Abstract
Background
Neonatal Opioid Withdrawal Syndrome (NOWS) is a consequence of in-utero exposure to prenatal maternal opioids, resulting in the manifestation of symptoms like irritability, feeding problems, tremors, and withdrawal signs. Opioid use disorder (OUD) during pregnancy can profoundly impact both mother and fetus, disrupting fetal brain neurotransmission and potentially leading to long-term neurological, behavioral, and vision issues, and increased infant mortality. Drug resistance complicates OUD and NOWS treatment, with protein kinase regulation of drug transporters not fully understood.
Methods
DNA methylation levels of ATP-binding cassette (ABC) and solute carrier (SLC) drug transporters, along with protein kinase C (PKC) genes, were assessed in 96 placental samples using the Illumina Infinium MethylationEPIC array (850K). Samples were collected from three distinct groups: 32 mothers with infants prenatally exposed to opioids who needed pharmacological intervention for NOWS, 32 mothers with prenatally opioid-exposed infants who did not necessitate NOWS treatment, and 32 mothers who were not exposed to opioids during pregnancy.
Results
We identified 69 significantly differentially methylated SLCs, with 24 hypermethylated and 34 hypomethylated, and 11 exhibiting both types of methylation changes including SLC13A3, SLC15A2, SLC16A11, SLC16A3, SLC19A2, and SLC26A1. We identified methylation changes in 11 ABC drug transporters (ABCA1, ABCA12, ABCA2, ABCB10, ABCB5, ABCC12, ABCC2, ABCC9, ABCE1, ABCC7, ABCB3): 3 showed hypermethylation, 3 hypomethylation, and 5 exhibited both. Additionally, 7 PKC family genes (PRKCQ, PRKAA1, PRKCA, PRKCB, PRKCH, PRKCI, and PRKCZ) showed methylation changes. These genes are associated with 13 pathways involved in NOWS, including ABC transporters, bile secretion, pancreatic secretion, insulin resistance, glutamatergic synapse, and gastric acid secretion.
Conclusion
We report epigenetic changes in PKC-related regulation of drug transporters, which could improve our understanding of clinical outcomes like drug resistance, pharmacokinetics, drug-drug interactions, and drug toxicity, leading to maternal relapse and severe NOWS. Novel drugs targeting PKC pathways and transporters may improve treatment outcomes for OUD in pregnancy and NOWS.
Keywords: drug transporters, protein kinases C, biomarker, opioid use, neonatal opioid withdrawal syndrome SLC transporters ABC transporters, bile secretion, pancreatic secretion, insulin resistance
Introduction
Opioid use disorder (OUD) represents a significant global health challenge. Maternal opioid misuse during pregnancy can result in Neonatal Opioid Withdrawal Syndrome (NOWS), which poses severe risks to newborns, including irritability, feeding difficulties, tremors, and withdrawal symptoms. These effects may extend into later life, impacting neurodevelopment, behavior, mental health, and potentially vision-related issues (Anbalagan and Mendez, 2023). Genetic and epigenetic variations in opioid receptors, metabolic enzymes, regulatory proteins, and transporters significantly influence susceptibility to NOWS (Metpally et al., 2019; Radhakrishna et al., 2021a,b, 2023a,b). Despite effective treatments for OUD and NOWS, drug resistance persists as a significant challenge (Wang X. et al., 2019). The roles of transporter proteins and signal transduction enzymes in drug resistance related to OUD and NOWS are not well understood.
Transporters are essential for pharmacokinetics and pharmacodynamics, impacting drug interactions, adverse effects, and body homeostasis by facilitating the transfer of ions, amino acids, sugars, and drugs across cell membranes (Zhang, 2018; Peng et al., 2020; Carbo and Rodriguez, 2023). Drug resistance arises from various mechanisms, with drug transporters and metabolizers significantly impacting drug efficacy (Mansoori et al., 2017; Kawano et al., 2022). Dysregulation in these genes can alter neurotransmitter levels and signaling pathways, contributing to opioid dependence and addiction (Mistry et al., 2014). Transporters, commonly situated on the plasma membrane, can be divided into solute carriers (SLC) and ATP-binding cassette (ABC) transporters. Both solute carrier (SLC) and ATP-binding cassette (ABC) drug transporters can be regulated by PKCs-related signaling pathways (Mayati et al., 2017; Puris et al., 2023). The SLC transporters include over 400 members across 52 families (Hediger et al., 2013; Baril et al., 2023). Most SLC transporters are responsible for the uptake of small molecules (including nutrients and xenobiotics), but a few SLCs act as both influx and efflux transporters. They are expressed in organs like the intestines, liver, and kidneys, which are involved in drug absorption, metabolism, and elimination (Brecht et al., 2020).
ABC transporters encompass a diverse group of 48 known proteins categorized into seven primary types: ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, and ABCG (Dean et al., 2001). ABC transporters use ATP hydrolysis to move a variety of substances across cell membranes. ABC transporters in the human placenta transport endogenous compounds and protect the fetus from exogenous substances like therapeutic agents, drugs of abuse, and other xenobiotics (Joshi et al., 2016). These transporters are linked to diseases like cystic fibrosis (ABCC7/CFTR), Tangier disease, cardiovascular disease (ABCA1), retinitis pigmentosa (ABCA4), and more (Vasiliou et al., 2009).
Protein Kinase C (PKC) is a family of serine/threonine kinases that play crucial roles in the proliferation, differentiation, survival, migration, invasion, apoptosis, and anticancer drug resistance of cancer cells (Kawano et al., 2022). Dysregulated PKC can cause abnormal phosphorylation and misregulation of SLC and ABC transporters, disrupting their activity and gene expression. This affects drug absorption, potentially leading to resistance or toxicity, and interferes with transporter trafficking and protein interactions, further disrupting cellular transport processes (Alves et al., 2022).
Previous reports highlight the significant roles of transporter genes such as SLC6A3 (DAT1), which regulates dopamine reuptake affecting reward pathways crucial for addiction development, and SLC6A4 (SERT), which controls serotonin levels impacting mood and emotional stability, in neurotransmitter regulation linked to addiction development and responses to opioids (Grover et al., 2020; Yuferov et al., 2021). PKC-related signaling pathways can regulate both SLC and ABC drug transporters. In the context of OUD, PKCs play a significant role by influencing addiction, tolerance, dependence, withdrawal, and drug-seeking behavior (Lee and Messing, 2008).
Our study used genome-wide methylation analysis to explore whether epigenetic modifications of placental PKC and drug transporters in infants exposed to prenatal opioids could predict NOWS. We found significant methylation changes in multiple PKCs and drug transporter genes involved in NOWS development. Targeting these transporters and PKC could lead to new therapeutic approaches for treating opioid addiction and managing NOWS.
Materials and methods
The research study received approval from the Institutional Review Board of Beaumont Health System, Royal Oak, MI, USA (HIC#: 2019-086). Pregnant women were identified retrospectively through chart review at William Beaumont Hospital, Royal Oak, MI. Informed consent was not required for this study as it solely involved the collection of discarded placental tissues from the subjects, along with obtaining limited de-identified, data from the hospital medical records. We collected demographic and clinical-pathological data, including age, sex, ethnicity, gestational age, and history of drug exposure (Radhakrishna et al., 2021b). Patients were diagnosed according to the assessment criteria outlined in the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) (Hasin et al., 2013).
