Abstract
Objective
We developed a few-shot learning (FSL) framework for the diagnosis of osteopenia and osteoporosis in knee X-ray images.
Methods
Computer vision models containing deep convolutional neural networks were fine-tuned to enable generalization from natural images (ImageNet) to chest X-ray images (normal vs. pneumonia, base images). Then, a series of automated machine learning classifiers based on the Euclidean distances of base images were developed to make predictions for novel images (normal vs. osteopenia vs. osteoporosis). The performance of the FSL framework was compared with that of junior and senior radiologists. In addition, the gradient-weighted class activation mapping algorithm was used for visual interpretation.
Results
In Cohort #1, the mean accuracy (0.728) and sensitivity (0.774) of the FSL models were higher than those of the radiologists (0.512 and 0.448). A diagnostic pipeline of FSL model (first)–radiologists (second) achieved better performance (0.653 accuracy, 0.582 sensitivity, and 0.816 specificity) than radiologists alone. In Cohort #2, the diagnostic pipeline also showed improved performance.
Conclusions
The FSL framework yielded practical performance with respect to the diagnosis of osteopenia and osteoporosis in comparison with radiologists. This retrospective study supports the use of promising FSL methods in computer-aided diagnosis tasks involving limited samples.
Keywords: Few-shot learning, osteopenia, osteoporosis, X-ray, deep learning, diagnosis
Introduction
Osteoporosis is a public health concern that affects populations worldwide. 1 The condition is characterized by low bone mass and microarchitectural bone tissue deterioration, resulting in increased bone fragility and a subsequent rise in fracture risk. 2 Bone mineral density (BMD) is a key diagnostic indicator for osteoporosis and is used to predict fracture risk. Dual X-ray absorptiometry (DXA) is the most commonly used tool for measuring BMD in clinical practice. 3 However, the cost and availability of the BMD technique limit the application of DXA-based BMD assessment for the clinical management of osteoporosis.4,5
X-ray imaging is the most widely used imaging technique for the diagnosis of bone pathologies.6,7 X-rays have been in use for over a century and are commonly used to visualize bones in various parts of the body, including the wrists, knees, shoulders, pelvis, and spine. X-ray imaging is useful for the diagnosis of fractures, joint dislocations, bone injuries, abnormal bone growth, arthritis, and even infections. 8 Despite its widespread use, X-ray imaging has limitations, particularly regarding the detection of early-stage osteoporosis, i.e., osteopenia. 9 Osteoporosis may not be visible on X-ray imaging until a substantial amount of bone loss has occurred, making it important to use additional imaging modalities and diagnostic tests for the early detection and management of the disease.10,11
In recent years, deep learning (DL)-based convolutional neural networks (CNNs) have gained popularity for medical image analysis owing to their state-of-the-art results when detecting diseases, including brain tumors, breast cancer, and pneumonia, from various medical images.12–14 CNNs have demonstrated superior performance regarding the classification of medical images. Recently, we published a literature review concerning artificial intelligence (AI)-assisted radiologic diagnosis of osteoporosis, which listed the datasets used and proposed methods in previous studies. 15 However, a major challenge encountered when using CNN classifiers in the medical field is the requirement for large amounts of labeled training data. 16 Insufficiently large datasets limit the use of CNNs for the detection and diagnosis of osteoporosis.
In response to the challenges faced by standard DL methods that require large datasets, few-shot learning (FSL) has emerged as a promising alternative. 17 As its name suggests, FSL enables the rapid and accurate completion of classification tasks by learning using only a small number of samples. 18 In this paper, we propose an FSL framework comprising feature extractors based on fine-tuning and Euclidean distance-based classifiers for the diagnosis of osteopenia and osteoporosis using knee X-ray images in two cohorts.
Methods
Chest x-ray images: base images for fine-tuning
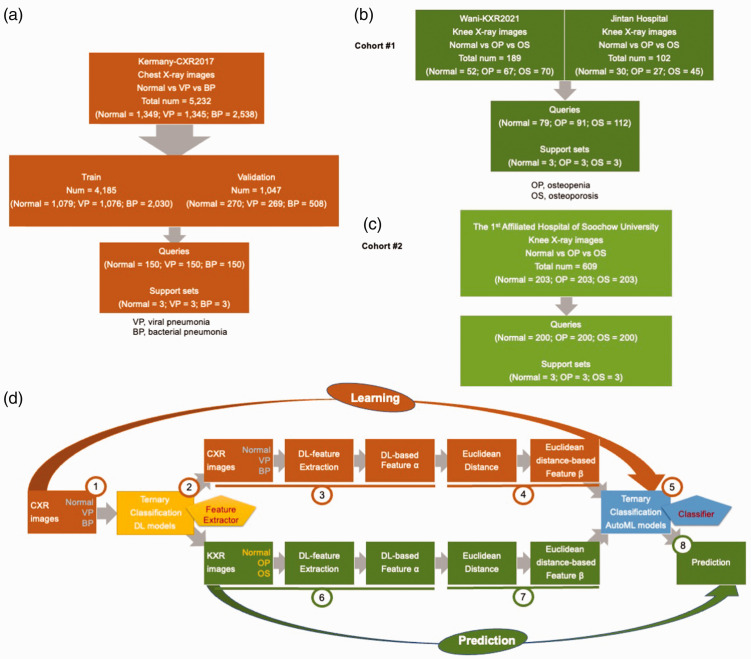
As shown in Figure 1(a), chest X-ray images acquired from the anteroposterior view were obtained from an open-access database, which comprised retrospective cohorts of pediatric patients aged 1 to 5 years from Guangzhou Women and Children’s Medical Center, Guangzhou, China. 19 All chest X-ray images were obtained as part of patients’ routine clinical care. Based on their clinical evaluation, the images were labeled normal (n = 1583) versus viral pneumonia (n = 1493) versus bacterial pneumonia (n = 2780). A high-resolution JPEG chest X-ray image dataset was also deposited into the public Mendeley database (https://doi.org/10.17632/rscbjbr9sj.3).
Figure 1.
Study flowchart and FSL framework. (a) Chest X-ray images: base images for fine-tuning; (b) knee X-ray images in Cohort #1: novel images for the target task; (c) knee X-ray images in Cohort #2: novel images for the target task and (d) FSL framework: learning and prediction parts. VP, viral pneumonia; BP, bacterial pneumonia; OP, osteopenia; OS, osteoporosis; FSL, few-shot learning framework; DL, deep learning; CXR, chest X-ray; KXR, knee X-ray.
Knee x-ray images: novel images for the target task
In Cohort #1 (Figure 1(b)), 52 normal knee X-ray images, 67 images of osteopenia, and 70 images of osteoporosis were obtained from a public database (Wani-KXR2021). 20 In addition, 30 normal, 27 osteopenia, and 45 osteoporosis knee X-ray images were obtained from Jintan Hospital Affiliated to Jiangsu University.
