equation,
| 2 |
where Vguest is the volume of the guest molecule obtained from its steric parameters (ref. 41 and Supplementary Table 5). The MOF unit cell VMOF of some of D-MIL-53⊃[DAT + guest] was estimated by PXRD peak positions using the program DICVOL56 (Supplementary Fig. 19and Supplementary Table 6). For D-MIL-53⊃[DAT + guest] containing THF, CHCl3, EtAc, h-Tol, EtOH, and AcNt as the guest, VMOF was estimated by using the unit cell volume of MIL-53 (Fe)⊃guest43, considering a linear relationship between the unit cell volumes of MIL-53 (Al)⊃guest and MIL-53 (Fe)⊃guest (Supplementary Fig. 20).
MD simulation
1. Models construction for MD simulations
To construct model structures, we referred to the reported experimental crystal structures of MIL-53 in degassed and H2O-solvated phases42, and optimized the infinite crystal structure using the Crystal17 program57,58 under periodic boundary conditions. For the optimization, we employed the pob-TZVP-rev2 basis set59 for all atoms and PBE functional60 together with Grimme D3 type dispersion correction61. The shrinking parameters were set to 2 and 2, and cell parameters were fixed to the experimental ones. Similarly, we optimized the MIL-53 in the BQ-solvated phase using the crystal structure of the desolvated phases after we changed the cell parameters to the estimated one in Supplementary Table 6. After the optimization of all three systems, we constructed each model structure of the 2 × 2 × 4 supercell (16 unit cells), where the 4 cells correspond to the pore direction. One DAT molecule was introduced into each model using the insert-molecule tool in GROMACS62. According to the experimental estimation of the amount of solvent molecules, 64 H2O molecules and 64 BQ molecules are added into each model, respectively, after the introduction of the DAT molecule.
2. Details of MD simulations
The force field parameters for the metal cluster of the MIL-53 were generated by using the Seminario methods63 in the MCPB.py modeling tool64 of AmberTools1865 with the general AMBER force field (GAFF)66. The Lennard-Jones parameters for the Al atoms and the O atoms coordinating to the Al atoms were taken from the universal force field67, and the Mulliken partial charges were taken for the literature68. The topologies files for the H2O, BQ, and DAT molecules were prepared as follows. The geometries of the three molecules were optimized using density functional theory at the B3LYP/6-311 + G(d, p) level of theory. The Mulliken atomic charges were obtained from the single point calculation at HF/6-31 G* level of theory. The optimization and charge evaluation were conducted using Gaussian 16 Rev. C. 0169. Using the obtained geometries and charges, the amber topology files were generated by the antechamber package70 with GAFF force field. The amber topology files were converted into a GROMACS format using the Python script, Acpype71.
For the equilibration of the solvents, first, energy minimization of the systems was performed based on the steepest descent algorithm with the threshold of the maximum force, 10.0 kJ/mol/nm. Second, NVT equilibration for 20 ns at 300 K were carried out for relaxation of the introduced solvent molecules within the pore, fixing the atomic positions of the MIL-53.
The main MD simulations with the canonical ensemble were performed without any restraints for 200 ns, and the snapshots were collected every 5 ps. To keep the temperature at 300 K, the velocity rescale thermostat was used72. Dynamics were propagated with a leapfrog integrator using a time step of 1 fs for the equilibration procedure and of 2 fs for the main MD simulation while preserving bond lengths using a linear constraint solver algorithm73. The geometric mixing rules were applied for cross-interactions. The particle-mesh Ewald scheme74 was used for calculating the long-range electrostatic interactions, while short-range contributions were computed with a cutoff distance of 11 Å. All simulations were carried out under periodic boundary conditions using GROMACS software version 2020.562.
To analyze the mobility of the DAT molecule in the MIL-53, we analyzed specific distances, as shown in Supplementary Figs. 15, 16, and Supplementary Table 4. Furthermore, to provide a comprehensive information about the spin relaxation, we conducted the analysis of dynamics of μ-OH (Supplementary Fig. 17).
Supplementary information
Acknowledgements
This work was partly supported by the JST-FOREST Program (JPMJFR201Y), the JST-CREST Program (JPMJCR23I6), JSPS KAKENHI (JP20H02713, JP22K19051, JP23H00304, JP23KJ1694 (A.Y.)), JST SPRING (JPMJSP2136) (A.Y.), the Innovation inspired by Nature Program of Sekisui Chemical Co. Ltd. Part of this work was conducted at Institute for Molecular Science, supported by Advanced Research Infrastructure for Materials and Nanotechnology (JPMXP1222MS0010), and by Nanotechnology Platform Program <Molecule and Material Synthesis> (JPMXP09S21MS0038), of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. We would like to thank Dr. Ryo Ohtani and Mr. Yudai Iwai for their help in the structural analysis of MIL-53. We appreciate Dr. Hiroshi Naka and Dr. Asuka Naraoka for the synthesis of deuterated DAT. We would like to thank Dr. Yusuke Nishiyama and Dr. Keiko Ideta for the measurement of solid-state NMR.
