Abstract
Bamboo plants are widely used in Asian traditional medicine for various health issues and exhibit therapeutic potential. Guadua species are renowned bamboos for their high phenolic compound content, including flavonoids and hydroxycinnamic acid derivatives, and possess noteworthy biological properties. Despite this, there is a notable scarcity of research on the chemical and biological aspects of Latin American bamboo leaf extracts (BLEs), especially concerning the Guadua genus. This study aimed to employ a metabolomics approach to integrate the phytochemical and activity profiles of BLEs to identify potential bioactive markers. We determined the metabolic fingerprints of 30 BLEs through HPTLC, HPLC-DAD, UHPLC-QTOF-MS, and 1H-NMR analyses and screened for antioxidant and cytotoxic activities using ABTS, DPPH, and MTT methods. Ultimately, correlation analyses were performed by using chemometric methods and molecular networking. Our findings present a comprehensive chemical characterization, encompassing 40 flavonoids and 9 cinnamic acid derivatives. Notably, most of these compounds have been reported for the first time within the genus, signifying novel discoveries. Additionally, certain compounds identified in other species of the subfamily Bambusoideae provide valuable comparative insights. These compounds demonstrated a significant correlation with antioxidant potential, with values exceeding 100 and 30 μmol of TE/g of extract for ABTS and DPPH, respectively, in the samples. Extracts from G. incana and G. angustifolia exhibited potent cytotoxic effects with IC50 values of 1.23 and 4.73 μg/mL against HCT-116 colon cancer cells, respectively. Notably, glycosylated flavones showed a strong correlation with cytotoxicity. These new findings significantly contribute to our understanding of the chemical composition and biological properties of these often overlooked bamboo species, providing them with important added value and alternative use.
1. Introduction
Bamboos, belonging to the subfamily Bambusoideae (Poaceae), comprise 1670 species distributed across 125 genera and divided into three tribes: herbaceous bamboos (Olyreae), tropical woody bamboos (Bambuseae), and temperate woody bamboos (Arundinarieae). They are distributed across various geographic regions, predominantly in Asia and Latin America.1,2 These plants, known as “a God’s gift” or “the plant of thousand uses”, have traditionally been used in Asia for their medicinal properties to address various health conditions including allergies, respiratory diseases, acute infections, dermatitis, diabetes, hair growth promotion, acne alleviation, hypertension management, fever reduction, detoxification assistance, and potential treatment for certain forms of cancer.3−11
Attractively, the Neotropical woody bamboos of the Guadua genus, often referred to as the “vegetable steel” or the “green gold of the 21st century”, lack a history of traditional medicinal use. They are regarded as the most significant American bamboos, comprising 33 species distributed across tropical regions in Central and South America.2 Despite their prominence in Latin America, comprehensive knowledge of their chemical composition and biological potential remains scarce. Preliminary phytochemical studies indicate the presence of flavonoids, phenolic acid derivatives, terpenes, saponins, and alkaloids.12−15 Subsequent investigations have focused on investigations into the influence of altitude on the chemical composition of bamboos have revealed intriguing patterns with higher altitude species exhibiting increased concentrations of flavonoids.16−20 These revealed phytochemicals have been reported to possess different biological activities, e.g., antimicrobial,13 antioxidant,17,21 anti-inflammatory,22,23 and anticancer effects.8,24 Despite these promising initial insights, a significant knowledge gap exists concerning the detailed relationship between the chemical compositions of Guadua species and their biological activities. Our study seeks to bridge this gap by conducting an in-depth investigation, utilizing a metabolomic approach to establish a correlation between chemical fingerprinting and biological activity in these bamboos. The primary objective is to identify chemical markers with antioxidant and cytotoxic potential, providing a more comprehensive and applicable understanding of these species in the realms of medicine and beyond. It is noteworthy that metabolomic research on Guadua species, particularly concerning compounds responsible for antioxidant and cytotoxic activities, is notably absent in the current scientific literature. Our study represents a significant step toward filling this void, offering valuable insights and laying the groundwork for future applications. This research endeavor not only contributes to the scientific community’s understanding of bamboos but also opens new perspectives for their utilization in various fields.
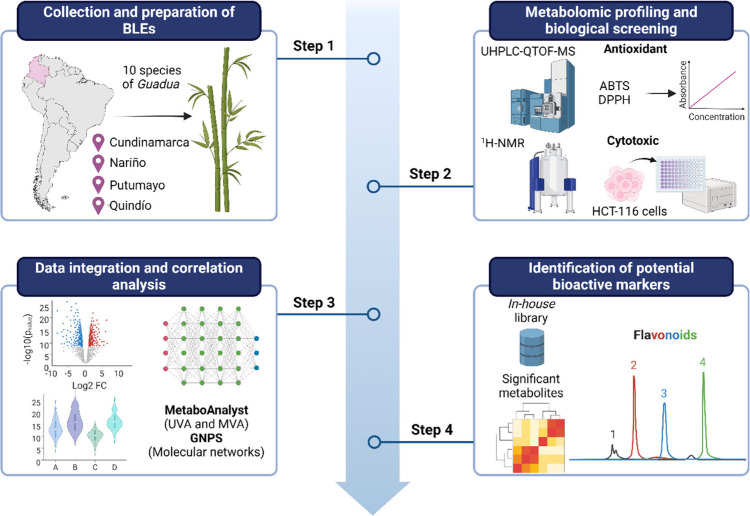
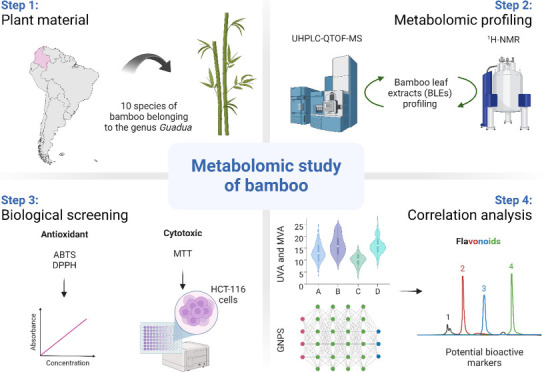
Therefore, this study aimed to integrate the chemical and bioactivity profiles of 30 BLEs based on a metabolomics approach for the identification of potential bioactive markers. Figure 1 illustrates the workflow used in this research.
Figure 1.
Workflow of the metabolomics strategy implemented for the identification of candidate bioactive compounds. Adapted from “T Cell Culture Workflow”, by BioRender.com. Retrieved from https://app.biorender.com/biorender-templates/figures/all/t-62ccc6c3da5217036d07073b-t-cell-culture-workflow.
2. Results
2.1. HPTLC and HPLC-DAD Phytochemical Profiling Analysis
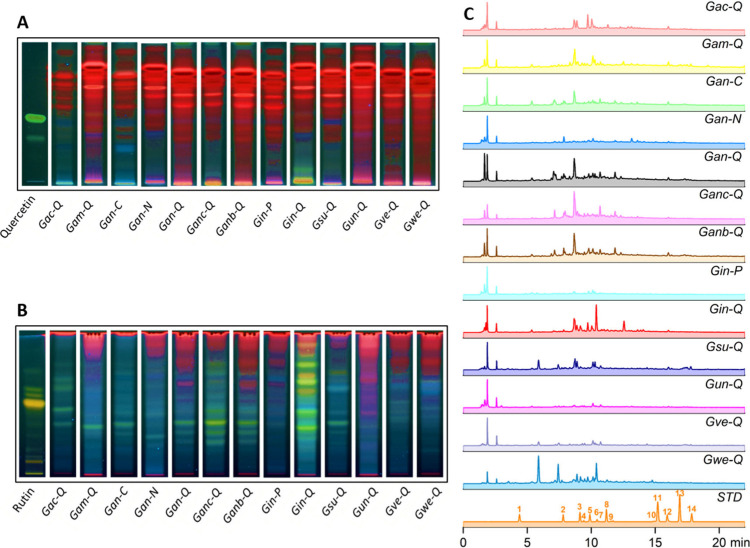
A previous study conducted on bamboos belonging to the genus Guadua, using an untargeted metabolomics approach to determine the changes in chemical composition under the influence of altitudinal variation, showed that these bamboos have a high chemical composition, specifically of phenolic compounds, encompassing flavonoids in both their aglycone and glycosidic forms, accounting for 48% of their total chemical makeup.16 These findings enabled us to conduct targeted chemical profiling in this study, aiming to dereplication for known compounds using specific commercial chemical standards related to flavonoids and phenolic acid derivatives (1–14) via HPTLC and HPLC-DAD. Figure 2A and B show the chromatographic profiles obtained by HPTLC for the BLEs collected at 13 distinct locations, as detailed in Table 2.
Figure 2.
HPTLC chromatographic profiles for (A) flavonoid aglycones and (B) flavonoid glycosides. HPLC-PDA chromatograms of the BLEs were expanded from 0 to 23 min at (C) 274 nm. Chemical standards (STD): Gallic acid (1, tR = 4.36 min), Chlorogenic acid (2, tR = 7.72 min), Caffeic acid (3, tR = 9.08 min), Isoorientin (4, tR = 9.39 min), Ampelopsin (5, tR = 9.88 min), Rutin (6, tR = 10.42 min), Vitexin (7, tR = 10.56 min), p-Coumaric acid (8, tR = 11.15 min), Sinapinic acid (9, tR = 11.55 min), Morin (10, tR = 14.79 min), Coumarin (11, tR = 15.19 min), Quercetin (12, tR = 15.86 min), Cinnamic acid (13, tR = 16.84 min), and Naringenin (14, tR = 17.75 min). Column: Luna C18 column (150 mm × 4.6 mm, 5 μm, 100 Å); mobile phase: (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile, 5–40% B (0–23 min); flow rate: 1 mL/min; detection wavelength: 274 nm. UV spectra were obtained via DAD detection. Chemical profiles were plotted by selecting one sample for each collection site and the different species (n = 13) according to the coding in Table 2.
Table 2. Information of Guadua Species.
| Species | Collection placea | Samples code | Voucher number (HPUJ) | Number of samplesb |
|---|---|---|---|---|
| Guadua aculeata E. Fourn. | Quindío | Gac-Q (1) | HPUJ-30736 | 1 |
| Guadua amplexifolia J. Pressl | Quindío | Gam-Q (1 to 3) | HPUJ-30733 | 3 |
| Guadua angustifolia Kunth | Cundinamarca | Gan-C (1 to 7) | HPUJ-30740 | 7 |
| Nariño | Gan-N (1 to 6) | HPUJ-30722 | 6 | |
| Quindío | Gan-Q (1 to 2) | HPUJ-30731 | 2 | |
| Guadua angustifolia Kunth biotype San Calixto | Quindío | Ganc-Q (1) | HPUJ-30737 | 1 |
| Guadua angustifolia var. bicolor Londoño | Quindío | Ganb-Q (1) | HPUJ-30735 | 1 |
| Guadua incana Londoño | Putumayo | Gin-P (1) | HPUJ-30723 | 1 |
| Quindío | Gin-Q (1 to 2) | HPUJ-30723 | 2 | |
| Guadua superba Huber | Quindío | Gsu-Q (1) | HPUJ-30734 | 1 |
| Guadua uncinata Londoño and L. G. Clark | Quindío | Gun-Q (1 to 3) | HPUJ-30730 | 3 |
| Guadua venezuelae Munro | Quindío | Gve-Q (1) | HPUJ-30729 | 1 |
| Guadua weberbaueri Pilg. | Quindío | Gwe-Q (1) | HPUJ-30738 | 1 |
Information regarding collection sites is provided in Supporting Information Table S2.
A total of 30 BLEs were prepared.
Figure 2A presents the chemical profile of the flavonoid aglycones, while Figure 2B presents the flavonoid glycosides alongside the reference standards Quercetin (12) and Rutin (6), respectively. Moreover, the plates were eluted based on the polarity of these compounds, then sprayed with the natural products-polyethyleneglycol reagent (NP/PEG) and developed at a wavelength of UV-365 nm.25 From the results, it is evident that the extracts contain a limited number of flavonoid aglycones, while a higher quantity of flavonoid glycosides is emphasized, as confirmed by the presence of yellow and orange fluorescent bands. The retardation factor (Rf) values for reference standards 12 and 6 were 0.58 and 0.62, respectively. G. incana Londoño (Gin-Q) is the species that exhibits a significant number of flavonoid glycosides compared to the other species, which are found in lower proportions. In this study, it was observed that flavonoid glycosides exhibit considerable variability among different extracts and are distinguished based on collection sites. This finding is consistent with earlier preliminary studies that revealed a higher accumulation of these compounds in these bamboos considered as giant grasses.12
Figure 2C shows the chromatographic profiles obtained by HPLC-DAD for the BLEs. We emphasize that between 5 and 12 min, a significant portion of the chemical composition of these extracts is observed, primarily comprising flavonoid glycosides and phenolic acid derivatives. The identification of these compounds was confirmed through ultraviolet spectra analysis (Supporting Information Figure S1). Most of the peaks exhibited two distinctive absorption bands characteristic of flavonoids, corresponding to band I (240–285 nm) and band II (300–560 nm).26,27 The presence of phenolic acid derivatives was also determined by the characteristic band with a shoulder (sh) between 290 and 310 nm.28 From the dereplication study of BLEs, chlorogenic acid (CGA) (2) was identified in the G. weberbaueri Pilg. species with a retention time (tR = 7.72 min) and an ultraviolet spectrum with absorption at 326 nm and a shoulder peak at 292 nm, which was compared with the commercial chemical standard and reports in the literature.29 This compound is reported for the first time for this species. However, it has been previously reported in other bamboo species.30,31 The remaining reference compounds were not found in any of the other species. Nevertheless, it can be inferred that these species may contain compounds related to flavonoid glycoside and phenolic acid derivative scaffolds based on the absorption bands observed in the ultraviolet spectra (Supporting Information Figure S1).
2.2. UHPLC-QTOF-MS-Based Metabolic Fingerprinting Analysis
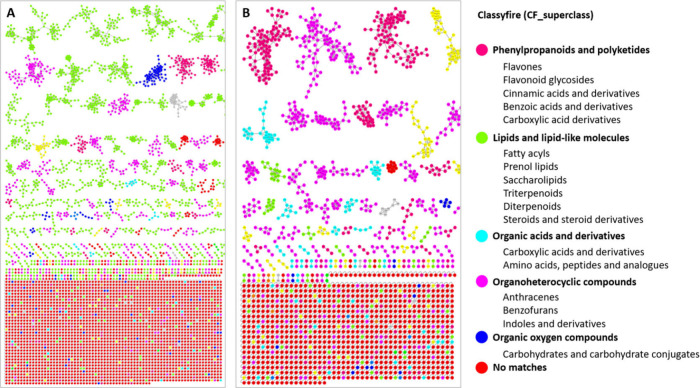
A comprehensive metabolic fingerprinting was conducted on the BLEs using UHPLC-QTOF-MS in both negative and positive modes. Initially with the MS2 data, an analysis was conducted on the GNPS platform to visualize the chemical composition of the global metabolome of BLEs. The comprehensive analysis in Figure 3A and B highlight the presence of molecular families (MFs), mainly corresponding to phenylpropanoids and polyketides, lipids and lipid-like molecules, organic acids and derivatives, organoheterocyclic compounds, and organic oxygen compounds for both ionization modes. Notably, in positive ionization mode, the MFs of phenylpropanoids and polyketides, including flavonoids and derivatives of cinnamic, benzoic, and carboxylic acids, are most abundant. In contrast, in the negative ionization mode, the MFs of lipids and related molecules predominate to a greater extent. However, the prevalence of phenylpropanoid and polyketide MFs also stands out.
