Highlights
-
•
Successfully developed a lactylation-based Riskscore model, which can accurately predict the prognosis of patients with diffuse large B-cell lymphoma.
-
•
Lactylation not only affects the prognosis of lymphoma but also regulates tumor immune function and drug resistance.
-
•
HNRNPH1, as a lactylation-related gene, plays a significant role in lymphoma.
Keywords: Diffuse large B-cell lymphoma, Lactylation, Riskscore, Machine learning, HNRNPH1
Abstract
Background
Lactylation, a newly discovered PTM involving lactic acid, is linked to solid tumor proliferation and metastasis. Lymphoma patients exhibit high lactic acid levels, yet lactylation's role in lymphoma is underexplored. This study aimed to identify lactylation-related genes in lymphoma using tumor databases and assess their predictive value in patient prognosis through cell experiments and clinical specimens.
Methods
Using TCGA and GEO datasets, we analyzed the expression levels of lactylation-related genes in diffuse large B-cell lymphoma patients. We also evaluated the prognostic significance of lactylation gene risk scores, exploring their impact on drug sensitivity and tumor immune function. Key lactylation-affecting genes were identified and functionally validated through cell experiments and mouse in vivo experiments. Additionally, the relationship between lactylation and lymphoma prognosis was examined in clinical specimens.
Results
We identified 70 genes linked to diffuse large B-cell lymphoma prognosis from the lactylation-related gene set. Using clinical data and a COX regression algorithm, we developed an optimized lactylation Riskscore model. This model significantly correlated with prognosis and showed differences in immune cell infiltration, particularly macrophages. High-risk patients showed resistance to chemotherapy drugs but responded well to immunotherapy. HNRNPH1, a lactylation-related gene, influenced patient prognosis, apoptosis, cell cycle distribution in lymphoma cells, and tumor volume in mice. In lymphoma specimens, lactylation levels correlated with Bcl-2, C-myc, and P53 levels.
Conclusions
Lactylation impacts diffuse large B-cell lymphoma prognosis, tumor immune function, and drug resistance. Our lactylation-based Riskscore model aids in patient stratification and treatment selection. HNRNPH1 regulates lactylation, thereby affecting patient prognosis.
Background
Lactylation is a newly discovered type of post-translational modification of proteins in recent years. It is a protein modification process where lactoyl groups covalently couple with protein lysine residues, thereby promoting gene regulation. Therefore, it is also known as lysine lactylation (Kla) [1]. In 2019, Professor Zhao from the University of Chicago discovered that histones within human and mouse cells can undergo lactylation, and that histone lactylation can affect chromatin gene expression regulation. This discovery opened a new chapter in the study of lactylation and pointed out a new direction for metabolism-epigenetic research [2].
The level of lactylation can be influenced by various factors. Professor Zhao found that lactylation is derived from lactic acid, and the level of lysine lactylation increases with the elevation of intracellular lactic acid concentration. Promoting cellular glycolysis can increase lactic acid content and histone lactylation levels [2]. Lactate dehydrogenase (LDH) is one of the important enzymes in glycolysis, capable of converting pyruvate into lactic acid. The level of LDH is closely related to lactic acid concentration and lactylation.
Lymphoma is a malignancy derived from lymphatic tissue, mainly including Hodgkin's lymphoma and non-Hodgkin's lymphoma, with non-Hodgkin's lymphoma accounting for the majority. Most non-Hodgkin's lymphomas are of the B-cell type, accounting for approximately 90% of the total [3]. LDH values are involved in the IPI prognostic scoring system for non-Hodgkin's lymphoma [4]. At the same time, it is not uncommon for lymphoma patients to have high lactic acid levels in clinical practice. lactylation may participate in the lymphoma process and affect the prognosis of lymphoma. However, there are very few studies on the relationship between lactylation and lymphoma, and the connection and underlying mechanisms between them remain to be further explored. This study selected diffuse large B-cell lymphoma, the most common type of non-Hodgkin's lymphoma, and screened lactylation-related genes through machine analysis of multiple clinical datasets. Combined with cell and animal experiments, we analyzed the role of lactylation in the progression of diffuse large B-cell lymphoma.
Methods
Data sources and preprocessing
The gene expression level data (FPKM expression level data) and clinical follow-up data of TCGA-DLBC were downloaded from the TCGA database (http://gdc.cancer.gov). The preprocessing of the data was as follows: (1) Samples with missing survival time or survival time of 0 were excluded, and only samples with prognostic information from TCGA patients were retained. (2) Genes with missing values or unexpressed genes exceeding 50% of the total sequenced genes were excluded. (3) Samples and genes with more than 50% of non-expressed genes were excluded. (4) All expression values were logarithmically transformed using log2(X + 1). Finally, 47 TCGA samples were retained. Additionally, clinical information-rich datasets with accession numbers GSE87371 (sequencing platform: GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array) and GSE32918 (sequencing platform: GPL8432 Illumina HumanRef-8 WG-DASL v3.0) were downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/). After excluding samples with overall survival time of 0 or missing survival time, 221 and 244 samples of diffuse large B-cell lymphoma were retained, respectively. Due to the limited number of TCGA data samples and the availability of clinical information such as staging in GSE87371, this dataset was selected as the subsequent analysis training set (main analysis data). The original data has been uploaded to Materials 1–3.
Acquisition of prognostic lactylation genes
The lactylation gene set in this study was obtained from previous research(Materials 4) [5]. Univariate Cox regression analysis was then performed using the survival package Version 2.41–1 (http://bioconductor.org/packages/survivalr/) in R language [6]. A threshold of p < 0.05 was selected to screen for specific genes significantly associated with survival prognosis in terms of expression levels for subsequent analysis. Subsequently, the expression correlation between prognostic genes was calculated using Pearson correlation. The STRING database (Version: 11.0, http://string-db.org/) was used to search for interactions between protein products related to prognostic lactylation genes, and an interaction network was constructed.
Construction of a prognostic risk prediction model
Survival regression analysis using the LASSO—COX algorithm from the glmnet package (https://cran.r-project.org/web/packages/glmnet/index.html) Version 1.2 in R language was performed. The penalty parameters were adjusted through 10-fold cross-validation [7] by selecting the lambda value that yielded the simplest model within a minimum variance range, and 1se provided a model with excellent performance and the minimum number of independent variables, where the non-zero coefficient gene variables were selected as key genes. The Risk score (risk prediction model) was constructed using stepwise Cox regression analysis from the survminer package (https://cran.rstudio.com/web/packages/survminer/index.html) Version 0.4.9. The Risk score formula was established based on the regression coefficients of each gene and the expression levels of the model genes, as follows
Note: In this formula, β represents the regression coefficient, ho(t) is the baseline hazard rate, and h(t,X) is the hazard rate associated with covariate X at time t.
Using the Risk score calculation formula, we separately computed the Risk score value for each sample in both the training set and the validation set. Subsequently, the samples were divided into two groups: High (with Risk scores higher than the median Risk score) and Low (with Risk scores lower than or equal to the median Risk score), using the median Risk score as the cutoff. The Kaplan–Meier curve method from the survival package was then employed to evaluate the correlation between the High and Low groupings and the actual survival prognosis information.
