Dear Editor,
We read with much interest the paper titled “Three Cases of False-positive Multiplex Ligation-dependent Probe Amplification of BRCA1” by Kim, et al. [1], published in your journal. We would like to supplement this study with the description of an additional case of a false-positive BRCA1 multiplex ligation-dependent probe amplification (MLPA) result identified in an Italian patient with high-grade serous ovarian cancer (HGSOC).
HGSOC is characterized by chromosomal instability attributed to homologous recombination deficiency (HRD) [2]. BRCA1/2 (BRCA) pathogenic variants (PVs) are well-known causes of HRD [3]. Poly (ADP-ribose) polymerase inhibitors have been recently approved for treating patients with HGSOC and BRCA PVs or HRD status, revolutionizing the ovarian cancer treatment landscape in both first-line and recurrent disease management. Therefore, assessing HRD status has become crucial for patient management [3].
In clinical trials, HRD status predominantly relies on centralized next-generation sequencing (NGS)-based assays. Recently, HRD assays, including Oncomine Comprehensive Assay Plus (Thermo Fisher Scientific, Waltham, MA, USA), SOPHiA DDM HRD Solution (SOPHiA Genetics, Rolle, Switzerland), and AmoyDx HRD Focus Assay (Amoy Diagnostics, Xiamen, China) have been marketed to facilitate in-house HRD testing in diagnostic laboratories equipped with high-throughput NGS platforms.
Our patient, a 50-yr-old woman diagnosed with HGSOC in March 2023, was started on neoadjuvant chemotherapy and considered eligible for HRD assay. Consequently, in-house HRD evaluation and sequencing were performed using SOPHiA DDM HRD Solution and the NextSeq550Dx platform (Illumina, San Diego, CA, USA), respectively. NGS data were analyzed using the SOPHiA DDM software, version 4.2.
SOPHiA DDM HRD Solution, employed to assess the NGS data obtained from a formalin-fixed paraffin-embedded HGSOC biopsy, analyzes copy number variations (CNVs) by measuring and normalizing the coverage for all exons within the genes covered by the panel. In its current version, the algorithm aggregates the coverage signal measured at the exon level for each gene analyzed and applies a threshold to identify gene amplification. In this study, this approach was complemented with an algorithm (proprietary of SOPHiA Genetics) utilizing a hidden Markov model to segment exon-level coverage data and identify exon-level CNVs.
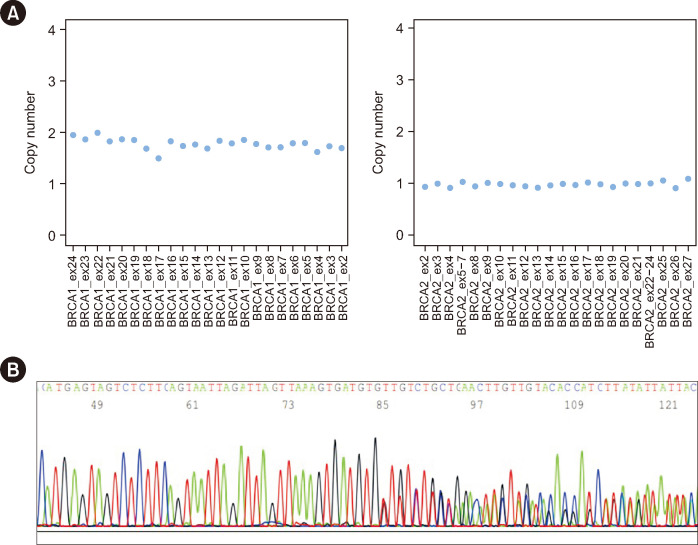
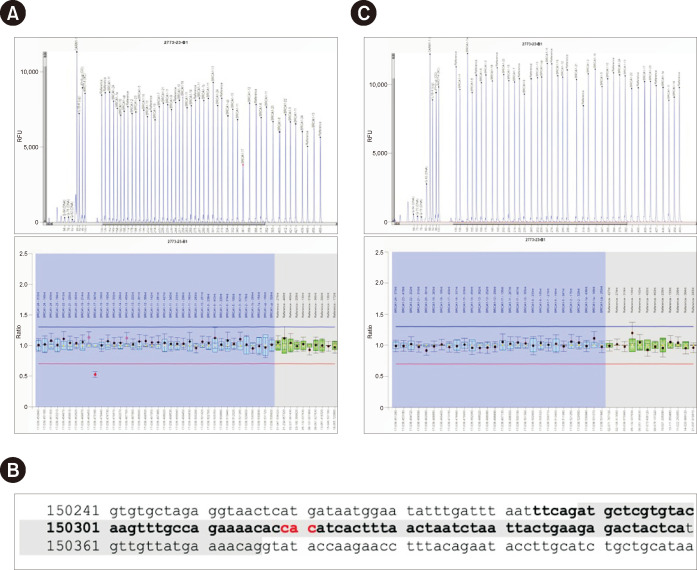
An HRD status was obtained, revealing the presence of both the c.5017_5019del (rs80358343) p.(His1673del) variant in exon 16 of BRCA1 (LRG_292t1) and the loss of the entire BRCA2 (LRG_293t1) gene (Fig. 1A). To evaluate the germline BRCA status, targeted Sanger sequencing and MLPA were performed on blood samples, as previously reported [4-6]. While Sanger sequencing confirmed the germline origin of the BRCA1 c.5017_5019del variant (Fig. 1B), SALSA MLPA Probemix P090 BRCA2 (MRC Holland, Amsterdam, the Netherlands) did not detect the loss of the entire BRCA2 gene, indicating a tissue-specific origin. Unexpectedly, MLPA identified a heterozygous deletion of BRCA1 in exon 16, with a final specific probe ratio of 0.53 (Fig. 2A). Owing to the discordance between the HRD CNV-calling and MLPA results, the tests were replicated using a fresh blood sample, yielding comparable results. Additionally, germinal-variant NGS analysis using a validated CNV bioinformatics algorithm [7] helped exclude any CNVs in BRCA1 (data not shown).
