Abstract
When equal volumes of two immiscible liquids are mixed (e.g., a hydrocarbon and a fluorocarbon), Janus droplets can form in an aqueous solution. In a gravity-aligned Janus droplet, the boundary between the two phases is flat and thus optically transparent when viewed from above. When tipped due to interactions with an analyte (i.e., agglutination), the resulting change in refraction and reflection yields an optical signal that can be detected and quantified. This study reports the detection and quantitation of interleukin-6 (IL-6) using emulsions functionalized at the hydrocarbon:aqueous interface with engineered proteins that bind IL-6 at high affinity and specificity. Hyperthermophilic affinity proteins (rcSso7d) are derived from thermophiles, giving them excellent thermal stability. Two rcSso7d affinity protein variants were synthesized with a noncanonical azide-functionalized amino acid to enable click chemistry to novel polymeric anchors embedded in the hydrocarbon phase. The two binding proteins recognize different epitopes, enabling the detection of both monomeric and dimeric IL-6 via agglutination. It is noteworthy that the rsSso7d protein variants, in addition to having superior thermal stability and facile recombinant synthesis in E. coli, show superior performance when compared to commercial antibodies for IL-6.
Introduction
Interleukin-6 (IL-6) is a multifunctional proinflammatory cytokine that plays an active role in immune responses and inflammation.1 IL-6 can be synthesized in a variety of cell types, and it induces the production of acute phase proteins, such as C-reactive protein (CRP).2 IL-6 levels in serum can be used as a diagnostic biomarker for sepsis and are a prognostic factor for different cancers, such as Hodgkin’s lymphoma,3 renal cell carcinoma,4 bladder carcinoma,5 ovarian cancer,6 and breast cancer.7−9 Additionally, serum IL-6 levels can be used for prognosticating severe cases of COVID-19.10−13 Common methods to measure IL-6 levels include enzyme-linked immunosorbent assay (ELISA)14,15 and electrochemical sensors16−18 with recent developments in point of care sensors such as paper-based assays10,19 and lateral flow assays (LFAs).20−22 Novel methods for the detection of IL-6 and other inflammatory biomarkers will be instrumental in the early diagnosis, monitoring, and treatment of a wide variety of diseases. While there are existing assays for IL-6, here we describe an agglutination assay for IL-6 as a proof-of-principle that sensors that exploit droplet morphology can be used to detect a single protein.
Dynamic complex droplets have traditionally been used for drug delivery23−27 and in the food industry,28−30 but have the potential to be leveraged as optical elements in the engineering of novel testing strategies. One type of complex colloidal droplets is composed of equal volumes of hydrocarbon and fluorocarbon oil, resulting in gravity-aligned droplets due to the higher density of the fluorous phase.31 When properly stabilized with surfactants, the biphasic droplets adopt a Janus morphology. The difference in refractive indices for the two phases results in directional optical properties that are sensitive to droplet alignment and morphology changes resulting from interfacial activity.32 Thus, Janus droplets, or double emulsions, represent a diverse platform for the generation of inexpensive optical sensing systems. In sensing schemes relying on droplet morphology changes in response to analyte, the directional output of emitted light from the interior of the droplets is highly sensitive to the droplet morphology. This method has been used to detect the presence of mannan or Salmonella,32 hydrogen peroxide,33 and per- and polyfluorinated substances.34 Alternatively, when recognition elements are attached to the hydrocarbon phase of the Janus droplets, the droplets can agglutinate around their target, wherein the droplets tip toward each other causing optical scattering from the agglutinated clusters; previously, E. coli was detected using this mechanism.35 Additionally, emissive droplet agglutination assays have also been used to detect Zika virus,36Listeria,37 and SARS-CoV-2.38
Similar to other biosensor schemes, droplet agglutination assays often rely on antibodies as the recognition elements. Although they are powerful recognition elements, antibodies are expensive, variable in quality, fragile, and not always readily available in monoclonal form. A viable alternative are variants of the hyperthermophilic protein, reduced-charge Sso7d (rcSso7d).39 The Sso7d protein was originally a DNA binding histone protein isolated from the hyperthermophilic archaeon Sulfolobus solfataricus. It has proved to be an ideal scaffold for the engineering of high-affinity binders to epitopes, due to its small size, high thermal stability (∼98 °C), ease of expression in both yeast and E. coli, and lack of cysteines or post-translational modifications.40 The native protein was modified to reduce the charge on its binding face, generating rcSso7d. This protein was used to generate a yeast display library of ∼1.4 × 109 variants which could be used to identify high affinity (low nM) binders against an epitope of choice. A sandwich pair of two high affinity (low nM KD) IL-6 binders were identified from the yeast display library using the Rapid Affinity Pair Identification via Directed Selection (RAPIDS) method, as previously described.41 Briefly, the library diversity of 1.4 × 109 variants was reduced via multiple rounds of selection for high affinity binders. This is achieved by attaching magnetic beads to IL-6 (via a HIS tag) and pulling down yeast expressing rcSso7d variants that bind with a high affinity to IL-6. Subsequently, the library is further refined by labeling IL-6 with a fluorescent tag (via primary and secondary antibodies) to enable fluorescence-activated cell sorting (FACS) of high affinity binders until a reasonable number of unique clones (∼100) can be sequenced. Analogous to a sandwich ELISA, we also set out to create binders that bind to independent epitopes. This was accomplished by replacing the primary and secondary antibodies with an isolated rcSso7d variant labeled with a fluorophore so that FACS could be used to identify variants that bind to a different epitope.41 The resulting sandwich pair binds to two different epitopes on IL-6 (termed E1 and E2). E1 and/or E2 were used to functionalize the hydrocarbon phase of Janus droplets.
We report an emulsion-based agglutination assay for the selective detection of IL-6 made possible by using click chemistry to attach high affinity rcSso7d binders to polymers embedded in the hydrocarbon phase. Specifically, 4-azidophenylalanaine (4AZP)-modified rcSso7d binders were used as recognition elements. We then developed amphiphilic surfactant polymers with click handles that localize at the hydrocarbon and water interface of our biphasic Janus droplets. Bioconjugation of the rcSso7d binders to the droplet surface occurs through a strain promoted alkyne–azide cycloaddition (SPAAC). Droplets containing immobilized rcSso7d binders bind to IL-6; the binding of two or more droplets to IL-6 yields agglutination such that droplets tip toward each other. In the absence of IL-6, the droplets remain gravity-aligned due to density-driven alignment. The deviation from a vertical alignment caused by agglutination produces a strong optical signal for detection, since the interface between the hydrocarbon and fluorocarbon phases reflects and refracts light. IL-6 exists both as a monomer and a dimer. Thus, agglutination can occur with droplets conjugated with a single rcSso7d variant (in the case of IL-6 being a dimer) or with a 1:1 mixture of the two different rcSso7d-functionalized droplets (analogous to a sandwich ELISA that is able to bind to IL-6 monomers). These clusters can be viewed under a microscope and counting of agglutinated droplets gives a quantitative measure of the amount of IL-6 present in a sample. Here we show that IL-6 levels at 50 ng/mL can be detected using the agglutination assay. While not yet effective for physiological levels of IL-6 (<10 pg/mL),42 this work serves as a proof-of-principle that an agglutination assay using Janus droplets can be used to detect a single protein, and furthermore points to the possibility of refinements for detecting IL-6 at biologically relevant levels.