Author contributions
N. Y. conceived the project. A. Y. and N. Y. designed the experiments. A. Y. and S. F. prepared and characterized the samples, with the input of N. K. and N. Y. A. Y., M. A., T. F., and T. N. conducted the EPR measurements. J. P. and Y. H. performed MD simulations. A. Y. and N. Y. wrote the manuscript with contributions from all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The processed data generated in this study have been deposited in the figshare 10.6084/m9.figshare.20586825 and are available from the corresponding authors upon request75.
Competing interests
All authors declare that they have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51715-w.
References
- 1.Ladd, T. D. et al. Quantum computers. Nature464, 45–53 (2010). 10.1038/nature08812 [DOI] [PubMed] [Google Scholar]
- 2.Arute, F. et al. Quantum supremacy using a programmable superconducting processor. Nature574, 505–510 (2019). 10.1038/s41586-019-1666-5 [DOI] [PubMed] [Google Scholar]
- 3.Muralidharan, S. et al. Optimal architectures for long distance quantum communication. Sci. Rep.6, 20463 (2016). 10.1038/srep20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaskar, M. K. et al. Experimental demonstration of memory-enhanced quantum communication. Nature580, 60–64 (2020). 10.1038/s41586-020-2103-5 [DOI] [PubMed] [Google Scholar]
- 5.Degen, C. L., Reinhard, F. & Cappellaro, P. Quantum sensing. Rev. Mod. Phys. 89, 035002 (2017).
- 6.Guo, X. et al. Distributed quantum sensing in a continuous-variable entangled network. Nat. Phys.16, 281–284 (2019). 10.1038/s41567-019-0743-x [DOI] [Google Scholar]
- 7.Jing, M. et al. Atomic superheterodyne receiver based on microwave-dressed Rydberg spectroscopy. Nat. Phys.16, 911–915 (2020). 10.1038/s41567-020-0918-5 [DOI] [Google Scholar]
- 8.Maze, J. R. et al. Nanoscale magnetic sensing with an individual electronic spin in diamond. Nature455, 644–647 (2008). 10.1038/nature07279 [DOI] [PubMed] [Google Scholar]
- 9.Üngör, O., Ozvat, T. M., Ni, Z. & Zadrozny, J. M. Record chemical-shift temperature sensitivity in a series of trinuclear cobalt complexes. J. Am. Chem. Soc.144, 9132–9137 (2022). 10.1021/jacs.2c03115 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, T. et al. Toward quantitative bio-sensing with nitrogen-vacancy center in diamond. ACS Sens.6, 2077–2107 (2021). 10.1021/acssensors.1c00415 [DOI] [PubMed] [Google Scholar]
- 11.Hendrickx, N. W. et al. A single-hole spin qubit. Nat. Commun.11, 3478 (2020). 10.1038/s41467-020-17211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gertler, J. M. et al. Protecting a bosonic qubit with autonomous quantum error correction. Nature590, 243–248 (2021). 10.1038/s41586-021-03257-0 [DOI] [PubMed] [Google Scholar]
- 13.Jackson, C. E., Moseley, I. P., Martinez, R., Sung, S. & Zadrozny, J. M. A reaction-coordinate perspective of magnetic relaxation. Chem. Soc. Rev.50, 6684–6699 (2021). 10.1039/D1CS00001B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasielewski, M. R. et al. Exploiting chemistry and molecular systems for quantum information science. Nat. Rev. Chem.4, 490–504 (2020). 10.1038/s41570-020-0200-5 [DOI] [PubMed] [Google Scholar]
- 15.Bader, K., Winkler, M. & van Slageren, J. Tuning of molecular qubits: very long coherence and spin-lattice relaxation times. Chem. Commun.52, 3623–3626 (2016). 10.1039/C6CC00300A [DOI] [PubMed] [Google Scholar]
- 16.Giménez-Santamarina, S., Cardona-Serra, S., Clemente-Juan, J. M., Gaita-Arino, A. & Coronado, E. Exploiting clock transitions for the chemical design of resilient molecular spin qubits. Chem. Sci.11, 10718–10728 (2020). 10.1039/D0SC01187H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita, M. Next generation multifunctional nano-science of advanced metal complexes with quantum effect and nonlinearity. Bull. Chem. Soc. Jpn.94, 209–264 (2021). 10.1246/bcsj.20200257 [DOI] [Google Scholar]
- 18.von Kugelgen, S. et al. Spectral addressability in a modular two qubit system. J. Am. Chem. Soc.143, 8069–8077 (2021). 10.1021/jacs.1c02417 [DOI] [PubMed] [Google Scholar]