Figure 3.
Representative MolNetEnhancer mass spectral molecular networking of consortium showing the chemical CF_superclasses in (A) negative and (B) positive electrospray ionization modes of BLEs based on UHPLC-QTOF-MS. This annotated GNPS library matches with network annotation (NAP) and DEREPLICATOR outputs.
The structures of the compounds representing the major detected peaks are discussed below. The identification of the detected compounds was based on a search for the main molecular ions (MS) and on some of the informative fragmentations observed (MS2), along with the use of standards when available, retention data, UV absorption, comparison with literature data, the GNPS web platform, and public databases.
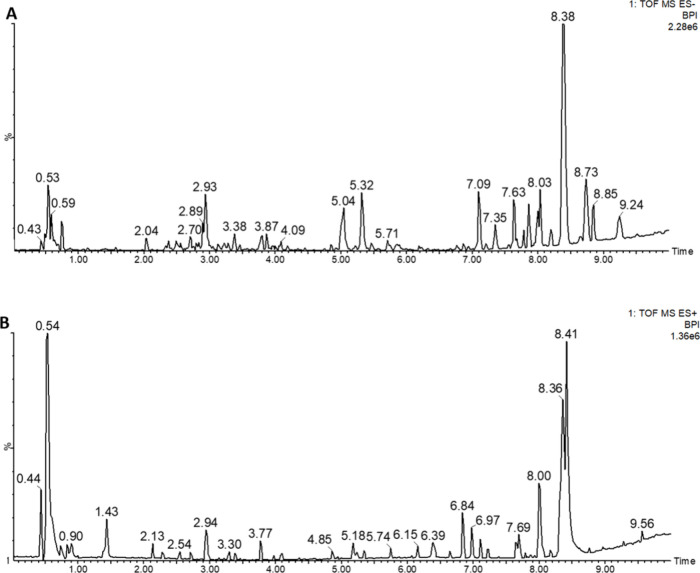
In the present work, 49 phytochemical compounds (2, 3, 8, and 15–60) have been tentatively characterized in the BLEs. Notably, there is the presence of 40 flavonoids and 9 cinnamic acid derivatives, listed in Supporting Information Table S1 and represented with the retention time in the base peak intensity (BPI) chromatograms of the quality control (QC) samples of the extracts, presented in Figure 4A and B for the negative and positive ionization modes, respectively. Importantly, many of these compounds are being reported for the first time within the genus, marking novel discoveries. Furthermore, certain compounds have been previously identified in other species of the subfamily Bambusoideae, as described in Supporting Information Table S1. The structures of the tentatively characterized compounds are presented in Supporting Information Figure S2, where the MS2 spectrum of several of them were determined based on the data obtained from UHPLC-QTOF-MS analysis of the studied extract, as illustrated in Supporting Information Figure S3. An extensive study of some of the compounds characterized according to their families is described below.
Figure 4.
Representative UHPLC-QTOF-MS base peak intensity chromatograms of the QC samples of the BLEs in (A) negative and (B) positive ions. The retention times of the compounds that could be annotated in the BLEs are listed in Table S1.
2.2.1. Flavonoids
We report the identification of 40 flavonoids in several of the BLEs. Of the identified scaffolds, five groups were found corresponding to 12 with apigenin skeleton (23, 25, 29, 30, 31, 32, 34, 35, 42, 43, 48, and 54), 5 with kaempferol skeleton (36, 37, 38, 57, and 58), 3 with luteolin skeleton (21, 44, and 49), 2 with quercetin skeleton (39 and 40), 2 with tricin skeleton (51 and 60), and 2 with naringenin skeleton (52 and 53). Most of them had C-glycosidized substituents at the C6 and C8 positions on the A-ring of the aglycone, while others had O-glycosidized substituents at the C7 and C3 positions of the corresponding aglycone. Flavonoids 33, 41, 45, 46, and 47 also presented C- and O-glycosidic substituents corresponding to other scaffolds, some of them with glycosidic substituents on the B-ring of the aglycone. The presence of three aglycones corresponding to one flavone 50 and 2 flavonols 55 and 59 was also identified. The presence of isoflavone 56 was also determined. Additionally, 5 compounds (22, 24, 26, 27, and 28) were found to be classified as unknown flavone glycosides based on their UV spectra and fragmentation patterns. These results were corroborated with the ultraviolet spectra obtained and compared with reports in literature where the range of flavones (λmax ≈ 275 and 335 nm), flavonols (λmax ≈ 275 and 355 nm), and flavanones (λmax ≈ 275 and 340 nm) is highlighted.32
Compound 31 exhibited a precursor ion at m/z 563.1406 [M-H]− and MS2 fragments at m/z 473 [M-H-90]− derived from the possible loss of five water molecules, 443 [M-H-120]−, 383 [M-H-180]−, 353 [M-H-210]−, 325 [M-H-238]−, and 297 [M-H-266]− derived from cross-ring cleavages of the respective sugars (hexoside and pentoside) present. Additionally, it displayed a precursor ion at m/z 565.1563 [M + H]+ with MS2 fragments at m/z 547 [M+H-18]+, 529 [M+H-36]+, 499 [M+H-66]+, 457 [M+H-108]+, 427 [M+H-138]+, and 379 [M+H-186]+. These results were consistent with those reported by Jiang et al.,30 and the compound was identified as isoschaftoside. This compound is a type of flavone C-glucoside previously reported in species such as Phyllostachys pracecox, Phyllostachys edulis, Olyra glaberrina, Parodiolyra micrantha, Aulonemia aristulata, Filgueirasia arenicola, Filgueirasia cannavieira, and Merostachys pluriflora.32
Compound 48 was identified as Apigenin 6-C-hexoside 7-O-hexoside with m/z = 593.1511 [M-H]−. The MS2 fragment at m/z 473 [M-H-120]− indicates a typical loss of C-linked hexose, and the base peak at m/z 431 [M-H-162]− indicates the loss of a hexose, suggesting O-glycosylation. These fragments confirm the presence of two hexose units, signifying an apigenin scaffold. Furthermore, fragments at m/z 353, 341, 311, and 283 resulting from cross-ring cleavages of the flavonoid glycosyl moiety’s hexoside part and the loss of water molecules, respectively, contribute additional structural insights. For bamboo species, this compound has been reported for the first time. This compound is a C,O-glycosidic flavone type reported in a study conducted for leaf extracts of Passiflora incarnata, Passiflora caerulea, and Passiflora alata species. In this study, they demonstrated the potent cytotoxic effect of P. alata and P. incarnata extracts against human acute lymphoblastic leukemia CCRF-CEM, mainly attributed to the content of phenolic compounds, specifically to the high content of flavones C-glycosides with scaffold related to apigenin and luteolin.33 According to the flavonoids identified and because the majority correspond to flavone C-glycosides, Supporting Information Table S1 details the corresponding MS2 fragment ions that for most of the molecules present the occurrence of the characteristic ions, e.g., [M-H-150]−, [M-H-120]−, [M-H-90]−, [M-H-66]−, [M-H-60]−, and [M-H-36]−, corresponding to the cross-ring cleavages of the hexoside and pentoside part of the flavonoid glycosyl moiety and characteristics of C-glycosides.
Compound 54 possesses a precursor ion m/z 269.0455 [M-H]− with MS2 fragments m/z 151 [M-H-188]−, 117 [M-H-152]−, and 107 [M-H-151-CO2]− corresponding to retro-Diels–Alder (RDA) cleavage accompanied by loss of CO2. These results were consistent with those reported by Troalen et al.,34 and the compound was identified as apigenin. The quercetin scaffold derived from flavone C-glucosides presented the product ion m/z 301 [M-H]− mainly presents five MS2 fragment ions with m/z 273 [M-H-28]−, 179 [M-H-122]−, 151 [M-H-150]−, 121 [M-H-180]−, and 107 [M-H-194]− being a product of RDA cleavage and CO and CO2 loss, respectively. Some quercetin derivatives have been reported for bamboo species such as G. angustifolia.16,35 Bamboo species are known to accumulate phenolic compounds, mainly glycosylated flavones, whose aglycones normally are apigenin, luteolin, and tricin. The most common flavonoids found in Asian bamboo species are the flavone C-glucosides.3,6,36
2.2.2. Phenolic Acid Derivatives
Nine phenolic acids were identified in several of the BLEs. Two phenolic acids including chlorogenic acid (CGA) (2), caffeic acid (3), and p-coumaric acid (8) were identified by comparison with authentic standards. Additionally, based on literature reports, three other phenolic acid derivatives were also identified. Compounds 18, 19, and 20 were recognized as derivatives of caffeoylquinic acid (CQAs) and were named O-Feruloylquinic acid, Dicaffeoylquinic acid, and O-Coumaroylquinic acid, respectively. Compounds 16 and 17 were identified to the chemical class designated as not identified. Finally, compound 15 identified as quinic acid (QA) was present in all species. QA has been recently reported in G. angustifolia as being one of the metabolites that showed a significant increase at high altitudes in these species.16
Compound 2 presented the precursor ion M-H at m/z 353.0531 [M-H]− and showed MS2 fragments at m/z 191 [M-H-162]−, formed by the cleavage of the ester bond between the quinic acid and caffeic acid residues, and the ion m/z 179 [M-H-174]− which loses a portion of carbon dioxide to produce the ion m/z 135 [M-H-44]−. This report is consistent with the identification based on the MS2 spectrum (Supporting Information Figure S3) of the CGA pattern, together with that reported by Reis Luz et al.37 Although CGA is a new compound reported for several species in this study, it has been found in other bamboo species such as Sasa argenteastriatus, Phyllostachys nigra var. henonis, and P. pubescens.38−40
2.3. NMR-Based Metabolomic Fingerprinting Analysis
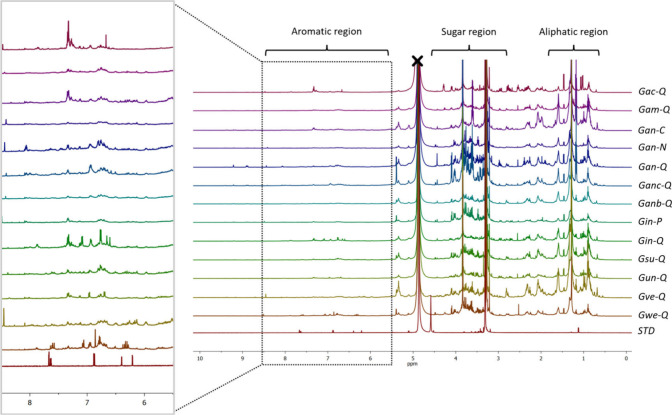
Representative 1H-NMR spectra obtained from BLEs are shown in Figure 5. The analysis of the 1H-NMR spectral data set led to the identification of representative metabolite signals in the samples. Fingerprinting analysis revealed that BLEs mainly contained compounds related to three specific regions, corresponding to aliphatic compounds, glycosidized compounds, and aromatic compounds. Signals in the region between δH 0.90 and 1.50 ppm were assigned to aliphatic compounds, specifically fatty acids and related molecules, determined by methyl (−CH3) and methylene (−CH2) proton signals, as confirmed by the allylic region of the aliphatic chains between δH 2.29 and 2.75 ppm.
Figure 5.
Stacked representative 1H-NMR spectra of the BLEs are presented. The panels display the partial assignment of aliphatic (δH 0.90–1.50 ppm), sugar (δH 3.00–5.50 ppm), and aromatic (δH 6.50–8.55 ppm) regions, respectively. An expanded spectral region from δH 5.50 to 8.50 ppm is shown for aromatic compounds. Assignments were established by extracting signals with reference standards (Quercetin and Rutin) and based on literature reports on the NMR spectra.
The signals between δH 3.00 and 5.50 ppm assigned to glucoside compounds were primarily determined by the anomeric signals of the sugar compounds. Mainly, the anomeric protons of glucose in the β position with δH 4.58 (d, J = 7.6 Hz) and in the α position with δH 5.15 (d, J = 3.5 Hz) were extracted and assigned. According to our results, this sugar is present in most of the flavonoids previously found and described in Supporting Information Figure S2, confirming the respective structure annotations. It is important to note that, due to the nature of an extract, many of the signals overlap in the spectrum. However, some of the signals extracted in this study were confirmed and compared with available reference compounds. The 1H-NMR spectra in the aromatic region between δH 6.85 and 8.05 ppm revealed the presence of phenolic compounds, primarily related to flavonoids and phenolic acids. The presence of aromatic protons at δH 6.26 (d, J = 2.20 Hz, 1H), 6.44 (d, J = 2.22 Hz, 1H), 6.96 (d, J = 8.90 Hz, 2H), and 7.84 (d, J = 8.90 Hz, 2H), distinctive signals corresponding to a flavonoid-type scaffold derived from apigenin with substitutions at the C6 and C8 positions, particularly stands out. Representative 1H-NMR signals were assigned as described in Table 1.
Table 1. Significantly Differential Major Metabolites Correlating with Cytotoxic Activity Determined by the OPLS-DA Model for UHPLC-QTOF-MS and 1H-NMR Analysisa.