Experimental Section
Materials
All reagents and solvents were purchased from commercial suppliers and used without further purification unless otherwise stated. PPFP-PPEG-b-PBn, PDBCO-PPEG-b-PBn, PN3-PPEG-b-PBn, the azido-modified anti IL-6 antibody, and the DBCO-modified anti-IL-6 antibody were synthesized as described in the Supporting Information. Additional information on the materials and their purification, including experimental details on cell culture and protein purification, is provided in the Supporting Information.
General Procedure for the Preparation of Janus Emulsions
Janus emulsions were fabricated via bulk emulsification, which generates polydisperse droplets with highly uniform morphology and composition. The disperse and continuous phases consists of 1 mg/mL of PDBCO–PPEG-b-PBn or PN3-PPEG-b-PBn in DEB and HFE-7500 combined in a 1:1 volume ratio and 0.1 wt % Tween 20:0.1 wt % Zonyl in 1× PBS (10 mM, pH = 7.4), respectively. The disperse phase was heated above the upper critical solution temperature (42 °C) until the two phases become miscible. The miscible disperse phase (30 μL) was rapidly added to 500 μL of warm continuous phase and subsequently emulsified via a vortex mixer at 3500 rpm for five seconds. A 4:6 volume ratio of 0.1 wt % CTAB:0.1 wt % Zonyl in 1× PBS was used as the continuous phase for droplets that will be further modified with DNA.
General Procedure for Droplet Bioconjugation
rcSso7d protein or antibody was added to droplet solutions consisting of 30 μL of droplets and 500 μL of continuous phase and incubated overnight on an orbital shaker (100 rpm). After incubation, the continuous phase was solvent exchanged five times with fresh continuous phase solution to remove unreacted rcSso7d protein or antibody. For droplets with additional PEG subunits, poly(ethylene glycol) methyl ether azide (PEG-N3) in 1× PBS (10 mM) was added to the rcSso7d-conjugated droplet solution and incubated overnight on an orbital shaker (100 rpm). The continuous phase was then solvent exchanged five times with a fresh solution to remove unreacted PEG-N3. Optimal functionalization schemes include reactions with ∼1 nmol rcSso7d protein for 30 μL of droplets in 500 μL overnight and passivation of the droplets with 25 nmol of poly(ethylene glycol) methyl ether azide.
General Procedure for Agglutination Assays
For each sensing experiment, the glass surface of a Thermo-Fisher Scientific Invitrogen Attofluor Cell Chamber was wetted with surfactant solution prior to depositing the droplets. For single droplet variant agglutination studies, 14 μL of droplets was used. For two droplet variant studies, 7 μL of each type of droplet variant was used. Once the droplets were deposited onto the imaging dish, they were spread out across the dish. Depending on the experiment, different volumes of analyte (IL-6 or CRP) were added to the experimental setup (total continuous phase volume = 500 μL). Specifically, 10 μL of human serum and 104 μL of 2.4 mg/mL CRP were added as analyte for the corresponding control experiments; a 0.1 w/v% solution of BSA in the continuous phase was prepared for the 1 mg/mL BSA control experiment. If necessary, analyte solutions were diluted with 1× PBS prior to addition. The droplets were incubated for specified amounts of time, and orientational changes were recorded using an inverted microscope.
Results and Discussion
Polymer Synthesis and Characterization
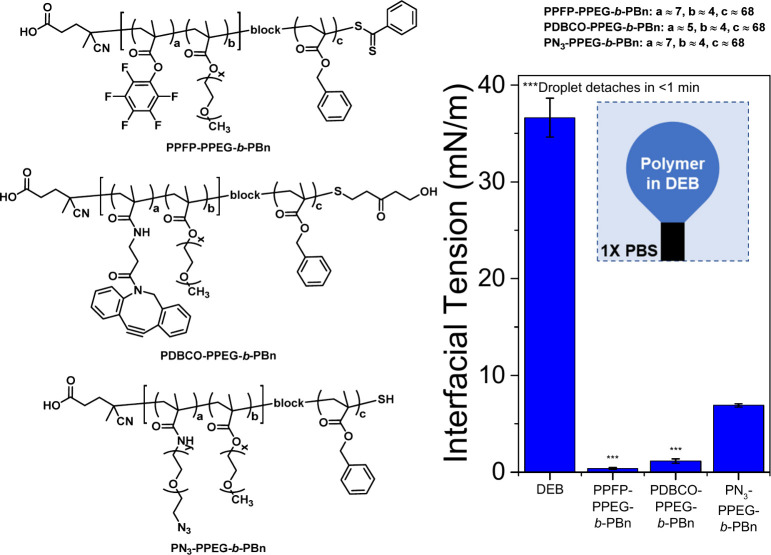
To functionalize and stabilize the hydrocarbon–water interface of the Janus droplets, a suite of block copolymer surfactants was synthesized with a hydrophobic block to anchor the polymer to the hydrocarbon phase and a hydrophilic bioconjugation block that is localized at the surface. An amphiphilic, hydrophilic-b-hydrophobic diblock copolymer, PPFP-PPEG-b-PBn, was synthesized via reversible addition–fragmentation chain transfer (RAFT) polymerization. PPFP-PPEG-b-PBn allows for easy access to polymers with different bioconjugation handles since the pentafluorophenyl (PFP) substituents allows for postpolymerization modification with amino-terminated functional groups enabling attachment of alkynes for click chemistry. To obtain polymers capable of undergoing SPAAC, PPFP-PPEG-b-PBn was reacted with either dibenzocyclooctyne (DBCO)-amine or azido-PEG-amine to obtain PDBCO-PPEG-b-PBn and PN3-PPEG-b-Pbn, respectively (Figure 1 and Schemes S1–S2.) The hydrophobic block consists of benzyl methacrylate subunits that anchor the polymer to the hydrocarbon phase of the Janus droplets. The hydrophilic block is a random copolymer of PEG methyl ether methacrylate (Mn = 500) and DBCO or azido-PEG methacrylate. PEG methyl ether methacrylate was chosen for its hydrophilicity, whereas DBCO and azido-PEG methacrylate were chosen to enable SPAAC with azido- and DBCO-functionalized recognition proteins, respectively. 19F NMR analysis indicates that approximately 80% of the original PFP subunits were successfully converted to DBCO-subunits whereas the azido-PEG-amine substitution is quantitative (Figure S5). Substitution of the PFP subunits of PPFP-PPEG-b-PBn with the azido-PEG-amine was further confirmed by FTIR (Figure S7).
Figure 1.
Chemical structures of the atactic amphiphilic diblock copolymers and pendant drop analysis of the polymers in diethylbenzene (DEB). An inverted pendant drop is utilized, because DEB is less dense than the buffer. Having copolymers dissolved in the DEB lowers the DEB-buffer interfacial tension, which indicates that the diblock copolymers are surface active.