In Cohort #2, 203 normal knee X-ray images, 203 images of osteopenia, and 203 images of osteoporosis were consecutively obtained from the First Affiliated Hospital of Soochow University (January 2020–December 2022), as shown in Figure 1(c). Patient details was de-identified such that they could not be identified in any way.
The Wani-KXR2021 images were collected from the BMD camp organized by the Unani and Panchkarma Hospital, Srinagar, J&K, India, which were deposited into a public Mendeley database (https://data.mendeley.com/datasets/fxjm8fb6mw/2). 20 The characteristics of participants could be obtained from the website. The BMD was measured just below the knee with the peripheral bone assessment QUS system known as the Sunlight Omnisense 7000S (Jacksonville, FL, USA) with simulation software from Pegasus Prestige (DMS Imaging, Gallargues-le-Montueux, France).
Participants who were evaluated at Jintan Hospital Affiliated to Jiangsu University and the First Affiliated Hospital of Soochow University between January 2020 and December 2022 were included. Participants received BMD scans of the femoral neck and lumbar spine (L2-L4) using DXA (Lunar Prodigy, General Electric Medical Systems, Aurora, OH, USA). At the three centers, the osteopenia and osteoporosis diagnoses were determined according to the BMD levels of patients on the basis of the T score values recommended by the World Health Organization. 21 Knee X-ray images from the anteroposterior view of each participant were obtained during the same visit at which their BMD was measured. Institutional Review Board approval was obtained from the Ethics Committee of The First Affiliated Hospital of Soochow University on 31 March 2022 (retrospective study review #2022098). The reporting of this study conforms to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines. 22 Written informed consent was obtained from all participants in this study. The characteristics of participants from the two centers are listed in Supplementary Table 1 (Jintan Hospital Affiliated to Jiangsu University) and Supplementary Table 2 (First Affiliated Hospital of Soochow University).
General design of the FSL framework
The FSL framework based on fine-tuning and Euclidean distance is presented in Figure 1(d).
The learning part (steps 1–5 in Figure 1(d)) was as follows.
VGG, ResNet, and Xception were pretrained on ImageNet, and then the three networks were fine-tuned on a ternary chest X-ray image classification task to learn X-ray features.
The fine-tuned DL models served as feature extractors (yellow).
A base three-way, three-shot set was constructed: three support sets (three images randomly selected from each class, i.e., normal vs. viral pneumonia vs. bacterial pneumonia) and three query sets (150 normal vs. 150 viral pneumonia vs. 150 bacterial pneumonia). The above chest X-ray images were transferred into feature vectors (feature α).
The Euclidean distance between the feature α of each query image and three support set images was calculated and transformed into feature β.
We used the H2O automated machine learning (AutoML) platform to develop a series of machine learning classifiers (blue) for the ternary classification task concerning feature β.
The prediction part (steps 6–8 in Figure 1(d)) was as follows.
6. A novel three-way, three-shot set was constructed: three support sets (three images randomly selected from each class in two cohorts, i.e., normal vs. osteopenia vs. osteoporosis) and three query sets (Cohort #1: 79 normal vs. 91 osteopenia vs. 112 osteoporosis; Cohort #2: 200 normal vs. 200 osteopenia vs. 200 osteoporosis). The above knee X-ray images were transferred into feature vectors (feature α) by the three feature extractors (yellow).
7. As in step 4, the Euclidean distance between the feature α of each query image and six support set images was calculated and transformed into feature β.
8. The trained AutoML classifiers from step 5 (blue) were used to perform prediction for the novel ternary classification task involving knee X-rays with respect to feature β.
Feature extractors formed by fine-tuning
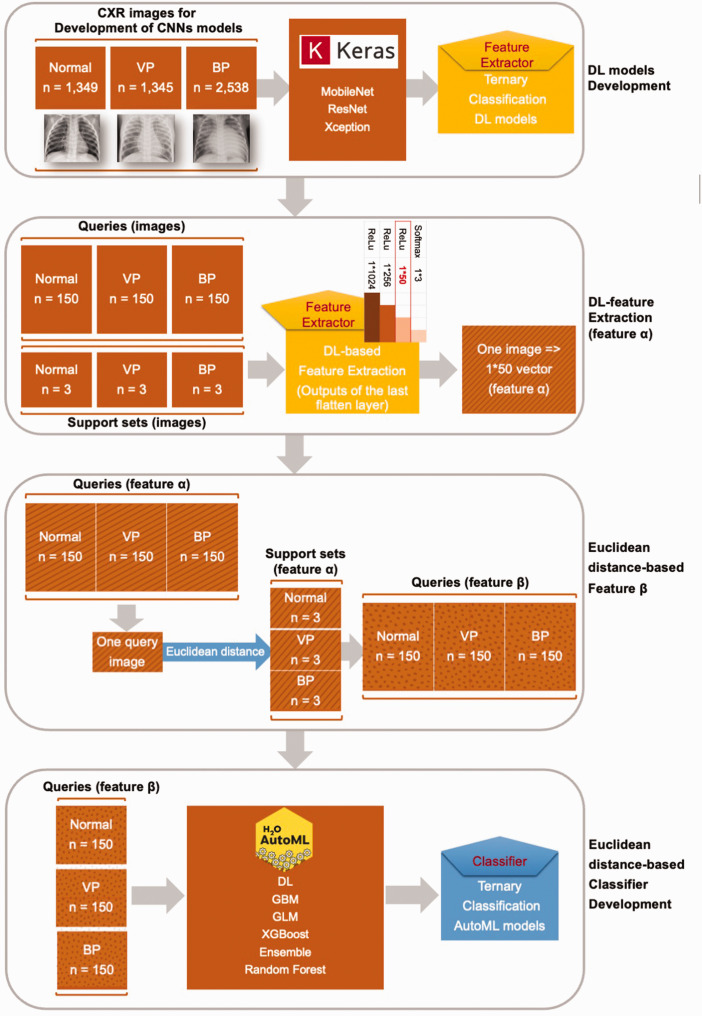
Generally, DL models trained on large datasets such as ImageNet are highly transferable to image classification and recognition tasks. 23 In this study, given that the images derived from the source dataset ImageNet (natural images) and the target task (X-rays) were of different types, fine-tuning was necessary to achieve a domain shift from a general natural view to X-ray imaging. 24 Thus, three classic DL models, i.e., VGG, ResNet, and Xception, were first pretrained on ImageNet, as shown in Figure 2. Then, the three models were fine-tuned in a base ternary chest X-ray image classification task (normal vs. viral pneumonia vs. bacterial pneumonia) to update the weights of the DL models and provide them with a better understanding of X-ray imaging. 25 After performing fine-tuning, the three DL models were regarded as feature extractors in the FSL framework. For each query chest X-ray image, a 1*50 feature vector α was obtained through the fine-tuned DL models, as well as each support set image.