Analysis of prognostic independence
By integrating clinical information data on diffuse large B-cell lymphoma, the correlation between Risk score and clinical factors (age and staging) was analyzed. To further investigate the prognostic independence between clinical prognostic factors and Risk score, clinical factors and Risk score from the diffuse large B-cell lymphoma data were included in both univariate and multivariate COX regression models. Using a threshold of p < 0.05, independent prognostic factors were screened.
Enrichment analysis
Using the R package limma (https://bioconductor.org/packages/release/bioc/html/limma.html) Version 3.34.7 [8], differentially expressed genes between the two groups defined by Risk score were identified based on the criteria of FDR < 0.05 and |log2 FC|≥1. Subsequently, GO functional and KEGG pathway enrichment analyses were conducted using the R package 'clusterProfiler' (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html, version 4.0.5) to explore the functional pathway entries involved by key genes [9]. Multiple testing corrections were applied using the Benjamini & Hochberg method to obtain adjusted p-values (adj.P.Value). Using an adj.P.Value threshold of less than 0.05 and a count threshold of greater than 2, the top 10 most significant enrichments were selected for presentation.
Gene Set Enrichment Analysis (GSEA) was used to assess the enrichment of glycolysis pathways [10]. Complete gene expression data for both high-risk and low-risk groups were obtained from previous procedures. The glycolysis gene set was retrieved from the GSEA algorithm at https://www.gsea-msigdb.org/gsea/. The number of permutations was set to 1000.
Comparison of immune microenvironments
Quantitative analysis of the enrichment of immune pathways in tumor samples was conducted using CIBERSORT (https://cibersort.stanford.edu/index.php) and single-sample gene set enrichment analysis (ssGSEA) (http://www.bioconductor.org/packages/release/bioc/html/ssGSEA.html) [[11], [12]]. The abundance level of immune cell infiltration was assessed using enrichment scores calculated based on normalized gene expression levels and empirical cumulative distribution functions.
Evaluation of therapeutic response
The evaluation of therapeutic response encompassed the analysis of drug sensitivity and immunotherapy efficacy. Drug sensitivity analysis was quantified using the R package pRRophetic (v.0.5) (https://github.com/paulgeeleher/pRRophetic) [13]. A statistical model was built using gene expression data obtained from a large number of cancer cell lines and then applied to the gene expression data of target samples to calculate IC50 values for each therapeutic agent in both groups. The differences in IC50 between different risk groups for 138 chemotherapy drugs were compared using the Wilcoxon test and displayed in box plots. Commonly used drugs for lymphoma were selected for graphical representation.
The immune dysfunction and exclusion (TIDE) score was calculated on the website (http://tide.dfci.harvard.edu/). Differences in TIDE scores between different risk groups were compared using the Wilcoxon test.
Single-Gene survival analysis and clinical correlation analysis
Using the R packages "survival" and "survminer," patients in the training set cohort were divided into two subgroups based on the expression level of a single gene. Subsequently, the correlation between clinical features and the expression level of the target gene was analyzed using the R package "ggpubr" (v0.4.0), and the results were displayed in a violin plot.
Cell culture
The diffuse large B-cell lymphoma cell line HBL-1 was donated by the Non-coding RNA Laboratory at Yangzhou University. Cells were cultured in a cell incubator at 37 °C with 5% carbon dioxide in a culture medium consisting of RMPI 1640, 10% fetal bovine serum, and 1% penicillin-streptomycin antibiotics.
Plasmid transfection
Log-phase growing cells were seeded into a 24-well plate. Plasmids and Lipofectamine 2000 (Thermo Fisher Scientific) were added to each well. The plate was then centrifuged horizontally at 30 °C and 2000 rpm for 4 h for infection. After centrifugation, 500 μl of medium was added, and the cells were allowed to continue growing. Seventy-two hours after infection, the fluorescence intensity was observed under a fluorescence microscope.
Cell apoptosis and cell cycle analysis
Log-phase growing cells were selected, and a cell suspension was collected. After centrifugation, the supernatant was discarded, and the cell pellet was resuspended in binding buffer. APC-ANNEXIN-V and Percp-7AAD were added, and the cells were incubated in the dark for 20 min before apoptosis was detected using a flow cytometer (BD, VERSE). For cell cycle analysis, log-phase growing cells were centrifuged, and the supernatant was discarded. The cell pellet was fixed with 75% ethanol at −20 °C for one week. After centrifugation and removal of the supernatant, the cells were washed twice with PBS and then stained with PI in the dark for 30 min. The cell cycle was then detected using a flow cytometer (BD, VERSE).
Detection of lactate content
The standard substances in the lactate detection kit (L-Lactic Acid Colorimetric Assay Kit, Elabscience, E-BC-K044-M) were diluted, and 5 μl of each was added to the corresponding wells. Similarly, 5 μl of the samples to be tested were added to the designated wells. Next, 100 μl of enzyme working solution and 20 μl of color reagent were added to each well. After incubation at 37 °C for 10 min, 180 μl of stop solution was added to each well. After shaking for 5 s, the OD values were measured using a microplate reader at 530 nm. A standard curve was plotted to quantify the lactate content in the samples.
Western blot
Cells were lysed, and the protein extracted and quantified. The protein samples were mixed with loading buffer, heated for denaturation, and then loaded onto an SDS-PAGE gel for electrophoresis. After electrophoresis, the proteins were transferred from the gel to a membrane through an electric field. The membrane was then blocked to prevent non-specific binding, and incubated overnight at 4 °C with primary antibodies (Bcl-2, BAX, Cleaved-caspase-3, and Bcl-xL were purchased from Cell Signaling Technology, pan Kla was purchased from PTM Biolabs Inc., and HNRNPH1 was purchased from ABclonal Technology). After washing, the membrane was incubated with secondary antibodies for two hours at room temperature. After further washing, a luminescent solution was applied, and the membrane was imaged using chemical luminescence (BIO RAD GelDoc XR). The resulting images were analyzed using ImageJ software to quantify the grayscale values and generate graphs.
Experimental animal keeping
Nude mice were purchased from the Comparative Medicine Center of Yangzhou University. Six-week-old nude mice were allowed to adapt in an SPF-grade animal facility for one week before being subcutaneously inoculated with tumor cells. HBL-1 cells in log-phase growth were suspended and injected subcutaneously into the nude mice at a concentration of 2 × 106 cells per 200 μl. We adhered to the guidelines of the Ethics Committee for Animal Research (NIH publication #85–23).
Immunohistochemical staining
Tissue sections were sliced and dried, followed by antigen retrieval through heating. The sections were then blocked with fetal bovine serum to prevent non-specific binding. Diluted specific primary antibodies were applied to the tissue sections, and the sections were incubated in a wet chamber at room temperature in the dark for 1 hour to allow the formation of antigen-antibody complexes. After washing, labeled secondary antibodies were applied to the tissue sections, and the sections were incubated in the wet chamber for 30 min. The secondary antibodies bind to the primary antibodies, and a chromogenic substrate was added for visualization. The distribution and expression of the target antigen in the tissue were observed under a microscope. This study has been approved by the Ethics Committee of the Clinical Medical College of Yangzhou University (Approval Number: 2023ky180).