| Compound number | RT (min) | δH (ppm) | Multiplicity, J in Hz | Type of hydrogen | CVb for QC (%) | FCc | VIPd | p-valuee |
|---|---|---|---|---|---|---|---|---|
| 2st | 2.46 | 7.05 | d, J = 2.0 Hz | Quinic acid derivatives aromatic, Ar–H | 7.25 | 0.35 ↓ | 1.86 | 3.67 × 10–02 |
| 6.82 | d, J = 7.6 Hz | |||||||
| 3st | 2.74 | 7.10 | d, J = 2.0 Hz | Phenolic acid aromatic, Ar–H | 1.97 | 0.52 ↓ | 1.67 | 3.67 × 10–02 |
| 6.75 | d, J = 7.6 Hz | |||||||
| 15 | 0.57 | 4.80 | s | Quinic acid derivatives α to oxygen, −CH–OH | 10.31 | 0.30 ↓ | 1.82 | 1.14 × 10–02 |
| 16 | 1.20 | 3.60 | 0.44 ↓ | 1.82 | 2.09 × 10–02 | |||
| 17 | 1.94 | 9.34 | 0.53 ↓ | 1.65 | 4.53 × 10–02 | |||
| 18 | 1.98 | 5.05 | s | Quinic acid derivatives α to oxygen, −CH–OH | 17.46 | 0.35 ↓ | 1.60 | 1.61 × 10–02 |
| 19 | 2.04 | 7.12 | d, J = 1.4 Hz | Quinic acid derivatives aromatic, Ar–H | 3.40 | 0.35 ↓ | 2.00 | 4.53 × 10–02 |
| 6.84 | dd, J = 7.5, 2.0 Hz | |||||||
| 20 | 2.33 | 5.05 | s | Quinic acid derivatives α to oxygen, −CH–OH | 17.79 | 0.42 ↓ | 1.75 | 2.64 × 10–02 |
| 21 | 2.36 | 7.47 | d, J = 2.0 Hz | Luteolin derivatives aromatic Ar–H b-ring | 4.81 | 2.40 ↑ | 1.72 | 1.13 × 10–02 |
| 6.92 | d, J = 7.5 Hz | |||||||
| 6.60 | s | Proton c-ring | ||||||
| 22 | 2.51 | 1.26 | 4.98 ↑ | 1.73 | 1.28 × 10–02 | |||
| 23 | 2.68 | 7.84 | m | Apigenin derivatives aromatic Ar–H b-ring | 4.50 | 2.44 ↑ | 1.41 | 1.45 × 10–02 |
| 6.96 | m | |||||||
| 6.68 | s | Proton c-ring | ||||||
| 25 | 2.71 | 7.83 | m | Apigenin derivatives aromatic Ar–H b-ring | 2.99 | 3.14 ↑ | 1.47 | 1.45 × 10–02 |
| 6.95 | m | |||||||
| 6.68 | s | Proton c-ring | ||||||
| 29 | 2.87 | 7.83 | m | Apigenin derivatives aromatic Ar–H b-ring | 4.37 | 5.13 ↑ | 1.50 | 4.08 × 10–02 |
| 6.95 | m | |||||||
| 6.68 | s | Proton c-ring | ||||||
| 6.28 | s | Proton a-ring | ||||||
| 30* | 2.88 | 7.80 | m | Apigenin derivatives aromatic Ar–H b-ring | 6.32 | 3.38 ↑ | 1.79 | 1.61 × 10–02 |
| 6.90 | m | |||||||
| 6.50 | s | Proton c-ring | ||||||
| 31* | 2.91 | 7.83 | m | Apigenin derivatives aromatic Ar–H b-ring | 12.60 | 4.70 ↑ | 2.26 | 4.27 × 10–03 |
| 6.95 | m | |||||||
| 6.68 | s | Proton c-ring | ||||||
| 32* | 3.13 | 8.89 | 6.73 ↑ | 1.55 | 2.64 × 10–02 | |||
| 33 | 3.16 | 13.65 | 3.52 ↑ | 1.41 | 6.18 × 10–03 | |||
| 34* | 3.18 | 7.87 | m | Apigenin derivatives aromatic Ar–H b-ring | 2.07 | 2.02 ↑ | 1.03 | 2.95 × 10–02 |
| 6.98 | m | |||||||
| 6.75 | s | Proton a-ring | ||||||
| 6.65 | s | Proton c-ring | ||||||
| 8st | 3.20 | 7.49 | m | Phenolic acid aromatic, Ar–H trans vinylic protons | 16.21 | 0.02 ↓ | 1.71 | 9.43 × 10–03 |
| 6.81 | m | |||||||
| 6.39 | d, J = 15.2 Hz | |||||||
| 35* | 3.23 | 7.85 | m | Apigenin derivatives aromatic Ar–H b-ring | 8.99 | 1.72 ↑ | 1.25 | 1.13 × 10–02 |
| 6.92 | m | |||||||
| 6.55 | s | Proton c-ring | ||||||
| 36 | 3.24 | 6.96 | m | Kaempferol derivatives aromatic Ar–H b-ring and a-ring | 13.97 | 2.61 ↑ | 1.34 | 2.48 × 10–02 |
| 6.73 | d, J = 1.6 Hz | |||||||
| 6.39 | d, J = 1.6 Hz | |||||||
| 38 | 3.28 | 9.34 | 2.51 ↑ | 1.00 | 4.53 × 10–02 | |||
| 42 | 3.54 | 10.33 | 2.14 ↑ | 1.34 | 1.86 × 10–02 | |||
| 43 | 3.56 | 4.94 | 1.96 ↑ | 1.08 | 3.67 × 10–02 | |||
| 46 | 3.61 | 6.39 | 1.36 ↑ | 1.68 | 8.64 × 10–03 | |||
| 48 | 3.73 | 7.85 | m | Apigenin derivatives aromatic Ar–H b-ring | 15.01 | 1.35 ↑ | 1.78 | 2.64 × 10–02 |
| 6.95 | m | |||||||
| 6.70 | s | Proton a-ring | ||||||
| 6.60 | s | Proton c-ring | ||||||
| 50 | 4.57 | 1.07 | 0.28 ↓ | 1.78 | 2.09 × 10–02 |
Direction of comparison: Active/Inactive. Statistical parameters: CV threshold <20% for QC; FC threshold >1 (increased activity ↑) and FC threshold <1 (decreased activity ↓); VIP threshold >1 and p-value threshold <0.05.
CV: coefficient of variation in the metabolites in the QC samples.
FC: fold change in the comparison (average in active samples/inactive samples).
VIP: variable importance in projection.
p-Value corresponding to the p-values calculated by the Benjamini-Hochberg (<0.05); st The identification of compounds has been verified by using authentic standards; * Isomers. The assignments of the 1H-NMR signals were established by extracting signals with reference standards (Quercetin and Rutin) and based on literature reports.
2.4. Antioxidant Capacity of BLEs
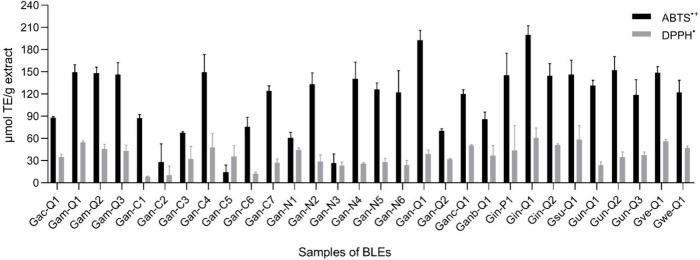
The results of the ABTS and DPPH assays are presented in Figure 6. Among the 30 BLEs tested, most exhibited significant antioxidant potential at the tested concentration of 1 mg/mL. It is evident that these extracts displayed robust antioxidant activity in both assays, which can be attributed to their high phenolic compound content. The activity was expressed as μmol of TE/g extract. Phenolic compounds constitute one of the main groups of compounds known to act as important antioxidants because they usually possess an aromatic ring that allows the stabilization and relocation of the unpaired electrons of their structure, thus facilitating the donation of hydrogen atoms and electrons from their hydroxyl groups.41,42 In our study, two methods were used to quantify the antioxidant capacity of 30 BLEs. When considering the antioxidant activity determined by ABTS, the species with the highest values were G. amplexifolia (Gam-Q1, Gam-Q2, and Gam-Q3), G. angustifolia (Gan-C4, Gan-C7, Gan-N2, Gan-N4, Gan-N5, Gan-N6, and Gan-Q1), G. incana (Gin-P1, Gin-Q1, and Gin-Q2), G. uncinata (Gun-Q2), and G. venezuelae (Gve-Q1), with 149.7, 148.5, 146.6, 149.7, 124.0, 133.5, 140.4, 126.4, 122.0, 192.8, 145.2, 200.2, 144.9, 152.6, and 149.1 μmol TE/g extract, respectively. Regarding the antioxidant activity determined by DPPH in each of the BLEs, the extracts with the highest antioxidant values were G. amplexifolia (Gam-Q1), G. angustifolia (Gan-C4 and Gan-N1), G. angustifolia (Ganc-Q1), G. incana (Gin-Q1), and G. superba (Gsu-Q1), with 54.5, 48.1, 44.4, 50.7, 61.1, and 58.5 μmol TE/g extract, respectively.
Figure 6.
Antioxidant activity is determined for BLEs. The ABTS and DPPH of the BLEs results were expressed as μmol Trolox equivalents per gram of extract (μmol TE/g extract).
2.5. Cytotoxic Activity of BLEs
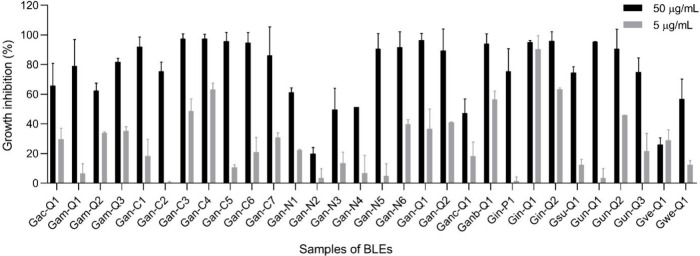
To determine the cytotoxic activity, two concentrations of 5 and 50 μg/mL of the extracts were tested against HCT-116 colon cancer cells. Figure 7 shows the percentage inhibition of the exposure of each of the extracts to the cell line. Out of the 30 extracts evaluated against the cell line, it is notable that 15 of them exhibited an inhibition percentage greater than 85%, meeting the criteria for activity established according to the National Cancer Institute (NCI) guidelines at the maximum evaluated concentration (50 μg/mL).43,44 It should be noted that G. angustifolia (Gan-C4) and G. incana (Gin-Q1) demonstrated the highest inhibition percentages in cell growth, with values of 97.7% and 96.1%, respectively.
Figure 7.
Percentage growth inhibition of BLEs evaluated at 50 and 5 μg/mL against HCT-116 (colorectal carcinoma) using MTT assay after 72 h of exposure. The percentage inhibition was calculated based on the growth of untreated cells. Data presented as mean ± SEM (standard error of the mean) from three independent duplicate experiments. All experiments utilized 0.05% DMSO (vehicle) and doxorubicin (DOXO) as negative and positive controls, respectively. Samples are considered active with an inhibition percentage higher than 85%. An extract was deemed active if it exhibited an inhibition of tumor cell lines >85% at 50 μg/mL.
Figure 8 displays the dose–response curves featuring the IC50 values of the extracts. Overall, it is evident that G. amplexifolia, G. angustifolia, and G. angustifolia var. bicolor, G. incana, and G. uncinata species exhibited a significant cytotoxic effect on HCT-116 colon cancer cell lines, with IC50 values below 50 μg/mL. Conversely, G. aculeata, G. angustifolia biotype San Calixto, G. superba, G. venezuelae, and G. weberbaueri did not demonstrate such an effect, displaying IC50 values above 50 μg/mL. According to the criteria established by the NCI, plant extracts are categorized into four groups: highly active (IC50 ≤ 20 μg/mL), moderately active (IC50 > 21–49 μg/mL), and weakly active (IC50 > 50 μg/mL). Cytotoxic analysis revealed that five extracts, corresponding to G. amplexifolia (Gam-Q3), G. angustifolia (Gan-C4 and Gan-Q2), and G. incana (Gin-Q1 and Gin-Q2), demonstrated a high cytotoxic effect with IC50 values of 14.1, 4.73, 11.34, 1.23, and 17.6 μg/mL, respectively. Additionally, ten extracts showed a moderate effect, including G. amplexifolia (Gam-Q1), G. angustifolia (Gan-C1, Gan-C3, Gan-C7, and Gan-N6), and G. angustifolia var. bicolor (Ganb-Q1), G. incana (Gin-P1), and G. uncinata (Gun-Q1, Gun-Q2, and Gun-Q3), with IC50 values of 41.6, 49.4, 27.8, 40.5, 32.2, 48.05, 44.9, 39.4, 36.1, and 48.6 μg/mL, respectively. The remaining 15 extracts exhibited a weak cytotoxic effect. This is the first report describing the cytotoxic activity of the BLEs.
Figure 8.
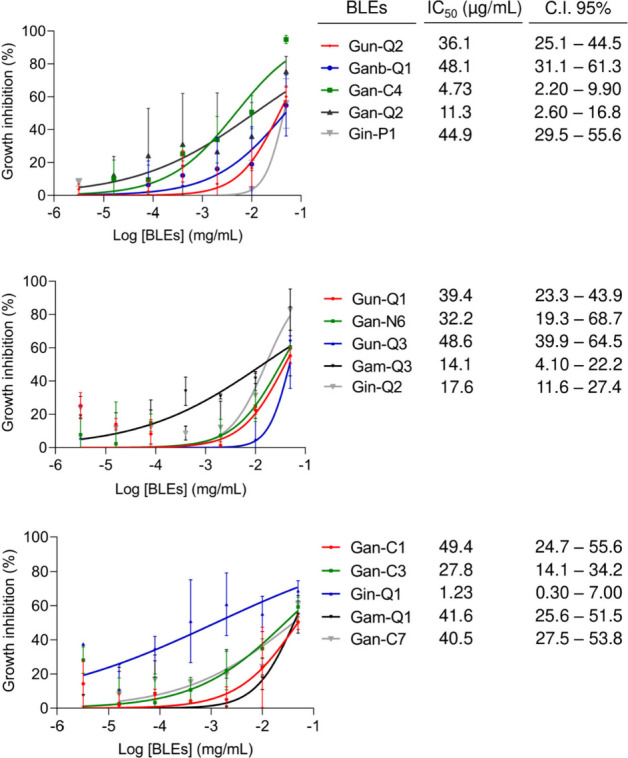
Dose–response curves of the cytotoxicity of HCT-116 cells after 72 h of treatment with the BLEs using concentrations of 3.2 × 10–6 to 5.0 × 10–2 mg/mL. The data represent the mean of three independent biological replicates with at least technical duplicates ± SEM (standard error of the mean). All experiments employed 0.05% DMSO (vehicle) and doxorubicin (DOXO) ranging from 0.00064 to 10 μM with IC50 = 0.2 μM (C.I 95% 0.14–0.25) as negative and positive controls, respectively.
2.6. Correlation-Based Untargeted Metabolomics Analysis
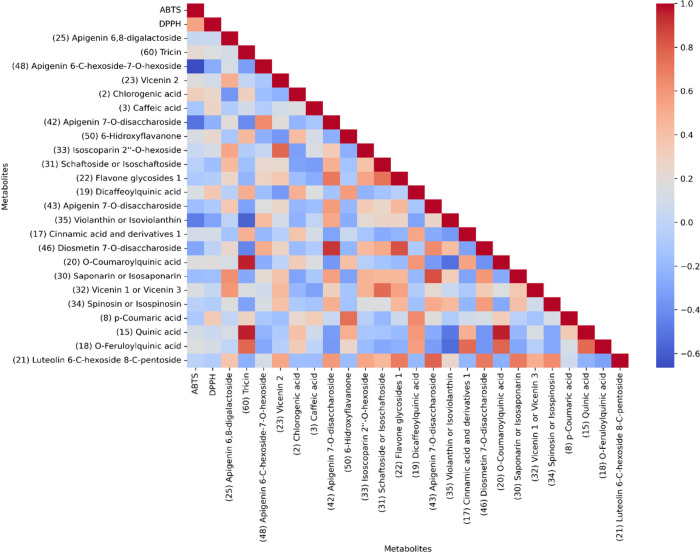
The correlation study based on the chemical composition of the extracts and antioxidant activity allowed us to determine that most of the BLEs showed a high antioxidant capacity, which was related to the high content of phenolic compounds present in these bamboos. Notably, the extracts that had a high content of flavonoids, as determined by HPTLC and HPLC-DAD analyses, were the ones that exhibited the highest antioxidant capacity, with G. incana and G. angustifolia species standing out. Figure 9 shows the relationship between antioxidant activity values and the chemical composition of the BLEs through Pearson’s simple correlation coefficients. We can observe a positive statistical relationship between the two activity variables concerning metabolites, with a correlation coefficient greater than 0.50 for most of them. We noticed a significant increase in both variables about the activity, which is closely related to the composition.
Figure 9.
Pearson’s simple correlation coefficients for the top 24 statistically differential metabolites and antioxidant activities monocation of ABTS and DPPH of BLEs. The interpretation of the r values corresponds to very weak or no correlation (−0.65 to 0.00 or 0.00 to 0.25); weak correlation (0.25 to 0.50); moderate correlation (0.50 to 0.75); and strong or perfect correlation (0.75 to 1.00).