Figure 2.
Detailed process of the learning part. VP, viral pneumonia; BP, bacterial pneumonia; CXR, chest X-ray; DL, deep learning; CNN, convolutional neural network; AutoML, automated machine learning; GBM, gradient boost machine; GLM, general linear regression; XGBoost, eXtreme gradient boosting.
Classifiers constructed using AutoML based on Euclidean distance
Euclidean distance is one of the most common distance metrics used to measure the absolute distance between two points in multidimensional space. 26 It can be used to develop an intuitive and traditional similarity algorithm. Given that every image had been transformed into a 1*50 feature vector α, the Euclidean distance (d) from one query image (q) to one support set image (s) was calculated as shown in Equation 1:
| (1) |
where d is the Euclidean distance, q indicates query, s is the support set, and 50 is 1*50 feature vector α.
Consequently, every query image was transformed into a 1*9 feature vector β based on the distance from one query to nine support set images (three images for each base class, i.e., normal vs. viral pneumonia vs. bacterial pneumonia).
H2O AutoML (https://www.h2o.ai) is a platform for automating the machine learning workflow, including automated training and tuning for a series of models. 27 It offers a variety of interpretation methods for variables and models. H2O AutoML supports six common algorithms: DL, gradient boost machines, general linear regression, eXtreme gradient boosting (XGBoost), ensemble models, and random forests. The code for the AutoML training process is available at https://osf.io/zjahc.
As shown in Figures 1(d) and 2, a series of AutoML models were developed based on the base ternary classification of feature vector β.
Prediction procedure
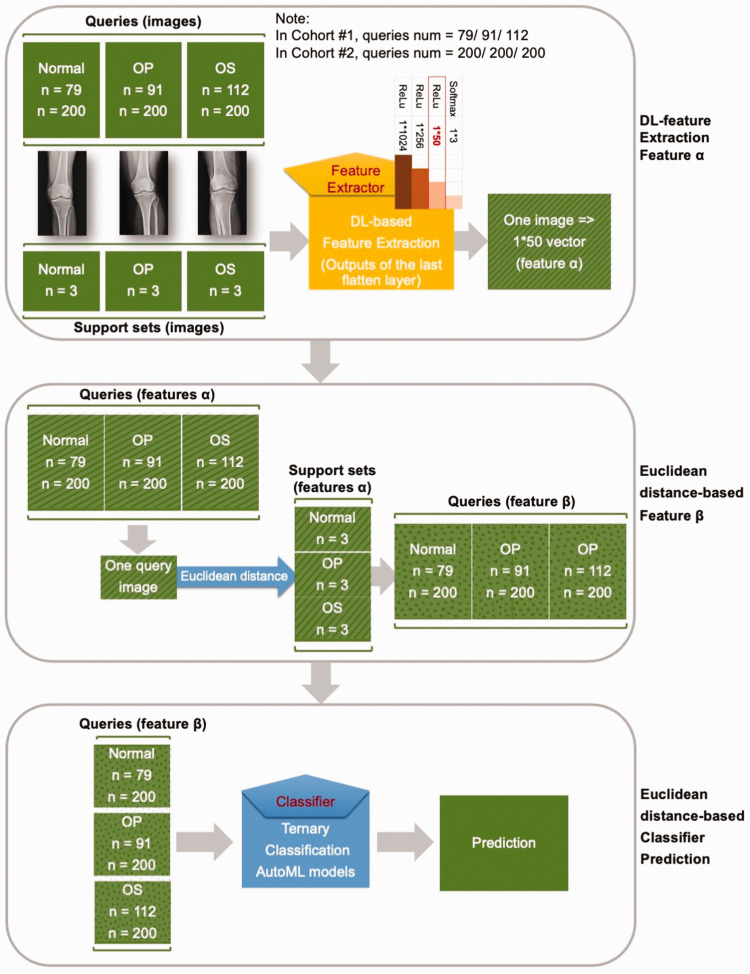
In this study, the novel task was the ternary classification of knee X-rays. In each round, a three-way, three-shot set was constructed: three support sets (three images randomly selected from each class in two cohorts, i.e., normal vs. osteopenia vs. osteoporosis) and three query sets (Cohort #1: 79 normal vs. 91 osteopenia vs. 112 osteoporosis; Cohort #2: 200 normal vs. 200 osteopenia vs. 200 osteoporosis).
As shown in Figure 3, every knee X-ray image was transferred into a feature vector (feature α) by the DL models, i.e., the feature extractors; then every query image was transformed into a 1*9 distance vector (feature β) based on the Euclidean distance. Finally, the osteopenia and osteoporosis predictions were processed by the trained classifier.
Figure 3.
Detailed prediction process. OP, osteopenia; OS, osteoporosis; DL, deep learning; AutoML, automated machine learning.
Visual interpretation
The gradient-weighted class activation mapping (Grad-CAM) algorithm was used to visually interpret the fine-tuned DL model. 28 Grad-CAM uses the gradients of any target concept, flowing into the final convolutional layer to produce a coarse localization map highlighting important regions in the image for predicting the concept. The results of Grad-CAM are presented in the form of heatmaps.
Model training and evaluation
The Keras Python (version 3.8.0) open access platform (Backbone: TensorFlow version 2.8.0; Google Inc., Santa Clara, CA, USA) was used to train the VGG16/ResNet50/Xception models. Each image was resized to 331 × 331 pixels and loaded into the DL models in the form of RGB channels. The random split-sample method was used to divide the images into training and validation datasets (8:2). The adaptive moment estimation optimizer and categorical cross-entropy cost function, with a learning rate of 0.0001 and a batch size of 32, were used to train the models with an early stopping approach. The training code for the DL models is available at https://osf.io/7aujc.
In Cohort #1, the whole FSL framework was executed in a three-round evaluation. The performance of the framework was compared with that of three junior radiologists (less than 5 years of experience) and three senior radiologists (more than 10 years of experience) who were blinded for the data collection and model development. In Cohort #2, the FSL framework (algorithm chosen based on the best model in Cohort #1) was applied in one round and compared with evaluation by one senior radiologist.
A confusion matrix was calculated using Equation 2 and used to evaluate the performance of the FSL model. TN, FN, TP, and FP indicate true negatives, false negatives, true positives, and false positives, respectively.
| (2) |
Accuracy indicates the proportion of samples that were classified correctly among all samples, as shown in Equation 3.
| (3) |
Sensitivity indicates the proportion of samples that were classified correctly out of the total number of actual true samples, as shown in Equation 4. In the study, positive events included osteopenia and osteoporosis; thus, the sensitivity values for these events and the mean were calculated separately.
| (4) |
Specificity denotes the proportion of samples that were completely classified correctly out of all actual false samples, as shown in Equation 5.
| (5) |
Cohen’s Kappa score and Matthews correlation coefficient (MCC) are statistical measures used to evaluate the performance of classification models. Cohen’s Kappa is a measure of inter-rater agreement for qualitative (categorical) items. It is generally used to compare the agreement between two raters or classifiers. The score is normalized and takes into account the possibility of the agreement occurring by chance. The MCC, another measure of classification, takes into account TNs, FNs, TPs, and FPs. The MCC is generally considered a balanced measure, which can be used even if the classes are imbalanced.