Statistical analysis
All statistical analyses were performed using R software version 4.1.2 and GraphPad Prism 9. The t-test was used for comparisons between groups, and statistical significance was indicated by asterisks (*). A two-sided p-value of less than 0.05 was considered statistically significant (p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Result
Establishment of lactate-related riskscore model
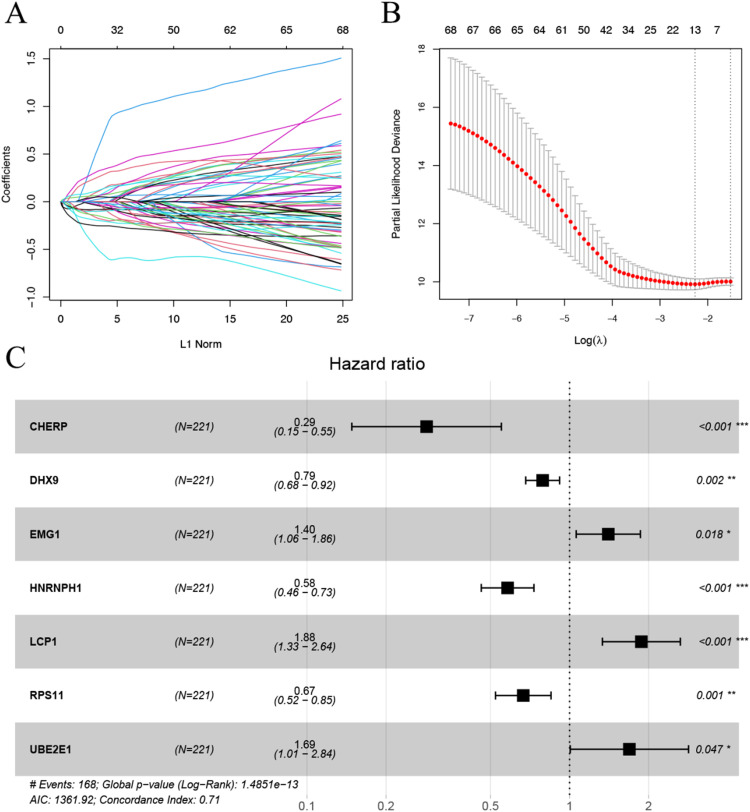
Using the univariate Cox regression analysis in the survival package, we screened 70 genes significantly associated with prognosis by setting the threshold of p < 0.05. The results are shown in Supplementary Figure 1A, and the patient list is detailed in the Supplementary Table. Supplementary Figure 1B presents the results of gene correlation analysis. The PPI network in Supplementary Figure 1C indicates that most of the proteins encoded by these genes are intricately connected in a complex manner. Thirteen key genes were then selected using the LASSO algorithm, as shown in Fig. 1A and 1B. Subsequently, the optimal gene combination was obtained using the stepwise COX regression algorithm, as presented in Table Fig. 1C, resulting in a final set of seven model genes. Finally, a Riskscore model was constructed based on the regression coefficients of the seven model genes and their expression levels in the training dataset.
Fig. 1.
A. Distribution of LASSO coefficients. B. Likelihood deviation of LASSO coefficient distribution. The two vertical dashed lines represent lambda.min (left line) and lambda.1se (right black line), respectively. C. Multivariate COX regression forest plot for the seven model genes.
Validation of the lactate-related Riskscore model efficacy
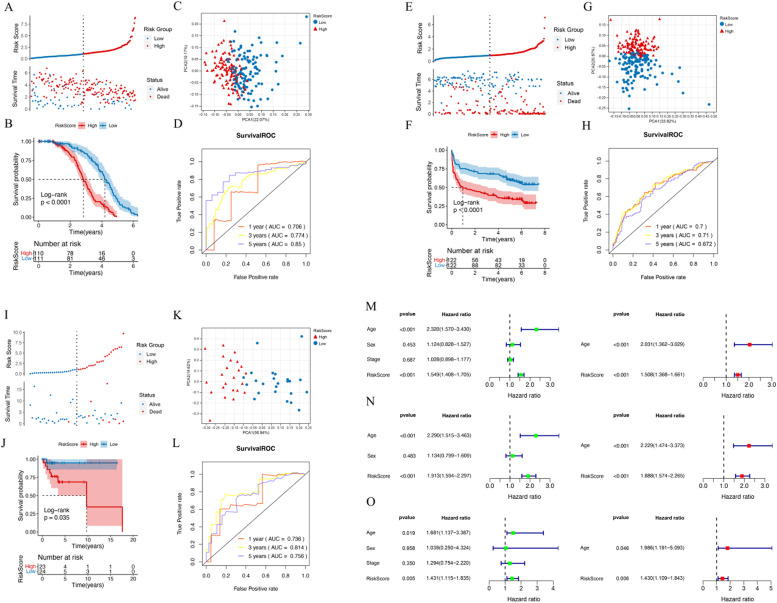
The Riskscore for each patient in the dataset was calculated, and the samples in the training set, TCGA, and GEO validation sets were divided into High (Riskscore higher than the median Riskscore) and Low (Riskscore equal to or lower than the median Riskscore) groups, respectively. The distribution of Riskscore values and survival time for each group are shown in Fig. 2A, E, and I. The KM curves for each dataset are presented in Fig. 2B, F, and J, respectively. The prognostic model was evaluated from multiple dimensions using principal component analysis (PCA), as shown in Fig. 2C, G, and K. The ROC curves for 1-year, 3-year, and 5-year prognoses based on gene prognostic features are presented in Fig. 2D, H, and L. The results showed that in the training set, TCGA, and GEO validation sets, there was a significant correlation between the different risk groups obtained by Riskscore model prediction and the actual prognosis. The age threshold was set at 65 years. Univariate Cox regression analysis was performed on the clinical indicators and RiskScore of the samples, and factors with P < 0.05 were selected for multivariate Cox regression to determine significant independent prognostic factors. The results are shown in Fig. 2M, N, and O.
Fig. 2.
A. Distribution of Riskscore (upper panel) and survival time status (lower panel) in the training set. B. KM curve showing the correlation between Riskscore prediction model and prognosis. C. PCA plot. D. ROC curves for 1-year, 3-year, and 5-year prognoses based on gene prognostic features. E. Distribution of Riskscore (upper panel) and survival time status (lower panel) in the TCGA validation set. F. KM curve based on Riskscore prediction model and prognosis. G. PCA plot. H. ROC curves for 1-year, 3-year, and 5-year prognoses based on gene prognostic features. I. Distribution of Riskscore (upper panel) and survival time status (lower panel) in the GEO validation set. J. KM curve based on Riskscore prediction model and prognosis. K. PCA plot. L. ROC curves for 1-year, 3-year, and 5-year prognoses based on gene prognostic features. M-O. Forest plots of univariate and multivariate Cox regression for clinical information.