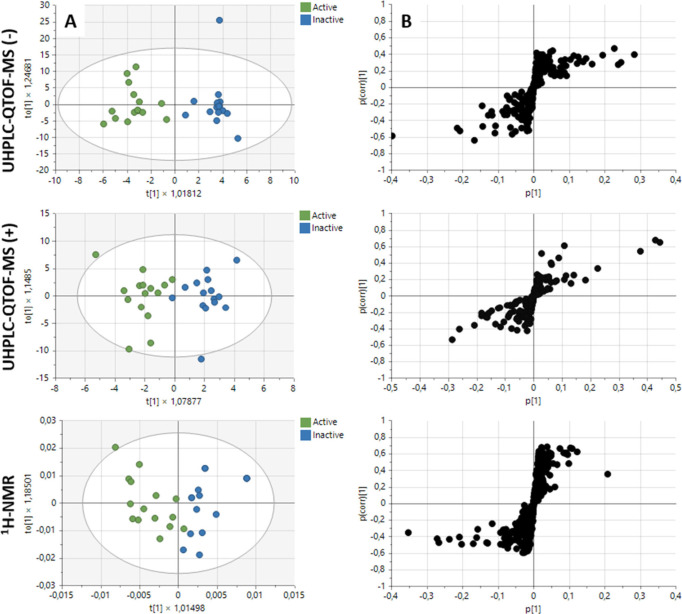
To identify the differential metabolites associated with cytotoxic potential, a multivariate analysis was conducted to correlate the bioactivity data with the metabolic fingerprinting obtained from UHPLC-QTOF-MS and 1H-NMR (Figure 10A-B). Initially, an unsupervised principal component analysis (PCA) was employed to assess the quality and stability of the instrumental platform. Supporting Information Figure S4 illustrates the clustering of the QC samples, demonstrating the reproducibility of the analysis. For a detailed investigation of the correlation between bioactivity and metabolites, a supervised orthogonal partial squares discriminant analysis (OPLS-DA) was applied, allowing for discrimination of activity variability among samples. Figure 10A represents the OPLS-DA plots for the UHPLC-QTOF-MS data in both negative and positive modes, as well as the 1H-NMR data. Notably, these plots highlight the discrimination between active and inactive sample groups within the specified activity range (>85% at 50 μg/mL with IC50 < 50 μg/mL). The established supervised models were validated through a permutation test, which assesses the quality of fit and predictive capability. In the case of cytotoxic activity against the colon cancer cell line (HCT-116), the R2X(cum), R2Y(cum), and Q2(cum) values obtained from OPLS-DA analysis were deemed acceptable, demonstrating excellent reproducibility and predictability on the analytical platforms used.
Figure 10.
(A) OPLS-DA score plot demonstrating a distinct separation between active (green) and inactive (blue) extracts against the colon cancer cell line (HCT-116). (B) S-loading plot depicting metabolites highly correlated with HCT-116 cytotoxicity. The prediction and quality values of the OPLS-DA model for each platform used were as follows: UHPLC-QTOF-MS (−) = R2X(cum): 0.382; R2Y(cum): 0.895; Q2(cum): 0.226; UHPLC-QTOF-MS (+) = R2X(cum): 0.216; R2Y(cum): 0.795; Q2(cum): 0.578, and 1H-NMR = R2X(cum): 0.815; R2Y(cum): 0.680; Q2(cum): 0.393. The corresponding loading plot displays metabolites on the right that contributed the most to activity and on the left the metabolites that contributed the least to activity.
The variable importance in projection (VIP) plot was generated to calculate the influence intensity and interpretability of each metabolite’s expression mode in distinguishing and classifying sample groups. In this case, this analysis facilitated the prediction of metabolites responsible for the activity based on statistically significant changes between active and inactive sample groups. The s-plot is presented in Figure 10B, highlighting variables with a VIP score >1.5, which serves as the prediction threshold. Additionally, we can observe regions where samples share a group of metabolites that contributed to cytotoxic activity to a lesser extent, and another region with metabolites contributing more significantly to cytotoxic activity. According to the above and from the OPLS-DA analysis, 27 differential metabolites were found between the active vs inactive BLEs, as summarized in Table 1. These findings are linked to the MFs identified in the analysis conducted by MolNetEnhancer. Of the metabolites that correlated with the cytotoxic potential, it was found that flavone glycosides were the ones that statistically contributed to the bioactivity in the platforms used. In the case of UHPLC-QTOF-MS, it was evident that the noted flavonoids were those that correlated positively with cytotoxicity, while for 1H-NMR, the signals present in the aromatic and sugar regions (δH 6.50–8.50 ppm and δH 3.00–5.50 ppm, respectively in the identified buckets) showed a positive correlation.
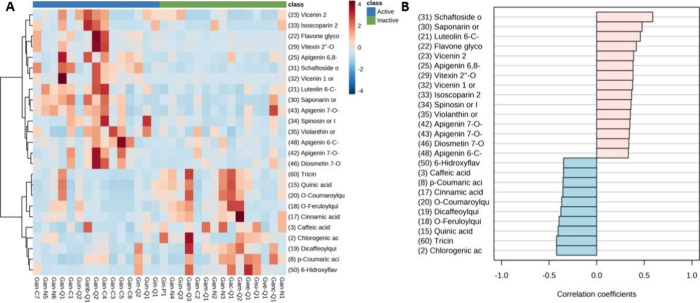
The heatmap shows the abundance of the main statistically significant metabolites correlated with cytotoxic activity (Figure 11A). Hierarchical clustering analysis clearly reveals two clusters showing the differential chemical classes of flavonoid glycosides and caffeoylquinic acid (CQAs) derivatives with respect to activity groups. In terms of abundance, it is observed that the flavonoid glycosides are present in the BLEs that presented a higher cytotoxic effect against the colon cancer cell line HCT-116, i.e., G. incana and G angustifolia species, respectively. Confirming these findings, we constructed a graph to search for patterns of metabolites correlated with cytotoxic activity (Figure 11B). It is observed that the group of flavonoid glycosides presented positive correlation coefficients, while the CQAs did not. Taken together, these data demonstrate that BLEs are good sources of new antioxidant and cytotoxic compounds.
Figure 11.
(A) Hierarchical clustering analysis (HCA) with heatmap illustrating the difference in the compounds annotated abundance between active and inactive samples. The x-axis shows the clustering of all the samples, and the y-axis shows the clustering of the 25 compounds significantly statistically. (B) Pattern search for correlation analysis of metabolites correlated with cytotoxic activity. Compounds annotated significantly statistically by UHPLC-QTOF-MS from BLEs comparing Active/Inactive groups (p-value threshold <0.05 and FC threshold >2).
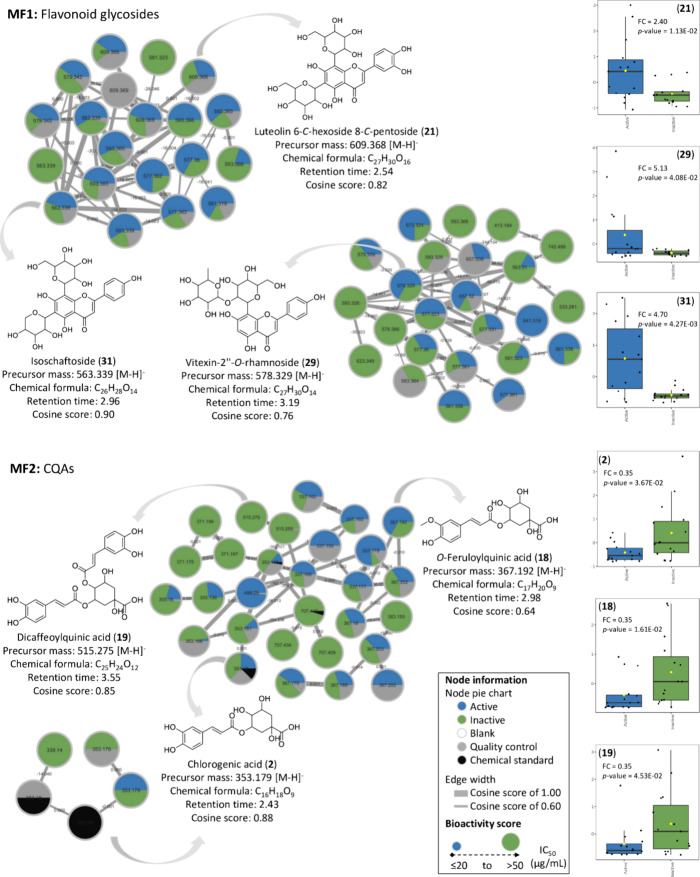
2.7. Bioactivity Score-Based Molecular Networking
To complement the correlation analysis, the effectiveness of the molecular networking (MN) was evaluated together with the bioactivity variable of the extracts by using the GNPS web platform. The MN containing the classification component of activity is represented in Figure 12. The constructed molecular networks show the two main molecular families (MFs) present in the BLEs. On one hand, the first molecular family (MF1) corresponds to flavonoid glycosides where three metabolites corresponding to Luteolin 6-C-hexoside 8-C-pentoside (21, FC = 2.40; p-value = 1.13 × 10–02), Vitexin-2″-O-rhamnoside (29, FC = 5.13; p-value = 4.08 × 10–02), and Schaftoside or Isoschaftoside (31, FC = 4.70; p-value = 4.27 × 10–03) are represented correlating positively with cytotoxicity, and the second molecular family (MF2) corresponds to CQAs including CGA (2, FC = 0.35; p-value = 3.67 × 10–02), O-Feruloylquinic acid (18, FC = 0.35; p-value = 1.61 × 10–02), and Dicaffeoylquinic acid (19, FC = 0.35; p-value = 4.53 × 10–02), which in this case did not exhibit a correlation with cytotoxic activity against the colon cancer cell line HCT-116.
Figure 12.
Bioactive molecular networking of the BLEs displays compounds that contribute most significantly to the activity. Particularly, flavonoid glycosides are highlighted with a high bioactivity score, while CQAs present a low bioactivity score against the colon cancer cell line HCT-116. The nodes were colored based on the classification of active and inactive groups, considering the IC50 value as a reference for the bioactivity score. Compounds 2, 18, 19, 21, 29, and 31, represented by the nodes, are the most representative according to the Pearson’s simple correlation coefficients determined by statistical analysis.
3. Discussion
In recent years, bamboo has found diverse applications, primarily in construction, agriculture, food production, and the manufacturing of paper and furniture.45−47 Despite its versatile utility, there is a notable paucity of comprehensive chemical and biological information about Guadua species. Specifically, the lack of studies employing metabolomic approaches for the identification of potential bioactive markers, correlating chemical composition and biological activity, hinders a deep understanding of the potential of these plants. Preliminary phytochemical investigations have provided insights into the chemical profile of Guadua species, revealing primarily the presence of flavonoid compounds and phenolic acid derivatives, etc.12,48 The studies reported to date provide a basis for comparing and confirming our findings regarding chemical composition and biological activity. Previously, several flavonoid compounds were identified in polar extracts of G. angustifolia leaves, including Quercetin, Kaempferol, Violanthin, and Kaempferol-7-O-neohesperidoside.35 Additional studies explored vinegar obtained from G. angustifolia, emphasizing its antioxidant and antimicrobial activity against Pseudomonas aeruginosa and Staphylococcus aureus, attributed to the high content of phenolic compounds.13,49 Another study focused on the essential oil from G. angustifolia and G. chacoensis (Rojas Acosta) Londoño and P.M. Peterson. The researchers analyzed the chemical composition and assessed antimicrobial activity against Aspergillus brasiliensis, Candida albicans, Escherichia coli, P. aeruginosa, and S. aureus. The majority of the essential oil composition in both essential oils primarily consisted of terpenes and compounds related to terpenes. In the essential oil of G. angustifolia, the main compounds were hexahydrofarnesyl acetone (23.1%) and (Z)-phytol (21.3%), while for G. chacoensis, the main compounds included (E)-β-ionone (8.8%), hexadecanoic acid (6.8%), hexadecenoic acid (6.5%), (Z)-phytol (5.3%), and (E)-α-ionone (5.0%). Some of these essential oils exhibited significant activity, with a MIC < 100 μg/mL, suggesting their potential as candidates for the development of new antimicrobial agents.50 Others studies, involving G. angustifolia var. bicolor, evaluated the antioxidant and antityrosinase potential of leaf and culm extracts. The n-butanol fraction exhibited the highest antioxidative activity, while the dichloromethane fraction showed tyrosinase inhibition, with inhibition percentages of 86.39% and 40.66%, respectively. Finally, the metabolites responsible for the evaluated activities were tentatively identified as p-coumaric acid, catechin, epicatechin, quercetagetin 7-O-glucoside, and genistein.51,52 In accordance with the above, several compounds identified in this research through chemical profiling had already been reported in BLEs, demonstrating antioxidant,48 antimicrobial,50 and antityrosinase51,52 potential. This knowledge of the activity of these species confirms our findings. Furthermore, these results are consistent with a recent study where untargeted metabolomics was applied to determine changes in chemical composition under the influence of altitude in 111 samples of bamboo plants collected in different regions of Colombia, confirming the presence of some of the MFs reported in this study. Additionally, the study highlights the prevalence of phenylpropanoid and polyketide MFs, indicating the presence of flavonoids and cinnamic acid derivatives, mainly.16 These compounds, widely distributed as secondary metabolites in most plants, are important due to their ability to act as antioxidants. Many phenolic compounds have been reported to possess potent antioxidant activity, which may also be associated with anticancer, antibacterial, antiviral, or anti-inflammatory properties to varying degrees.53−55
The findings of this work demonstrate that BLEs have antioxidant potential, which aligns with data collected for certain species of bamboo leaves such as G. angustifolia.14 In their study, different extraction methods were employed with solvents of varying polarity, including ethyl acetate, diethyl ether, and 50% ethanol, using three separate extraction techniques: reflux, Soxhlet, and ultrasound. Additionally, they evaluated the antioxidant capacity using the DPPH methodology in their extracts at a concentration of 50 mg/mL. It was highlighted that the extracts of medium and high polarity, corresponding to ethyl acetate and 50% ethanol, presented the highest antioxidant activity. This observation could indicate the presence of polyphenolic compounds, such as flavonoids, to which antioxidant activity has been attributed and that have been found in bamboo leaves, specifically Phyllostachys nigra var. henonis.56,57 It is important to highlight that in our study we used a concentration of 1 mg/mL to evaluate antioxidant activity through the ABTS and DPPH assays. Therefore, when compared with the previous study, our results for antioxidant activity indicate that most of the BLEs presented a substantial antioxidant potential, with values greater than 100 and 30 μmol of TE/g of extract for ABTS and DPPH, respectively. Another study assessed the antioxidant potential of bamboo vinegar obtained through the pyrolysis process of G. angustifolia. The antioxidant activity in vinegar was determined by both methods. In this study, the concentration of syringol was quantified in the different formulations obtained from the vinegar, revealing that this compound, along with others such as guaiacol, exhibited antioxidant capacity. Furthermore, typical compounds of phenolic acids with antioxidant properties were also identified. Free radical scavenging activity of 30.9 μmol/L TEAC was observed at a concentration of 0.23%. Notably, at a vinegar concentration of 10%, the most effective capture of the DPPH radical was achieved, with a value of 940.2 μmol/L TEAC.58
The highest antioxidant capacity of BLEs using the DPPH method was 61.1 μmol of Trolox/g of extract. These values were lower than those obtained by Mosquera et al.48 in ethanolic extracts evaluated at a concentration of 1 mg/mL in G. angustifolia, which presented an antioxidant activity of 80 μmol Trolox/g of extract. However, for the ABTS method, the highest antioxidant capacity of the extracts was 200.2 μmol Trolox/g of extract, which was higher compared to the results obtained by Mosquera et al.,48 who reported an antioxidant capacity of 180 μmol Trolox/g extract by ABTS. Studies conducted by Lozano-Puentes et al.19 reported the antioxidant activity of several ethanol extracts from optimized leaves of G. angustifolia obtained through different levels of sonication (low, medium, and high) evaluated at three times (10, 20, and 30 min) and three temperatures (20 °C, 35 °C, and 50 °C), where the antioxidant capacity was determined. It was found that extraction at 50 °C for 20 min favored the extraction of phenolic compounds and total flavonoids, presenting the highest antioxidant capacity with values of 144.76 and 209.23 μmol Trolox/g of extract for the ABTS and DPPH methods, respectively. This aligns with what was found in this study, given that many of the extracts were obtained under room temperature conditions and by the percolation method and an abundant presence of flavonoids was detected, as seen in the species G. incana with antioxidant activity values of 200.2 and 61.1 μmol Trolox/g of extract and in G. angustifolia with values of 140.4 and 26.1 μmol Trolox/g for ABTS and DPPH, respectively.