The sample details and methodology have been documented in our prior publication (Radhakrishna et al., 2021b). To summarize, ninety-six formalin-fixed, paraffin-embedded (FFPE) placental tissue biopsies were collected and processed. These tissue samples were categorized into three groups: Group 1 comprised 32 newborns prenatally exposed to opioids requiring treatment for Neonatal Opioid Withdrawal Syndrome (NOWS) (+Opioids/+NOWS), Group 2 included 32 newborns prenatally exposed to opioids not requiring treatment for NOWS (+Opioids/-NOWS), and Group-3 served as the control group consisting of newborns with no prenatal opioid exposure and no NOWS (-Opioids/-NOWS, control). The mean gestational age at delivery (in weeks) was 37.94 (SD = 3.16) for Group 1, 37.49 (SD = 2.96) for Group 2, and 38.09 (SD = 3.37) for Group 3. NOWS diagnosis (P96.1) was determined by neonatologists based on clinical criteria. Infants born to mothers with a history of opioid or illicit drug use were observed in the inpatient unit for 4–5 days to detect signs of NOWS. Scoring was conducted using the Finnegan Neonatal Abstinence Scoring Tool (FNAST). Postpartum nurses and/or NICU nurses conducted the scoring process. If the scores indicated a need for pharmacologic treatment according to set criteria, the infant was moved to the NICU for continued monitoring, scoring, and treatment. Parental involvement was encouraged to enhance non-pharmacologic interventions as the primary approach before and during treatment, regardless of whether the infant received pharmacologic treatment. The initiation of pharmacologic management with morphine was determined using the Finnegan Neonatal Abstinence Scoring Tool (FNAST). The analysis comprised four comparisons: I. (+Opioids/+NOWS) vs. (+Opioids/-NOWS); II. (+Opioids/+NOWS) + (+Opioids/-NOWS) vs. (-Opioids/-NOWS, control); III. (+Opioids/+NOWS) vs. (-Opioids/-NOWS, control); IV. (+Opioids/-NOWS) vs. (-Opioids/-NOWS, control), calculated for each unique differentially methylated CpG locus.
Methylation analysis
All participant mothers with OUD were of European-American ancestry. Placental specimens were collected from the maternal side, approximately 2 cm from the site of umbilical cord insertion. Generally, eight to ten 10 mm curls of formalin-fixed paraffin-embedded (FFPE) placental tissue from each block were used for DNA preparation. The extensive discussion of Illumina Infinium MethylationEPIC array BeadChip (850K) assay (Illumina, Inc., San Diego, CA, USA) has been previously referenced (Radhakrishna et al., 2021b). These state-of-the-art arrays boast coverage of over 850,000 CpG sites across the genome, offering unparalleled single-nucleotide precision. We obtained data on differentially methylated CG dinucleotides from previously unpublished DNA methylation datasets concerning SLC transporters, ABC drug transporters, and PKC family genes (Radhakrishna et al., 2021b). The information on SLC transporters was sourced from https://slc.bioparadigms.org/, ABC transporters from http://www.genenames.org, and the PKC gene family was obtained from https://rgd.mcw.edu/rgdweb/homepage/.
Statistical and bioinformatic analysis
Before analysis, CpG-probes with missing ß-values were excluded. Differential methylation was evaluated by comparing ß-values for cytosines at each CpG locus between NOWS and controls. Probes linked to sex chromosomes, non-specific probes, and those targeting CpG sites within 10 bp of SNPs were removed to mitigate confounding factors. SNPs with a minor allele frequency ≤ 0.05 were considered for further analysis (Liu et al., 2013; Wilhelm-Benartzi et al., 2013; Zhang et al., 2013).
The p-value for methylation differences between the case and control groups at each locus was computed as outlined previously (Vishweswaraiah et al., 2019; Radhakrishna et al., 2021b). CpG sites showing significant differential methylation between NOWS, and controls were identified using predefined cutoff criteria of FDR p < 0.05 and retained for further analysis. Raw and FDR-adjusted p-values for multiple testing (using the Benjamini-Hochberg test) were calculated. The area under the receiver operating characteristic curve (AUC-ROC) for combinations of loci was determined using the ‘ROCR’ package (v3.5.0) in the ‘R’ program, based on methylation levels at the most significantly differentially methylated CpG loci.
Network interaction analysis using STRING
Protein-protein interaction analysis was conducted using the STRING database (version 12.0), available at http://string-db.org, following the identification of differently methylated genes with an FDR p-value < 0.05. The database compiles known and predicted protein-protein interactions, including both physical and functional associations. For the analysis, all interaction sources were utilized: text mining from scientific literature, experimental data, aggregated information from curated databases, co-expression data, genomic context predictions (neighbourhood), evidence from gene fusion events, and phylogenetic tree-based co-occurrence. To ensure the interactions were significant while maintaining a comprehensive dataset, a medium confidence score threshold of 0.400 was applied. This method aimed to create a balanced protein interaction network, minimizing the inclusion of false positives yet allowing for the discovery of potentially relevant associations that may elucidate the complex mechanisms underlying NOWS.
Gene ontology (GO) and KEGG pathway analyses
We conducted gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses on the same pool of differentially methylated genes, identified with an FDR p-value < 0.05. This was aimed at unveiling their biological relevance and participation in dysregulated signaling pathways associated with drug resistance. For GO analysis, the identified genes were categorized into three main ontologies: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). This classification provided insights into the functional roles of the genes, their cellular localization, and the molecular activities they may influence. KEGG analysis was conducted to integrate the differently methylated genes into known genetic pathways, providing an understanding of how alterations in gene methylation could affect specific biological pathways and processes. This integration helps to identify which pathways are potentially altered in the context of NOWS. Both GO and KEGG analyses utilized ‘clusterProfiler’, an R package designed for statistical analysis and visualization of functional profiles for genes and gene clusters. It facilitates the comparison of biological themes among gene clusters, enhancing the interpretation of high-throughput genomics data.
Heatmaps
Differential methylation patterns of CpG sites, especially those associated with dysregulated transporters such as ABC genes, SLC genes, and PKC genes related to pain, were used to generate a heatmap using the ComplexHeatmap (v1.6.0) package in the R environment (v3.2.2). Sample hierarchical clustering was conducted using Ward distance (Gu, 2015).
Results
Analysis of transporters & PKCs in NOWS
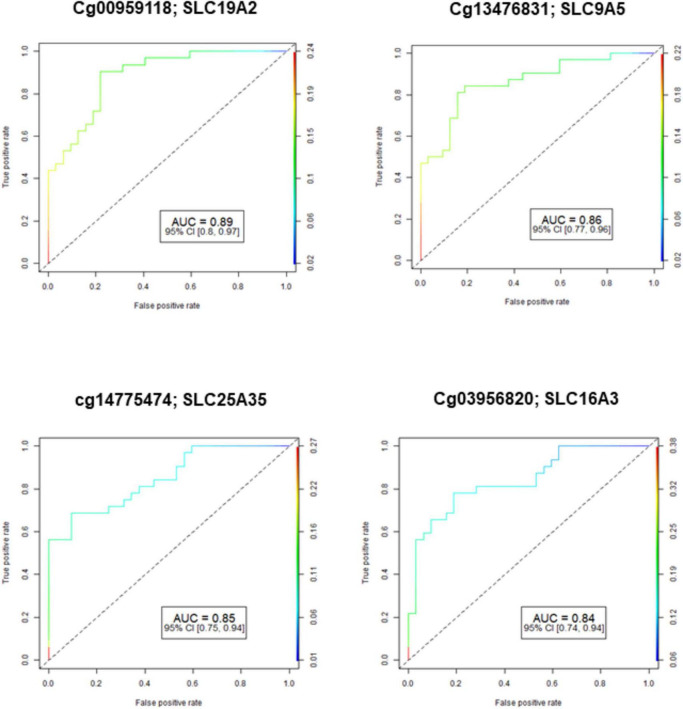
The demographic characteristics of both NOWS and control groups were examined, indicating no significant disparities. This data has been previously published (Radhakrishna et al., 2021a). Figure 1 shows the Receiver Operating Characteristic (ROC) curve analysis of four significantly differently methylated CpGs in ABC, SLC, and PKC genes (FDR p ≤ 0.05). The results of four different analyses showed that Analysis I identified 8 dysregulated ABC genes, 38 SLC genes, and 2 PKC genes when comparing individuals with (+Opioids/+NOWS) to those with (+Opioids/-NOWS) (Supplementary Table 1). Analysis II compared individuals with (+Opioids/+NOWS) and (+Opioids/-NOWS) against those with (-Opioids/-NOWS, control), identifying 2 ABC genes, 15 SLC genes, and 4 PKC genes (Supplementary Table 2). Analysis III showed differential regulation of 1 ABC gene, 29 SLC genes, and 1 PKC gene in individuals with (+Opioids/+NOWS) compared to those with (-Opioids/-NOWS, control) (Supplementary Table 3). Analysis IV revealed differential regulation of 6 ABC genes, 18 SLC genes, and 5 PKC genes in individuals with (+Opioids/-NOWS) compared to those with (-Opioids/-NOWS, control) (Supplementary Table 4).