- 19.Jellen, M. J., Ayodele, M. J., Cantu, A., Forbes, M. D. E. & Garcia-Garibay, M. A. 2D Arrays of organic qubit candidates embedded into a pillared-paddlewheel metal-organic framework. J. Am. Chem. Soc.142, 18513–18521 (2020). 10.1021/jacs.0c07251 [DOI] [PubMed] [Google Scholar]
- 20.Yu, C. J., von Kugelgen, S., Laorenza, D. W. & Freedman, D. E. A molecular approach to quantum sensing. ACS Cent. Sci.7, 712–723 (2021). 10.1021/acscentsci.0c00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zadrozny, J. M., Gallagher, A. T., Harris, T. D. & Freedman, D. E. A porous array of clock qubits. J. Am. Chem. Soc.139, 7089–7094 (2017). 10.1021/jacs.7b03123 [DOI] [PubMed] [Google Scholar]
- 22.Harvey, S. M. & Wasielewski, M. R. Photogenerated spin-correlated radical pairs: From photosynthetic energy transduction to quantum information science. J. Am. Chem. Soc.143, 15508–15529 (2021). 10.1021/jacs.1c07706 [DOI] [PubMed] [Google Scholar]
- 23.Bayliss, S. L. et al. Optically addressable molecular spins for quantum information processing. Science370, 1309–1312 (2020). 10.1126/science.abb9352 [DOI] [PubMed] [Google Scholar]
- 24.Kirk, M. L., Shultz, D. A., Hewitt, P., Chen, J. & van der Est, A. Excited state magneto-structural correlations related to photoinduced electron spin polarization. J. Am. Chem. Soc.144, 12781–12788 (2022). 10.1021/jacs.2c03490 [DOI] [PubMed] [Google Scholar]
- 25.Paquette, M. M., Plaul, D., Kurimoto, A., Patrick, B. O. & Frank, N. L. Opto-spintronics: photoisomerization-induced spin state switching at 300 K in photochrome cobalt-dioxolene thin films. J. Am. Chem. Soc.140, 14990–15000 (2018). 10.1021/jacs.8b09190 [DOI] [PubMed] [Google Scholar]
- 26.McGuire, J., Miras, H. N., Donahue, J. P., Richards, E. & Sproules, S. Ligand radicals as modular organic electron spin qubits. Chem. Eur. J.24, 17598–17605 (2018). 10.1002/chem.201804165 [DOI] [PubMed] [Google Scholar]
- 27.Christensen, J. A., Zhou, J., Tcyrulnikov, N. A., Krzyaniak, M. D. & Wasielewski, M. R. Spin-polarized molecular triplet states as qubits: Phosphorus hyperfine coupling in the triplet state of benzoisophosphinoline. J. Phys. Chem. Lett.11, 7569–7574 (2020). 10.1021/acs.jpclett.0c01912 [DOI] [PubMed] [Google Scholar]
- 28.Mayländer, M., Chen, S., Lorenzo, E. R., Wasielewski, M. R. & Richert, S. Exploring photogenerated molecular quartet states as spin qubits and qudits. J. Am. Chem. Soc.143, 7050–7058 (2021). 10.1021/jacs.1c01620 [DOI] [PubMed] [Google Scholar]
- 29.Kothe, G. et al. Initializing 214 pure 14-qubit entangled nuclear spin states in a hyperpolarized molecular solid. J. Phys. Chem. Lett.12, 3647–3654 (2021). 10.1021/acs.jpclett.1c00726 [DOI] [PubMed] [Google Scholar]
- 30.Jacobberger, R. M., Qiu, Y., Williams, M. L., Krzyaniak, M. D. & Wasielewski, M. R. Using molecular design to enhance the coherence time of quintet multiexcitons generated by singlet fission in single crystals. J. Am. Chem. Soc.144, 2276–2283 (2022). 10.1021/jacs.1c12414 [DOI] [PubMed] [Google Scholar]
- 31.Kirner, S. V. et al. On-off switch of charge-separated states of pyridine-vinylene-linked porphyrin-C60 conjugates detected by EPR. Chem. Sci.6, 5994–6007 (2015). 10.1039/C5SC02051D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yong, L. et al. Electron spin relaxation of triarylmethyl radicals in fluid solution. J. Magn. Reson.152, 156–161 (2001). 10.1006/jmre.2001.2379 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, J. et al. Embedded nano spin sensor for in situ probing of gas adsorption inside porous organic frameworks. Nat. Commun.14, 4922 (2023). 10.1038/s41467-023-40683-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, L. et al. Room-temperature quantitative quantum sensing of lithium ions with a radical-embedded metal-organic framework. J. Am. Chem. Soc.144, 19008–19016 (2022). 10.1021/jacs.2c07692 [DOI] [PubMed] [Google Scholar]