Considering that only previous reports have been found for the species of the genus Guadua, and to strengthen our discussion, a search was carried out at the Bambusoideae subfamily level for related studies to compare our results of chemical composition and biological activity. In this sense, it was found, particularly, that the dried leaves of Sasa veitchii bamboo, a species traditionally used in Asian folk medicine, reveal its use as a source of antioxidant food supplements in Japan. Additionally, the Chinese Food Additives Standardization Committee approved an extract of Phyllostachys nigra var. henonis known as antioxidant of bamboo leaves (AOB) as a novel kind of natural antioxidant due to its potent antioxidant capacity attributed to phenolic compounds.3,59 Products derived from bamboo extracts, coupled with the increased production of reactive oxygen species (ROS), are believed to play a significant role in preventing disease development and reducing the impact of oxidative stress when disease occurs.39,60
We highlight that according to our results, to date, the BLEs included in this research do not present any report of cytotoxic activity, this being the first report. Despite the absence of reports on cytotoxicity in these bamboos, it is striking that, in general, other bamboo species such as Phyllostachys heterocycla and Dendrocalamus asper have shown anticancer effects against various cell lines, including HepG2, MCF-7, HeLa, A549, and THP-1, which are directly associated with the high content of polyphenols.31,61 For example, in a previous study, the polar fraction of Phyllostachys heterocycla var. pubescens (Pradelle) Ohwi was shown to exhibit selectivity by inhibiting the epidermal growth factor receptor (EGFR) in HepG2 cells. In this study, the isolation of 7-hydroxy-5-methoxy-methylcinnamate showed IC50 values of 7.43 and 10.65 μM against HepG2 and MCF-7 cell lines, respectively. Moreover, it induced apoptotic cell death in HepG2 cells, with a total apoptotic rate of 58.6% compared to 4.71% in the control, achieved by arresting cell cycle progression in the G1 phase.9 Previous studies in other bamboo species have reported cytotoxic activity, e.g., in the crude methanol extract obtained from the skin of the shoots of bamboo Phyllostachys heterocycla. The crude extract was evaluated against five cell lines-four of them tumorigenic (HepG2, HeLa, A549, and MCF-7) and one nontumorigenic cell line. The results showed that the crude extract presented a moderate cytotoxic effect against HepG2 and MCF-7, with IC50 values of 48.4 and 38.9 μg/mL, respectively, while against HeLa it showed a weak effect with an IC50 value of 50 μg/mL.11 In a more recent study of P. heterocycla var. pubescens, the cytotoxic effect was evaluated against the crude methanol extract and the fractions obtained from n-hexane, ethyl acetate, and n-butanol, targeting five cell lines: HepG2, HeLa, A549, MCF-7, and THP-1. It was reported that the n-butanol fraction exhibited inhibition percentages exceeding 60% against all cell lines at the maximum concentration of 100 μg/mL. Notably, the n-butanol fraction demonstrated the highest inhibition at 94.31% against the HeLa cell line.9 In the broader context of our research on the chemical and biological properties of Guadua species, our study significantly expands the existing knowledge base, particularly concerning cytotoxic activity. Although previous reports have been scarce, our results of the activity of BLEs against HCT-116 cells represent a novel contribution to the understanding of the anticancer potential of these bamboos. However, there is still much to explore about the mechanisms of action that exert their effect on cells. The absence of previous reports on cytotoxicity in these plants, as highlighted in our study, underscores the limited exploration of their therapeutic effects. However, previous studies support our findings, confirming that these extracts may be promising candidates for bioactive compounds, specifically as anticancer agents.
Our interest in HCT-116 cells and the need for further research on the antioxidant and cytotoxic potential of Guadua species led us to establish a correlation between the chemical composition and the antioxidant and cytotoxic activities of BLEs to identify the bioactive compounds responsible for these effects. This holistic approach not only improves our understanding of BLEs but also lays the foundation for the development of new therapeutic agents with potential applications in cancer research and treatment. Antioxidant activity is intricately linked to fundamental cellular processes such as apoptosis, proliferation, lipid metabolism, cell differentiation, and immune response. In the course of cellular metabolism, the generation of reactive oxygen species (ROS) can induce cellular damage and contribute to diseases primarily through superoxide anions, nitric oxide (NO•) radicals, hydroxyl radicals, and hydrogen peroxide. The oxidation of oxygen free radicals has been identified as a pivotal factor in the onset and progression of various diseases, including cardiovascular diseases, neurodegenerative disorders, atherosclerosis, tumors, etc.62 Based on this context and taking into account that it is the first approach in bamboos, it is highlighted that the compounds corresponding to flavonoids and derivatives of phenolic acids correlated positively with the antioxidant potential, previously highlighted in the studies described. Particularly, in a study conducted by Ni et al. on the species Sasa argenteastriatus, it was found that they correlated the chemical composition with antioxidant activity, determining that the content of total flavonoids (TF) and total phenols (TP) obtained from the leaves of this ornamental bamboo exhibited a high correlation with antioxidant activity. Additionally, eight characteristic compounds of this bamboo were identified, including orientin, isoorientin, vitexin, homovitexin, p-coumaric acid, chlorogenic acid, caffeic acid, and ferulic acid. In general, it is highlighted that seasonal variation and the content of TF and TP influence the DPPH radical scavenging capacity. The correlation coefficient between TF, TP, and DPPH radical scavenging is TF > TP, indicating that TF in bamboo leaves plays a primary role in the DPPH radical scavenging capacity. The correlation coefficient between TF, TP, and antioxidant capacity obtained by the FRAP assay is TP > TF.38 According to the above, our correlation results of the chemical composition and antioxidant activity align with those of the previous study. Although the total flavonoid and total phenolic contents were not determined, it was established that about 50% of the chemical composition of the BLEs corresponds to flavonoids and derivatives of phenolic acids. On the other hand, compounds reported in this work, such as vicenin 2, chlorogenic acid, caffeic acid, schaftoside or isoschaftoside, saponarin or isosaponarin, etc., presented a higher correlation coefficient >0.5, demonstrating powerful antioxidant activity, in agreement with the findings reported and compared by Ni et al. Similarly, a recent study by Ko et al. on the effect of seasonal variations on phenolic compounds and the antioxidant and anti-inflammatory activity of S. quelpaertensis Nakai through a correlation study demonstrated that phenolic compounds (p-coumaric acid, tricine, TP, and TF content) in ethanol extracts of leaves (ESL) showed an excellent correlation with antioxidant activities, indicating that ESL contained the highest levels of antioxidant and anti-inflammatory substances.63 However, in our study, the flavone tricine showed correlation coefficients <0.5, indicating a negative correlation with antioxidant activity, unlike the findings reported by Ko et al. This allows us to determine that the effect is possibly attributed to the synergistic effect64,65 of all the compounds present in the BLEs or to other specific phenolic compounds such as p-coumaric acid and Luteolin 6-C-hexoside 8-C-pentoside, presenting a coefficient of correlation >0.7, indicating a potent antioxidant effect for both methods.
On the other hand, for cytotoxic activity, it was found that flavonoid glycosides presented a positive correlation with this effect. These results are consistent with those reported for a wide variety of flavonoid glycosides that exhibit cytotoxic activity against various cell lines, including MCF-7, HCT-116, HepG2, HeLa, etc. Some flavones, including apigenin, baicalein, chrysin, luteolin, nobiletin, tangeretin, and wogonin, have shown cytotoxic potential against various colon cancer cell lines, such as Caco-2, COL-2, COLO201, COLO205, COLO320, HCT-115, HCT-116, HT-29, etc., with IC50 values <100 μM.66 We found that glycosated flavones particularly showed a positive relationship with increased cytotoxic activity, whereas caffeoylquinic acids did not. Although there are no studies on the effect of BLEs against HCT-116 colon cancer cell lines, the effect of extracts of bamboo shoots (Dendrocalamus asper, Phyllostachys heterocycla var. Pubescens, and S. quelpaertensis) against MCF-7 breast cancer cells and HCT-116 colon cancer cells lines has been reported, showing cytotoxic, antiproliferative, proapoptotic, and proin-flammatory effects.7,67 A recent study investigated the role of NO• and inhibitors of apoptosis (IAPs) during the process of apoptosis induced by S. quelpaertensis leaf extracts (SQE) in p53 wild-type (WT) and p53 null HCT-116 colon carcinoma cells. SQE was shown to induce apoptosis independent of p53 status and was associated with modulation of endogenous NO• in HCT-116 cells.67
Sak reported that flavonoids, such as quercetin and baicalein, are recognized as beneficial agents for preventing and treating colon cancer. However, in contrast to their glycosides, rutin and baicalin, respectively, they did not exhibit growth inhibitory effects on colon cancer cells. This observation highlights the substantial impact of the sugar fraction on the bioactivity of flavonoids.66 Interestingly, our correlation study contradicts this finding, especially concerning glycosylated flavones identified as vicenin 1 or vicenin 3 (32, FC = 6.73; p-value = 2.64 × 10–02), Vitexin 2″-O-rhamnoside (29, FC = 5.13; p-value = 4.08 × 10–02), Flavone glycosides (22, FC = 4.98; p-value = 1.28 × 10–02), Schaftoside or Isoschaftoside (31, FC = 4.70; p-value = 4.27 × 10–03), and Saponarin or Isosaponarin (30) (FC = 3.38; p-value = 1.61 × 10–02), which demonstrated significant changes and are associated with cytotoxic potential in the most active BLEs. To validate these results, isolating potential bioactive markers from the most active extract and confirming their cytotoxic activity is essential. However, a recent study by Vukovic et al. evaluated the cytotoxicity of 11 flavonoids isolated from propolis against colon cancer cell lines (HCT-116) and breast cancer cells (MDA-MB-231). Six flavonoids induced cytotoxic effects in both cell lines. Luteolin exhibited cytotoxicity, especially in HCT-116 cells, with an IC50 value after 72 h of exposure of 66.86 μM. Myricetin demonstrated cytotoxicity with an IC50 value of 114.75 μM, inducing apoptosis in MDA-MB-231 cells.68 The preceding data, related to the correlation study and bioactivity of glycosylated flavones against the colon cancer cell line HCT-116, align consistently with our results. Thus, a straightforward identification of potential biomarkers was achieved prior to any fractionation or isolation efforts. However, studies focused on the isolation of cytotoxic compounds are required to confirm our hypothesis.
Molecular networking analysis has enabled the identification of bioactive molecules within BLEs, whose active constituents were previously unknown against colon cancer cells HCT-116. In the study, it was emphasized that compounds 21, 29, and 31 exhibited a positive correlation, affirming the earlier observations. The present study demonstrated that the application of a metabolomics approach, based on correlation of chemical composition and biological activity, together with the construction of integrated molecular networks with bioactivity scores, can accelerate the search for bioactive molecules and rationalize the isolation procedure in bioassay-based phytoconstituent discovery.69 This innovative tool offers the possibility to narrow the pool of candidate compounds for further isolation, thus minimizing the time required in this process.
Our study faced several limitations. First, the isolation of bioactive compounds is necessary to validate the correlation study. Second, due to the limited availability of individuals of certain species during the plant collection phase, we recommend incorporating a larger number of individuals in future studies. This approach will strengthen statistical analysis and ensure a more comprehensive exploration of the metabolome in bamboos. Third, given the limited biological information available for these species, it is crucial to evaluate additional biological activities that support our findings. Finally, to elucidate the mechanisms of action on HCT-116 cells and validate their therapeutic potential, more specific biological activity assays are needed.
4. Conclusion
The present study offers a comprehensive view of Guadua species, emphasizing their chemical and biological potential. Despite incomplete information on these plants, 49 compounds, including flavonoids and phenolic acid derivatives, have been identified, showing a strong correlation with antioxidant and cytotoxic activities. Remarkably, this study is the first to report the cytotoxic activity of BLEs against the colon cancer cell line HCT-116. Our findings underscore a direct link between the abundance of phenolic compounds, such as Vicenin 2, Chlorogenic acid, and Caffeic acid, and antioxidant activity. Additionally, glycosylated flavones are positively correlated with cytotoxicity, contrasting with the negligible impact of CQAs. Furthermore, the application of a metabolomic approach and molecular network analysis has enabled the identification of bioactive molecules within BLEs, streamlining the pursuit of compounds for future isolation studies. Nevertheless, further investigations are warranted to isolate and validate these bioactive compounds as well as to better understand the mechanisms of action associated with the antioxidant and cytotoxic activities observed in these species. A detailed understanding of the bioactive compounds present in bamboo and their potential impact on human health could lead to the development of innovative therapies for various diseases as well as the formulation of cosmetic and nutraceutical products. In addition, future research could explore the role of bamboo as a natural source of antioxidants and cytotoxic agents in the prevention and treatment of cancer diseases as well as its application in the food and health industry.
5. Materials and Methods
5.1. Reagents
1,1-Diphenyl-2-picrylhydrazyl (DPPH), 2,2-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), and 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox, Hoffman-La Roche) were purchased from Sigma-Aldrich Co., (St. Louis, MO, USA). Methanol, acetonitrile, and formic acid HPLC grade were acquired from Merck (Darmstadt, Germany). Ultrapure water was used throughout and was produced by a Milli-Q Ultrapure water system with a resistivity of 18.2 MΩ·cm at 25 °C (Millipore, Bedford, USA). For the NMR experiments, methanol-d4 (>99.8 atom % D) and deuterium oxide (>99.8 atom % D) were purchased from Sigma-Aldrich USA (St. Louis, MO, USA). Dimethyl sulfoxide (DMSO) was purchased from Panreac AppliChem. Gallic acid (purity: > 98%, CAS No. 149–91–7), Chlorogenic acid (purity: > 98%, CAS No. 202650–88–2), Caffeic acid (purity: > 98%, CAS No. 501–16–6), Isoorientin (purity: > 98%, CAS No. 4261–42–1), Ampelopsin (purity: > 98%, CAS No. 27200–12–0), Rutin (purity: > 98%, CAS No. 153–18–4), Vitexin (purity: > 98%, CAS No. 3681–93–4), p-Coumaric acid (purity: > 98%, CAS No. 501–98–4), Sinapic acid (purity: > 98%, CAS No. 7362–37–0), Morin (purity: > 98%, CAS No. 480–16–0), Coumarin (purity: > 98%, CAS No. 91–64–5), Quercetin (purity: > 98%, CAS No. 117–39–5), Cinnamic acid (purity: > 98%, CAS No. 140–10–3), and Naringenin (purity: > 98%, CAS No. 480–41–1) were obtained from Sigma-Aldrich (St. Louis, MO, USA). All these compounds were used as reference standards. RPMI-1640 medium, fetal bovine serum (FBS), and penicillin–streptomycin were obtained from Gibco BRL (Grand Island, NY, USA).