FIGURE 1.
Receiver operating characteristic (ROC) curve analysis of methylated CpGs in ABC, SLC, and PKC genes (FDR-p ≤ 0.05) that had good diagnostic accuracy with CpGs AUC ≥ 0.80-0.89). AUC: Area Under the Receiver Operating Characteristics Curve; 95% CI: 95% Confidence Interval. Confidence intervals (CI) in parentheses show bounds.
Across all four analyses, multiple genes were identified, totaling 87, showing significant methylation changes in genes linked to drug transporters and PKCs. Among them, 69 SLC transporters exhibit various methylation patterns: 24 are hypermethylated, 34 are hypomethylated, and 11 display both hypo- and hypermethylation. We observed 11 ABC drug transporters: ABCA1, ABCA12, ABCA2, ABCB10, ABCB5, ABCC12, ABCC2, ABCC9, ABCE1, CFTR (ABCC7), and TAP2 (ABCB3) that were differentially methylated. Among these, ABCA2, ABCC12, and ABCE1 were found to have hypomethylation, while ABCC2, ABCC9, and TAP2 (ABCB3) exhibited hypermethylation. Notably, ABCA1, ABCA12, ABCB10, ABCB5, and CFTR (ABCC7) genes showed both hypo- and hypermethylation tendencies. Furthermore, variations in methylation were noted in 7 genes of the protein kinase C family, including hypomethylated genes PRKAA1, PRKCB, and PRKCH, hypermethylated gene PRKCA, and genes PRKCI, PRKCQ, and PRKCZ displaying both hypo- and hypermethylation (Table 1).
TABLE 1.
A comprehensive detail of CpG targets that exhibit significant differential methylation in NOWS.
| Target ID | Genes | Location | p-Value | FDR p-Value |
% Methylation | AUC | CI | Gene detail |
|||
| Cases | Control | Change | Lower | Upper | |||||||
| cg10434274 | ABCA1 | 9q31.1 | 4.14763E-09 | 0.003587703 | 60.83 | 68.83 | −8.00 | 0.75 | 0.63 | 0.87 | ABC |
| cg14299235 | ABCA1 | 9q31.1 | 5.50932E-10 | 0.000476556 | 67.75 | 59.84 | 7.91 | 0.68 | 0.55 | 0.81 | ABC |
| cg00972111 | ABCA12 | 2q35 | 4.72015E-14 | 4.08293E-08 | 78.32 | 70.24 | 8.08 | 0.73 | 0.61 | 0.85 | ABC |
| cg02820283 | ABCA2 | 9q34.3 | 2.81873E-08 | 0.02438205 | 72.63 | 78.14 | −5.51 | 0.71 | 0.60 | 0.83 | ABC |
| cg21045171 | ABCB10 | 1q42.13 | 2.51239E-11 | 2.17322E-05 | 85.30 | 79.30 | 6.00 | 0.79 | 0.68 | 0.90 | ABC |
| cg21045171 | ABCB10 | 1q42.13 | 2.66472E-08 | 0.023049813 | 78.01 | 83.60 | −5.59 | 0.75 | 0.64 | 0.87 | ABC |
| cg08888968 | ABCB5 | 7p21.1 | 2.90402E-16 | 2.51198E-10 | 68.08 | 77.76 | −9.69 | 0.76 | 0.64 | 0.88 | ABC |
| cg08888968 | ABCB5 | 7p21.1 | 9.25029E-39 | 8.0015E-33 | 80.18 | 70.50 | 9.68 | 0.75 | 0.63 | 0.87 | ABC |
| cg21723907 | ABCC12 | 16q12.1 | 8.67446E-09 | 0.007503411 | 74.56 | 80.83 | −6.28 | 0.78 | 0.67 | 0.89 | ABC |
| cg01614760 | ABCC2 | 10q24.2 | 2.00132E-08 | 0.017311439 | 43.41 | 35.80 | 7.60 | 0.74 | 0.62 | 0.86 | ABC |
| cg01909678 | ABCC9 | 12p12.1 | 3.14029E-14 | 2.71635E-08 | 76.39 | 67.95 | 8.44 | 0.78 | 0.67 | 0.89 | ABC |
| cg26690672 | ABCE1 | 4q31.21 | 2.09819E-12 | 1.81494E-06 | 63.82 | 72.88 | −9.06 | 0.76 | 0.64 | 0.88 | ABC |
| cg26635219 | ABCC7/CFTR | 7q31.2 | 9.66969E-39 | 8.36428E-33 | 24.59 | 14.19 | 10.40 | 0.76 | 0.64 | 0.88 | ABC |
| cg22533025 | ABCC7/CFTR | 7q31.2 | 1.97867E-11 | 1.71155E-05 | 72.00 | 79.58 | −7.59 | 0.73 | 0.61 | 0.85 | ABC |
| cg00720839 | ABCB3/TAP2 | 6p21.32 | 6.28153E-09 | 0.005433526 | 80.38 | 74.32 | 6.06 | 0.64 | 0.50 | 0.78 | ABC |
| cg22826226 | SLC11A2 | 12q13.12 | 7.94208E-09 | 0.006869897 | 80.63 | 74.65 | 5.98 | 0.76 | 0.65 | 0.88 | SLC |
| cg10439765 | SLC12A5 | 20q13.12 | 2.61284E-08 | 0.022601057 | 15.53 | 21.81 | −6.27 | 0.70 | 0.57 | 0.82 | SLC |
| cg18756954 | SLC12A7 | 5p15.33 | 2.03828E-08 | 0.017631088 | 72.18 | 78.71 | −6.53 | 0.74 | 0.62 | 0.86 | SLC |
| cg15062310 | SLC12A9 | 7q22.1 | 5.60196E-08 | 0.048456948 | 14.05 | 9.28 | 4.77 | 0.78 | 0.66 | 0.89 | SLC |
| cg09192862 | SLC13A3 | 20q13.12 | 2.0586E-08 | 0.017806874 | 79.71 | 85.13 | −5.42 | 0.84 | 0.74 | 0.94 | SLC |
| cg02131891 | SLC15A2 | 3q13.33 | 2.47989E-09 | 0.002145104 | 78.88 | 84.68 | −5.80 | 0.83 | 0.73 | 0.94 | SLC |
| cg15639045 | SLC16A11 | 17p13.1 | 3.49989E-08 | 0.03027406 | 55.79 | 63.69 | −7.90 | 0.76 | 0.65 | 0.88 | SLC |
| cg03956820 | SLC16A3 | 17q25.3 | 4.63951E-09 | 0.004013176 | 17.56 | 12.01 | 5.55 | 0.84 | 0.74 | 0.94 | SLC |
| cg08296680 | SLC16A3 | 17q25.3 | 3.54873E-09 | 0.00306965 | 44.17 | 51.88 | −7.72 | 0.67 | 0.55 | 0.79 | SLC |
| cg13460167 | SLC17A5 | 6q13 | 9.83035E-10 | 0.000850325 | 56.82 | 65.43 | −8.61 | 0.78 | 0.67 | 0.90 | SLC |
| cg24865549 | SLC17A5 | 6q13 | 2.09993E-08 | 0.018164393 | 85.53 | 80.50 | 5.04 | 0.72 | 0.60 | 0.85 | SLC |
| cg02624701 | SLC17A7 | 19q13.33 | 1.26807E-08 | 0.010968811 | 30.60 | 23.62 | 6.98 | 0.71 | 0.58 | 0.84 | SLC |