The amphiphilic characteristics and surfactant effectiveness of all the polymers were evaluated via pendant drop tensiometry, a method that monitors the droplet’s dimension and profile characteristics and uses the Young–Laplace equation to determine interfacial tensions. When PDBCO-PPEG-b-PBn and PN3-PPEG-b-Pbn are dissolved in diethylbenzene (DEB), the DEB-buffer interfacial tension, γH, drops from ∼36 mN/m to <1.2 and 6.9 mN/m, respectively, indicating that the diblock copolymers are surface active and stabilize the interface by lowering the interfacial energy between DEB and the buffer. The initial polymer with the PPFP scaffold (PPFP-PPEG-b-PBn) also has high interfacial activity with γH < 1 mN/m (Figure 1). In contrast to polymers used in our previous studies that were synthesized by post polymerization modification of commercial polystyrene-b-poly(acrylic acid),36,38 the present synthetic approach allows for more precise control over the hydrophobicity and hydrophilicity, molecular weight, and ratio of PEG. Moreover, it provides an efficient path for bioconjugation by incorporation of reactive pentafluorophenyl esters. The synthesized polymers allow for the facile functionalization of Janus droplets with IL-6 binders derived from rcSso7d or commercially available antibodies.
Bioconjugation at the Interface of Janus Emulsions
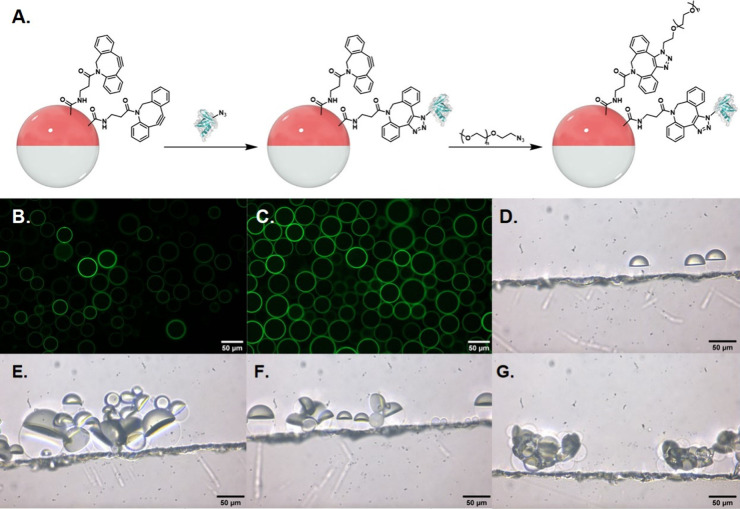
Polydisperse Janus droplets were obtained by combining a 1 mg/mL DEB solution of PDBCO-PPEG-b-PBn with an equal volume of 2-(trifluoromethyl)-3-ethyoxydodecafluorohexane (HFE-7500), a fluorocarbon solvent. The resulting mixture was heated above its upper critical solution temperature (42 °C) to create a single phase and then dispersed into a larger volume of an aqueous continuous phase solution composed of 1× Phosphate Buffered Saline (PBS) (10 mM, pH = 7.4) and surfactants (0.1 wt % Tween 20:0.1 wt % Zonyl, 1:1 (v/v)). Upon cooling, the polydisperse droplets phase separate into Janus morphology. Interfacial bioconjugation with hyperthermophilic affinity proteins via SPAAC was performed by adding the AZP-functionalized rcSso7d proteins (0.2–1 nmol) to DBCO-functionalized droplets (30 μL volume) in 500 μL of the aqueous continuous phase described above (Figure 2A). The mixture was allowed to react overnight at room temperature with mixing by an orbital shaker. The droplets have a net density higher than that of the aqueous phase and will settle at the bottom of a vial, thereby allowing for exchange of the continuous aqueous phase to remove unreacted azide-functionalized proteins. The AZP-functionalized rcSso7d proteins for bioconjugation were purified via a gravity column using HisPURE Ni2+ NTA resin unless otherwise stated. In the bioconjugation step, only one of the two IL-6-binding rcSso7d variants (E1 or E2) is added to the reaction mixture to produce two different functionalized emulsions. To confirm successful bioconjugation, E1 and E2 were labeled with NHS-FITC and used in the bioconjugation step. Confocal fluorescence imaging of the postconjugated droplets confirms successful bioconjugation of the rcSso7d proteins on the DEB-continuous phase interface (Figures 2B and 2C). When N3-functionalized droplets are used instead of the DBCO-functionalized ones, no fluorescence is observed after addition of FITC-labeled E2, which further confirms that the proteins are covalently bound to the droplet surface (Figure S8).
Figure 2.
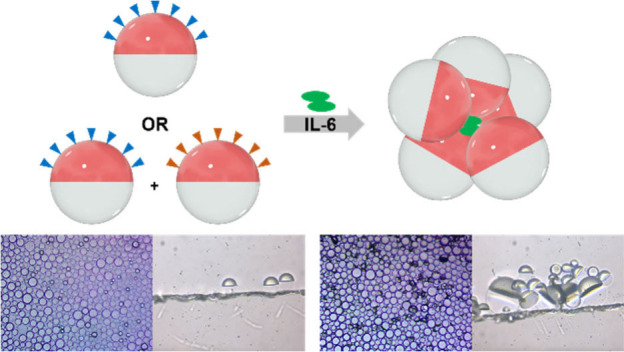
A. Droplet bioconjugation scheme. PDBCO-PPEG-b-PBn-functionalized droplets were sequentially reacted with azide-functionalized rcSso7d proteins, followed by poly(ethylene glycol) methyl ether azide. Images are not drawn to scale; droplets are orders of magnitude larger than protein components. Fluorescence microscopy images of droplets reacted with B. FITC-labeled E1 and C. FITC-labeled E2. Side-view images of D. E1+PEG-functionalized Janus droplets, E. and F. 1:1 E1+PEG- and E2+PEG-functionalized droplets incubated with 5 μg/mL of IL-6, and G. DNA-functionalized droplets in 0.1 wt % of 4:6 CTAB:Zonyl that are agglutinated due to electrostatic interactions between the DNA and CTAB. All images are of polydisperse droplets.
The bioconjugation step is not assumed to be quantitative, and when we push the reaction further by adding more than 8 nmol of rcSso7d binder, we observe some self-interaction (droplet aggregation) when no analyte is present. These self-interactions can be prevented by reacting the remaining residual DBCO moieties on the droplets with poly(ethylene glycol) methyl ether azide (Mn = 1000) overnight. The resulting PEG- and rcSso7d-functionalized droplets do not self-interact (Figure S9); the long PEG chains likely mitigate interactions between neighboring droplets. Despite having longer PEG chains on the surface than those present on the polymer, these functionalized droplets can still readily detect IL-6. Representative side-view images of droplets functionalized with E1/E2 further confirm IL-6 dependent agglutination (Figures 2D-F). Agglutination can also occur due to electrostatic interactions between cationic surfactants and DNA (Figure 2G). As a result, our optimal functionalization schemes include reactions with ∼1 nmol of rcSso7d protein for 30 μL of droplets in 500 μL of continuous phase overnight and passivation of the droplets with 25 nmol of poly(ethylene glycol) methyl ether azide.