5.2. Plant Material and Preparation of BLEs
Mature leaves obtained from the apical branches of 10 bamboo species belonging to the genus Guadua were collected from different locations in Colombia as described in Table 2. The voucher species were deposited in the Herbarium of Pontificia Universidad Javeriana (HPUJ) and determined by Néstor García and Ximena Londoño. The plant materials were dried for 96 h at 35 °C in an oven with forced air circulation, followed by being ground to a fine powder. The material obtained was subjected to a percolation process with ethanol (plant:solvent, 1:10 w/v) at room temperature for 24 h with solvent changes repeating four times. Subsequently, the extracted solutions obtained were concentrated on a rotary evaporator under reduced pressure at an average temperature of 35 °C, providing the corresponding BLEs and stored at room temperature until used.
5.3. Cleanup of the BLEs
The BLEs were solubilized in methanol:water 95:5 (v/v) and a cleanup step was performed for biological test and chromatographic analyses. The extracts underwent a cleanup step in a solid phase extraction cartridge (Strata X, C18). In this procedure, first the cartridge was packed with 6 volumes (∼5 mL) of HPLC grade methanol. Then the cartridge was equilibrated with 6 volumes of methanol:water solution at 95% and the extract obtained in the previous step was applied. The elution was carried out with 3 mL of 95% methanol:water. The extract obtained was dried by rotary evaporation and stored at room temperature until used for analysis.
5.4. Preparation of Chemical Standards and Quality Control (QC) Samples
Chemical standards solutions were prepared as 1 mg/mL solutions in methanol. Multiple QC samples were prepared by pooling and mixing equal volumes of each extracted sample to check the system performance and reproducibility in sample analysis. To assess the robustness of the instrument, pooled QC samples were injected prior to the sample analysis until system equilibration was achieved and after every ten randomized sample injections.
5.5. HTPLC Data Acquisition
The standard solutions were prepared by dissolving 1 mg of Quercetin and Rutin in 1 mL of methanol (1000 μg/mL), and the extracts were prepared by dissolving 10 mg in 1 mL of ethanol (10000 μg/mL) to seed 10 μL in each band of the plate and obtain a final amount of 10 and 100 μg, respectively. Each extract was filtered through a 0.22 mm polypropylene membrane filter for subsequent HPTLC analysis. Next, all samples were examined on HPTLC equipment (Camag, Muttenz, Switzerland), which included an autosampler (ATS 4), a developer (ADC), a derivatizer (DV), a development chamber, a visualization chamber (Visualizer 2, CAMAG), and VisionCATS software version 3.1.21109.3. HPTLC was performed on the Silica gel 60 F254 precoated TLC plates of 10 × 10 cm2 (Merck, Darmstadt, Germany). Ten microliter sampless of the extracts were applied to the plates in the form of 7 mm bands with the Camag automatic applicator equipped with a 10 mL syringe and operated with the following settings: band length 7 mm, application rate 10 s/mL, distance between 6 mm, distance from the lateral edge of the plate 2 cm, and distance from the bottom of the plate 2 cm. For each analysis, the factors that were kept constant were developing distance: 75 mm, developing time: 10 min, detection reagent: natural product reagent (1 g of 2-aminoethyl diphenyl borate in 100 mL of ethanol) (NP/PEG), postchromatographic derivatization time: 5 min. Plates were eluted with n-hexane/ethyl acetate/formic acid 10/6/1 (v/v/v) for flavonoid aglycones and ethyl acetate/formic acid/acetic acid/water 100/11/11/27 (v/v/v) for flavonoid glycosides in a chamber (20 cm × 10 cm, Camag) followed by air drying. For derivatization, the following reagents were applied by piezoelectric spraying. After development and derivatization of the bands, the plates were scanned at a wavelength of 365 nm by using the visualization chamber. The VisionCATS software controlled all of the modules of the equipment. The bands of the reference standards (Quercetin and Rutin) had their Rf measured for comparison with the bands of the extracts.
5.6. HPLC-DAD Data Acquisition
Samples were prepared by weighing 5 mg of each extract and solubilizing them with 1 mL of methanol. The chromatograph used was a Shimadzu UFLC system (Shimadzu, Duisburg, Germany) coupled to a diode array detector (DAD, VIS-Shimadzu 1520 PC-Diode Array), a pump model 6-AD Shimadzu SCL-10VP control system, an in-line degasser, and an autosampler. Chromatographic separation was performed on a Luna C18 column (150 × 4.6 mm2, 5 μm, 100 Å) was used to carry on the chromatographic separation at 30 °C. The mobile phase used was eluent A (Water 0.1% formic acid, v/v) and eluent B (Acetonitrile 0.1% formic acid, v/v) with flow of 1 mL/min. Gradient elution was performed as follows: 0 to 40 min, 5% to 100% B; 40 to 50 min, 100% B; 50 to 53 min, 100% to 5% B; and 53 to 63 min, 5% B. The wavelengths used were 274 and 365 nm, with spectra acquired in a range of 200 to 400 nm. Dereplication studies were performed to compare retention times and UV spectrum.
5.7. UHPLC-QTOF-MS Data Acquisition
For UHPLC-QTOF-MS analysis, the BLEs were dissolved in methanol at a concentration of 1 mg/mL. Samples were further diluted with methanol to obtain a final concentration of 500 ppm. Analyses were performed on an ACQUITY UPLC System (Waters, Milford, MA, USA) coupled with a Waters Xevo G2-XS QToF Mass Spectrometer. An ACQUITY UPLC HSS T3 C18 column (2.1 × 100 mm2, 1.8 μm) from Waters (Waters Corporation, Wexford, Ireland) was used to carry on the chromatographic separation at 30 °C. The mobile phases consisted of eluent A (Water 0.1% formic acid, v/v) and eluent B (Acetonitrile 0.1% formic acid, v/v) with a flow rate of 0.5 mL/min. The gradient was programmed as follows: 10–100% B from 0 to 10 min, 100% B from 10 to 14 min, 100–5% B from 14 to 15 min, 5% B from 15 to 20 min. Mass spectrometry was performed on a Xevo G2-XS QToF instrument using electrospray ionization (ESI). Data mass spectra were acquired in ionization negative (ESI−) and positive (ESI+) modes, in a mass range of m/z 100–1500 Da in data dependent acquisition (DDA) mode at a rate set to 0.1 s scans, followed by an MS2 scan of the most intense ions. For MS2 fragmentation, a low collision energy ramp ranging from 10 to 40 eV and a high collision energy ramp ranging from 50 to 80 eV were applied to acquire the data in centroid format. The QTOF instrument was operated at a frequency of 6.0 GHz and 76.0 μs utilizing high resolution calibrated with the reference mass correction for Leucine-enkephalin (C28H37N5O7) and was used as a lock mass compound in positive ion mode ([M + H]+ = 556.2771) and negative ion mode ([M-H]− = 554.2615), respectively. The spectrometer parameters were as follows: capillary voltage was set to 2.2 kV (ESI−) or 2.6 kV (ESI+), cone voltage was set to 40 V, source temperature was maintained at 120 °C, cone gas was set to 50 L/h, and desolvation gas was set to 800 L/h at 300 °C. Retention time data compared between HPLC-DAD and UHPLC-QTOF-MS analyses of the reference standards are detailed in Supporting Information Table S3.
5.8. 1H-NMR Data Acquisition
To the BLEs were added 500 μL of methanol-d4 (CD3OD) and 200 μL of phosphate buffer (KH2PO4, 99% anhydrous) in deuterium oxide (D2O) (pH 6.0) from Sigma-Aldrich USA Isotope with 99.8% purity; the solution was sonicated (frequency of 50 kHz/20 min) and centrifuged (10.000 rpm/5 min), and 700 μL of the supernatant was transferred to 5 mm NMR tubes for further 1D NMR analyses. 1H-NMR spectra were recorded at 25 °C on a Bruker Avance III HD 600 MHz spectrometer (Bruker, Karlsruhe, Germany) operating at a proton NMR frequency of 600.13 MHz. Each 1H-NMR spectrum consisted of 128 scans using the following parameters: 0.16 Hz/point, pulse width (PW) = 30 (11.3 μs), and relaxation delay (RD) = 1.5 s. Free induction decays (FIDs) were Fourier transformed with line broadening (LB) = 0.3 Hz. Deuterated methanol was used as the internal lock. Chemical shifts were expressed in δ (ppm), and coupling constants were reported in hertz. The 1H-NMR spectra of some reference standards were included in Supporting Information Figure S5 and Figure S6.
5.9. GNPS Classical Molecular MS2 Network
The MS2 data files in mzML format were submitted to the GNPS online platform (http://gnps.ucsd.edu). The molecular networking of BLEs was obtained using parameters provided in the Supporting Information, as shown in Table S4 and described by Aron et al.70 Classical molecular networks for both negative and positive ionization modes are accessible on the GNPS Web site at the following links:
Negative mode: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=d4d5f88562114bf0b1a6a8083ecfb166
Positive mode: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f6f3a908473a462eb8822d718a087d8b
Visualization of comprehensive chemical space was incorporated in the MolNetEnhancer tool. Chemical class annotations were performed using ClassyFire chemical ontology. Both negative and positive ionization modes are accessible on the GNPS Web site at the following links:
Negative mode: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=a4dd538e6fc845858df14c6308ff317a
Positive mode: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=2a2f566db5994ec8a05fa024c751f1ef
The attribute table of the generated nodes was visualized in Cytoscape version 3.7.2 software to analyze the molecular network. The data used for the analysis of molecular networks were deposited in the MassIVE Public GNPS database (http://massive.ucsd.edu) with the accession number MSV000093471 for negative mode and MSV000093474 for positive mode.
5.10. Antioxidant Activity
The assays were carried out in triplicate for all the samples using a single extract concentration of 1 mg/mL21,48 diluted in ethanol and determined by the azinobis(ethylbenzothiazoline-6-sulfonic acid) (ABTS•+) radical scavenging method and scavenging of 1,1-diphenyl-2-picryl-hydrazyl (DPPH•) radical assay.
5.11. ABTS•+ Free Radical Scavenging Ability Assay
The ABTS assay was conducted following the method proposed by Lozano-Puentes et al., with some modifications.19,71 Briefly, the radical was generated by oxidizing ABTS at 7 mM with potassium persulfate at 2.45 mM and incubated for 12 h in darkness until the reaction was completed and the absorbance was stable. To perform the test, 30 μL of the BLEs was taken at 1 mg/mL and mixed with 970 μL of the ABTS•+ solution previously prepared and adjusted to an approximate absorbance value of 0.70 ± 0.02 with ethanol. This mixture was allowed to react for 30 min at room temperature in the dark. The reduction of ABTS•+ was measured by using a Varioskan LUX Multimode Microplate Reader (Thermo Scientific, Vantaa, Finland) at 734 nm and compared against a standard curve of Trolox. As a negative control, the solvent in which the extracts were solubilized (ethanol) was used. A Trolox calibration curve (0–100 mg/L, R2 = 0.9705) was constructed to express the antioxidant capacity in μmol trolox equivalents per gram of extract (μmol TE/g extract) in Supporting Information Figure S7. Three independent experiments were performed in triplicate.
5.12. DPPH• Free Radical Scavenging Ability Assay
Antioxidant activity was measured using the DPPH• radical method described by Lozano-Puentes et al., with some modifications.19,72 The electron-capturing capacity was determined by reacting 280 μL of a DPPH• solution at 240 mg/L with 20 μL of BLEs at 1 mg/mL for 30 min under conditions of darkness and room temperature. Radical reduction was measured using a Varioskan LUX Multimode Microplate Reader (Thermo Scientific, Vantaa, Finland) at 517 nm and compared against a standard curve of Trolox. As a negative control, the solvent in which the extracts were solubilized (ethanol) was used. A calibration curve of trolox (0–100 mg/L, R2 = 0.9709) was conducted to express the antioxidant capacity in μmol trolox equivalents per gram of extract (μmol TE/g extract) in Supporting Information Figure S7. Three independent experiments were performed in triplicate.
5.13. Cytotoxic Activity
The cell cultures of the human colorectal carcinoma cell line HCT-116 were acquired from the American Type Culture Collection (ATCC, Rockville, MD, USA) and maintained under standard cell culture conditions (37 °C, 95% with 5% CO2 atmosphere) in RPMI-1640 medium supplemented with 10% FBS, 2 g/L NaHCO3, and 1% antibiotics (penicillin and streptomycin).
5.14. MTT Cell Viability Assay
Colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to evaluate metabolic activity.73 Approximately 1 × 104 cells per well were seeded in 96-well plates. Twenty-four hours after seeding, the cells were then treated with DMSO 0.05% (vehicle), doxorubicin 10 μM (control), and BLEs at two concentrations of 5 and 50 μg/mL (treatments) diluted in DMSO and incubated under standard cell culture conditions for 72 h. Subsequently, MTT solution (150 μL at 10%) was added to each control and treatment and incubated again for 3 h. Then the medium was removed and the formazan crystal was solubilized with 200 μL of DMSO. The Bio-Tek Synergy HTX multimode reader (BioTeck Instruments) was applied to read the absorbance at 570 nm. Percent growth inhibition was calculated with eq 1:
| 1 |
To determine the dose–response curves (percentage cell survival versus concentration) and to obtain the IC50 value (the minimum concentration of each ethanol extract that provides 50% survival of HCT-116 cells), extracts that exceeded 85% inhibition of cell growth were prepared over a range of concentrations from 3.2 × 10–6 to 5.0 × 10–2 mg/mL, following the same procedure explained above. Data were determined in three independent biological replicates, each with technical quadruplicates. IC50 curves and extract values were calculated using GraphPad Prism version 8.0.2 software (San Diego, CA, USA) using sigmoidal dose–response curve fitting (variable slope, four parameters).
5.15. Data Processing and Biochemometric Analysis
The raw UHPLC-QTOF-MS data files were converted into mzML format using the MSConvert tool from ProteoWizard software version 3.0 (Palo Alto, CA, USA) to separate negative and positive ionization modes. The files were processed in MZmine version 2.53 (http://mzmine.github.io/download.html), including feature detection, chromatogram builder, deconvolution, deisotopes, alignment, filtering, and gap filling whose parameters summarized in Supporting Information Table S5. The list of data information, including tR-m/z (characterize the detected ions) and normalized peak area were exported as .csv files and manually inspected to eliminate noise, and presence filter was applied. The features that were present in 100% of the samples and had a coefficient of variation in the QC of less than 20% were used for statistical analysis.
The raw 1H-NMR spectra were imported into NMRProcFlow version 1.4.20 (https://www.nmrprocflow.org) for ppm calibration, baseline correction, alignment, spectra bucketing, and data normalization whose parameters summarized in Supporting Information Table S6. Regions at δH 4.85–4.95 and δH 3.25–3.35 ppm were removed from the analysis due to residual solvents signals of D2O and CD3OD, respectively. Spectra bucketing utilized the intelligent bucketing method and variable size bucketing, encompassing the full range of δH 0.40–10.0 ppm.
Statistical analysis was performed with SIMCA-P+ software, version 18.0 (Umetrics, Umeå, Sweden). The multivariate analysis allowed the generation of PCA and OPLS-DA models. The OPLS-DA model was validated by cross-validation less than 0.05, and quality was evaluated based on parameters R2X, R2Y, and Q2. The groups were compared to determine the significantly differential metabolites by calculating the variable importance in the projection (VIP) combined with FC > 2.0 or FC < 0.5. Annotated metabolites were organized in a table (.csv) containing the peak area were uploaded at MetaboAnalyst 5.0 software–statistical, functional, and integrative analysis of metabolomics data (https://www.metaboanalyst.ca/) was used visualization for heatmap clustering,74 normalized by Pareto scaling. Univariate analysis was performed to determine the p-value features.