Fig. 1. Bioinformatic prediction of CNVs in BRCA1 and BRCA2 and validation using Sanger sequencing. (A) Bioinformatics prediction of CNVs in BRCA1 and BRCA2 genes using the prototype algorithm of SOPHiA DDM® tool. The CNV analysis indicated a wild-type status for the BRCA1 gene (left) and complete deletion of the BRCA2 gene (right). (B) Sanger sequencing result confirming the heterozygous status of the BRCA1 c.5017_5019del variant.
Fig. 2. MLPA results obtained using the SALSA MLPA probemix and detailed probe sequences for BRCA1. (A) MLPA results from comparative analysis experiment obtained using Coffalyser.NET Software with SALSA MLPA probemix P002-D1 BRCA1. The final ratio of BRCA1 exon 17 probes indicates a heterozygous deletion. (B) Nucleotide sequence of BRCA1 exon 17 is highlighted in gray, with the MLPA sequence probe denoted in bold font. The three deleted nucleotides of the variant p.(His1673del) are highlighted in red. (C) MLPA results from comparative analysis experiment obtained using Coffalyser.NET Software using SALSA MLPA probemix P087-D1 BRCA1, where the final ratio of BRCA1 exon 17 probes showed no heterozygous deletion. The final ratio (FR) of each reference probe in the patient’s sample should fall between 0.80 and 1.20 for the diploid normal copy number. The following FR cut-off values were used: heterozygous deletion, 0.40<FR< 0.65; heterozygous duplication, 1.30<FR<1.65. Please note that the BRCA1 exon numbering used in both SALSA MLPA probemix product descriptions follows the traditional exon numbering, wherein no exon 4 is present. Notably, BRCA1 exon 17 reported in the MLPA kit corresponds to BRCA1 exon 16 of LRG_292.
Abbreviation: RFU, relative fluorescent units.
SNVs located within the target sequence of a probe can potentially influence probe hybridization and/or ligation. However, upon reviewing the datasheet of the SALSA MLPA Probemix P002-D1 BRCA1, we observed no evidence of possible interference caused by the BRCA1 c.5017_5019del variant. To investigate the likelihood of a false-positive MLPA result attributable to the presence of the in-frame small deletion within BRCA1 exon 16, we examined the nucleotide sequence of BRCA1 probe 18031-L23028 covering exon 16 and confirmed that the 3′ position of the right probe oligo aligned adjacent to a microsatellite variant. This led to an apparent deletion of BRCA1 exon 16 as an artifact (Fig. 2B). The MLPA BRCA1 P087-D1 confirmation kit did not validate the deletion, as the 21496-L30761 probe, specific for BRCA1 exon 16, exhibited a wild-type final ratio of 0.97 (Fig. 2C). This interference has been reported to MRC Holland, the manufacturer of the SALSA MLPA kit.