Functional annotation of the lactate-related Riskscore model
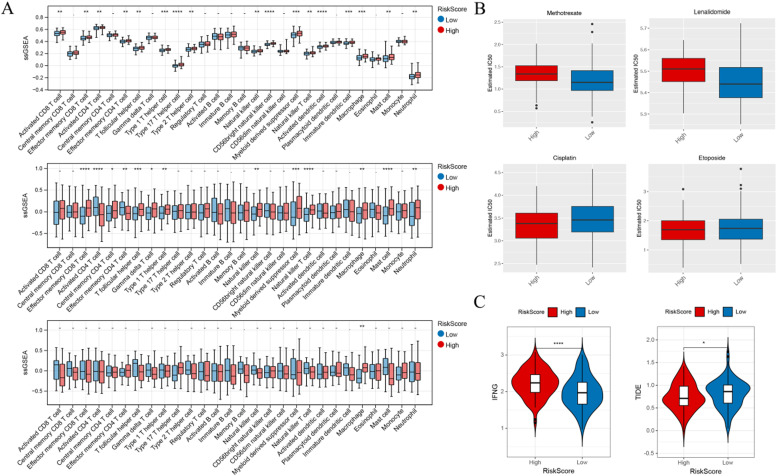
We conducted GO annotation and KEGG analysis in the training set and validation set GSE32918 data, as shown in Supplementary Figure 2. The results suggest a significant enrichment of glycolytic pathways in the tissues of high-risk patients compared to low-risk patients. Given that the enrichment analysis revealed the presence of many tumor immune microenvironment-related pathways, we performed ssGSEA to assess immune cell infiltration and immune function in the training and validation sets. Fig. 3A demonstrates that most immune-related cells are upregulated in the high-risk group, particularly macrophages, which showed a significant increase across all three datasets.
Fig. 3.
A. Box plots of differences in 28 immune cells in the training set, TCGA, and GEO validation sets, showing a significant increase in Macrophage in high-risk patients. B. High-risk patients exhibited an increase in IC50 for Methotrexate and Lenalidomide, but a decrease in IC50 for Cisplatin and Etoposide. C. High-risk patients showed a significant decrease in the responsiveness to immunotherapy, represented by TIDE, and a significant increase in the activation level of the interferon-γ (IFN-γ) signaling pathway, represented by IFNG.
To explore whether the lactate-related prognostic model can be used to guide the treatment of patients with diffuse large B-cell lymphoma, we analyzed a large number of chemical and targeted drugs. We compared the differences in IC50 levels of 138 chemotherapy drugs, and Fig. 3B showcases four drugs. High-risk patients exhibited increased resistance to two commonly used drugs, Methotrexate and Lenalidomide. Additionally, compared to the low-risk group, the high-risk group showed a significantly lower TIDE score for immunotherapy responsiveness and a significantly higher activation level of the interferon-γ (IFN-γ) signaling pathway, as indicated by IFNG (Fig. 3C).
Efficacy of lactate-related Riskscore model and prognosis of lymphoma
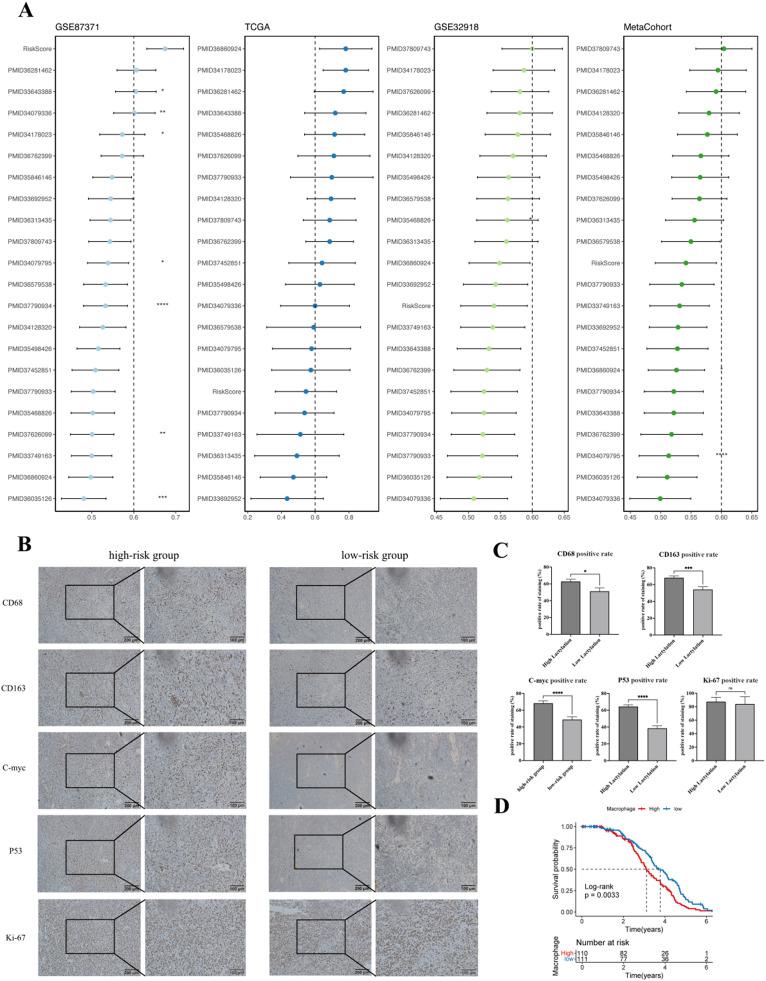
We compared the C-index of RiskScore with the features in other article models. Notably, RiskScore performed well in the training dataset and a few external datasets, but its performance was relatively weak in the TCGA dataset (Fig. 4A). We scored 71 lymphoma patient samples collected from the sample bank of Subei People's Hospital and found that patients in the high-risk group had a higher positive rate of macrophage phenotypes CD68 and CD163, as well as high expression of C-myc and P53(Fig. 4B-C). In evaluating the correlation between macrophage grouping in the dataset and the actual prognosis information of patients, we found that a high macrophage score was closely related to significantly poor prognosis (Fig. 4D).
Fig. 4.
A. Comparison of prognostic features based on gene expression in diffuse large B-cell lymphoma in published literature. (B-C). The expression levels of macrophage markers CD68 and CD163, as well as the expression of prognostic-related markers C-myc, Ki67, and P53, in patients with different groupings. D. KM curve diagram of macrophage prognosis-related factors in the dataset.
Impact of lactate-related gene HNRNPH1 on the progression of diffuse large B-cell lymphoma
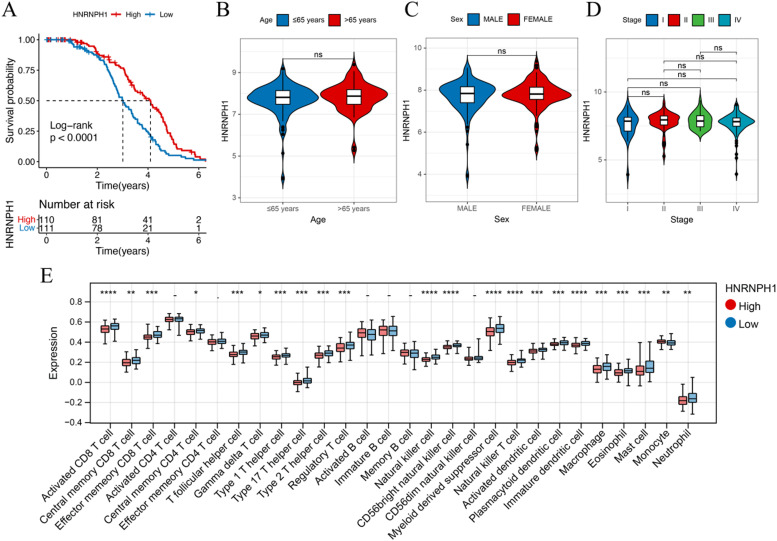
HNRNPH1, a prognostic gene associated with lactate metabolism, is a component of the genetic marker. Based on its expression level, patients in the training set GSE87371 cohort were divided into a high-risk group (N = 110) and a low-risk group (N = 111) (Fig. 5A), indicating that low expression of HNRNPH1 is associated with a poorer prognosis. The expression of HNRNPH1 was found to be unrelated to patient age (with a threshold of 65 years), gender, and staging (Fig. 5B-D). Analysis of immune cell infiltration also revealed differences in macrophages and regulatory T cells between the high and low expression subgroups of HNRNPH1 (Fig. 5E).