| cg12285003 | SLC17A9 | 20q13.33 | 3.10992E-09 | 0.002690084 | 71.71 | 78.63 | −6.91 | 0.83 | 0.73 | 0.93 | SLC |
| cg12285003 | SLC17A9 | 20q13.33 | 1.08447E-08 | 0.00938070 | 73.03 | 78.63 | −5.60 | 0.78 | 0.67 | 0.88 | SLC |
| cg00959118 | SLC19A2 | 1q24.2 | 7.31371E-10 | 0.000632636 | 17.10 | 11.39 | 5.71 | 0.89 | 0.80 | 0.97 | SLC |
| cg09726804 | SLC19A3 | 2q36.3 | 8.28861E-09 | 0.007169646 | 73.34 | 66.44 | 6.90 | 0.73 | 0.61 | 0.86 | SLC |
| cg03989758 | SLC1A3 | 5p13.2 | 1.52389E-11 | 1.31816E-05 | 69.13 | 77.23 | −8.10 | 0.76 | 0.65 | 0.88 | SLC |
| cg06960901 | SLC1A3 | 5p13.2 | 4.06686E-09 | 0.003517831 | 27.60 | 20.80 | 6.80 | 0.70 | 0.58 | 0.83 | SLC |
| cg05414613 | SLC1A7 | 1p32.3 | 1.97336E-11 | 1.70696E-05 | 62.49 | 71.39 | −8.90 | 0.82 | 0.72 | 0.93 | SLC |
| cg17949403 | SLC22A23 | 6p25.2 | 1.62173E-08 | 0.014027981 | 63.91 | 56.38 | 7.53 | 0.70 | 0.57 | 0.83 | SLC |
| cg11713788 | SLC22A23 | 6p25.2 | 1.14405E-09 | 0.00098960 | 15.53 | 21.56 | −6.03 | 0.55 | 0.43 | 0.68 | SLC |
| cg01739295 | SLC22A3 | 6q25.3 | 3.72641E-09 | 0.003223343 | 34.96 | 27.49 | 7.47 | 0.73 | 0.60 | 0.85 | SLC |
| cg17978727 | SLC23A2 | 20p13 | 3.2701E-08 | 0.028286374 | 35.05 | 27.91 | 7.14 | 0.86 | 0.77 | 0.95 | SLC |
| cg13282929 | SLC23A2 | 20p13 | 1.05376E-11 | 9.11502E-06 | 38.36 | 48.43 | −10.07 | 0.78 | 0.66 | 0.89 | SLC |
| cg13223777 | SLC24A5 | 15q21.1 | 1.26703E-14 | 1.09598E-08 | 72.94 | 81.33 | −8.39 | 0.85 | 0.76 | 0.95 | SLC |
| cg12030923 | SLC25A13 | 7q21.3 | 3.85551E-10 | 0.000333502 | 64.94 | 72.99 | −8.04 | 0.73 | 0.61 | 0.85 | SLC |
| cg14710071 | SLC25A16 | 10q21.3 | 4.75308E-08 | 0.041114139 | 80.54 | 85.65 | −5.11 | 0.78 | 0.66 | 0.89 | SLC |
| cg23045610 | SLC25A24 | 1p13.3 | 1.71658E-09 | 0.001484844 | 64.20 | 56.25 | 7.95 | 0.77 | 0.65 | 0.88 | SLC |
| cg00298230 | SLC25A26 | 3p14.1 | 4.97067E-11 | 4.29963E-05 | 79.20 | 85.45 | −6.25 | 0.85 | 0.75 | 0.94 | SLC |
| cg10343071 | SLC25A26 | 3p14.1 | 5.46218E-11 | 4.72479E-05 | 73.11 | 65.32 | 7.79 | 0.78 | 0.66 | 0.89 | SLC |
| cg25217269 | SLC25A27 | 6p12.3 | 1.95076E-11 | 1.68741E-05 | 29.90 | 38.17 | −8.27 | 0.73 | 0.62 | 0.84 | SLC |
| cg25217269 | SLC25A27 | 6p12.3 | 2.82907E-09 | 0.002447147 | 29.73 | 38.17 | −8.44 | 0.71 | 0.59 | 0.84 | SLC |
| cg27155504 | SLC25A3 | 12q23.1 | 3.46705E-09 | 0.002999001 | 80.86 | 74.78 | 6.08 | 0.77 | 0.65 | 0.88 | SLC |
| cg14775474 | SLC25A35 | 17p13.1 | 1.44284E-08 | 0.01248054 | 11.75 | 7.23 | 4.52 | 0.85 | 0.75 | 0.94 | SLC |
| cg18270394 | SLC25A36 | 3q23 | 1.46857E-11 | 1.27031E-05 | 77.30 | 84.04 | −6.74 | 0.75 | 0.63 | 0.87 | SLC |
| cg03084648 | SLC25A37 | 8p21.2 | 5.46944E-09 | 0.00473107 | 77.37 | 83.36 | −5.99 | 0.77 | 0.65 | 0.88 | SLC |
| cg16405055 | SLC25A44 | 1q22 | 1.02268E-10 | 8.84615E-05 | 7.03 | 11.86 | −4.83 | 0.65 | 0.53 | 0.77 | SLC |
| cg09153458 | SLC26A1 | 4p16.3 | 4.46293E-13 | 3.86043E-07 | 55.56 | 45.90 | 9.66 | 0.77 | 0.65 | 0.88 | SLC |
| cg26303603 | SLC26A2 | 5q32 | 2.80194E-11 | 2.42368E-05 | 59.10 | 50.17 | 8.93 | 0.78 | 0.67 | 0.89 | SLC |
| cg26303603 | SLC26A2 | 5q32 | 1.71203E-08 | 0.014809033 | 47.20 | 55.60 | −8.40 | 0.75 | 0.63 | 0.87 | SLC |
| cg12588047 | SLC28A3 | 9q21.32-q21.33 | 2.14091E-14 | 1.85189E-08 | 77.39 | 69.01 | 8.38 | 0.75 | 0.63 | 0.87 | SLC |
| cg12588047 | SLC28A3 | 9q21.32-q21.33 | 8.78757E-10 | 0.000760125 | 67.46 | 75.07 | −7.60 | 0.71 | 0.59 | 0.84 | SLC |
| cg00309135 | SLC2A12 | 6q23.2 | 1.74595E-09 | 0.001510246 | 71.49 | 78.46 | −6.97 | 0.77 | 0.66 | 0.89 | SLC |
| cg07645864 | SLC2A13 | 12q12 | 4.80512E-08 | 0.041564282 | 81.05 | 75.43 | 5.62 | 0.73 | 0.61 | 0.86 | SLC |
| cg07645864 | SLC2A13 | 12q12 | 4.7902E-11 | 4.14352E-05 | 71.55 | 79.07 | −7.53 | 0.73 | 0.61 | 0.85 | SLC |
| cg19132526 | SLC2A2 | 3q26.2 | 8.64142E-12 | 7.47483E-06 | 51.94 | 61.80 | −9.86 | 0.78 | 0.67 | 0.89 | SLC |
| cg20566657 | SLC2A9 | 4p16.1 | 3.53496E-09 | 0.003057739 | 64.98 | 72.69 | −7.71 | 0.82 | 0.72 | 0.92 | SLC |
| cg08789022 | SLC30A3 | 2p23.3 | 2.09826E-38 | 1.815E-32 | 24.84 | 15.24 | 9.60 | 0.75 | 0.63 | 0.87 | SLC |
| cg16989032 | SLC30A4 | 15q21.1 | 3.04023E-12 | 2.6298E-06 | 61.32 | 70.61 | −9.29 | 0.76 | 0.64 | 0.88 | SLC |
| cg16989032 | SLC30A4 | 15q21.1 | 1.74464E-11 | 1.50912E-05 | 73.48 | 65.55 | 7.93 | 0.70 | 0.57 | 0.83 | SLC |
| cg02530515 | SLC30A7 | 1p21.2 | 1.18431E-08 | 0.010244289 | 69.42 | 76.32 | −6.90 | 0.76 | 0.65 | 0.88 | SLC |
| cg11178666 | SLC33A1 | 3q25.31 | 2.61279E-09 | 0.002260067 | 56.99 | 65.38 | −8.40 | 0.76 | 0.64 | 0.87 | SLC |
| cg02272859 | SLC34A2 | 4p15.2 | 1.42711E-08 | 0.012344479 | 58.59 | 66.60 | −8.01 | 0.78 | 0.67 | 0.89 | SLC |