Results
Model evaluation in three rounds in Cohort #1
The performance of the FSL models and radiologists in round #1 is summarized in Table 1. The best FSL model (VGG16+XGBoost) achieved higher accuracy (0.745) and higher mean sensitivity (0.748) than those of the junior radiologist (0.454 and 0.372, respectively) and senior radiologist (0.585 and 0.529, respectively). However, the specificity of the model (0.709) was equal to that of the senior radiologist, which was still higher than that of the junior radiologist (0.658).
Table 1.
Performance of few-shot learning framework and radiologists in Cohort #1, round #1.
| Round #1 | Confusion matrix | Metrics | |||||
|---|---|---|---|---|---|---|---|
|
VGG16+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.745 | ||
| Actual | Normal | 56 | 5 | 18 | 79 | Sensitivity (OP) | 0.648 |
| OP | 6 | 59 | 26 | 91 | Sensitivity (OS) | 0.848 | |
| OS | 8 | 9 | 95 | 112 | Sensitivity (mean) | 0.748 | |
| Total | 70 | 73 | 139 | 282 | Specificity | 0.709 | |
|
ResNet50+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.610 | ||
| Actual | Normal | 36 | 14 | 29 | 79 | Sensitivity (OP) | 0.538 |
| OP | 13 | 49 | 29 | 91 | Sensitivity (OS) | 0.777 | |
| OS | 7 | 18 | 87 | 112 | Sensitivity (mean) | 0.658 | |
| Total | 56 | 81 | 145 | 282 | Specificity | 0.456 | |
|
Xception+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.716 | ||
| Actual | Normal | 49 | 7 | 23 | 79 | Sensitivity (OP) | 0.626 |
| OP | 12 | 57 | 22 | 91 | Sensitivity (OS) | 0.857 | |
| OS | 10 | 6 | 96 | 112 | Sensitivity (mean) | 0.742 | |
| Total | 71 | 70 | 141 | 282 | Specificity | 0.620 | |
|
Junior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.454 | ||
| Actual | Normal | 52 | 9 | 18 | 79 | Sensitivity (OP) | 0.352 |
| OP | 33 | 32 | 26 | 91 | Sensitivity (OS) | 0.393 | |
| OS | 27 | 41 | 44 | 112 | Sensitivity (mean) | 0.372 | |
| Total | 112 | 82 | 88 | 282 | Specificity | 0.658 | |
|
Senior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.585 | ||
| Actual | Normal | 56 | 10 | 13 | 79 | Sensitivity (OP) | 0.451 |
| OP | 29 | 41 | 21 | 91 | Sensitivity (OS) | 0.607 | |
| OS | 12 | 32 | 68 | 112 | Sensitivity (mean) | 0.529 | |
| Total | 97 | 83 | 102 | 282 | Specificity | 0.709 | |
Bold figures indicate the highest numeric values.
OP, osteopenia; OS, osteoporosis; XGBoost, eXtreme gradient boosting.
In rounds #2 and #3 (Tables 2 and 3), the best FSL model (Xception+XGBoost) yielded higher accuracy (0.709 and 0.730, respectively) and mean sensitivity (0.800 and 0.775, respectively) values than those of the two radiologists. However, the specificity of the model in theses rounds (0.456 and 0.582, respectively) was lower than those of the two senior radiologists (0.658 and 0.709, respectively) and two junior radiologists (0.595 and 0.646, respectively).
Table 2.
Performance of few-shot learning models and radiologists in Cohort #1, round #2.
| Round #2 | Confusion Matrix | Metrics | |||||
|---|---|---|---|---|---|---|---|
|
VGG16+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.688 | ||
| Actual | Normal | 39 | 11 | 29 | 79 | Sensitivity (OP) | 0.648 |
| OP | 6 | 59 | 26 | 91 | Sensitivity (OS) | 0.857 | |
| OS | 5 | 11 | 96 | 112 | Sensitivity (mean) | 0.753 | |
| Total | 50 | 81 | 151 | 282 | Specificity | 0.494 | |
|
ResNet50+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.585 | ||
| Actual | Normal | 22 | 24 | 33 | 79 | Sensitivity (OP) | 0.670 |
| OP | 5 | 61 | 25 | 91 | Sensitivity (OS) | 0.732 | |
| OS | 4 | 26 | 82 | 112 | Sensitivity (mean) | 0.701 | |
| Total | 31 | 111 | 140 | 282 | Specificity | 0.278 | |
|
Xception+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.709 | ||
| Actual | Normal | 36 | 18 | 25 | 79 | Sensitivity (OP) | 0.725 |
| OP | 3 | 66 | 22 | 91 | Sensitivity (OS) | 0.875 | |
| OS | 5 | 9 | 98 | 112 | Sensitivity (mean) | 0.800 | |
| Total | 44 | 93 | 145 | 282 | Specificity | 0.456 | |
|
Junior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.436 | ||
| Actual | Normal | 47 | 19 | 13 | 79 | Sensitivity (OP) | 0.330 |
| OP | 30 | 30 | 31 | 91 | Sensitivity (OS) | 0.411 | |
| OS | 26 | 40 | 46 | 112 | Sensitivity (mean) | 0.370 | |
| Total | 103 | 89 | 90 | 282 | Specificity | 0.595 | |
|
Senior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.504 | ||
| Actual | Normal | 52 | 16 | 11 | 79 | Sensitivity (OP) | 0.363 |
| OP | 33 | 33 | 25 | 91 | Sensitivity (OS) | 0.509 | |
| OS | 14 | 41 | 57 | 112 | Sensitivity (mean) | 0.436 | |
| Total | 99 | 90 | 93 | 282 | Specificity | 0.658 | |
Bold figures indicate the highest numeric values.
OP, osteopenia; OS, osteoporosis; XGBoost, eXtreme gradient boosting.
Table 3.