Fig. 5.
A. Correlation between OS and HNRNPH1 expression level in the training cohort. (B-D) Correlation between clinical information such as patient age, gender, staging, and HNRNPH1 expression level. E. Differences in immune cells between the high and low expression subgroups of HNRNPH1.
Impact of lactate-related gene HNRNPH1 on diffuse large B-cell lymphoma cells
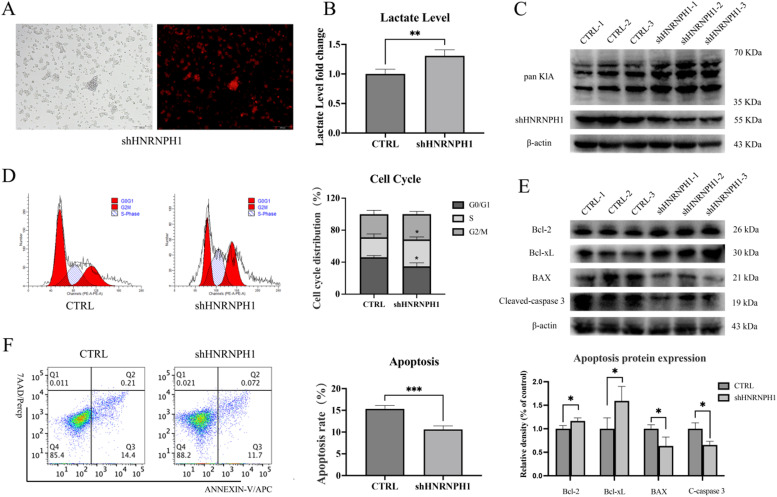
In addition to demonstrating the function of HNRNPH1 in the dataset, we also conducted relevant research in vitro cell experiments. We knocked down the HNRNPH1 gene in the diffuse large B-cell lymphoma HBL-1 cell line (Fig. 6A). The lymphoma cells with knockdown exhibited robust proliferation, and the lactate level in the cell supernatant significantly increased (Fig. 6B). Collecting the cells, we found that the total Lactylation level in the knockdown cells significantly rose (Fig. 6C). The cell cycle distribution of the knockdown cells was abnormal, mainly reflected in the increase of the S phase, indicating active cell division (Fig. 6D). The expression of apoptosis-inhibiting proteins Bcl-2 and Bcl-xL was upregulated in the knockdown lymphoma cells, while the expression of apoptosis-promoting proteins BAX and Cleaved-caspase3 was downregulated. Correspondingly, ANNEXIN-V/7AAD apoptosis staining also suggested reduced apoptosis in the knockdown lymphoma cells (Fig. 6E-F).
Fig. 6.
A. Knockdown of HNRNPH1 in diffuse large B-cell lymphoma HBL-1 cells. B. Compared with the control group, the lactate level in the supernatant of HBL-1 cells in the HNRNPH1 knockdown group significantly increased. C. Compared with the control group, the total Lactylation level in HBL-1 cells in the HNRNPH1 knockdown group significantly rose. D. The S phase of HBL-1 cells in the HNRNPH1 knockdown group significantly increased. E. In the HNRNPH1 knockdown group, the expression of apoptosis-inhibiting proteins Bcl-2 and Bcl-xL was upregulated, while the expression of apoptosis-promoting proteins BAX and Cleaved-caspase3 was downregulated. F. The apoptosis rate of HBL-1 cells in the HNRNPH1 knockdown group decreased.
Impact of lactate-related gene HNRNPH1 on tumor-bearing mice with diffuse large B-cell lymphoma
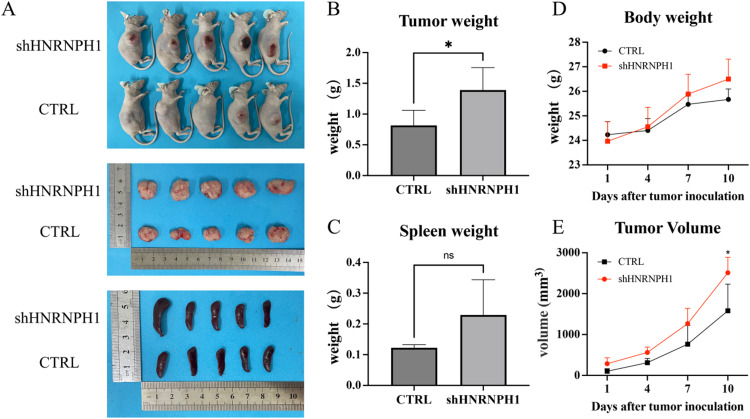
To validate the function of HNRNPH1 in vivo, we inoculated HBL-1 cells with knocked down HNRNPH1 gene in the subcutaneous tissue of nude mice during the logarithmic growth phase. The tumor volume and weight in the knockdown group were significantly greater than those in the control group, but there was no significant difference in spleen weight (Fig. 7A-C). After tumor formation, the body weight and tumor volume of the tumor-bearing mice were regularly monitored. Although the body weight of the tumor-bearing mice in the knockdown group showed an upward trend, there was no significant difference compared with the control group. However, the tumor volume of the tumor-bearing mice in the knockdown group was significantly higher than that in the control group (Fig. 7d-E).
Fig. 7.
A. Tumor and spleen images of tumor-bearing mice with diffuse large B-cell lymphoma in the HNRNPH1 knockdown group and the control group. B. The tumor size in the knockdown group was significantly larger than that in the control group. C. There was no significant difference in spleen size between the knockdown group and the control group. D. Compared with the control group, the body weight of the tumor-bearing mice in the knockdown group increased, but there was no significant difference. E. Compared with the control group, the tumor volume of the tumor-bearing mice in the knockdown group was significantly larger, with statistically significant differences.