| cg16584327 | SLC35B4 | 7q33 | 3.26095E-11 | 2.82072E-05 | 69.14 | 77.07 | −7.93 | 0.75 | 0.63 | 0.87 | SLC |
| cg26453171 | SLC35F2 | 11q22.3 | 4.69575E-12 | 4.06182E-06 | 13.57 | 20.97 | −7.40 | 0.77 | 0.65 | 0.88 | SLC |
| cg14469376 | SLC37A2 | 11q24.2 | 4.6003E-09 | 0.003979261 | 9.77 | 5.46 | 4.31 | 0.78 | 0.67 | 0.89 | SLC |
| cg02765475 | SLC38A9 | 5q11.2 | 4.08508E-09 | 0.003533594 | 75.04 | 81.37 | −6.33 | 0.75 | 0.64 | 0.87 | SLC |
| cg00003999 | SLC39A10 | 2q32.3 | 6.45175E-09 | 0.005580765 | 79.37 | 73.18 | 6.20 | 0.74 | 0.61 | 0.86 | SLC |
| cg14228592 | SLC39A4 | 8q24.3 | 8.63426E-11 | 7.46864E-05 | 21.89 | 15.16 | 6.74 | 0.70 | 0.57 | 0.83 | SLC |
| cg17926678 | SLC39A9 | 14q24.1 | 1.62919E-38 | 1.40925E-32 | 34.99 | 24.49 | 10.50 | 0.77 | 0.65 | 0.88 | SLC |
| cg25287207 | SLC41A2 | 12q23.3 | 2.1923E-09 | 0.001896343 | 71.73 | 78.63 | −6.90 | 0.80 | 0.69 | 0.91 | SLC |
| cg25287207 | SLC41A2 | 12q23.3 | 6.23886E-11 | 5.39661E-05 | 80.33 | 73.56 | 6.77 | 0.79 | 0.68 | 0.90 | SLC |
| cg07291744 | SLC43A1 | 11q12.1 | 2.60106E-39 | 2.24991E-33 | 56.96 | 45.99 | 10.97 | 0.58 | 0.44 | 0.72 | SLC |
| cg19272348 | SLC43A2 | 17p13.3 | 2.95112E-10 | 0.000255272 | 73.67 | 80.66 | −6.99 | 0.76 | 0.65 | 0.88 | SLC |
| cg22521553 | SLC44A1 | 9q31.1-q31.2 | 1.22738E-12 | 1.06169E-06 | 73.51 | 65.18 | 8.33 | 0.71 | 0.58 | 0.83 | SLC |
| cg02426178 | SLC44A2 | 19p13.2 | 2.01983E-08 | 0.01747157 | 80.20 | 85.49 | −5.29 | 0.70 | 0.57 | 0.83 | SLC |
| cg08203794 | SLC45A4 | 8q24.3 | 2.12648E-11 | 1.8394E-05 | 83.88 | 77.62 | 6.26 | 0.81 | 0.70 | 0.92 | SLC |
| cg17156227 | SLC4A11 | 20p13 | 8.58965E-12 | 7.43005E-06 | 65.62 | 74.28 | −8.66 | 0.79 | 0.67 | 0.90 | SLC |
| cg09988421 | SLC4A2 | 7q36.1 | 1.9444E-09 | 0.001681904 | 26.37 | 19.51 | 6.86 | 0.78 | 0.67 | 0.90 | SLC |
| cg20078681 | SLC4A3 | 2q35 | 1.86189E-08 | 0.016105309 | 84.71 | 89.24 | −4.53 | 0.85 | 0.76 | 0.95 | SLC |
| cg13250541 | SLC5A6 | 2p23.3 | 2.56509E-14 | 2.2188E-08 | 13.99 | 7.73 | 6.26 | 0.85 | 0.75 | 0.95 | SLC |
| cg26758670 | SLC6A12 | 12p13.33 | 4.31442E-10 | 0.000373197 | 18.40 | 12.37 | 6.03 | 0.79 | 0.68 | 0.90 | SLC |
| cg04394707 | SLC6A15 | 12q21.31 | 2.35189E-08 | 0.020343858 | 60.54 | 68.20 | −7.66 | 0.72 | 0.60 | 0.85 | SLC |
| cg16321159 | SLC6A17 | 1p13.3 | 3.57318E-08 | 0.030907997 | 81.60 | 86.58 | −4.98 | 0.76 | 0.65 | 0.88 | SLC |
| cg17277001 | SLC6A18 | 5p15.33 | 8.83477E-09 | 0.007642076 | 28.79 | 36.89 | −8.10 | 0.71 | 0.58 | 0.84 | SLC |
| cg26339753 | SLC6A20 | 3p21.31 | 1.3871E-09 | 0.001199845 | 58.65 | 67.14 | −8.49 | 0.73 | 0.61 | 0.86 | SLC |
| cg06617455 | SLC6A6 | 3p25.1 | 1.60204E-08 | 0.013857662 | 51.42 | 59.71 | −8.29 | 0.76 | 0.65 | 0.88 | SLC |
| cg22367705 | SLC9A1 | 1p36.11 | 1.4567E-14 | 1.26004E-08 | 18.10 | 11.05 | 7.05 | 0.78 | 0.66 | 0.89 | SLC |
| cg13476831 | SLC9A5 | 16q22.1 | 1.00782E-09 | 0.000871768 | 15.73 | 10.21 | 5.52 | 0.88 | 0.79 | 0.96 | SLC |
| cg25268697 | SLCO1B3 | 12p12.2 | 6.83856E-12 | 5.91536E-06 | 35.62 | 44.35 | −8.74 | 0.66 | 0.54 | 0.78 | SLC |
| cg22822824 | SLCO2A1 | 3q22.1-q22.2 | 3.31677E-08 | 0.02869003 | 6.18 | 10.14 | −3.96 | 0.61 | 0.48 | 0.73 | SLC |
| cg12878682 | SLCO5A1 | 8q13.3 | 2.47908E-12 | 2.14441E-06 | 68.47 | 76.89 | −8.42 | 0.74 | 0.62 | 0.86 | SLC |
| cg26294217 | PRKAA1 | 5p13.1 | 6.80901E-09 | 0.005889795 | 5.54 | 10.08 | −4.54 | 0.76 | 0.65 | 0.88 | PKC |
| cg11899080 | PRKCA | 17q24.2 | 5.88738E-39 | 5.09258E-33 | 65.29 | 55.15 | 10.15 | 0.82 | 0.71 | 0.92 | PKC |
| cg13127598 | PRKCB | 16p12.2-p12.1 | 1.76442E-08 | 0.01526223 | 18.23 | 24.17 | −5.94 | 0.57 | 0.45 | 0.70 | PKC |
| cg18417061 | PRKCH | 14q23.1 | 6.21097E-10 | 0.00053725 | 76.05 | 81.62 | −5.57 | 0.81 | 0.71 | 0.91 | PKC |
| cg05878107 | PRKCI | 3q26.2 | 7.36142E-39 | 6.36763E-33 | 72.02 | 62.10 | 9.92 | 0.77 | 0.66 | 0.89 | PKC |
| cg08532220 | PRKCI | 3q26.2 | 2.61116E-10 | 0.000225865 | 57.36 | 66.20 | −8.84 | 0.74 | 0.62 | 0.86 | PKC |
| cg00105154 | PRKCQ | 10p15.1 | 9.19432E-14 | 7.95308E-08 | 21.46 | 14.05 | 7.42 | 0.75 | 0.63 | 0.87 | PKC |
| cg00105154 | PRKCQ | 10p15.1 | 5.72985E-13 | 4.95632E-07 | 14.86 | 22.82 | −7.97 | 0.70 | 0.57 | 0.83 | PKC |
| cg16269144 | PRKCZ | 1p36.33 | 3.26129E-10 | 0.000282101 | 80.02 | 73.44 | 6.58 | 0.77 | 0.65 | 0.89 | PKC |
| cg07836663 | PRKCZ | 1p36.33 | 8.27326E-13 | 7.15637E-07 | 66.22 | 73.87 | −7.65 | 0.76 | 0.66 | 0.87 | PKC |
These include CpG sites with Target ID, ABC, SLC, and PKC Genes ID, chromosome location, p-value, FDR p-value, and percentage of methylation change. The bold values indicate markers both hyper and hypo methylated.