Agglutination Assay with IL-6
Sensing experiments were performed with rcSso7d+PEG-functionalized droplets in cell chambers by adding different concentrations of IL-6 to the continuous phase at room temperature. For the droplets to tilt from their equilibrium gravity (density) alignment, the rcSso7d-conjugated droplets need multiple interactions between the droplets and the analyte (Figures 2E-F). Orientational changes in the droplets were recorded for quantitation using an inverted microscope with incubation times varied from 1 h to overnight.
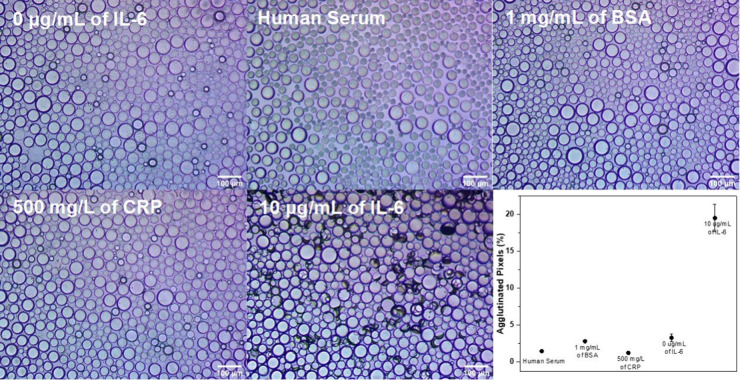
To test for specificity, the droplets were challenged with other purified proteins (albumin and CRP), as well as complex mixtures (human serum). Control experiments with rcSso7d+PEG-functionalized droplets without IL-6 in 1× PBS and in 1× PBS with human serum showed no agglutination (Figures 3 and S10). Additionally, no agglutination was observed in control experiments performed with 1 mg/mL of bovine serum albumin (BSA) and 500 mg/L of CRP, which is 50 times higher than the healthy physiological CRP levels (Figures 3 and S10). These results indicate high specificity in relevant mixtures. Control experiments were performed with a 1:1 ratio of each variant of rcSso7d+PEG-functionalized droplets. Both rcSso7d proteins purified using a Ni2+ resin and proteins that underwent additional purification via size exclusion chromatography (SEC) were used for the previously described control experiments. These results indicate that our sensing scheme is selective for IL-6 and that there are no nonspecific interactions between different rcSso7d-conjugated droplets. Furthermore, the small impurities that may be present in the rcSso7d proteins only purified with Ni2+ resin do not affect the agglutination study. This shows that a simple, inexpensive, fast, single step purification is sufficient for these assays, thereby making these proteins accessible to laboratories that do not have access to expensive fast protein liquid chromatography (FPLC) machinery.
Figure 3.
Inverted microscope images of 1:1 E1+PEG- and E2+PEG-functionalized droplets incubated under different conditions overnight (0 μg/mL of IL-6, human serum, 1 mg/mL of BSA, 500 mg/L of CRP, and 10 μg/mL of IL-6). The agglutinated droplets appear on their side and create darkened spots in the images as a result of their efficient scattering of transmitted light. At the bottom right is a plot of the percent of agglutinated pixels according to image processing for each control relative to the 10 μg/mL of IL-6 assay.
In principle, it is possible for agglutination to occur with droplets functionalized with a single rcSso7d variant because IL-6 exists as both a monomer and dimer under physiological conditions.43,44 If IL-6 is in its dimeric form, there will be a binding site for the same rcSso7d variant on each monomer (Scheme S3). To determine the effect of a single or multiple binding event(s), we performed three types of agglutination assays that comprise (1) E1+PEG-functionalized droplets, (2) E2+PEG-functionalized droplets, and (3) 1:1 ratio of E1+PEG- and E2+PEG-functionalized droplets with rcSso7d proteins purified via Ni2+ resin. At high IL-6 concentrations (10 μg/mL), we observe agglutination after only 1 h of incubation in all three types of assays (Figure S11). This suggests that using just one rcSso7d variant is sufficient to detect IL-6. The extent of agglutination intensifies with incubation times, which means that the assay’s sensitivity increases with time. Additionally, the extent of agglutination also decreases with analyte concentration, allowing for the creation of a quantitative IL-6 assay.
Quantification of the Extent of Agglutination
Previously, an image processing program was implemented to calculate the percentage of area covered by agglutinated Janus droplets by evaluating the differences in the optical intensity of the images before and after analyte exposure. The program used the adaptive thresholding algorithm to distinguish regions of higher transparency (pristine droplets) from opaque regions (agglutinated droplets). However, the program has limitations when applied to Janus emulsion assays composed of polydisperse droplets since an average radius is required as an input in order to properly use the adaptive thresholding algorithm.35,37 To address the incompatibility of polydisperse droplets with the previous software, we established an alternate image processing work flow using the free ImageJ software that is compatible with polydisperse droplets. Briefly, each optical image was split into its red, green, and blue components, and pixel counting was performed with ImageJ’s built-in histogram function on the blue channel image. The pixel thresholds for agglutination and baseline correction were determined using optical images taken prior to analyte addition. The percent agglutination level was calculated by summing the number of pixels below the threshold and dividing by the total number of pixels. This process was applied to all images for a given time point, but the top quintile of images or the three images with the highest level of agglutination, whichever was greater, were used in the final response curve. We note that the extent of agglutination is not homogeneous across the entire imaging dish. The agglutination is dependent on the droplet packing density. Therefore, image analysis was performed only on regions that had at least ∼90% coverage, which resulted in consistent determinations of the amount of agglutination for a given amount of IL-6. Additionally, images were taken across the entire dish to account for the inhomogeneity. A full description of the image processing work flow is provided in the Supporting Information. The level of agglutination was also determined for the different control assays (human serum, 1 mg/L of BSA, and 500 mg/L of CRP). The percent of agglutinated pixels was minimal and comparable to that of assays with no analyte (Figure 3).
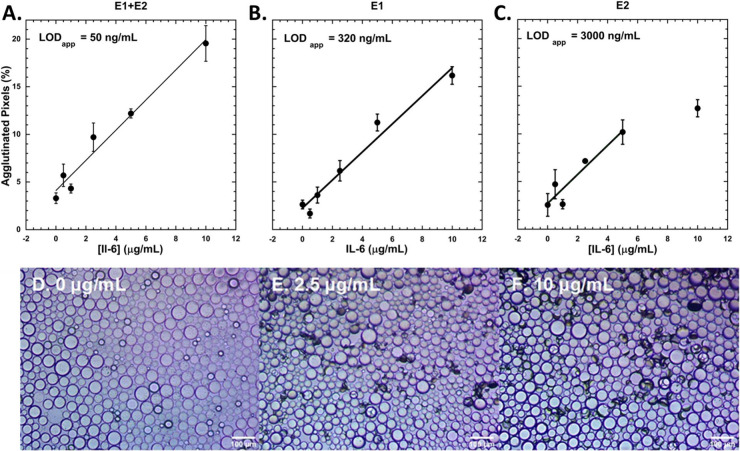
Response curves for the three types of agglutination assays after overnight incubation with IL-6 were determined and are shown in Figure 4. The data were fit to linear regressions, and the full set of data (unaveraged) is provided in the Supporting Information (Figure S16). The limit of detection (LOD) of assays performed with droplets functionalized with E1 only, E2 only, E1 and E2 are 320, 3000, and 50 ng/mL, respectively. The sandwich assay utilizing 1:1 E1+PEG- and E2+PEG-functionalized droplets unsurprisingly has the lowest LOD. This is expected because when both rcSso7d variants (E1 and E2) are present, both monomeric and dimeric forms of IL-6 can trigger agglutination, whereas only dimeric IL-6 is capable of triggering agglutination with a single variant (E1 or E2). Additionally, a single binder (E1 or E2) can bind to one IL-6 dimer at two locations at most, whereas the sandwich can bind at four locations of a single IL-6 dimer. A response curve for the sandwich assay after incubation for 5 h was also determined (Figure S17). The response curve shows that IL-6 levels can be quantified without overnight incubation with a LOD of 3275 ng/mL. Furthermore, the kinetics of the agglutination for the sandwich assay were monitored at 1 h intervals for the first 5 h of the assay. The kinetics for the sandwich assay for different IL-6 concentrations is shown in Figure 5. This data shows qualitatively that the rate of agglutination increases with increasing IL-6 concentration.