Expression of lactate-related gene HNRNPH1 in tumor tissues of lymphoma patients
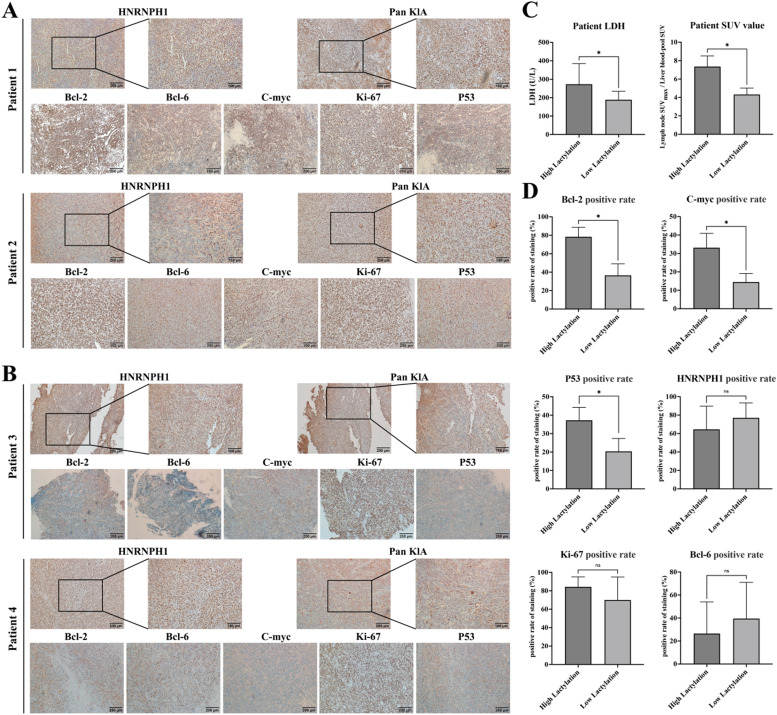
To validate the function of lactate modification and HNRNPH1 in lymphoma patients using clinical specimens, we retrieved 20 cases of diffuse large B-cell lymphoma specimens from our center and performed immunohistochemical staining for pan Kla and HNRNPH1 on their tissues. Detailed patient information is provided in the supplementary table. Immunohistochemical staining for pan Kla and HNRNPH1 was performed on the 20 patient specimens. Patients with high HNRNPH1 expression had low pan Kla levels, accompanied by low expression of Bcl-2, Bcl-6, C-myc, Ki-67, and P53 (Fig. 8A-B). The 20 patients were divided into a high lactate modification group and a low lactate modification group based on their pan Kla expression levels. The serum LDH levels and SUVmax of tumor lymph nodes in PET-CT were significantly elevated in the high lactate modification group (Fig. 8C). Additionally, patients in the high lactate modification group had higher positive expressions of Bcl-2, C-myc, and P53 in their tumors. However, unexpectedly, there was no significant difference in HNRNPH1 expression between the two groups, possibly due to the small sample size and large numerical deviations (Fig. 8D). There were no significant differences in Bcl-6, Ki-67 expression, patient age, blood routine values, tumor staging, and grouping between the two groups (Supplementary Figure 3).
Fig. 8.
A. Patients with low HNRNPH1 expression exhibited an increased level of Pan Kla, accompanied by high expressions of Bcl-2, Bcl-6, C-myc, Ki-67, and P53. B. In patients with high HNRNPH1 expression, the level of Pan Kla decreased, along with decreased expressions of Bcl-2, Bcl-6, C-myc, Ki-67, and P53. C. The 20 patients were divided into high and low Pan Kla groups. The serum LDH levels and SUVmax of tumor lymph nodes in PET-CT were significantly elevated in the high lactate modification group. D. In the high lactate modification group, the positive expression rates of Bcl-2, C-myc, and P53 in the tumors increased, but there were no significant differences in the positive expression rates of HNRNPH1, Bcl-6, and Ki-67.
Discussion
The classic Warburg theory demonstrates that tumor cells absorb glucose more efficiently than other cells, preferring glycolysis even under non-hypoxic conditions, releasing lactate during glucose absorption [[14], [15]]. A large amount of lactate is produced in the bodies of cancer patients, but it was previously considered a metabolic waste until the theory of lactylation was proposed. Lactylation can influence tumor progression, such as promoting tumor angiogenesis [[16], [17]], and lactate can suppress the killing function of immune cells in the tumor microenvironment [[18], [19], [20]]. Lactate can also act as a signaling molecule mediating intracellular or intercellular communication [[21], [22]]. This study explored the role of lactylation in lymphoma progression. lactylation affects the lymphoma process, and we can use the level of lactylation to score the prognosis of lymphoma and assess its prognsis early.
The TCGA and GEO databases provide rich genomic and gene expression data. We screened the lactylation-related gene sets in these datasets and identified seven prognostic lactylation-related genes through the LASSO algorithm and stepwise COX regression: CHERP, DHX9, EMG1, HNRNPH1, LCP1, RPS11, and UBE2E1. The lactylation Riskscore model constructed based on the expression levels of these genes was proven to have predictive value for lymphoma prognosis through comparison with clinical data. In the immune cell infiltration analysis, we found an interesting link between lactylation modification and macrophage levels in lymphoma patients. In Professor Zhao's research, he discovered that macrophage genomic histones undergo lactylation modification under the influence of lactate, promoting the transformation of macrophages from the proinflammatory and anticancer M1 type to the anti-inflammatory and cancer-promoting M2 type. Lymphoma lactylation may have similar macrophage changes. Through the analysis of lymphoma samples from our hospital, we found that patients in the high-risk group indeed had higher expression of macrophages. Many previous studies have demonstrated that macrophage infiltration in diffuse large B-cell lymphoma is associated with poor prognosis[[23], [24]]. But which gene in macrophages undergoes lactylation modification, what changes have occurred in its function, and whether it has a synergistic effect with tumor cells are all issues that needs to address in future experiments. The alteration of macrophages may affect the immunotherapy response in lymphoma patients, so we evaluated the TIDE scores of lymphoma patients with different lactylation modifications. The TIDE score is associated with tumor immune dysfunction and used to assess the possibility of tumor immune evasion in the gene expression profile of tumor samples. It can predict the response to immune checkpoint blockade therapy, and a higher TIDE score is generally associated with poorer immunotherapy outcomes [[25], [26]]. The IFNG score is a scoring system related to interferon-gamma (IFNG), a protein with various biological activities. The IFNG score may be associated with patient prognosis, and a higher IFNG score may indicate a better prognosis [[27], [28]]. Lymphoma patients with high-risk lactylation modification who have low TIDE scores and high IFNG scores are predicted to have better immunotherapy outcomes, which may be related to abnormally expressed macrophages. This suggests that immunotherapy may be a viable first-line treatment option for such patients.
In addition to predicting prognosis, the lactylation risk score can also guide the choice of medication for lymphoma patients. We analyzed the IC50 levels of 138 commonly used chemotherapy drugs and found patients with high-risk lactylation lymphoma may not be suitable for chemotherapy regimens containing methotrexate and lenalidomide as first-line treatment. However, the sensitivity to Cisplatin and Etoposide increased, indicating that these patients may be more suitable for the ESHAP protocol (Etoposide + Methylprednisolone + High-dose Cytarabine + Cisplatin). We have analyzed the correlation between R-IPI and RiskScore in our dataset. Unfortunately, while there is a trend, the differences in RiskScore across different R-IPI categories are not very significant(Supplementary Figure 4). To further clarify the diagnostic role of the lactylation risk score gene set, we randomly selected the key gene HNRNPH1 from the gene set. HNRNPH1 is a member of the widely expressed heterogeneous nuclear ribonucleoprotein (hnRNPs) subfamily, and its expression level has been observed to increase in various cancers and tumors [[29], [30]]. For example, the expression level of HNRNPH1 is significantly higher in glioblastoma tissues compared to normal tissues [[31], [32]]. In diffuse large B-cell lymphoma, our analysis of the data revealed that patients with high HNRNPH1 expression had a better prognosis, and there was no significant correlation between HNRNPH1 expression and patient gender, age, or stage. Additionally, HNRNPH1 expression was associated with the number of various immune cells.