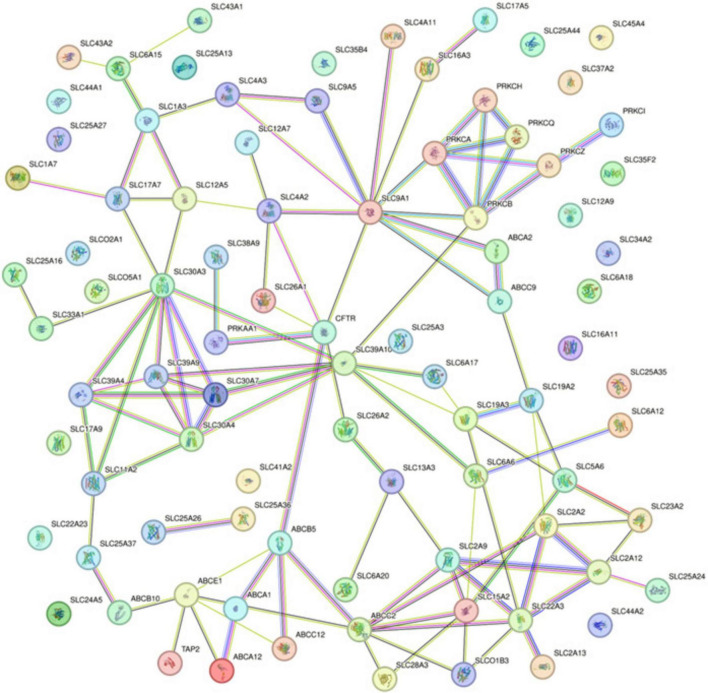
Protein-protein interaction network
The Protein-Protein Interaction (PPI) network analysis, utilizing the STRING database, delineated the complex interaction landscape among the 87 differentially methylated genes (Figure 2). The generated network comprised 87 nodes, each representing an individual gene product, connected by 110 edges that signified the putative protein-protein interactions. With an average node degree of 2.53, the network demonstrated a modest level of connectivity, where, on average, a protein was associated with approximately two and a half other proteins. The network’s average local clustering coefficient stood at 0.402, indicating a moderate propensity for proteins to form clusters, suggesting the presence of functional groupings within the network. The number of edges observed in the network significantly exceeded what would be expected in a random set of proteins, with an actual edge count of 110 compared to an expected count of 10. This substantial difference, validated by a PPI enrichment p-value < 1.0e–16, implies that the interactions are statistically significant and likely to be biologically relevant, rather than occurring by mere chance.
FIGURE 2.
Protein-Protein Interaction Network from STRING Analysis for Differentially Methylated Genes. Interactions are depicted as lines connecting nodes, which represent individual proteins. Line colors correspond to the type of evidence supporting the interaction: red for gene fusion; green for the neighborhood; blue for co-occurrence; purple for experimental; yellow for text mining; light blue for database evidence; and black for co-expression.
Specifically, noteworthy interactions were observed between kinase and transporter proteins. PRKCB showed connections with SLC39A10 and SLC9A1, while PRKCA was linked to SLC9A1. Additionally, PRKAA1 exhibited interactions with CFTR (ABCC7) and SLC38A9. These kinase and transporter protein interactions underscore potential regulatory points critical to the molecular mechanisms of NOWS.
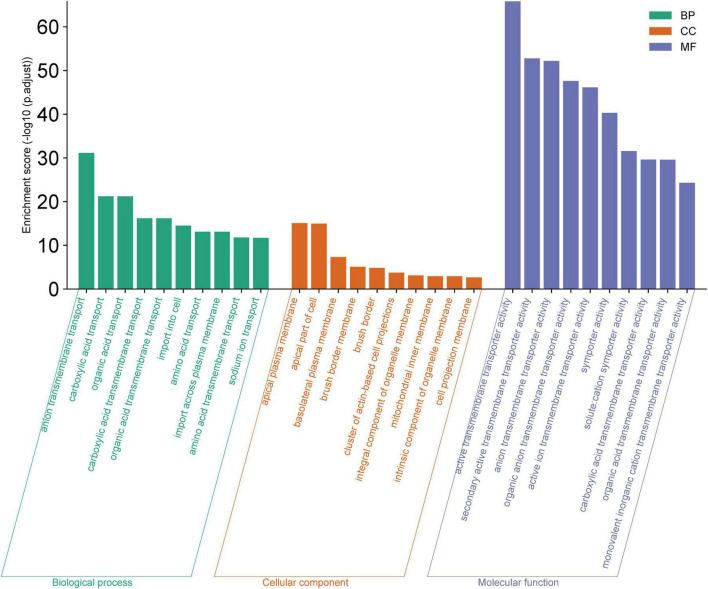
GO and KEGG pathway enrichment
The GO analysis highlighted significant enrichments in biological processes (BP) primarily related to various substance transport activities, particularly anion and carboxylic acid transport. The cellular components (CC) most represented were those associated with the cell membrane, including the apical plasma membrane and basolateral plasma membrane. Molecular functions (MF) predominantly involved active transport activities, with a focus on transmembrane transporter activities (Figure 3). The GO analysis enrichment scores and related details are present in Supplementary Tables 5–7.
FIGURE 3.
Gene Ontology (GO) Analysis for Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The bar chart displays the enrichment scores (−log10 (p.adjust) for the top ten GO terms within each category. Elevated enrichment scores indicate a higher level of significance, suggesting that these GO terms are notably overrepresented in the dataset, and may be integral to the underlying mechanisms of the biological system being investigated.
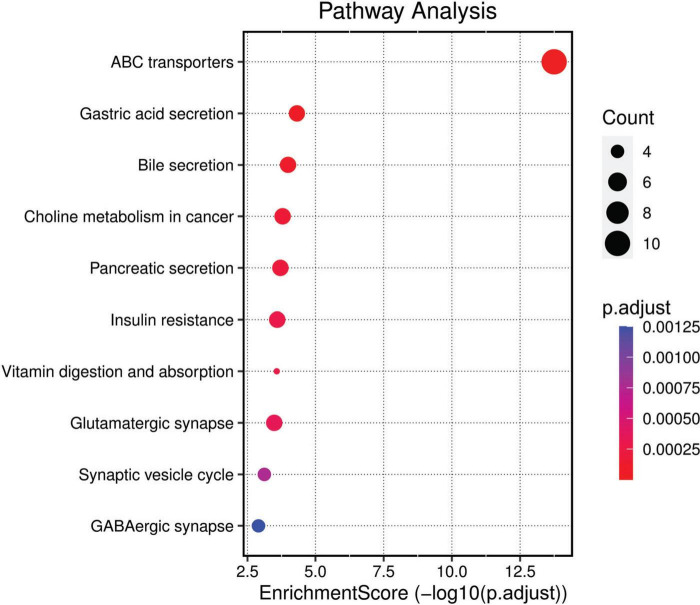
KEGG pathway analysis revealed that the differently methylated genes were significantly associated with 13 different pathways including ABC transporters, Gastric acid secretion, Bile secretion, Choline metabolism in cancer, Pancreatic secretion, Insulin resistance, Vitamin digestion, and absorption, Glutamatergic synapse, Synaptic vesicle cycle, GABAergic synapse, Salivary secretion, Inflammatory mediator regulation of TRP channels and Mineral absorption. Among them, the ‘ABC transporters’ pathway was prominent, suggesting a potential role in the transport of a wide range of substrates across extra- and intracellular membranes. Other notable pathways included those related to digestive system functions, metabolism, and synaptic neurotransmission, which could be linked to the biological underpinnings of NOWS. The visual representation and distribution of the enrichment scores across these pathways highlight the key biological processes potentially disrupted in NOWS (Figure 4). Complete details regarding the KEGG pathway analysis are provided in Supplementary Table 8.
FIGURE 4.