Performance of few-shot learning models and radiologists in Cohort #1, round #3.
| Round #3 | Confusion Matrix | Metrics | |||||
|---|---|---|---|---|---|---|---|
|
VGG16+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.660 | ||
| Actual | Normal | 39 | 10 | 30 | 79 | Sensitivity (OP) | 0.659 |
| OP | 4 | 60 | 27 | 91 | Sensitivity (OS) | 0.777 | |
| OS | 8 | 17 | 87 | 112 | Sensitivity (mean) | 0.718 | |
| Total | 51 | 87 | 144 | 282 | Specificity | 0.494 | |
|
ResNet50+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.589 | ||
| Actual | Normal | 33 | 21 | 25 | 79 | Sensitivity (OP) | 0.560 |
| OP | 10 | 51 | 30 | 91 | Sensitivity (OS) | 0.732 | |
| OS | 14 | 16 | 82 | 112 | Sensitivity (mean) | 0.646 | |
| Total | 57 | 88 | 137 | 282 | Specificity | 0.418 | |
|
Xception+XGBoost |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.730 | ||
| Actual | Normal | 46 | 11 | 22 | 79 | Sensitivity (OP) | 0.648 |
| OP | 7 | 59 | 25 | 91 | Sensitivity (OS) | 0.902 | |
| OS | 7 | 4 | 101 | 112 | Sensitivity (mean) | 0.775 | |
| Total | 60 | 74 | 148 | 282 | Specificity | 0.582 | |
|
Junior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.496 | ||
| Actual | Normal | 51 | 14 | 14 | 79 | Sensitivity (OP) | 0.418 |
| OP | 23 | 38 | 30 | 91 | Sensitivity (OS) | 0.455 | |
| OS | 28 | 33 | 51 | 112 | Sensitivity (mean) | 0.436 | |
| Total | 102 | 85 | 95 | 282 | Specificity | 0.646 | |
|
Senior radiologist |
|||||||
| Normal | OP | OS | Total | Accuracy | 0.596 | ||
| Actual | Normal | 56 | 10 | 13 | 79 | Sensitivity (OP) | 0.451 |
| OP | 19 | 41 | 31 | 91 | Sensitivity (OS) | 0.634 | |
| OS | 9 | 32 | 71 | 112 | Sensitivity (mean) | 0.542 | |
| Total | 84 | 83 | 115 | 282 | Specificity | 0.709 | |
Bold figures indicate the highest numeric values.
OP, osteopenia; OS, osteoporosis; XGBoost, eXtreme gradient boosting.
General model evaluation in Cohort #1
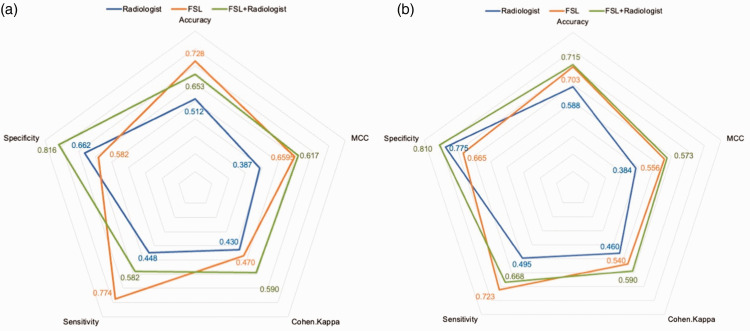
As shown in Figure 4(a), the mean accuracy (0.728) and sensitivity (0.774) of the best FSL models in the three rounds were higher than those of the radiologists (0.512 and 0.448) in Cohort #1. The radiologists (0.662) only demonstrated an advantage over the models (0.582) in terms of the mean specificity metric.
Figure 4.
Performance of models, radiologists, and diagnostic pipeline in (a) Cohort #1 and (b) Cohort #2.
FSL, few-shot learning framework; MCC, Matthews correlation coefficient.
A diagnostic pipeline of FSL model (first)–radiologists (second) achieved improved performance (0.653 mean accuracy, 0.582 mean sensitivity, and 0.816 mean specificity) over that of the radiologists. The MCC was improved from 0.387 to 0.617, and the Cohen’s Kappa was improved from 0.430 to 0.590.
General model evaluation in Cohort #2
In Figure 4(b), the accuracy (0.703) and sensitivity (0.723) of the FSL model (Xception+XGBoost) were higher than those of the senior radiologist (0.588 and 0.495, respectively) in Cohort #2. The radiologists (0.662) only showed an advantage (0.775) in specificity.
The diagnostic pipeline of FSL model (first)–radiologist (second) also achieved improved performance (0.715 accuracy, 0.668 sensitivity, and 0.810 specificity) over that of the senior radiologist. The MCC was improved from 0.384 to 0.573, and the Cohen’s Kappa was improved from 0.460 to 0.590.
Model interpretation
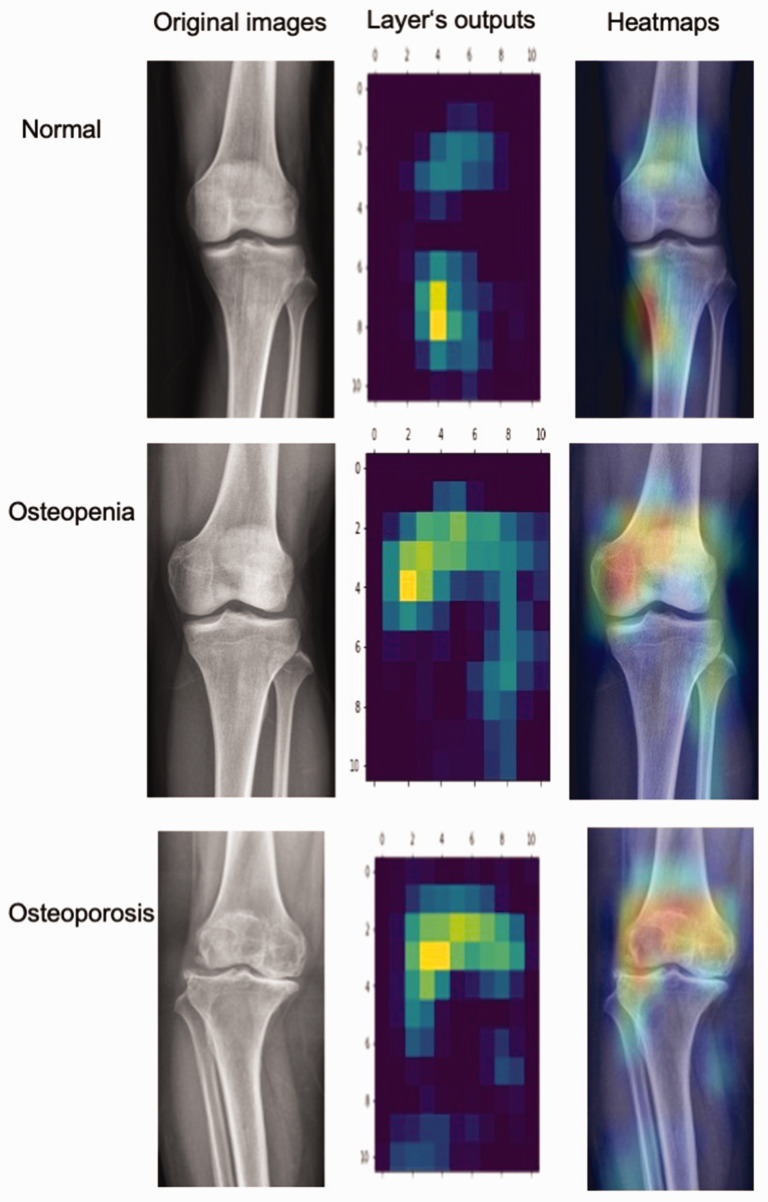
The heatmaps produced by Grad-CAM are plotted in Figure 5. One knee X-ray image was selected from each novel classification. The gradient difference between the fully connected layer and the final output of the fine-tuned DL model (Xception) was calculated to plot the class activation map. The highlighted regions in the heatmaps are the important areas for prediction considered by the model.