We found that knockdown of HNRNPH1 in diffuse large B-cell lymphoma cells led to increased lactate production and a corresponding rise in lactylation levels. Additionally, the S-phase distribution of cells increased, indicating robust cell proliferation and reduced apoptosis. Using a tumor-bearing mouse model constructed with the HNRNPH1 knockdown cell line, we observed a significant increase in tumor volume, suggesting that HNRNPH1 may inhibit the proliferation of diffuse large B-cell lymphoma cells. Based on this, we analyzed 20 tissue samples from patients with diffuse large B-cell lymphoma and found that patients with low HNRNPH1 expression had elevated Pan Kla levels, accompanied by high expression of classical disease progression and prognostic indicators such as Bcl-2, Bcl-6, C-myc, Ki-67, and P53. This suggests that HNRNPH1 may affect cell proliferation by regulating the expression of cell cycle regulatory factors, growth factor receptors, signal transduction molecules, and other genes. HNRNPH1 binds to the mRNA of these genes and regulates their expression levels by affecting their splicing or stability.
Finally, we divided the 20 patients into high and low Pan-Kla expression groups. By comparing clinical indicators between the two groups, we found that patients in the high lactylation group had significantly elevated serum LDH levels and SUVmax values in PET-CT for tumor lymph nodes, indicating a close correlation between lactylation levels and glucose metabolism in lymphoma patients. Additionally, the positive expression rates of Bcl-2, C-myc, and P53 were increased in the tumors of patients in the high lactylation group, confirming that lactylation is involved in the tumorigenesis of lymphoma. However, the malignancy of diffuse large B-cell lymphoma is high, and Ki-67 levels are generally elevated, which may explain the lack of significant differences. The high dispersion of HNRNPH1 data may have contributed to the lack of significant differences observed. Future studies with increased sample sizes are needed for further analysis.
Conclusion
In summary, we have investigated the role of lactylation in patients with diffuse large B-cell lymphoma, screening for genes that exhibit differences in lactylation levels. Based on these genes, we have constructed a lactylation gene risk score, which has been validated through three datasets to confirm its predictive power. Our findings have clarified that lactylation affects macrophage immune infiltration in diffuse large B-cell lymphoma and reduces sensitivity to commonly used drugs such as Methotrexate and Lenalidomide. Additionally, both in vitro and in vivo experiments, combined with patient tissue biopsies, have demonstrated that HNRNPH1 can regulate lactylation to promote lymphoma cell proliferation.
Ethics approval and participant consent
The ethical approval for sample collection from participants in this study was granted by the Clinical Medicine Approval Committee of Yangzhou University, with approval number 2023ky180. Written informed consent was obtained from all participants. This study adhered to the principles outlined in the Declaration of Helsinki.
Consent for publication
Not applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102102.
Contributor Information
Miao Zhu, Email: realzhumiao@163.com.
Xiaoyan Xie, Email: xxylw8848@163.com.
Mei Sun, Email: hematologysunmei@163.com.
Appendix. Supplementary materials
References
- 1.Yu J., Chai P., Xie M., Ge S., Ruan J., Fan X., Jia R. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021 Mar 16;22(1):85. doi: 10.1186/s13059-021-02308-z. PMID: 33726814; PMCID: PMC7962360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M., Ding J., Czyz D., Hu R., Ye Z., He M., Zheng Y.G., Shuman H.A., Dai L., Ren B., Roeder R.G., Becker L., Zhao Y. Metabolic regulation of gene expression by histone lactylation. Nature. 2019 Oct;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. Epub 2019 Oct 23. PMID: 31645732; PMCID: PMC6818755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage J.O., Gascoyne R.D., Lunning M.A., Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017 Jul 15;390(10091):298–310. doi: 10.1016/S0140-6736(16)32407-2. Epub 2017 Jan 31. PMID: 28153383. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Barta S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019 May;94(5):604–616. doi: 10.1002/ajh.25460. PMID: 30859597. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Z., Huang H., Li M., Liang X., Tan Y., Chen Y. Lactylation-related gene signature effectively predicts prognosis and treatment responsiveness in hepatocellular carcinoma. Pharmaceuticals. (Basel). 2023 Apr 25;16(5):644. 10.3390/ph16050644. PMID: 37242427; PMCID: PMC10221268. [DOI] [PMC free article] [PubMed]
- 6.Rizvi A.A., Karaesmen E., Morgan M., Preus L., Wang J., Sovic M., Hahn T., Sucheston-Campbell L.E. gwasurvivr: an R package for genome-wide survival analysis. Bioinformatics. 2019 Jun 1;35(11):1968–1970. doi: 10.1093/bioinformatics/bty920. PMID: 30395168; PMCID: PMC7963072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tibshirani R. The lasso method for variable selection in the Cox model. Stat. Med. 1997 Feb 28;16(4):385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. PMID: 9044528. [DOI] [PubMed] [Google Scholar]
- 8.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids. Res. 2015 Apr 20;43(7):e47. doi: 10.1093/nar/gkv007. Epub 2015 Jan 20. PMID: 25605792; PMCID: PMC4402510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012 May;16(5):284–287. doi: 10.1089/omi.2011.0118. Epub 2012 Mar 28. PMID: 22455463; PMCID: PMC3339379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., Merico D., Bader G.D. Pathway enrichment analysis and visualization of omics data using g:profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019 Feb;14(2):482–517. doi: 10.1038/s41596-018-0103-9. PMID: 30664679; PMCID: PMC6607905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. PMID: 29344893; PMCID: PMC5895181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao B., Liu L., Li A., Xiang C., Wang P., Li H., Xiao T. Identification and verification of immune-related gene prognostic signature based on ssGSEA for osteosarcoma. Front. Oncol. 2020 Dec 15;10:607622. 10.3389/fonc.2020.607622. PMID: 33384961; PMCID: PMC7771722. [DOI] [PMC free article] [PubMed]
- 13.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS. One. 2014 Sep 17;9(9) doi: 10.1371/journal.pone.0107468. PMID: 25229481; PMCID: PMC4167990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaupel P., Multhoff G. Revisiting the Warburg effect: historical dogma versus current understanding. J. Physiol. 2021 Mar;599(6):1745–1757. doi: 10.1113/JP278810. Epub 2021 Jan 4. PMID: 33347611. [DOI] [PubMed] [Google Scholar]