Figure 4.
Dose response curves for A. 1:1 ratio of E1+PEG- and E2+PEG- functionalized Janus droplets (k = 1.591, R = 0.98), B. E1+PEG-functionalized droplets (k = 1.445, R = 0.99), and C. E2+PEG-functionalized droplets (k = 1.559, R = 0.95) after overnight incubation with IL-6. Data are fit to linear responses, and standard errors are reported. The last data point for C. is excluded from the linear fit. Data represent n ≥ 3 replicates. Representative images of agglutinated droplets (1:1 ratio of E1+PEG- and E2+PEG- functionalized) after incubation with D. 0 μg/mL, E. 2.5 μg/mL, and F. 10 μg/mL of IL-6.
Figure 5.
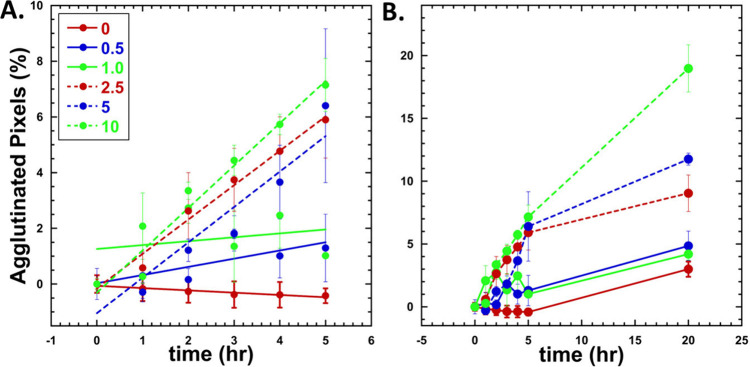
Kinetics of agglutination at various concentrations (0–10 ug/mL) A. First 5 h after addition of IL-6. Lines indicate linear fit of data (0 μg/mL = −0.082, R = 0.95, 0.5 μg/mL = 0.29, R = 0.76, 2.5 μg/mL = 1.23, R = 0.99, 5 μg/mL = 1.27, R = 0.94, and 10 μg/mL = 1.52, R = 0.98. 1.0 μg/mL did not fit well to a linear (R = 0.255) but fit is still shown. B. Line graph of data collected overnight indicated increase in agglutination over time as a function of concentration. Standard error is displayed. Data represent n ≥ 3 replicates.
We have significantly improved the sensitivity and cost-effectiveness of agglutination assays that use rcSso7d proteins. Previously, we reported an emulsion agglutination assay that utilized rcSso7d proteins to detect the hexameric Zika NS1 protein and achieved a LOD of 100 nM,36 which is 2 orders of magnitude higher than the LOD of 50 ng/mL (∼1.1–2.2 nM) that we achieved for IL-6. Unlike the Zika NS1 assay, which relied on an optical detection scheme such as backscattering and inner filter effect, the IL-6 agglutination assay does not require additional fluorescent dyes in the hydrocarbon or fluorocarbon phase or an optical fiber detection setup, thus making it more cost-effective. The LOD of the IL-6 assay is comparable to that of an agglutination assay for anti-SARS-CoV-2 spike IgG antibody (0.2 μg/mL or ∼1.3 nM) that utilizes antibodies instead of rcSso7d proteins as the biorecognition element. The anti-SARS-CoV-2 spike IgG assay takes advantage of fluorescent dyes and thus requires an optical detection setup. We note that adding fluorescent dyes and changing the detection setup can potentially lower the LOD and incubation time of our IL-6 assay; however, changing the detection scheme will increase the overall cost of the assay. Additionally, the anti-SARS-CoV-2 spike IgG assay utilizes monodisperse droplets produced using a microfluidics device, whereas the IL-6 assay leverages polydisperse droplets that are more cost-effective to produce. Furthermore, there are significant advantages to utilizing rcSso7d over commercial antibodies as biorecognition elements (vide infra). Compared to the Zika NS1 protein and the anti-SARS-CoV-2 spike IgG protein, IL-6, which is 21–28 kDa,43 is a much smaller analyte and thus more challenging to detect. Although the Zika NS1 protein is 48 kDa and comparable in size to dimeric IL-6,45 it exists as a hexameric lipoprotein particle in infected individuals, and thus, there are six binding sites instead of the two to four sites present for monomeric/dimeric IL-6.45 With respect to the kinetics of the system, the IL-6 assay reaches its nM LOD overnight whereas the anti-SARS-CoV-2 spike IgG assays reaches a similar LOD in 2 h.38 One potential explanation for the difference in the kinetics is the size of the analytes. Since droplets functionalized with rcSso7d recognition elements are relatively large compared to IL-6, a second interaction between a droplet and IL-6 becomes more difficult after the initial binding event.
Comparison with Commercial Antibodies
In previous investigations, we have employed commercial antibodies in droplet agglutination assays.37,38 In order to functionalize droplets with antibodies, the antibodies first need to be functionalized with the appropriate bioconjugation handle via a reaction with the appropriate NHS ester. The NHS-functionalization step results in variability in the position and number of lysines that react. In the rcSso7d proteins, we chose the position of 4AZP to not affect the protein binding face and guarantee that each droplet has only one bioconjugation handle. Nevertheless, antibodies typically retain their activity provided that standard NHS functionalization protocols are followed, and we seek to determine the effectiveness of using the rcSso7d proteins relative to commercial antibodies.