Figure 5.
Grad-CAM heatmaps. The left column presents the original images. The middle column shows the heatmaps based on the output of the feature extractor’s last layer of CNNs. The right column shows the Grad-CAM heatmaps covering the original images. Grad-CAM; gradient-weighted class activation mapping; CNN, convolutional neural network.
Discussion
For the task of diagnosing osteopenia and osteoporosis in knee X-ray images, we developed an FSL framework comprising chest X-ray-based fine-tuned feature extractors and Euclidean distance-based AutoML classifiers. In both cohorts, compared with radiologists, the FSL model achieved better accuracy and sensitivity in a total of four rounds of evaluation. This showed that the FSL model could improve the clinical diagnosis results obtained with the FSL model (first)–radiologist (second) pipeline.
The prevalence of osteoporosis-related fractures is increasing in women aged 55 years and older and men aged 65 years and older, leading to significant bone-associated morbidity, mortality, and health care costs.4,5 Advances in research have enabled more precise assessments of fracture risk and have expanded the range of therapeutic options that are available to prevent fractures. In clinical practice, fracture risk algorithms that incorporate clinical risk factors and BMD are commonly used to identify high-risk individuals. 29 Osteopenia, a precursor of osteoporosis, is also a significant risk factor for fragility fractures. Research has shown that most women who experience fragility fractures have previously been diagnosed with osteopenia. 30 Unfortunately, many cases of osteoporosis and osteopenia remain undiagnosed until a fracture occurs, increasing the likelihood of complications and mortality. Therefore, the early detection of osteoporosis and osteopenia is crucial for disease prevention and management, which can reduce the incidence of osteoporotic fractures and alleviate the burden of this disease.
It has been widely accepted that BMD is a reliable marker for the early detection of osteoporosis and osteopenia. 31 Various tools, e.g., DXA and quantitative ultrasound, have been used for BMD measurement worldwide. However, their application is limited owing to inaccessibility, a lack of screening knowledge, and high cost, with only a few developing countries having access to DXA and quantitative ultrasound. 32 Moreover, DXA-based BMD measures only account for two-dimensional cortical and cancellous bone structures and cannot fully explain bone geometries, sizes, and microstructures. Thus, it is important to explore alternative, effective, safe, and cost-effective methods to improve this situation.
To address the limitations described above when detecting osteopenia and osteoporosis, researchers have turned to recent advancements in imaging technology and AI algorithms to develop computer-aided diagnostic systems. 33 Using medical images and applying advanced algorithms, these systems can provide cost-effective, readily available, and accurate means of detecting osteopenia, osteoporosis, and other medical conditions. 34
FSL is characterized by its ability to rapidly generalize to new tasks with only a few training examples, making it ideal for scenarios where training data are limited. 35 Generally, FSL is a K-way, N-shot task, where K represents the number of novel classes and N is the number of support set images for each class. In addition, K query sets contain M images selected randomly from the remaining data. The primary objective of FSL is to develop the ability to accurately classify images and extract features from the training dataset. Through this process, the model can learn to recognize and generalize patterns from a small set of examples, making it well suited for use in situations where only a limited number of labeled data are available. By incorporating FSL techniques, researchers can potentially improve the accuracy and efficiency of image classification and feature extraction methods in a wide range of applications. 36 Thus, for various medical datasets, FSL could be a powerful extension of standard DL and has emerged as a promising technique for disease recognition and classification. 37
A series of previous studies have demonstrated the application of DL algorithms for building osteoporosis diagnosis models. 34 In 2018, Naoufami and colleagues 38 proposed a DL model to detect osteoporotic vertebral fractures based on vertebral computed tomography (CT) images. Logical imaging features were extracted for system building, which achieved practical results. In 2019, another DL study concerning CT scans of vertebrae was reported by Krishnaraj et al. 39 to distinguish osteoporosis from normal conditions. Those authors used U-Net CNNs for the segmentation of radiological images and achieved good accuracy. In 2020, Lee et al. 40 used spine X-ray images to extract imaging features with CNN architecture and then loaded the features into a machine learning model for classification purposes. Using VGG as the feature extractor and a random forest as the classifier, 0.71 accuracy was achieved in the binary classification task. Yasaka and colleagues 41 developed a DL-based model to predict the BMD of lumbar vertebrae in CT images of the abdomen. A good correlation was observed between the predicted BMD values and the actual DXA BMD values. Most previous reports have used spine images in the AI-assisted radiological diagnosis of osteoporosis. He et al. 42 collected anteroposterior knee X-ray images and T scores from the DXA scans of 361 patients. They measured two radiographic parameters, i.e., cortical bone thickness and distal femoral cortex, which exhibited significant correlations with the BMD and T score. Wani et al. 20 selected four CNNs (AlexNet, VGG16, ResNet, and VGG19) to conduct a ternary classification task involving normal, osteopenia, and osteoporosis images in a set of 381 knee X-ray images. The models attained good accuracy, even though there was no independent test set.
Our study comprised several unique features. First, we applied FSL to the challenging task of diagnosing osteopenia and osteoporosis based on knee X-rays, which are the most widely used radiographic images. To the best of our knowledge, this is the first report on FSL in this field. Second, we designed a fine-tuning strategy that enables generalization from natural images (ImageNet) to X-ray images (pneumonia). In addition, a series of AutoML classifiers based on the Euclidean distance features derived from base images were developed to make predictions for novel images. The multistep FSL framework achieved practical performance in comparisons with radiologists.
This study also has several limitations. First, only knee X-ray images were used in the study, which may limit the generalizability of the FSL model. According to previous studies, further investigation involving spine X-rays or vertebral CT images are required. Clinical baselines and laboratory data are also valuable for disease prediction. Thus, it is worth conducting multimodal FSL modeling on the fusion of radiological data and structured covariates.
Conclusions
In conclusion, we developed a three-way, three-shot FSL framework for the diagnosis of osteopenia and osteoporosis in knee X-ray images. The FSL framework, based on fine-tuned feature extractors and Euclidean distance-based classifiers, achieved practical performance in a three-round evaluation when compared with radiologists. This framework supports the development of promising FSL methods for computer-aided diagnoses involving limited samples.