KEGG Pathway Analysis. This bubble plot visualizes the enriched pathways among the differently methylated genes based on the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database. Pathways are listed on the y-axis and are ordered by the enrichment score (−log10(p.adjust)), plotted on the x-axis. The size of each bubble corresponds to the gene count within the pathway, and the color gradient represents the adjusted p-value, with darker hues indicating higher significance.
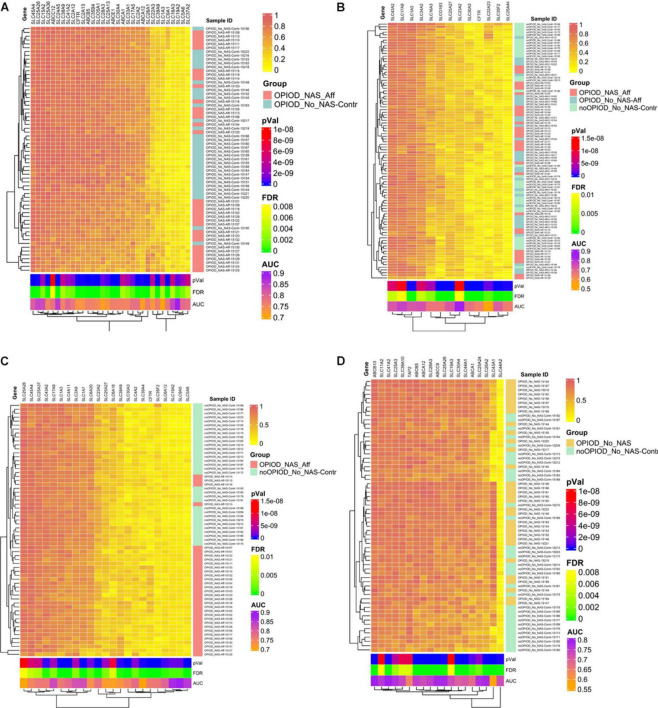
Evaluation of heatmaps
The heatmap, driven by CpG methylation markers associated with transporters and PKCs, clearly delineates distinct clusters: one for NOWS and the other for the control group. This compelling evidence underscores the reliability of these methylation markers in distinguishing between NOWS-affected patients and unaffected individuals, as demonstrated across all four distinct analyses presented in Figures 5A–D. In essence, our findings corroborate the accuracy and efficacy of these methylation markers in accurately discriminating between the two study groups.
FIGURE 5.
Heatmap displaying normalized beta values representing the top differentially methylated CpG sites in subjects exposed to prenatal opioids, along with CpG sites associated with transporters and protein kinase C (PKCs). Beta values for CpG sites with an unadjusted p < 0.005 and |Δβ| ≥ 0.05 are shown. Each row contains individual samples (affected and controls), and the columns pertain to the beta value for a CpG site color-coded from 0-1 (see color key, top right). The analysis was done in four combinations. Hierarchical sorting was performed by the CpG-associated genes site (columns), and the dendrogram (left) indicates similarities in methylation trends across all CpG sites. (A) Analysis I was conducted to distinguish NOWS from prenatal opioid exposure without NOWS symptoms. Heatmap comparing +Opioids/+NOWS and +Opioids/-NOWS reveals significant differences in methylation status among SLC and ABC drug transporter genes, alongside PKCs family members linked to NOWS. (B) Analysis II was conducted to distinguish prenatal opioid abuse from normal controls (OUD detection). Heatmap analyzing (+Opioids/+NOWS), (+Opioids/-NOWS), versus (-Opioids/-NOWS, control). (C) Analysis III was conducted to distinguish NOWS from unexposed controls. Heatmap displaying a comparison of (+Opioids/+NOWS) versus (-Opioids/-NOWS, control). (D) Analysis IV was conducted to distinguish opioid-induced epigenetic changes. Heatmap displaying analysis of (+Opioids/-NOWS), versus (-Opioids/-NOWS, control).
Discussion
Treating NOWS effectively remains challenging due to the paradoxical risks associated with unknown molecular and long-term developmental consequences. While short-term symptoms are observable soon after birth, uncertainties about long-term effects underscore the need for extensive studies. The detailed dysregulation of significant methylation changes in SLC, ABC transporters, and PKC genes associated with NOWS is described below.
ABCA1 is involved in cholesterol and lipid transport (Oram, 2003). Variations in ABCA1 can cause Tangier disease, characterized by impaired lipid efflux from macrophages leading to early atherosclerosis and low HDL levels (Peters et al., 2022).
ABCA12 transports lipids like glucosylceramides across cell membranes to create the skin’s protective outer layer, the stratum corneum, shielding against environmental damage and preventing water loss (Akiyama, 2011). Variations in ABCA12 can cause autosomal recessive congenital ichthyosis, marked by abnormal skin scaling and dryness (Hotz et al., 2023). Dry skin is common among opioid users or those addicted to opioids.
ABCA2 plays a role in regulating cholesterol homeostasis in the brain, facilitating lipid transport, and impacting drug resistance in cancer cells, notably tumor stem cells. Variations in this gene are associated with early-onset Alzheimer’s disease (AD) (Mack et al., 2008). ABCA2 also exhibits resistance to compounds like estradiol and mitoxantrone (Mack et al., 2007).
ABCB10 plays a crucial role in transporting porphyrins, necessary for heme biosynthesis and mitochondrial function (Yamamoto et al., 2014), its variations can impact heme metabolism and mitochondrial function, potentially influencing cellular responses to oxidative stress or drug-induced toxicity (Martinez et al., 2020).
ABCB5 plays a key role in multidrug resistance in cancer cells, exporting chemotherapy drugs and reducing their effectiveness (Muriithi et al., 2020).
ABCC12, a multidrug resistance protein (MRP), removes drugs, toxins, and metabolites from cells, playing a crucial role in drug resistance. Variations in ABCC12 are linked to breast cancer, liver hepatocellular carcinoma (Meng et al., 2022), bile duct paucity, cholestatic liver disease (Pham et al., 2021), and may also affect spermatid development and sperm function (Ono et al., 2007).
ABCC2 and ABCC9 genes, encoding multidrug resistance-associated proteins (MRPs), play crucial roles in drug resistance by regulating drug efflux, particularly in chemotherapy and antiretroviral therapy (Sodani et al., 2012). ABCC2 expels glucuronidated metabolites and bilirubin, impacting liver function and inflammation (Choudhuri and Klaassen, 2006). ABCC2 gene variants contribute to susceptibility to nonalcoholic fatty liver disease (NAFLD) (Sookoian et al., 2009), which is prevalent in cases of NAFLD with cirrhosis, high BMI, and psychiatric disorders involving opioid use (Moon et al., 2021). ABCC9 expressed in the cardiac, smooth muscle, and brain, influences drug responses in cardiovascular and neurological contexts, thereby impacting overall drug efficacy and resistance (Nelson et al., 2015) and often causing sudden cardiac death (Subbotina et al., 2019). Infants born to opioid-addicted mothers have experienced sudden and unexpected deaths (Pierson et al., 1972).
ABCC7 (CFTR) gene encodes a chloride channel critical for regulating ion transport and maintaining salt and water balance in tissues like the lungs, pancreas, and intestines (Lukasiak and Zajac, 2021). Dysfunction of this protein is linked to cystic fibrosis and can contribute to multidrug resistance in specific cell types. Variations in CFTR disrupt glucose homeostasis, leading to drug resistance through complex metabolic and cellular mechanisms (Ntimbane et al., 2009).
ABCE1 is a multifunctional protein involved in viral replication and cellular antiviral responses. In HIV pathogenesis, it aids in viral core disassembly and HIV-1 capsid formation (Ramnani et al., 2021). ABCE1 is also associated with chemotherapy and broader drug resistance mechanisms (Tian et al., 2012).
ABCB3 is crucial for antigen presentation and contributes to multidrug resistance (Fan et al., 2023). Variations in ABCB3 affect susceptibility to viral infections, autoimmune diseases, autoinflammatory diseases, and certain cancers (Mantel et al., 2022).