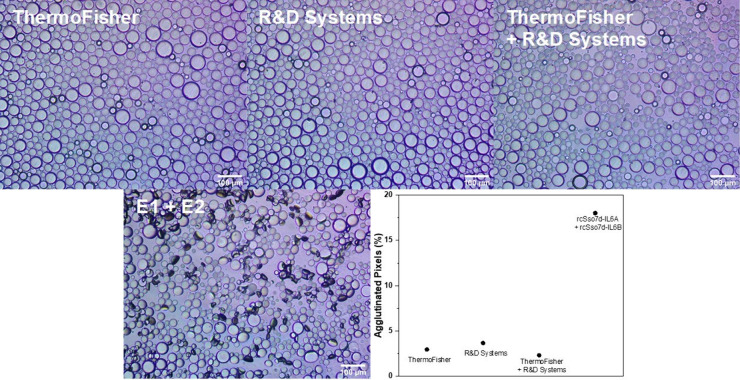
Two commercial anti-IL-6 antibodies from ThermoFisher and R&D Systems that are typically utilized in ELISA assays were functionalized with either azidobutyric acid NHS ester or DBCO NHS ester, respectively. A full description of the functionalization protocol is provided in the Supporting Information. Briefly, a 1–4 mg/mL solution of antibody was reacted with 8-fold molar excess of the corresponding NHS ester in 20 mM HEPES (pH 8.0) and 100 mM NaCl for 4 h. The resulting anti-IL-6 antibodies were then conjugated onto either PDBCO-PPEG-b-PBn- or PPN3-PPEG-b-PBn-functionalized droplets (0.2 nmol of antibody for 30 μL of droplets overnight, with mixing); after the initial bioconjugation, droplets made with PDBCO-PPEG-b-PBn underwent subsequent functionalization with PEG-N3 by the same procedure as used for the rsSso7d-functionalized droplets. Successful bioconjugation was confirmed by using dye-labeled secondary antibodies (Figure S12). Different combinations of the antibody-functionalized droplets were incubated overnight in 5 μg/mL of IL-6 in 1× PBS, and no agglutination was observed (Figure 6). Since less (0.2 nmol) of anti-IL-6 antibody was used during the bioconjugation step compared to our conventional procedure (1 nmol), we performed new assays with droplets functionalized with 0.2 nmol of FPLC purified rcSso7d proteins. We again observed agglutination at 5 μg/mL of IL-6, thus confirming that the rcSso7d proteins are superior to antibodies at detecting IL-6 in agglutination assays.
Figure 6.
Inverted microscope images of anti-IL-6 antibody+PEG-functionalized droplets and 1:1 E1+PEG- and E2+PEG-functionalized droplets incubated with 5 μg/mL of IL-6 overnight. Droplets with antibodies from ThermoFisher and R&D Systems and rcSso7d proteins purified via Ni2+ resin and subsequent FPLC are shown. Plot of the percent of agglutinated pixels according to image processing for each antibody-based assay relative to the 5 μg/mL of IL-6 assay.
Conclusions
We report a new method of detection of IL-6 by using polydisperse Janus emulsions, amphiphilic polymers, and AZP-functionalized rcSso7d proteins. In contrast to previous agglutination assays that utilize monodisperse Janus droplets fabricated using microfluidic equipment, we have shown that polydisperse droplets that can be readily produced without specialized equipment are effective for IL-6 detection and quantitation. An image processing workflow using an open-source program was developed to quantify the degree of agglutination. The rcSso7d proteins are proven to be an attractive alternative to antibodies, as they are thermally stable and precisely functionalized away from the recognition site. Furthermore, the AZP-functionalized rcSso7d proteins outperformed commercial anti-IL-6 antibodies, which displayed no agglutination response at 5 μg/mL of IL-6. Only one rcSso7d variant needs to present to produce a response; however, sandwich assays with a 1:1 ratio of E1+PEG- and E2+PEG-functionalized droplets perform better than the single variant counterparts with a LOD of 50 ng/mL. Although the LOD is not physiologically relevant, the amphiphilic polymers and rcSso7d variants used in the assays are promising materials that can be incorporated into other types of sensors such as lateral flow assays, which have LODs as low as 93 fg/mL for IL-6.20 Specifically, the amphiphilic polymers can be utilized to anchor recognition elements onto supports, and the rcSso7d variants can replace antibodies as recognition elements, allowing for more shelf-stable assays. Furthermore, the amphiphilic polymers in this study have the potential to be leveraged in multiplex assays. Future studies will investigate appending multiple recognition elements onto Janus droplets to develop multiplex assays to ensure diagnostic reliability. Additionally, previous successful assays were performed with much larger objects for detection (microbes, viruses). Remarkably, much smaller objects (i.e. IL-6) can tip droplets if their affinity to their corresponding binder is sufficiently high, despite the fact that the droplets are several orders of magnitude larger than the analyte. This work demonstrates the versatility of this assay from very large to comparably small objects. In summary, the agglutination assay is generalizable for a wide host of analytes by changing the biomolecular recognition element on the surface of the Janus droplet and thus has considerable utility in the creation of diverse bioassays.
Acknowledgments
We are appreciative of the support from the National Cancer Institute Innovative Molecular Analysis program (R21 CA256081-02) and from the US Army through the MIT Institute for Soldier Nanotechnologies. This material is based upon work sponsored (in part) by the U.S. Army DEVCOM ARL Army Research Office through the MIT Institute for Soldier Nanotechnologies under Cooperative Agreement number W911NF-23-2-0121.
Data Availability Statement
All original data from this study are available at https://fairdomhub.org/studies/1241.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c03959.
Additional materials, instruments, experimental procedures, and supplementary figures including Figures S1–S18 and Schemes S1–S3 (PDF)
Author Present Address
# Department of Chemistry, Wesleyan University, Middletown, Connecticut 06459, United States
Author Contributions
§ M.C. and E.I.C. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Kishimoto T. The Biology of Interleukin-6. Blood 1989, 74 (1), 1–10. 10.1182/blood.V74.1.1.1. [DOI] [PubMed] [Google Scholar]
- Li S.-P.; Goldman N. D. Regulation of Human C-Reactive Protein Gene Expression by Two Synergistic IL-6 Responsive Elements. Biochemistry 1996, 35 (28), 9060–9068. 10.1021/bi953033d. [DOI] [PubMed] [Google Scholar]
- Seymour J.F; Talpaz M; Hagemeister F.B; Cabanillas F; Kurzrock R Clinical Correlates of Elevated Serum Levels of Interleukin-6 in Patients with Untreated Hodgkin’s Disease. Am. J. Med. 1997, 102 (1), 21–28. 10.1016/S0002-9343(96)00352-X. [DOI] [PubMed] [Google Scholar]