Supplemental Material
Supplemental material, sj-pdf-1-imr-10.1177_03000605241274576 for A few-shot learning framework for the diagnosis of osteopenia and osteoporosis using knee X-ray images by Hua Xie, Chenqi Gu, Wenchao Zhang, Jiacheng Zhu, Jin He, Zhou Huang, Jinzhou Zhu and Zhonghua Xu in Journal of International Medical Research
Supplemental material, sj-pdf-2-imr-10.1177_03000605241274576 for A few-shot learning framework for the diagnosis of osteopenia and osteoporosis using knee X-ray images by Hua Xie, Chenqi Gu, Wenchao Zhang, Jiacheng Zhu, Jin He, Zhou Huang, Jinzhou Zhu and Zhonghua Xu in Journal of International Medical Research
Acknowledgements
We thank all radiologists and postgraduate students in this project.
Authors’ contributions: JZ and ZX conceived and designed the study. HX, CG, ZH, and WZ acquired the data. HX, JZ, and JH performed analyses and drafted the manuscript. CG provided statistical assistance. All authors contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content. All authors read and approved the final manuscript. JZ and ZX are the guarantors and take responsibility for the integrity of the data and the accuracy of the data analysis.
The authors declare that there is no conflict of interest.
Funding: This study was supported by the National Natural Science Foundation of China (82000540), Changzhou Science and Technology Program (CJ20210003, CJ20210005), Suzhou Health Committee Program (KJXW2019001), and Jiangsu Health Committee Program (Z2022077). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iD: Jinzhou Zhu https://orcid.org/0000-0003-0544-9248
Data availability
All the code used to extract features, generate models, and evaluate model performance can be found in an open-accessed website (deep learning with Python [https://osf.io/7aujc] and automated machine learning with R [https://osf.io/zjahc]).
Supplementary material
Supplemental material for this article is available online.
References
- 1.Compston JE, McClung MR, Leslie WD. Osteoporosis. Lancet 2019; 393: 364–376. 2019/01/31. DOI: 10.1016/S0140-6736(18)32112-3. [DOI] [PubMed] [Google Scholar]
- 2.Akesson K, Marsh D, Mitchell PJ, et al. Capture the Fracture: a Best Practice Framework and global campaign to break the fragility fracture cycle. Osteoporos Int 2013; 24: 2135–2152. 2013/04/17. DOI: 10.1007/s00198-013-2348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ensrud KE, Crandall CJ. Osteoporosis. Ann Intern Med 2017; 167: ITC17–ITC32. 2017/08/02. DOI: 10.7326/AITC201708010. [DOI] [PubMed] [Google Scholar]
- 4.Anam AK, Insogna K. Update on Osteoporosis Screening and Management. Med Clin North Am 2021; 105: 1117–1134. 2021/10/25. DOI: 10.1016/j.mcna.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 5.LeBoff MS, Greenspan SL, Insogna KL, et al. The clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int 2022; 33: 2049–2102. 2022/04/29. DOI: 10.1007/s00198-021-05900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Yu K, Ning Z, et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone 2020; 140: 115561. 2020/07/31. DOI: 10.1016/j.bone.2020.115561. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal S, Das SK, Agarwal GG, et al. X-ray knee as a screening tool for osteoporosis. J Clin Densitom 2012; 15: 362–365. 2012/04/24. DOI: 10.1016/j.jocd.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Oehme F, Kremo V, Veelen NV, et al. Routine X-Rays after the Osteosynthesis of Distal Radius and Ankle Fractures. Dtsch Arztebl Int 2022; 119: 279–284. 2022/02/11. DOI: 10.3238/arztebl.m2022.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalili C, Kazemi M, Taheri E, et al. Exposure to heavy metals and the risk of osteopenia or osteoporosis: a systematic review and meta-analysis. Osteoporos Int 2020; 31: 1671–1682. 2020/05/04. DOI: 10.1007/s00198-020-05429-6. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh CI, Zheng K, Lin C, et al. Automated bone mineral density prediction and fracture risk assessment using plain radiographs via deep learning. Nat Commun 2021; 12: 5472. 2021/09/18. DOI: 10.1038/s41467-021-25779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang M, Kim M, Bae SJ, et al. Opportunistic Osteoporosis Screening Using Chest Radiographs With Deep Learning: Development and External Validation With a Cohort Dataset. J Bone Miner Res 2022; 37: 369–377. 2021/11/24. DOI: 10.1002/jbmr.4477. [DOI] [PubMed] [Google Scholar]
- 12.Ge H, Zhou X, Wang Y, et al. Development and Validation of Deep Learning Models for the Multiclassification of Reflux Esophagitis Based on the Los Angeles Classification. J Healthc Eng 2023; 2023: 7023731. 20230218. DOI: 10.1155/2023/7023731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Hong Y, Wang Y, et al. Automated Multimodal Machine Learning for Esophageal Variceal Bleeding Prediction Based on Endoscopy and Structured Data. J Digit Imaging 2023; 36: 326–338. 20221024. DOI: 10.1007/s10278-022-00724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin M, Liang X, Wang Z, et al. Identification of Asymptomatic COVID-19 Patients on Chest CT Images Using Transformer-Based or Convolutional Neural Network-Based Deep Learning Models. J Digit Imaging 2023: 1–10. 2023/01/04. DOI: 10.1007/s10278-022-00754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Lin J, Zhu S, et al. Deep learning in the radiologic diagnosis of osteoporosis: a literature review. J Int Med Res 2024; 52: 3000605241244754. DOI: 10.1177/03000605241244754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Fu Y, Chen J, et al. Generalized Meta-FDMixup: Cross-Domain Few-Shot Learning Guided by Labeled Target Data. IEEE Trans Image Process 2022; 31: 7078–7090. 2022/11/09. DOI: 10.1109/TIP.2022.3219237. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Chen Y, Zhang Y, et al. Diagnosis of arrhythmias with few abnormal ECG samples using metric-based meta learning. Comput Biol Med 2023; 153: 106465. 2023/01/08. DOI: 10.1016/j.compbiomed.2022.106465. [DOI] [PubMed] [Google Scholar]
- 18.Fei-Fei L, Fergus R, Perona P. One-shot learning of object categories. IEEE Trans Pattern Anal Mach Intell 2006; 28: 594–611. 2006/03/29. DOI: 10.1109/TPAMI.2006.79. [DOI] [PubMed] [Google Scholar]