- 15.Koppenol W.H., Bounds P.L., Otto Dang CV. Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011 May;11(5):325–337. doi: 10.1038/nrc3038. Epub 2011 Apr 14. Erratum in: Nat Rev Cancer. 2011 Aug;11(8):618. PMID: 21508971. [DOI] [PubMed] [Google Scholar]
- 16.Porporato P.E., Payen V.L., De Saedeleer C.J., Préat V., Thissen J.P., Feron O., Sonveaux P. Lactate stimulates angiogenesis and accelerates the healing of superficial and ischemic wounds in mice. Angiogenesis. 2012 Dec;15(4):581–592. doi: 10.1007/s10456-012-9282-0. Epub 2012 Jun 3. PMID: 22660894. [DOI] [PubMed] [Google Scholar]
- 17.Végran F., Boidot R., Michiels C., Sonveaux P., Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011 Apr 1;71(7):2550–2560. doi: 10.1158/0008-5472.CAN-10-2828. Epub 2011 Feb 7. PMID: 21300765. [DOI] [PubMed] [Google Scholar]
- 18.Fischer K., Hoffmann P., Voelkl S., Meidenbauer N., Ammer J., Edinger M., Gottfried E., Schwarz S., Rothe G., Hoves S., Renner K., Timischl B., Mackensen A., Kunz-Schughart L., Andreesen R., Krause S.W., Kreutz M. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007 May 1;109(9):3812–9. 10.1182/blood-2006-07-035972. Epub 2007 Jan 25. PMID: 17255361. [DOI] [PubMed]
- 19.Gottfried E., Kunz-Schughart L.A., Ebner S., Mueller-Klieser W., Hoves S., Andreesen R., Mackensen A., Kreutz M. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006 Mar 1;107(5):2013–2021. doi: 10.1182/blood-2005-05-1795. Epub 2005 Nov 8. PMID: 16278308. [DOI] [PubMed] [Google Scholar]
- 20.Brand A., Singer K., Koehl G.E., Kolitzus M., Schoenhammer G., Thiel A., Matos C., Bruss C., Klobuch S., Peter K., Kastenberger M., Bogdan C., Schleicher U., Mackensen A., Ullrich E., Fichtner-Feigl S., Kesselring R., Mack M., Ritter U., Schmid M., Blank C., Dettmer K., Oefner P.J., Hoffmann P., Walenta S., Geissler E.K., Pouyssegur J., Villunger A., Steven A., Seliger B., Schreml S., Haferkamp S., Kohl E., Karrer S., Berneburg M., Herr W., Mueller-Klieser W., Renner K., Kreutz M. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK Cells. Cell Metab. 2016 Nov 8;24(5):657–671. doi: 10.1016/j.cmet.2016.08.011. Epub 2016 Sep 15. PMID: 27641098. [DOI] [PubMed] [Google Scholar]
- 21.Brooks G.A. The science and translation of lactate shuttle theory. Cell Metab. 2018 Apr 3;27(4):757–785. doi: 10.1016/j.cmet.2018.03.008. PMID: 29617642. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Wang G., Xu Z.G., Tu H., Hu F., Dai J., Chang Y., Chen Y., Lu Y., Zeng H., Cai Z., Han F., Xu C., Jin G., Sun L., Pan B.S., Lai S.W., Hsu C.C., Xu J., Chen Z.Z., Li H.Y., Seth P., Hu J., Zhang X., Li H., Lin H.K. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019 Jun 27;178(1):176–189.e15. doi: 10.1016/j.cell.2019.05.003. Epub 2019 May 30. PMID: 31155231; PMCID: PMC6625351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai L., Fan G., Xie T., Li L., Tang L., Chen H., Shi Y., Han X. Single-cell and spatial transcriptomics reveal a high glycolysis B cell and tumor-associated macrophages cluster correlated with poor prognosis and exhausted immune microenvironment in diffuse large B-cell lymphoma. Biomark. Res. 2024 Jun 5;12(1):58. doi: 10.1186/s40364-024-00605-w. PMID: 38840205; PMCID: PMC11155084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koudouna A., Gkioka A.I., Gkiokas A., Tryfou T.M., Papadatou M., Alexandropoulos A., Bartzi V., Kafasi N., Kyrtsonis M.C. Serum-soluble CD163 levels as a prognostic biomarker in patients with diffuse large B-cell lymphoma treated with chemoimmunotherapy. Int. J. Mol. Sci. 2024 Mar 1;25(5):2862. doi: 10.3390/ijms25052862. PMID: 38474108; PMCID: PMC10931688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Z., Huang H., Li M., Liang X., Tan Y., Chen Y. Lactylation-related gene signature effectively predicts prognosis and treatment responsiveness in hepatocellular carcinoma. Pharmaceuticals. (Basel) 2023 Apr 25;16(5):644. doi: 10.3390/ph16050644. PMID: 37242427; PMCID: PMC10221268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S., Wang L., Ding W., Ye B., Cheng C., Shao J., Liu J., Zhou H. Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework. Front. Endocrinol. (Lausanne) 2023 Apr 19;14 doi: 10.3389/fendo.2023.1180404. PMID: 37152941; PMCID: PMC10154596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozeman E.A., Hoefsmit E.P., Reijers I.L.M., Saw R.P.M., Versluis J.M., Krijgsman O., Dimitriadis P., Sikorska K., van de Wiel B.A., Eriksson H., Gonzalez M., Torres Acosta A., Grijpink-Ongering L.G., Shannon K., Haanen J.B.A.G., Stretch J., Ch'ng S., Nieweg O.E., Mallo H.A., Adriaansz S., Kerkhoven R.M., Cornelissen S., Broeks A., Klop W.M.C., Zuur C.L., van Houdt W.J., Peeper D.S., Spillane A.J., van Akkooi A.C.J., Scolyer R.A., Schumacher T.N.M., Menzies A.M., Long G.V., Blank C.U. Survival and biomarker analyses from the OpACIN-neo and OpACIN neoadjuvant immunotherapy trials in stage III melanoma. Nat. Med. 2021 Feb;27(2):256–263. doi: 10.1038/s41591-020-01211-7. Epub 2021 Feb 8. PMID: 33558721. [DOI] [PubMed] [Google Scholar]
- 28.Quan Q., Xiong X., Wu S., Yu M. Identification of immune-related key genes in ovarian cancer based on WGCNA. Front. Genet. 2021 Nov 15;12:760225. 10.3389/fgene.2021.760225. PMID: 34868239; PMCID: PMC8634599. [DOI] [PMC free article] [PubMed]
- 29.Feng S., Li J., Wen H., Liu K., Gui Y., Wen Y., Wang X., Yuan S. hnRNPH1 recruits PTBP2 and SRSF3 to modulate alternative splicing in germ cells. Nat. Commun. 2022 Jun 23;13(1):3588. doi: 10.1038/s41467-022-31364-7. PMID: 35739118; PMCID: PMC9226075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fultz E.K., Coelho M.A., Lieberman D., Jimenez-Chavez C.L., Bryant C.D., Szumlinski K.K. Hnrnph1 is a novel regulator of alcohol reward. Drug Alcohol Depend. 2021 Mar 1;220:108518. 10.1016/j.drugalcdep.2021.108518. Epub 2021 Jan 10. PMID: 33454624; PMCID: PMC7899125. [DOI] [PMC free article] [PubMed]
- 31.Xi S., Cai H., Lu J., Zhang Y., Yu Y., Chen F., Huang Q., Wang F., Chen Z. The pseudogene PRELID1P6 promotes glioma progression via the hnHNPH1-Akt/mTOR axis. Oncogene. 2021 Jul;40(26):4453–4467. doi: 10.1038/s41388-021-01854-x. Epub 2021 Jun 9. PMID: 34108621; PMCID: PMC8249232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Z., Xia M., Shi W., Zhao C., Wang J., Xin D., Dong X., Xiong Y., Zhang F., Berry K., Ogurek S., Liu X., Rao R., Xing R., Wu L.M.N., Cui S., Xu L., Lin Y., Ma W., Tian S., Xie Q., Zhang L., Xin M., Wang X., Yue F., Zheng H., Liu Y., Stevenson C.B., de Blank P., Perentesis J.P., Gilbertson R.J., Li H., Ma J., Zhou W., Taylor M.D., Lu Q.R. Human fetal cerebellar cell atlas informs medulloblastoma origin and oncogenesis. Nature. 2022 Dec;612(7941):787–794. doi: 10.1038/s41586-022-05487-2. Epub 2022 Nov 30. PMID: 36450980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.