- Blay J.-Y.; Negrier S.; Combaret V.; Attali S.; Goillot E.; Merrouche Y.; Mercatello A.; Ravault A.; Tourani J.-M.; Moskovtchenko J.-F.; Philip T.; Favrot M. Serum Level of Interleukin 6 as a Prognosis Factor in Metastatic Renal Cell Carcinoma1. Cancer Res. 1992, 52 (12), 3317–3322. [PubMed] [Google Scholar]
- Andrews B.; Shariat S. F.; Kim J.-H.; Wheeler T. M.; Slawin K. M.; Lerner S. P. Preoperative Plasma Levels of Interleukin-6 and Its Ssoluble Receptor Predict Disease Recurrence and Survival of Patients with Bladder Cancer. J. Urol. 2002, 167 (3), 1475–1481. 10.1016/S0022-5347(05)65348-7. [DOI] [PubMed] [Google Scholar]
- Tempfer C.; Zeisler H.; Sliutz G.; Haeusler G.; Hanzal E.; Kainz C. Serum Evaluation of Interleukin 6 in Ovarian Cancer Patients. Gynecol. Oncol. 1997, 66 (1), 27–30. 10.1006/gyno.1997.4726. [DOI] [PubMed] [Google Scholar]
- Haverty A. A.; Harmey J. H.; Redmond H. P.; Bouchier-Hayes D. J. Interleukin-6 Upregulates GP96 Expression in Breast Cancer. J. Surg. Res. 1997, 69 (1), 145–149. 10.1006/jsre.1997.5036. [DOI] [PubMed] [Google Scholar]
- Yokoe T.; Iino Y.; Takei H.; Horiguchi J.; Koibuchi Y.; Maemura M.; Ohwada S.; Morishita Y. Changes of Cytokines and Thyroid Function in Patients with Recurrent Breast Cancer. Anticancer Res. 1997, 17 (1B), 695–699. [PubMed] [Google Scholar]
- Jiang X.-P.; Yang D. C.; Elliott R. L.; Head J. F. Down-Regulation of Expression of Interleukin-6 and Its Receptor Results in Growth Inhibition of MCF-7 Breast Cancer Cells. Anticancer Res. 2011, 31 (9), 2899 LP–2906. [PubMed] [Google Scholar]
- Adrover-Jaume C.; Alba-Patiño A.; Clemente A.; Santopolo G.; Vaquer A.; Russell S. M.; Barón E.; González del Campo M. del M.; Ferrer J. M.; Berman-Riu M.; García-Gasalla M.; Aranda M.; Borges M.; de la Rica R. Paper Biosensors for Detecting Elevated IL-6 Levels in Blood and Respiratory Samples from COVID-19 Patients. Sensors Actuators B Chem. 2021, 330, 129333 10.1016/j.snb.2020.129333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.; Ma Q.; Li C.; Liu R.; Zhao L.; Wang W.; Zhang P.; Liu X.; Gao G.; Liu F.; Jiang Y.; Cheng X.; Zhu C.; Xia Y. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 Are Disease Severity Predictors. Emerg. Microbes Infect. 2020, 9 (1), 1123–1130. 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D.; Nilsson-Payant B. E.; Liu W.-C.; Uhl S.; Hoagland D.; Møller R.; Jordan T. X.; Oishi K.; Panis M.; Sachs D.; Wang T. T.; Schwartz R. E.; Lim J. K.; Albrecht R. A.; tenOever B. R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181 (5), 1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucourant C.; Filipovic I.; Ponzetta A.; Aleman S.; Cornillet M.; Hertwig L.; Strunz B.; Lentini A.; Reinius B.; Brownlie D.; Cuapio A.; Ask E. H.; Hull R. M.; Haroun-Izquierdo A.; Schaffer M.; Klingström J.; Folkesson E.; Buggert M.; Sandberg J. K.; Eriksson L. I.; Rooyackers O.; Ljunggren H.-G.; Malmberg K.-J.; Michaëlsson J.; Marquardt N.; Hammer Q.; Strålin K.; Björkström N. K. Natural Killer Cell Immunotypes Related to COVID-19 Disease Severity. Sci. Immunol. 2020, 5 (50), eabd6832 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle M.; Boeije L.; de Groot E.; de Vos A.; Aarden L. Sensitive ELISA for Interleukin-6: Detection of IL-6 in Biological Fluids: Synovial Fluids and Sera. J. Immunol. Methods 1991, 138 (1), 47–56. 10.1016/0022-1759(91)90063-L. [DOI] [PubMed] [Google Scholar]
- Usami S.; Motoyama S.; Koyota S.; Wang J.; Hayashi-Shibuya K.; Maruyama K.; Takahashi N.; Saito H.; Minamiya Y.; Takasawa S.; Ogawa J.; Sugiyama T. Regenerating Gene I Regulates Interleukin-6 Production in Squamous Esophageal Cancer Cells. Biochem. Biophys. Res. Commun. 2010, 392 (1), 4–8. 10.1016/j.bbrc.2009.12.129. [DOI] [PubMed] [Google Scholar]
- Malhotra R.; Patel V.; Vaqué J. P.; Gutkind J. S.; Rusling J. F. Ultrasensitive Electrochemical Immunosensor for Oral Cancer Biomarker IL-6 Using Carbon Nanotube Forest Electrodes and Multilabel Amplification. Anal. Chem. 2010, 82 (8), 3118–3123. 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Yang M. Electrochemical Sensor Utilizing Ferrocene Loaded Porous Polyelectrolyte Nanoparticles as Label for the Detection of Protein Biomarker IL-6. Sensors Actuators B Chem. 2011, 158 (1), 361–365. 10.1016/j.snb.2011.06.035. [DOI] [Google Scholar]
- Wei H.; Ni S.; Cao C.; Yang G.; Liu G. Graphene Oxide Signal Reporter Based Multifunctional Immunosensing Platform for Amperometric Profiling of Multiple Cytokines in Serum. ACS Sensors 2018, 3 (8), 1553–1561. 10.1021/acssensors.8b00365. [DOI] [PubMed] [Google Scholar]
- Alba-Patiño A.; Russell S. M.; Borges M.; Pazos-Pérez N.; Álvarez-Puebla R. A.; de la Rica R. Nanoparticle-Based Mobile Biosensors for the Rapid Detection of Sepsis Biomarkers in Whole Blood. Nanoscale Adv. 2020, 2 (3), 1253–1260. 10.1039/D0NA00026D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Gupta P.; Wang S.; Melnykov A.; Jiang Q.; Seth A.; Wang Z.; Morrissey J. J.; George I.; Gandra S.; Sinha P.; Storch G. A.; Parikh B. A.; Genin G. M.; Singamaneni S. Ultrasensitive Lateral-Flow Assays via Plasmonically Active Antibody-Conjugated Fluorescent Nanoparticles. Nat. Biomed. Eng. 2023, 7, 1556. 10.1038/s41551-022-01001-1. [DOI] [PubMed] [Google Scholar]
- Huang D.; Ying H.; Jiang D.; Liu F.; Tian Y.; Du C.; Zhang L.; Pu X. Rapid and Sensitive Detection of Interleukin-6 in Serum via Time-Resolved Lateral Flow Immunoassay. Anal. Biochem. 2020, 588, 113468 10.1016/j.ab.2019.113468. [DOI] [PubMed] [Google Scholar]