5.16. Metabolite Identification
Differential metabolite identification involved mass accuracy (with a maximum error 10 ppm), isotopic pattern distribution (to generate of molecular formulas), and adduct formation. After that, we utilized the CEU Mass Mediator tool (https://ceumass.eps.uspceu.es/) to search for potential metabolites candidates through different public online databases such as METLIN (http://metlin.scripps.edu), KEGG (https://www.genome.jp/kegg/), HMDB (https://hmdb.ca/), PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ChEBI (https://www.ebi.ac.uk/chebi/). We confirmed the identity of the metabolites through MS2 analysis, employing tools such as MS-DIAL 4.80 (http://prime.psc.riken.jp/compms/msdial/main.html), MS-FINDER 3.52 (http://prime.psc.riken.jp/compms/msfinder/main.html), SIRIUS 5.5.7 (https://bio.informatik.uni-jena.de/software/sirius/), CFM-ID 4.0 (https://cfmid.wishartlab.com/) for in silico mass spectral fragmentation. This process also included the use of the GNPS web platform (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp) and manual interpretation with the MassLynx software version 4.01 (Waters Company, Milford, MA, USA). Furthermore, an in-house library was developed using metabolites previously isolated from the bamboo species subfamily (Subfamily: Bambusoideae) within the research group, facilitating the confirmation of metabolite identities. Finally, identification level were assigned for each metabolite accordance with Metabolomics Standards Initiative (MSI).75
Acknowledgments
The authors would like to acknowledge Pontificia Universidad Javeriana, al Ministerio de Ciencia, Tecnología e Innovación, al Ministerio de Educación Nacional, al Ministerio de Industria, Comercio y Turismo e ICETEX, 2a Convocatoria Ecosistema Científico - Colombia Científica 792-2017, Programa “Generación de alternativas terapéuticas en cáncer a partir de plantas a través de procesos de investigación y desarrollo traslacional, articulados en sistemas de valor sostenibles ambiental y económicamente” (Contrato no. FP44842-221-2018) and INCTBioNat CNPq 23038.000776/2017-54. We also extend our gratitude to the Ministerio de Ambiente y Desarrollo Sostenible for granting permission to use genetic resources and derived products (Contract number 212/2018; Resolution 210/2020), and the producers linked to the Federación Nacional de Cafeteros and Fundación Suyusama, El Paraiso del Bambú y la Guadua and Fundación Yarumo Jardín Botánico Forestal de Cundinamarca for providing the raw materials from different Guadua species. Furthermore, we are thankful to Dra. Isabel Duarte Coutinho, CEO and Co-founder of NATCROM, for the collaboration in providing chemical standards. Additionally, the authors would like to thank Dr. João Bronzel for their assistance in acquiring the UHPLC-QTOF-MS raw data and Dr. Uenifer Rodrigues Couto and the NMR Instrumental Platform of the Instituto de Química of the Universidade Estadual Paulista “Julio de Mesquita Filho” for their assistance in acquiring the NMR experiments. Biorender that was used for the elaboration of the graphical abstract. Finally, Luis Carlos Chitiva expresses appreciation for the approval of the cotutelle agreement (Doctoral Thesis and Double Degree) between Pontificia Universidad Javeriana Bogotá and Universidade Estadual Paulista “Júlio de Mesquita Filho”, signed on June 21st, 2022.
Glossary
Abbreviations
- HPTLC
High-Performance Thin-Layer Chromatography
- HPLC-DAD
High-Performance Liquid Chromatography coupled with Diode Array Detector
- UHPLC-QTOF-MS
Ultra-High Performance Liquid Chromatography coupled with Quadrupole Time-of-Flight Mass Spectrometry
- 1H-NMR
Proton Nuclear Magnetic Resonance
- MIC
Minimal Inhibitory Concentration
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c09114.
UV spectra of the major peaks found in the BLEs obtained by HPLC-DAD; Phytochemicals tentatively identified in BLEs using UHPLC-QTOF-MS; Chemical structures of the metabolites were found in BLEs; MS2 spectrum match feature of GNPS showing the similarity of fragments patterns of the experimental and library data; PCA score plot including Quality Controls (QCs) and all samples from UHPLC-QTOF-MS analysis; List and information on the environmental variables of collection of the Guadua species used for the study; Comparison of retention times of reference standards between HPLC-DAD and UHPLC-QTOF-MS analyses; 1H-NMR spectra obtained for the reference standards (Quercetin and Rutin); Calibration curves obtained for the determination of antioxidant capacity by ABTS and DPPH; Parameters used in the GNPS Classical Molecular Networking platform, MZmine version 2.53, and NMRProcFlow software for processing the data obtained by UHPLC-QTOF-MS, and 1H-NMR (PDF)
Author Contributions
L.C.C. wrote the original manuscript; L.C.C., H.S.L.-P., P.R.-T., and L.V.C.-L. performed the experiments and analyzed the data; P.R.-T. conducted cytotoxicity assay; X.L. conducted taxonomy, classification, description, and supply of bamboo samples; L.C.C., T.F.L., and I.C.-G. conducted molecular networking analysis; P.R.-T., X.L., T.F.L., L.A.D.-A., L.V.C.-L., J.A.P.-R., G.M.C., and I.C.-G. performed writing, review, and editing of the manuscript. G.M.C. and I.C.-G. supervised the project. All authors have read and agreed to the published version of the manuscript.
The Article Processing Charge for the publication of this research was funded by the Coordination for the Improvement of Higher Education Personnel - CAPES (ROR identifier: 00x0ma614).
The authors declare no competing financial interest.
Supplementary Material
References
- Vorontsova M. S.; Clark L. G.; Dransfield J.; Govaerts R.; Baker W. J.. World Checklist of Bamboos and Rattans; International Network of Bamboo and Rattan & the Board of Trustees of the Royal Botanic Gardens, Kew: Beijing, 2016. [Google Scholar]
- Ruiz-Sanchez E.; Tyrrell C. D.; Londoño X.; Oliveira R. P.; Clark L. G. Diversity, Distribution, and Classification of Neotropical Woody Bamboos (Poaceae: Bambusoideae) in the 21St/s Century. Botanical Sciences. 2021, 99, 198–228. 10.17129/botsci.2722. [DOI] [Google Scholar]
- Van Hoyweghen L.; Karalic I.; Van Calenbergh S.; Deforce D.; Heyerick A. Antioxidant Flavone Glycosides from the Leaves of Fargesia Robusta. J. Nat. Prod. 2010, 73 (9), 1573–1577. 10.1021/np100220g. [DOI] [PubMed] [Google Scholar]
- Wróblewska K. B.; Oliveira D. C. S.; Grombone-Guaratini M. T.; Moreno P. R.. Medicinal Properties of Bamboos. In Pharmacognosy-medicinal plants; Perveen S., Ed.; IntechOpen: Saudi Arabia, 2018; pp 1–18. [Google Scholar]
- Panee J. Bamboo Extract in the Prevention of Diabetes and Breast Cancer. Complementary and Alternative Therapies and the Aging Population: An Evidence-Based Approach 2009, 159–177. 10.1016/B978-0-12-374228-5.00009-3. [DOI] [Google Scholar]
- Grombone-Guaratini M. T.; Furlan C. M.; Lopes P. S.; Barsalobra K. P.; Leite e Silva V. R.; Moreno P. R. H. Antioxidant and Photoprotective Properties of Neotropical Bamboo Species. Plant Antioxidants and Health 2021, 1–35. 10.1007/978-3-030-45299-5_33-1. [DOI] [Google Scholar]
- Yansen I. A.; Budhy T. I.; Adam D.; Fahria F. Potential of Bamboo Shoots against Breast Cancer: A Review. Malaysian J. Med. Heal. Sci. 2023, 19 (SUPP3), 144–148. [Google Scholar]
- Abdelhameed R. F. A.; Nafie M. S.; Ibrahim A. K.; Yamada K.; Abdel-Kader M. S.; Ibrahim A. K.; Ahmed S. A.; Badr J. M.; Habib E. S. Cytotoxic, Apoptosis-Inducing Activities, and Molecular Docking of a New Sterol from Bamboo Shoot Skin Phyllostachys Heterocycla Var. Pubescens. Molecules 2020, 25 (23), 5650. 10.3390/molecules25235650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhameed R. F. A.; Habib E. S.; Ibrahim A. K.; Yamada K.; Abdel-Kader M. S.; Ahmed S. A.; Ibrahim A. K.; Badr J. M.; Nafie M. S. Chemical Constituent Profiling of Phyllostachys Heterocycla Var. Pubescens with Selective Cytotoxic Polar Fraction through EGFR Inhibition in HepG2 Cells. Molecules 2021, 26 (4), 940. 10.3390/molecules26040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A.; Abdelhameed R.; Habib E.; Ahmed S.; Badr J. Biological Activities of Different Species of the Genus Phyllostachys. Rec. Pharm. Biomed. Sci. 2021, 5 (1), 64–73. 10.21608/rpbs.2021.61559.1092. [DOI] [Google Scholar]
- Abdelhameed R. F. A.; Habib E. S.; Ibrahim A. K.; Yamada K.; Abdel-Kader M. S.; Ibrahim A. K.; Ahmed S. A.; Badr J. M.; Nafie M. S. Chemical Profiling, Cytotoxic Activities through Apoptosis Induction in MCF-7 Cells and Molecular Docking of Phyllostachys Heterocycla Bark Nonpolar Extract. J. Biomol. Struct. Dyn. 2022, 40 (20), 9636–9647. 10.1080/07391102.2021.1932599. [DOI] [PubMed] [Google Scholar]
- Mosquera Martínez O. M.; González Cadavid L. M.; Cortes Ossa Y. J.; Camargo García J. C. Caracterización Fitoquímica de Los Extractos de Acetona y Contenido de Lignina En Culmos de Guadua Angustifolia. Recur. Nat. y Ambient. 2012, 65 (65), 10–15. [Google Scholar]
- Mejía Gallón A.; Cadavid Torres E.; Gallardo Cabrera C. Actividad Antiséptica de Vinagre de Guadua Angustifolia Kunth. Rev. Cuba. plantas Med. 2011, 16 (3), 244–252. [Google Scholar]
- Valencia M.; Durango S.; Pinillos J. F.; Mejía C. A.; Gallardo-Cabrera C. C. Extracción de Fracciones Con Actividad Antioxidante En Hojas de Guadua Angustifolia Kunth. Rev. Cuba. Plantas Med. 2011, 16 (4), 364–373. [Google Scholar]
- Álvarez E. S. D.; Cabrera C. G.; Contreras A. C.; Durango Álvarez E. S.; Gallardo Cabrera C.; Contreras Contreras A. Estudios Para El Aprovechamiento Potencial de Hojas de Guadua Angustifolia Kunth (Poaceae), Para El Sector Cosmético. Rev. Cuba. Farm. 2015, 49 (3), 535–542. [Google Scholar]
- Chitiva L. C.; Lozano-Puentes H. S.; Londoño X.; Leão T. F.; Cala M. P.; Ruiz-Sanchez E.; Díaz-Ariza L. A.; Prieto-Rodríguez J. A.; Castro-Gamboa I.; Costa G. M. Untargeted Metabolomics Approach and Molecular Networking Analysis Reveal Changes in Chemical Composition under the Influence of Altitudinal Variation in Bamboo Species. Front. Mol. Biosci. 2023, 10, 1192088 10.3389/fmolb.2023.1192088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano Puentes H. S.; Modesti Costa G.; Díaz Ariza L. A. PPV-4 Determinación de La Composición de Flavonoides de Hojas de G. Angustifolia Kunth En Guaduales Naturales Del Departamento de Nariño. Rev. Prod. Nat. 2022, 5 (2), 176–178. 10.3407/rpn.v5i2.6927. [DOI] [Google Scholar]
- Villamarin-Raad D. A.; Lozano-Puentes H. S.; Chitiva L. C.; Costa G. M.; Diaz-Gallo S. A.; Diaz-Ariza L. A. Changes in Phenolic Profile and Total Phenol and Total Flavonoid Contents of Guadua Angustifolia Kunth Plants under Organic and Conventional Fertilization. ACS Omega 2023, 8, 41223–41231. 10.1021/acsomega.3c04579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Puentes H. S.; Sánchez-Matiz J. J.; Ruiz-Sanchez E.; Costa G. M.; Díaz-Ariza L. A. Guadua Angustifolia Kunth Leaves as a Source for Bioactive Phenolic Compounds: Optimization of Ultrasound-Assisted Extraction Using Response Surface Methodology and Antioxidant Activities. Heliyon 2023, 9 (12), e22445 10.1016/j.heliyon.2023.e22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Matiz J. J.; Lozano-Puentes H. S.; Villamarín-Raad D. A.; Díaz-Gallo S. A.; Díaz-Ariza L. A. Dynamic of Phenolic Compounds in Guadua Angustifolia Kunth under Chemical, Organic, and Biological Fertilization. Agronomy 2023, 13 (11), 2782. 10.3390/agronomy13112782. [DOI] [Google Scholar]
- Lozano-Puentes S. H.; Sánchez-Matiz J.; Ruiz-Sanchez E.; Costa G. M.; Díaz-Ariza L. A. Guadua Angustifolia Kunth Leaves as a Source for Bioactive Phenolic Compounds: Optimization of Ultrasound-Assisted Extraction Using Response Surface Methodology and Antioxidant Activities. SSRN Heliyon 2023, 9, e22445. 10.1016/j.heliyon.2023.e22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey W.; Dasi J.; Rao N.; Gottumukkala K. Anti-Inflammatory Activity of Methanolic Extract of Bambusa Vulgaris Leaves. Int. J. Green Pharm. 2009, 3 (3), 234–238. 10.4103/0973-8258.56282. [DOI] [Google Scholar]
- Tundis R.; Augimeri G.; Vivacqua A.; Romeo R.; Sicari V.; Bonofiglio D.; Loizzo M. R. Anti-Inflammatory and Antioxidant Effects of Leaves and Sheath from Bamboo (Phyllostacys Edulis J. Houz). Antioxidants 2023, 12 (6), 1239. 10.3390/antiox12061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitiva L. C.; Santamaría-Torres M. A.; Rezende-Teixeira P.; Borlot J. R. P. de O.; Romagna R. de A.; Londoño X.; Kitagawa R. R.; Costa-Lotufo L. V.; Prieto-Rodríguez J. A.; Castro-Gamboa I.; Costa G. M. Uncovering Metabolic Alterations in HCT-116 Colon Cancer Cells upon Exposure to Bamboo Leaf Extract Obtained from Guadua Incana Londoño. Molecules 2024, 29 (13), 2985. 10.3390/molecules29132985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H.; Blandt S.; Zgainski E.; Springer-Verlang. Plant Drug Analysis: A Thin Layer Chromatography Atlas, 1st ed.; Springer Science & Business Media: Munchen, Germany, 1996. [Google Scholar]
- Ma J.; Yin Y.-M.; Liu H.-L.; Xie M.-X. Interactions of Flavonoids with Biomacromolecules. Curr. Org. Chem. 2011, 15 (15), 2627–2640. 10.2174/138527211796367345. [DOI] [Google Scholar]