- 19.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018; 172: 1122–1131 e1129. 2018/02/24. DOI: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Wani IM, Arora S. Osteoporosis diagnosis in knee X-rays by transfer learning based on convolution neural network. Multimed Tools Appl 2022: 1–25. 2022/10/04. DOI: 10.1007/s11042-022-13911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimai HP. Use of dual-energy X-ray absorptiometry (DXA) for diagnosis and fracture risk assessment; WHO-criteria, T- and Z-score, and reference databases. Bone 2017; 104: 39–43. 2017/01/04. DOI: 10.1016/j.bone.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007; 147: 573–577. DOI: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 23.Kaur N, Mittal A. CADxReport: Chest x-ray report generation using co-attention mechanism and reinforcement learning. Comput Biol Med 2022; 145: 105498. 2022/05/20. DOI: 10.1016/j.compbiomed.2022.105498. [DOI] [PubMed] [Google Scholar]
- 24.Ho CS, Chen YP, Fan TY, et al. Application of deep learning neural network in predicting bone mineral density from plain X-ray radiography. Arch Osteoporos 2021; 16: 153. 2021/10/10. DOI: 10.1007/s11657-021-00985-8. [DOI] [PubMed] [Google Scholar]
- 25.Suh B, Yu H, Kim H, et al. Interpretable Deep-Learning Approaches for Osteoporosis Risk Screening and Individualized Feature Analysis Using Large Population-Based Data: Model Development and Performance Evaluation. J Med Internet Res 2023; 25: e40179. 2022/12/10. DOI: 10.2196/40179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das D, Lee CSG. A Two-Stage Approach to Few-Shot Learning for Image Recognition. IEEE Trans Image Process 2020; 29: 3336–3350. DOI: 10.1109/TIP.2019.2959254. [DOI] [PubMed] [Google Scholar]
- 27.Yu C, Li Y, Yin M, et al. Automated Machine Learning in Predicting 30-Day Mortality in Patients with Non-Cholestatic Cirrhosis. J Pers Med 2022; 12: 1930. 2022/11/25. DOI: 10.3390/jpm12111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, Xu J, Shi R, et al. A Multi-Label Deep Learning Model with Interpretable Grad-CAM for Diabetic Retinopathy Classification. Annu Int Conf IEEE Eng Med Biol Soc 2020; 2020: 1560–1563. DOI: 10.1109/EMBC44109.2020.9175884. [DOI] [PubMed] [Google Scholar]
- 29.US Preventive Services Task Force, Curry SJ, Krist AH, Owens DK, et al. Screening for Osteoporosis to Prevent Fractures: US Preventive Services Task Force Recommendation Statement. JAMA 2018; 319: 2521–2531. 2018/06/28. DOI: 10.1001/jama.2018.7498. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Chen Q, Nie B, et al. Trends in Bone Mineral Density, Osteoporosis, and Osteopenia Among U.S. Adults With Prediabetes, 2005-2014. Diabetes Care 2020; 43: 1008–1015. 2020/03/08. DOI: 10.2337/dc19-1807. [DOI] [PubMed] [Google Scholar]
- 31.Bruyere O, Reginster JY. Monitoring of osteoporosis therapy. Best Pract Res Clin Endocrinol Metab 2014; 28: 835–841. 2014/11/30. DOI: 10.1016/j.beem.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Reid IR. Monitoring Osteoporosis Therapy. J Bone Miner Res 2021; 36: 1423–1424. 2021/06/17. DOI: 10.1002/jbmr.4393. [DOI] [PubMed] [Google Scholar]
- 33.Kolanu N, Silverstone EJ, Ho BH, et al. Clinical Utility of Computer-Aided Diagnosis of Vertebral Fractures From Computed Tomography Images. J Bone Miner Res 2020; 35: 2307–2312. 2020/08/05. DOI: 10.1002/jbmr.4146. [DOI] [PubMed] [Google Scholar]
- 34.Dimai HP. New Horizons: Artificial Intelligence Tools for Managing Osteoporosis. J Clin Endocrinol Metab 2023; 108: 775–783. 2022/12/09. DOI: 10.1210/clinem/dgac702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drori I, Zhang S, Shuttleworth R, et al. A neural network solves, explains, and generates university math problems by program synthesis and few-shot learning at human level. Proc Natl Acad Sci U S A 2022; 119: e2123433119. 2022/08/03. DOI: 10.1073/pnas.2123433119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, Gao M, Li H, et al. Multi-Learner Based Deep Meta-Learning for Few-Shot Medical Image Classification. IEEE J Biomed Health Inform 2022; 27: 17–28. PP 2022/10/18. DOI: 10.1109/JBHI.2022.3215147. [DOI] [PubMed] [Google Scholar]
- 37.Paul A, Tang YX, Shen TC, et al. Discriminative ensemble learning for few-shot chest x-ray diagnosis. Med Image Anal 2021; 68: 101911. 2020/12/03. DOI: 10.1016/j.media.2020.101911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita N, Cheung YY, Hassanpour S. Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Comput Biol Med 2018; 98: 8–15. 2018/05/15. DOI: 10.1016/j.compbiomed.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Krishnaraj A, Barrett S, Bregman-Amitai O, et al. Simulating Dual-Energy X-Ray Absorptiometry in CT Using Deep-Learning Segmentation Cascade. J Am Coll Radiol 2019; 16: 1473–1479. 2019/04/16. DOI: 10.1016/j.jacr.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Choe EK, Kang HY, et al. The exploration of feature extraction and machine learning for predicting bone density from simple spine X-ray images in a Korean population. Skeletal Radiol 2020; 49: 613–618. 2019/11/25. DOI: 10.1007/s00256-019-03342-6. [DOI] [PubMed] [Google Scholar]
- 41.Yasaka K, Akai H, Kunimatsu A, et al. Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol 2020; 30: 3549–3557. 2020/02/16. DOI: 10.1007/s00330-020-06677-0. [DOI] [PubMed] [Google Scholar]
- 42.He QF, Sun H, Shu LY, et al. Radiographic predictors for bone mineral loss: Cortical thickness and index of the distal femur. Bone Joint Res 2018; 7: 468–475. 2018/08/21. DOI: 10.1302/2046-3758.77.BJR-2017-0332.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-imr-10.1177_03000605241274576 for A few-shot learning framework for the diagnosis of osteopenia and osteoporosis using knee X-ray images by Hua Xie, Chenqi Gu, Wenchao Zhang, Jiacheng Zhu, Jin He, Zhou Huang, Jinzhou Zhu and Zhonghua Xu in Journal of International Medical Research
Supplemental material, sj-pdf-2-imr-10.1177_03000605241274576 for A few-shot learning framework for the diagnosis of osteopenia and osteoporosis using knee X-ray images by Hua Xie, Chenqi Gu, Wenchao Zhang, Jiacheng Zhu, Jin He, Zhou Huang, Jinzhou Zhu and Zhonghua Xu in Journal of International Medical Research
Data Availability Statement
All the code used to extract features, generate models, and evaluate model performance can be found in an open-accessed website (deep learning with Python [https://osf.io/7aujc] and automated machine learning with R [https://osf.io/zjahc]).