The c.5017_5019del variant is classified as pathogenic according to reports by Parsons, et al. [8]. Additionally, this variant is extremely rare in population-based databases but has been documented in multiple individuals and families with breast and ovarian cancers in northern Italy, suggesting a founder effect in this geographical region [8, 9].
Although in-house HRD testing may aid in bridging the gap between molecular testing and routine clinical/diagnostic requirements, we emphasize that the standardization of analytical pipelines, simplification of complex technical procedures, and implementation of different workflows for data analysis are essential to successfully integrate in-house HRD testing into routine clinical practice.
Finally, the integration of an exon-level CNV bioinformatics pipeline into our HRD analysis and a comprehensive evaluation of the molecular results facilitated the resolution of the unexpected MLPA result. This highlights the importance of data exchange among seasoned professionals, organizations, and clinicians, particularly within complex diagnostic frameworks.
ACKNOWLEDGEMENTS
We thank Pierluigi Iapicca and Christian Pozzorini for participating as field experts and their valuable contributions.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, Concolino P, De Paolis E, and Minucci A; methodology, Rinelli M, Maneri G, and Brisighelli F; software, Concolino P, Giacò L, and Preziosi A; validation, Minucci A, Concolino P, De Paolis E, and Nero C; investigation, De Paolis E, Concolino P, Rinelli M, and Maneri G; data curation, Trozzi R, Duranti S, and Panfili A; writing—original draft preparation, Concolino P, De Paolis E, Minucci A, and Piane M; writing—review and editing, Concolino P, De Paolis E, Minucci A, and De Bonis M; supervision, Minucci A and Scambia G.
CONFLICTS OF INTEREST
None declared.
RESEARCH FUNDING
None declared.
References
- 1.Kim KB, Park S, Ha JS, Ryoo N, Kim DH. Three cases of false-positive multiplex ligation-dependent probe amplification of BRCA1. Ann Lab Med. 2022;42:497–9. doi: 10.3343/alm.2022.42.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network, author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–54. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giacò L, Palluzzi F, Guido D, Nero C, Giacomini F, Duranti S, et al. A computational framework for comprehensive genomic profiling in solid cancers: the analytical performance of a high-throughput assay for small and copy number variants. Cancers (Basel) 2022;14:6152. doi: 10.3390/cancers14246152.5fc382c2d0e5420191465cbf17292b1a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minucci A, Scambia G, Santonocito C, Concolino P, Canu G, Mignone F, et al. Clinical impact on ovarian cancer patients of massive parallel sequencing for BRCA mutation detection: the experience at Gemelli hospital and a literature review. Expert Rev Mol Diagn. 2015;15:1383–403. doi: 10.1586/14737159.2015.1081059. [DOI] [PubMed] [Google Scholar]
- 6.Concolino P, Mello E, Minucci A, Santonocito C, Scambia G, Giardina B, Capoluongo E. Advanced tools for BRCA1/2 mutational screening: comparison between two methods for large genomic rearrangements (LGRs) detection. Clin Chem Lab Med. 2014;52:1119–27. doi: 10.1515/cclm-2013-1114. [DOI] [PubMed] [Google Scholar]
- 7.Minucci A, Scambia G, Santonocito C, Concolino P, Urbani A. BRCA testing in a genomic diagnostics referral center during the COVID-19 pandemic. Mol Biol Rep. 2020;47:4857–60. doi: 10.1007/s11033-020-05479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons MT, Tudini E, Li H, Hahnen E, Wappenschmidt B, Feliubadaló L, et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum Mutat. 2019;40:1557–78. doi: 10.1002/humu.23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuntini R, Cortesi L, Calistri D, Pippucci T, Martelli PL, Casadio R, et al. BRCA1 p.His1673del is a pathogenic mutation associated with a predominant ovarian cancer phenotype. Oncotarget. 2014;8:22640–8. doi: 10.18632/oncotarget.15151. [DOI] [PMC free article] [PubMed] [Google Scholar]