- Falcone Ferreyra M. L.; Rius S. P.; Casati P. Flavonoids: Biosynthesis, Biological Functions, and Biotechnological Applications. Front. Plant Sci. 2012, 3 (SEP), 222. 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holser R. A. Principal Component Analysis of Phenolic Acid Spectra. ISRN Spectrosc. 2012, 2012, 1–5. 10.5402/2012/493203. [DOI] [Google Scholar]
- Belay A.; Gholap A. V. Characterization and Determination of Chlorogenic Acids (CGA) in Coffee Beans by UV-Vis Spectroscopy. African J. Pure Appl. Chem. 2009, 3 (11), 234–240. [Google Scholar]
- Jiang L.; Belwal T.; Huang H.; Ge Z.; Limwachiranon J.; Zhao Y.; Li L.; Ren G.; Luo Z. Extraction and Characterization of Phenolic Compounds from Bamboo Shoot Shell under Optimized Ultrasonic-Assisted Conditions: A Potential Source of Nutraceutical Compounds. Food Bioprocess Technol. 2019, 12, 1741–1755. 10.1007/s11947-019-02321-y. [DOI] [Google Scholar]
- Cheng Y.; Wan S.; Yao L.; Lin D.; Wu T.; Chen Y.; Zhang A.; Lu C. Bamboo Leaf: A Review of Traditional Medicinal Property, Phytochemistry, Pharmacology, and Purification Technology. J. Ethnopharmacol. 2023, 306, 116166 10.1016/j.jep.2023.116166. [DOI] [PubMed] [Google Scholar]
- Gagliano J.; Altenhofen S.; Nabinger D. D.; Gusso D.; Kuhl-Silva J. M.; Anselmo-Moreira F.; Yamaguchi L. F.; Kato M. J.; Bonan C. D.; Furlan C. M. Are Brazilian Bamboo Species Helpful for Cognition and Memory?. Phytomedicine Plus 2022, 2 (1), 100183 10.1016/j.phyplu.2021.100183. [DOI] [Google Scholar]
- Ozarowski M.; Piasecka A.; Paszel-Jaworska A.; Chaves D. S. de A.; Romaniuk A.; Rybczynska M.; Gryszczynska A.; Sawikowska A.; Kachlicki P.; Mikolajczak P. L.; Seremak-Mrozikiewicz A.; Klejewski A.; Thiem B. Comparison of Bioactive Compounds Content in Leaf Extracts of Passiflora Incarnata, P. Caerulea and P. Alata and in Vitro Cytotoxic Potential on Leukemia Cell Lines. Rev. Bras. Farmacogn. 2018, 28 (2), 179–191. 10.1016/j.bjp.2018.01.006. [DOI] [Google Scholar]
- Troalen L. G.; Phillips A. S.; Peggie D. A.; Barran P. E.; Hulme A. N. Historical Textile Dyeing with Genista Tinctoria L.: A Comprehensive Study by UPLC-MS/MS Analysis. Anal. Methods 2014, 6 (22), 8915–8923. 10.1039/C4AY01509F. [DOI] [Google Scholar]
- Morales Noreña N.; Sanchez Vallejo L.. Contribución Al Estudio Fitoquímico de Las Hojas de Guadua Angustifolia Kunth; Universidad del Quindío, 2008. [Google Scholar]
- Van Hoyweghen L.; Beer T. D.; Deforce D.; Heyerick A. Phenolic Compounds and Anti-oxidant Capacity of Twelve Morphologically Heterogeneous Bamboo Species. Phytochem. Anal. 2012, 23 (5), 433–443. 10.1002/pca.1377. [DOI] [PubMed] [Google Scholar]
- Dos Reis Luz L.; Porto D. D.; Castro C. B.; Silva M. F. S.; de Godoy Alves Filho E.; Canuto K. M.; de Brito E. S.; Becker H.; do Ó; Pessoa C.; Zocolo G. J.; Dos Reis Luz L.; Porto D. D.; Castro C. B.; Silva M. F. S.; de Godoy Alves Filho E.; Canuto K. M.; de Brito E. S.; Becker H.; do Ó; Pessoa C.; Zocolo G. J.; et al. Metabolomic Profile of Schinopsis Brasiliensis via UPLC-QTOF-MS for Identification of Biomarkers and Evaluation of Its Cytotoxic Potential. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1099 (April), 97–109. 10.1016/j.jchromb.2018.09.019. [DOI] [PubMed] [Google Scholar]
- Ni Q.; Xu G.; Wang Z.; Gao Q.; Wang S.; Zhang Y. Seasonal Variations of the Antioxidant Composition in Ground Bamboo Sasa Argenteastriatus Leaves. Int. J. Mol. Sci. 2012, 13 (2), 2249–2262. 10.3390/ijms13022249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. H.; Jo H. G.; Yang J. H.; Ki S. H.; Shin H. J. Antioxidative and Anti-Melanogenic Activities of Bamboo Stems (Phyllostachys Nigra Variety Henosis) via PKA/CREB-Mediated MITF Downregulation in B16F10 Melanoma Cells. Int. J. Mol. Sci. 2018, 19 (2), 409. 10.3390/ijms19020409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A.; Zhu Q.; Tan H.; Horiba H.; Ohnuki K.; Mori Y.; Yamauchi R.; Ishikawa H.; Iwamoto A.; Kawahara H.; Shimizu K. Biological Activities and Phytochemical Profiles of Extracts from Different Parts of Bamboo (Phyllostachys Pubescens). Molecules 2014, 19 (6), 8238–8260. 10.3390/molecules19068238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer M.; Kiese S.; Tybussek T.; Herfellner T.; Schneider F.; Schweiggert-Weisz U.; Eisner P. Radical Scavenging Mechanisms of Phenolic Compounds: A Quantitative Structure-Property Relationship (QSPR) Study. Front. Nutr. 2022, 9, 882458. 10.3389/fnut.2022.882458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeb A.Concept, Mechanism, and Applications of Phenolic Antioxidants in Foods. J. Food Biochem. 2020, 44 ( (9), ). 10.1111/jfbc.13394. [DOI] [PubMed] [Google Scholar]
- Nordin M. L.; Abdul Kadir A.; Zakaria Z. A.; Abdullah R.; Abdullah M. N. H. In Vitro Investigation of Cytotoxic and Antioxidative Activities of Ardisia Crispa against Breast Cancer Cell Lines, MCF-7 and MDA-MB-231. BMC Complement. Altern. Med. 2018, 18 (1), 1–10. 10.1186/s12906-018-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharum Z.; Akim A. M.; Taufiq-Yap Y. H.; Hamid R. A.; Kasran R. In Vitro Antioxidant and Antiproliferative Activities of Methanolic Plant Part Extracts of Theobroma Cacao. Molecules 2014, 19 (11), 18317–18331. 10.3390/molecules191118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Zhu R.; Wu B.; Hu Y.; Yu W. Fabrication, Material Properties, and Application of Bamboo Scrimber. Wood Sci. Technol. 2015, 49 (1), 83–98. 10.1007/s00226-014-0683-7. [DOI] [Google Scholar]
- Akinlabi E. T.; Anane-Fenin K.; Akwada D. R.. Applications of Bamboo. In Bamboo: the multipurpose plant; Springer International Publishing, 2017; pp 179–219. 10.1007/978-3-319-56808-9_5. [DOI] [Google Scholar]
- Emamverdian A.; Ding Y.; Ranaei F.; Ahmad Z. Application of Bamboo Plants in Nine Aspects. Sci. World J. 2020, 2020, 1–9. 10.1155/2020/7284203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera O. M.; González L. M.; Cortés Y. J.; Camargo J. C. Caracterización Fitoquímica, Determinación Del Contenido de Lignina y La Actividad Antioxidante de Los Culmos de Guadua Angustifolia Kunth. Rev. Fac. Ciencias Básicas 2015, 11 (2), 124–135. 10.18359/rfcb.1301. [DOI] [Google Scholar]
- Mejía Gallón A.; Ramírez López G.; Palacio Torres H.; López C. Identificación de Compuestos Volátiles Del Vinagre de Guadua Angustifolia Kuth. Rev. Cuba. Plantas Med. 2011, 2 (16), 190–201. [Google Scholar]
- Moreno P. R. H.; De Oliveira D. C. S.; Santos F. T.; Nunes F. A.; Bílek T.; Tereza Grombone-Guaratini M. PP11. Chemical Composition and Antimicrobial Properties of the Essential Oils of Two Guadua Kunth Species (Poaceae-Bambusoideae). Chemistry and Technology 2018, 16, 75. [Google Scholar]
- Corrêa J. K. C.; Moreno P. R. H. Atividade Antioxidante e Anti-Tirosinase Nas Folhas de Guadua Angustifolia Var. Bicolor Londoño (Bambusoideae:Poaceae). 10o Workshop do Mestrado Profissional – Universidade de São Paulo, Instituto de Química 2022, 42–49. 10.5151/9786555502084-07. [DOI] [Google Scholar]
- Caetano Corrêa J. K.; Hrihorowitsch Moreno P. R. Isolamento e Identificação de Compostos Com Atividade Antioxidante e Anti-Tirosinase Em Extratos de Guadua Angustifolia Var. Bicolor Londoño. 29o SIICUSP 2020, 1–2. [Google Scholar]
- Panche A. N.; Diwan A. D.; Chandra S. R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, 1–15. 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Baker D. H. An Ethnopharmacological Review on the Therapeutical Properties of Flavonoids and Their Mechanisms of Actions: A Comprehensive Review Based on up to Date Knowledge. Toxicol. Reports 2022, 9, 445–469. 10.1016/j.toxrep.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Pandey A. K.. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013. 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N.; Guo J.; Chen S.; Yuan X.; Zhang T.; Ding Y. Antioxidant and Compositional HPLC Analysis of Three Common Bamboo Leaves. Molecules 2020, 25 (2), 409. 10.3390/molecules25020409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S.; Lee H. C.; Jo J. S.; Jung J. Y.; Ha Y. L.; Yang J. K. Evaluation of Antioxidant and Anticancer Activity of Steam Extract from the Bamboo Species. J. Korean Wood Sci. Technol. 2014, 42 (5), 543–554. 10.5658/WOOD.2014.42.5.543. [DOI] [Google Scholar]
- Arboleda Echavarría C.; Jaramillo Yepes F.; Palacio Torres H. Determinación Del Potencial Antioxidante En Extractos de Vinagre Guadua Angustifolia Kunth Para Aplicaciones Alimenticias. Rev. Cuba. plantas Med. 2012, 17 (4), 330–342. [Google Scholar]
- Van Hoyweghen L.; Deforce D.; Heyerick A. Antioxidant Capacity and Secondary Metabolites of 12 Bamboo Species. I International Symposium on Genetic Resources of Bamboos and Palms and III International Symposium on Ornamental Palms 1003 2013, 107–112. 10.17660/ActaHortic.2013.1003.13. [DOI] [Google Scholar]
- Yu Y.; Li Z.; Cao G.; Huang S.; Yang H. Bamboo Leaf Flavonoids Extracts Alleviate Oxidative Stress in HepG2 Cells via Naturally Modulating Reactive Oxygen Species Production and Nrf2-Mediated Antioxidant Defense Responses. J. Food Sci. 2019, 84 (6), 1609–1620. 10.1111/1750-3841.14609. [DOI] [PubMed] [Google Scholar]
- Gagliano J.; Anselmo-Moreira F.; Sala-Carvalho W. R.; Furlan C. M. What Is Known about the Medicinal Potential of Bamboo?. Adv. Tradit. Med. 2022, 22 (3), 467–495. 10.1007/s13596-020-00536-5. [DOI] [Google Scholar]
- Kamble S.; Gacche R. Evaluation of Anti-Breast Cancer, Anti-Angiogenic and Antioxidant Properties of Selected Medicinal Plants. Eur. J. Integr. Med. 2019, 25, 13–19. 10.1016/j.eujim.2018.11.006. [DOI] [Google Scholar]
- Ko H. C.; Lee J. Y.; Jang M. G.; Song H.; Kim S. J. Seasonal Variations in the Phenolic Compounds and Antioxidant Activity of Sasa Quelpaertensis. Ind. Crops Prod. 2018, 122, 506–512. 10.1016/j.indcrop.2018.06.031. [DOI] [Google Scholar]
- Crespo Y. A.; Bravo Sánchez L. R.; Quintana Y. G.; Cabrera A. S. T.; Bermúdez del Sol A.; Mayancha D. M. G.. Evaluation of the Synergistic Effects of Antioxidant Activity on Mixtures of the Essential Oil from Apium Graveolens L.; Thymus Vulgaris L.; Coriandrum Sativum L.. Using Simplex-Lattice Design. Heliyon 2019, 5 ( (6), ). 10.1016/j.heliyon.2019.e01942. [DOI] [PMC free article] [PubMed]
- Zhang L.; Zhu C.; Liu X.; Su E.; Cao F.; Zhao L. Study on Synergistic Antioxidant Effect of Typical Functional Components of Hydroethanolic Leaf Extract from Ginkgo Biloba In Vitro. Molecules 2022, 27 (2), 439. 10.3390/molecules27020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sak K. Cytotoxicity of Dietary Flavonoids on Different Human Cancer Types. Pharmacogn. Rev. 2014, 8 (16), 122–146. 10.4103/0973-7847.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. Y. Sasa Quelpaertensis Nakai Extract Induces P53-Independent Apoptosis via the Elevation of Nitric Oxide Production in Human HCT116 Colon Cancer Cells. Oncol. Lett. 2020, 19 (4), 3027–3034. 10.3892/ol.2020.11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukovic N. L.; Obradovic A. D.; Vukic M. D.; Jovanovic D.; Djurdjevic P. M. Cytotoxic, Proapoptotic and Antioxidative Potential of Flavonoids Isolated from Propolis against Colon (HCT-116) and Breast (MDA-MB-231) Cancer Cell Lines. Food Res. Int. 2018, 106, 71–80. 10.1016/j.foodres.2017.12.056. [DOI] [PubMed] [Google Scholar]
- Tahir A. T.; Fatmi Q.; Nosheen A.; Imtiaz M.; Khan S. Metabolomic Approaches in Plant Research. Essentials of Bioinformatics, Volume III: In Silico Life Sciences: Agriculture 2019, 109–140. 10.1007/978-3-030-19318-8_7. [DOI] [Google Scholar]
- Aron A. T.; Gentry E. C.; McPhail K. L.; Nothias L. F.; Nothias-Esposito M.; Bouslimani A.; Petras D.; Gauglitz J. M.; Sikora N.; Vargas F.; van der Hooft J. J. J.; Ernst M.; Kang K. B.; Aceves C. M.; Caraballo-Rodríguez A. M.; Koester I.; Weldon K. C.; Bertrand S.; Roullier C.; Sun K.; Tehan R. M.; Boya P C. A.; Christian M. H.; Gutiérrez M.; Ulloa A. M.; Tejeda Mora J. A.; Mojica-Flores R.; Lakey-Beitia J.; Vásquez-Chaves V.; Zhang Y.; Calderón A. I.; Tayler N.; Keyzers R. A.; Tugizimana F.; Ndlovu N.; Aksenov A. A.; Jarmusch A. K.; Schmid R.; Truman A. W.; Bandeira N.; Wang M.; Dorrestein P. C. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15 (6), 1954–1991. 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- Re R.; Pellegrini N.; Proteggente A.; Pannala A.; Yang M.; Rice-Evans C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free Radic. Biol. Med. 1999, 26 (9–10), 1231. 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Saeed N.; Khan M. R.; Shabbir M. Antioxidant Activity, Total Phenolic and Total Flavonoid Contents of Whole Plant Extracts Torilis Leptophylla L. BMC Complement. Altern. Med. 2012, 12, 221. 10.1186/1472-6882-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65 (1–2), 55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Pang Z.; Zhou G.; Ewald J.; Chang L.; Hacariz O.; Basu N.; Xia J. Using MetaboAnalyst 5.0 for LC–HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. Nat. Protoc. 2022, 17 (8), 1735–1761. 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- Blaženović I.; Kind T.; Ji J.; Fiehn O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.