- 35.Mirzoyan, R., Kazmierczak, N. P. & Hadt, R. G. Deconvolving contributions to decoherence in molecular electron spin qubits: A dynamic ligand field approach. Chem. Eur. J.27, 9482–9494 (2021). 10.1002/chem.202100845 [DOI] [PubMed] [Google Scholar]
- 36.Kouno, H. et al. Nonpentacene polarizing agents with improved air stability for triplet dynamic nuclear polarization at room temperature. J. Phys. Chem. Lett.10, 2208–2213 (2019). 10.1021/acs.jpclett.9b00480 [DOI] [PubMed] [Google Scholar]
- 37.Sloop, D. J., Yu, H. L., Lin, T. S. & Weissman, S. I. Electron spin echoes of a photoexcited triplet: Pentacene in p-terphenyl crystals. J. Chem. Phys.75, 3746–3757 (1981). 10.1063/1.442520 [DOI] [Google Scholar]
- 38.Yamauchi, S. Recent developments in studies of electronic excited states by means of electron paramagnetic resonance spectroscopy. Bull. Chem. Soc. Jpn.77, 1255–1268 (2004). 10.1246/bcsj.77.1255 [DOI] [Google Scholar]
- 39.Oxborrow, M., Breeze, J. D. & Alford, N. M. Room-temperature solid-state maser. Nature488, 353–356 (2012). 10.1038/nature11339 [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi, S. et al. The lowest photoexcited triplet state of subphthalocyanine in solid and fluid environments. Time-resolved electron paramagnetic resonance studies. J. Phys. Chem. A107, 1478–1485 (2003). 10.1021/jp0258210 [DOI] [Google Scholar]
- 41.Férey, G. & Serre, C. Large breathing effects in three-dimensional porous hybrid matter: facts, analyses, rules and consequences. Chem. Soc. Rev.38, 1380–1399 (2009). 10.1039/b804302g [DOI] [PubMed] [Google Scholar]
- 42.Loiseau, T. et al. A rationale for the large breathing of the porous aluminum terephthalate (MIL-53) upon hydration. Chem. Eur. J.10, 1373–1382 (2004). 10.1002/chem.200305413 [DOI] [PubMed] [Google Scholar]
- 43.Millange, F., Serre, C., Guillou, N., Férey, G. & Walton, R. I. Structural effects of solvents on the breathing of metal-organic frameworks: an in situ diffraction study. Angew. Chem. Int. Ed. Engl.47, 4100–4105 (2008). 10.1002/anie.200705607 [DOI] [PubMed] [Google Scholar]
- 44.Chandler, B. D. et al. Mechanical gas capture and release in a network solid via multiple single-crystalline transformations. Nat. Mater.7, 229–235 (2008). 10.1038/nmat2101 [DOI] [PubMed] [Google Scholar]
- 45.Horike, S., Shimomura, S. & Kitagawa, S. Soft porous crystals. Nat. Chem.1, 695–704 (2009). 10.1038/nchem.444 [DOI] [PubMed] [Google Scholar]
- 46.Schneemann, A. et al. Flexible metal-organic frameworks. Chem. Soc. Rev.43, 6062–6096 (2014). 10.1039/C4CS00101J [DOI] [PubMed] [Google Scholar]
- 47.Fujiwara, S. et al. Triplet dynamic nuclear polarization of guest molecules through induced fit in a flexible metal–organic framework. Angew. Chem. Int. Ed. Engl.61, e202115792 (2022). 10.1002/anie.202115792 [DOI] [PubMed] [Google Scholar]
- 48.Dai, Y. Z. et al. Chemical modification toward long spin lifetimes in organic conjugated radicals. Chemphyschem19, 2972–2977 (2018). 10.1002/cphc.201800742 [DOI] [PubMed] [Google Scholar]
- 49.Fujisawa, J.-I., Ohba, Y. & Yamauchi, S. A time-resolved electron paramagnetic resonance study of excited triplet porphyrins in fluid solution. J. Am. Chem. Soc.119, 8736–8737 (1997). 10.1021/ja970507i [DOI] [Google Scholar]
- 50.Stoll, S. & Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson.178, 42–55 (2006). 10.1016/j.jmr.2005.08.013 [DOI] [PubMed] [Google Scholar]
- 51.Sakamoto, K. et al. Polarizing agents beyond pentacene for efficient triplet dynamic nuclear polarization in glass matrices. Proc. Natl. Acad. Sci. USA120, e2307926120 (2023). 10.1073/pnas.2307926120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrachtrup, J., Borczyskowski, C. V., Bernard, J., Orrit, M. & Brown, R. Optical detection of magnetic resonance in a single molecule. Nature363, 244–245 (1993). 10.1038/363244a0 [DOI] [Google Scholar]
- 53.Mena, A. et al. Room-temperature optically detected coherent control of molecular spins. Preprint at arXiv10.48550/arXiv.2402.07572 (2024).