- Bradley Z.; Coleman P. A.; Courtney M. A.; Fishlock S.; McGrath J.; Uniacke-Lowe T.; Bhalla N.; McLaughlin J. A.; Hogan J.; Hanrahan J. P.; Yan K.-T.; McKee P. Effect of Selenium Nanoparticle Size on IL-6 Detection Sensitivity in a Lateral Flow Device. ACS Omega 2023, 8 (9), 8407–8414. 10.1021/acsomega.2c07297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S.-H.; Chen S.-Y.; Gao X. Multifunctional Nanocapsules for Simultaneous Encapsulation of Hydrophilic and Hydrophobic Compounds and On-Demand Release. ACS Nano 2012, 6 (3), 2558–2565. 10.1021/nn205023w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.; Ren Y.; Hou L.; Liu W.; Jiang T.; Deng X.; Tao Y.; Jiang H. Electrically Controlled Rapid Release of Actives Encapsulated in Double-Emulsion Droplets. Lab Chip 2018, 18 (7), 1121–1129. 10.1039/C7LC01387F. [DOI] [PubMed] [Google Scholar]
- Weinberg G.; Ripper R.; Feinstein D. L.; Hoffman W. Lipid Emulsion Infusion Rescues Dogs from Bupivacaine-Induced Cardiac Toxicity. Reg. Anesth. Pain Med. 2003, 28 (3), 198–202. 10.1053/rapm.2003.50041. [DOI] [PubMed] [Google Scholar]
- Wadhwa J.; Nair A.; Kumria R. Emulsion Forming Drug Delivery System for Lipophilic Drugs. Acta Polym. Pharm. 2012, 69 (2), 179–191. [PubMed] [Google Scholar]
- Sadurní N.; Solans C.; Azemar N.; García-Celma M. J. Studies on the Formation of O/W Nano-Emulsions, by Low-Energy Emulsification Methods, Suitable for Pharmaceutical Applications. Eur. J. Pharm. Sci. 2005, 26 (5), 438–445. 10.1016/j.ejps.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Tavernier I.; Wijaya W.; Van der Meeren P.; Dewettinck K.; Patel A. R. Food-Grade Particles for Emulsion Stabilization. Trends Food Sci. Technol. 2016, 50, 159–174. 10.1016/j.tifs.2016.01.023. [DOI] [Google Scholar]
- Galus S.; Kadzińska J. Food Applications of Emulsion-Based Edible Films and Coatings. Trends Food Sci. Technol. 2015, 45 (2), 273–283. 10.1016/j.tifs.2015.07.011. [DOI] [Google Scholar]
- Augustin M. A.; Hemar Y. Nano- and Micro-Structured Assemblies for Encapsulation of Food Ingredients. Chem. Soc. Rev. 2009, 38 (4), 902–912. 10.1039/B801739P. [DOI] [PubMed] [Google Scholar]
- Zarzar L. D.; Sresht V.; Sletten E. M.; Kalow J. A.; Blankschtein D.; Swager T. M. Dynamically Reconfigurable Complex Emulsions via Tunable Interfacial Tensions. Nat. (London, United Kingdom) 2015, 518 (7540), 520–524. 10.1038/nature14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeininger L.; Nagelberg S.; Harvey K. S.; Savagatrup S.; Herbert M. B.; Yoshinaga K.; Capobianco J. A.; Kolle M.; Swager T. M. Rapid Detection of Salmonella Enterica via Directional Emission from Carbohydrate-Functionalized Dynamic Double Emulsions. ACS Cent. Sci. 2019, 5 (5), 789–795. 10.1021/acscentsci.9b00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D.; Swager T. M. Trace Detection of Hydrogen Peroxide via Dynamic Double Emulsions. J. Am. Chem. Soc. 2021, 143 (11), 4397–4404. 10.1021/jacs.1c00683. [DOI] [PubMed] [Google Scholar]
- Trinh V.; Malloy C. S.; Durkin T. J.; Gadh A.; Savagatrup S. Detection of PFAS and Fluorinated Surfactants Using Differential Behaviors at Interfaces of Complex Droplets. ACS Sensors 2022, 7 (5), 1514–1523. 10.1021/acssensors.2c00257. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Savagatrup S.; Kaplonek P.; Seeberger P. H.; Swager T. M. Janus Emulsions for the Detection of Bacteria. ACS Cent. Sci. 2017, 3 (4), 309–313. 10.1021/acscentsci.7b00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Zeininger L.; Sung K.-J.; Miller E. A.; Yoshinaga K.; Sikes H. D.; Swager T. M. Emulsion Agglutination Assay for the Detection of Protein-Protein Interactions: An Optical Sensor for Zika Virus. ACS Sens. 2019, 4 (1), 180–184. 10.1021/acssensors.8b01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Savagatrup S.; Nelson Z.; Yoshinaga K.; Swager T. M. Fluorescent Janus Emulsions for Biosensing of Listeria Monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (22), 11923–11930. 10.1073/pnas.2002623117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Concellón A.; Yoshinaga K.; Nelson Z.; He Q.; Swager T. M. Janus Emulsion Biosensors for Anti-SARS-CoV-2 Spike Antibody. ACS Cent. Sci. 2021, 7 (7), 1166–1175. 10.1021/acscentsci.1c00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung K. J.; Miller E. A.; Sikes H. D. Engineering Hyperthermostable RcSso7d as Reporter Molecule for in Vitro Diagnostic Tests. Mol. Syst. Des. Eng. 2018, 3 (6), 877–882. 10.1039/C8ME00049B. [DOI] [Google Scholar]
- Traxlmayr M. W.; Kiefer J. D.; Srinivas R. R.; Lobner E.; Tisdale A. W.; Mehta N. K.; Yang N. J.; Tidor B.; Wittrup K. D. Strong Enrichment of Aromatic Residues in Binding Sites from a Charge-Neutralized Hyperthermostable Sso7D Scaffold Library. J. Biol. Chem. 2016, 291 (43), 22496–22508. 10.1074/jbc.M116.741314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. A.; Sung K. J.; Kongsuphol P.; Baniya S.; Aw-Yong H. Q.; Tay V.; Tan Y.; Kabir F. M.; Pang-Yeo K.; Kaspriskie I. G.; Sikes H. D. Beyond Epitope Binning: Directed in Vitro Selection of Complementary Pairs of Binding Proteins. ACS Comb. Sci. 2020, 22 (1), 49–60. 10.1021/acscombsci.9b00176. [DOI] [PubMed] [Google Scholar]
- Song M.; Kellum J. A. Interleukin-6. Crit. Care Med. 2005, 33 (12), S463. 10.1097/01.CCM.0000186784.62662.A1. [DOI] [PubMed] [Google Scholar]
- Moreno A.; Villar M. L.; Cámara C.; Luque R.; Cespón C.; González-Porqué P.; Roy G.; López-Jiménez J.; Bootello A.; Santiago E. R. Interleukin-6 Dimers Produced by Endothelial Cells Inhibit Apoptosis of B-Chronic Lymphocytic Leukemia Cells. Blood 2001, 97 (1), 242–249. 10.1182/blood.V97.1.242. [DOI] [PubMed] [Google Scholar]
- Ward L. D.; Hammacher A.; Howlett G. J.; Matthews J. M.; Fabri L.; Moritz R. L.; Nice E. C.; Weinstock J.; Simpson R. J. Influence of Interleukin-6 (IL-6) Dimerization on Formation of the High Affinity Hexameric IL-6·receptor Complex. J. Biol. Chem. 1996, 271 (33), 20138–20144. 10.1074/jbc.271.33.20138. [DOI] [PubMed] [Google Scholar]
- Brown W. C.; Akey D. L.; Konwerski J. R.; Tarrasch J. T.; Skiniotis G.; Kuhn R. J.; Smith J. L. Extended Surface for Membrane Association in Zika Virus NS1 Structure. Nat. Struct. Mol. Biol. 2016, 23 (9), 865–867. 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data from this study are available at https://fairdomhub.org/studies/1241.