- 54.Singh, H. et al. Room-temperature quantum sensing with photoexcited triplet electrons in organic crystals. Preprint at arXiv10.48550/arXiv.2402.13898 (2024).
- 55.Yanai, N. et al. Gas detection by structural variations of fluorescent guest molecules in a flexible porous coordination polymer. Nat. Mater.10, 787–793 (2011). 10.1038/nmat3104 [DOI] [PubMed] [Google Scholar]
- 56.Boultif, A. & Louër, D. Indexing of powder diffraction patterns for low-symmetry lattices by the successive dichotomy method. J. Appl. Cryst.24, 987–993 (1991). 10.1107/S0021889891006441 [DOI] [Google Scholar]
- 57.R. Dovesi et al. CRYSTAL17 User’s Manual (University of Torino, Torino, 2017).
- 58.Dovesi, R. et al. Quantum-mechanical condensed matter simulations with CRYSTAL. WIREs Comput. Mol. Sci.8, e1360 (2018). 10.1002/wcms.1360 [DOI] [Google Scholar]
- 59.Vilela Oliveira, D., Laun, J., Peintinger, M. F. & Bredow, T. BSSE-correction scheme for consistent gaussian basis sets of double- and triple-zeta valence with polarization quality for solid-state calculations. J. Comput. Chem.40, 2364–2376 (2019). 10.1002/jcc.26013 [DOI] [PubMed] [Google Scholar]
- 60.Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett.77, 3865–3868 (1996). 10.1103/PhysRevLett.77.3865 [DOI] [PubMed] [Google Scholar]
- 61.Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem.27, 1787–1799 (2006). 10.1002/jcc.20495 [DOI] [PubMed] [Google Scholar]
- 62.Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX1-2, 19–25 (2015). 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 63.Seminario, J. M. Calculation of intramolecular force fields from second-derivative tensors. Int. J. Quantum Chem.60, 1271–1277 (1996). [DOI] [Google Scholar]
- 64.Li, P. & Merz, K. M. Jr. MCPB.py: A Python based metal center parameter builder. J. Chem. Inf. Model.56, 599–604 (2016). 10.1021/acs.jcim.5b00674 [DOI] [PubMed] [Google Scholar]
- 65.D. A. Case et al. AMBER 2018 (University of California, San Francisco, 2018).
- 66.Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem.25, 1157–1174 (2004). 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- 67.Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A. III & Skiff, W. M. UFF, a Full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc.114, 10024–10035 (1992). 10.1021/ja00051a040 [DOI] [Google Scholar]
- 68.Ramsahye, N. A. et al. Charge distribution in metal organic framework materials: transferability to a preliminary molecular simulation study of the CO2 adsorption in the MIL-53 (Al) system. Phys. Chem. Chem. Phys.9, 1059–1063 (2007). 10.1039/B613378A [DOI] [PubMed] [Google Scholar]
- 69.M. J. Frisch et al. Gaussian 16, Revision C.01 (Gaussian, Inc., Wallingford CT, 2016).
- 70.Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model.25, 247–260 (2006). 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 71.Silva, A. W. S. D. & Vranken, W. F. ACPYPE - Antechamber PYthon parser interfacE. BMC Res. Notes5, 367 (2012). 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys.126, 014101 (2007). 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 73.Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem.18, 1463–1472 (1997). [DOI] [Google Scholar]
- 74.Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys.98, 10089–10092 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- 75.Yamauchi, A. Modulation of triplet quantum coherence by guest-induced structural changes in a flexible metal-organic framework. Data sets. figshare 10.6084/m9.figshare.20586825 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data generated in this study have been deposited in the figshare 10.6084/m9.figshare.20586825 and are available from the corresponding authors upon request75.