The SLC transporters
Telomere-associated genes
We identified nine dysregulated genes involved in telomere maintenance.1 (ABCC12, ABCC2, ABCC9, CFTR, SLC25A36, SLC39A10, SLC6A12, PRKCB, PRKCQ) Accelerated telomere shortening in maternal cells may result in increased cellular aging, reduced regenerative capacity, and heightened susceptibility to age-related pathologies (Bar and Blasco, 2016). Compromised telomere maintenance during fetal development can lead to abnormalities and a higher risk of congenital defects due to its role in genomic stability (Tardat and Dejardin, 2018). Maternal opioid use increases the risk of telomere shortening (Rahimi Mehdi Abad et al., 2021).
Glucose metabolism-related genes
Maternal opioid exposure can modify glucose metabolism and insulin sensitivity, potentially influencing gene expression and the function of glucose transport pathways critical for drug resistance (Toorie et al., 2022). This disruption can affect fetal nutrient supply and development, potentially predisposing offspring to future metabolic issues (Toorie et al., 2021). We identified six key genes (SLC16A3, SLC19A2, SLC25A13, SLC2A2, SLC37A2, and PRKAA1) crucial for glucose metabolism.
Circadian rhythm dysregulation
Circadian rhythms are 24-h cycles regulating physiological processes like sleep, hormone release, and metabolism. Ultradian rhythms, occurring within a day, include REM and non-REM sleep, feeding, and hormone release. Sleep insufficiency and circadian disruptions correlate with obesity, cardiovascular disease, and cognitive impairment, though mechanisms are unclear (Chaput et al., 2023). We identified nine genes associated with circadian rhythm: ABCA1, SLC12A9, SLC22A23, SLC25A37, SLC43A1, SLC45A4, SLC4A2, SLC6A12, and SLC6A6. These genes are crucial for lipid transport (ABCA1) (Phillips, 2018), electrolyte balance (SLC12A9) (Levin-Konigsberg et al., 2023), and neurotransmitter transport (SLC6A12, SLC6A6) (Pramod et al., 2013). PRKCA, regulates circadian and ultradian rhythms, particularly sleep and hormonal cycles (Moller-Levet et al., 2013; Hu et al., 2023). Genes associated with ultradian rhythms include SLC23A2, SLC24A5, SLC25A37, and SLC6A6, influencing vitamin C transport (SLC23A2) (Kobayashi et al., 2024) and calcium transport (SLC24A5) (Quillen and Shriver, 2008).
Suicide-associated genes
The risk of suicide death is significantly higher in individuals with opioid use disorders (Rizk et al., 2021). We identified dysregulations in four PKC genes—PRKCA, PRKCB, PRKCH, and PRKCI—known for their roles in mood regulation and stress response, crucial for neurodevelopment and linked to psychiatric disorders (Choi et al., 2011; Coon et al., 2020; Sokolowski and Wasserman, 2020; Pandey et al., 2021). Additionally, the SLC genes SLC19A2, SLC1A3, SLC4A2, and SLC4A3 are implicated in suicide through their roles in neural function and neurotransmitter regulation: SLC19A2 encodes a thiamine transporter essential for brain metabolism; thiamine deficiencies can lead to mood dysregulation and suicidal behavior (Lutz et al., 2017). SLC1A3 encodes a glutamate transporter, and its dysregulation is linked to depression and anxiety, significant risk factors for suicide (Murphy et al., 2011). SLC4A2 regulates brain pH balance and is linked to mood disorders, observed in individuals with suicidal tendencies (Coon et al., 2020; Mirza et al., 2024). SLC4A3 encodes a bicarbonate transporter; its dysfunction can disturb the brain’s acid-base balance, possibly contributing to neurological and psychiatric disorders, noted for significant variability in individuals who have died by suicide (Punzi et al., 2022).
Metal ion transporters
Zinc, cobalt, manganese, and magnesium transporters support infants’ cellular balance, neuronal development, and drug resistance. Zinc is crucial for growth, brain development, and neuronal function (Li et al., 2022). Dysfunctional zinc transporters that hinder zinc uptake in the brain can disrupt neuronal signaling and synaptic plasticity, potentially impairing cognitive development and increasing the risk of neurological disorders later in life (Wang et al., 2023). The six SLC transporters (SLC30A3, SLC30A4, SLC30A7, SLC39A10, SLC39A4, SLC39A9) are crucial for zinc balance, affecting immune function, growth, and neuronal development. Other metal-related genes like SLC12A5 (cobalt carrier), SLC11A2 (manganese transporter), and SLC41A2 (magnesium transporter) significantly affect the immediate and long-term health of infants exposed to opioids in utero. Cobalt is vital for vitamin B12 production, hematopoiesis, immune responses, and antibacterial activities. Disruptions in cobalt levels can cause anemia, neurological issues, impaired hypoxia response, and drug resistance (Ma et al., 2022). Increased SLC11A2 activity can lead to excessive manganese uptake, contributing to neurodegenerative disorders such as Parkinson’s disease, impacting immune function, and potentially causing severe toxicity from manganese accumulation (Chen et al., 2015). Magnesium deficiency linked to dysregulated SLC41A2 can lead to neurological symptoms like seizures and muscle spasms, affecting neural function and overall health (Al Alawi et al., 2018).
Oxidative stress-related genes
We identified PRKAA1 (PKC gene) and five SLC genes (SLC17A5, SLC1A3, SLC23A2, SLC25A27, SLC6A6) linked to oxidative stress. PRKAA1, a regulator of cellular energy homeostasis activated under stress conditions, enhances antioxidant defenses and cellular repair in opioid exposure. However, chronic activation may alter metabolic states, impacting drug metabolism and resistance. SLC17A5 disruption by oxidative stress affects lysosomal function, impairing drug metabolism (Ruivo et al., 2009). SLC1A3 dysfunction due to oxidative stress causes neuronal damage and affects drug response (Ayka and Sehirli, 2020). SLC23A2, crucial for vitamin C uptake, combats oxidative stress; impaired function reduces antioxidant capacity (Teafatiller et al., 2021). SLC25A27 (UCP4), involved in mitochondrial ROS reduction, influences drug metabolism under chronic oxidative stress (Zhang et al., 2024). SLC6A6 alteration in taurine transport affects cellular resilience, potentially influencing drug resistance mechanisms (Baliou et al., 2021).
Opioid use is known to increase impulsivity
Impulsivity is common in opioid addiction cases (Tolomeo et al., 2021). We identified three genes linked to impulsivity—SLCO5A1, PRKCA, and PRKCH—in infants born to mothers with opioid use disorders.
SLCO5A1 regulates drug transport across cell membranes, influencing neurodevelopmental processes linked to impulsivity (Roshandel et al., 2023). PRKCA regulates signal transduction, influencing neurotransmitter signaling and synaptic plasticity, thereby increasing susceptibility to impulsive behaviors while PRKCH is critical for neuronal signaling and brain development; its dysregulation may disrupt brain development, potentially increasing impulsivity (Khadka et al., 2014). Animal studies similarly indicate that prenatal opioid exposure results in long-term cognitive deficits and impulsivity (Alaee et al., 2021).
Protein Kinase C family (PKC)
PKC family members play crucial roles in cell signaling and serve as therapeutic targets for various conditions, including diabetes, cancer, cardiovascular issues, dermatological conditions, psychiatric disorders, neurological diseases, and immune-mediated ailments (Mochly-Rosen et al., 2012). Dysregulation of the seven PKCs—PRKCA, PRKCB, PRKCH, PRKCI, PRKCQ, and PRKAA1—is linked to specific diseases and disorders. Both PRKCA and PRKCH are associated with impulsivity, as previously explained. Variations in both PRKCB and PRKCQ play significant roles in certain cancers and are considered excellent predictive biomarkers for these diseases (Abdelatty et al., 2021). Chronic opioid therapy increases the risk of cancer in noncancer patients with chronic pain (Oh and Song, 2020).
Pathways
We found significant epigenetic changes in multiple genes across 13 pathways, outlining important dysregulated pathways linked to NOWS and OUD.
ABC Transporters: ABC transporters are pivotal in drug resistance as they actively expel drugs from cells, diminishing their therapeutic efficacy. Moreover, these transporters can influence the pharmacokinetics of drugs of abuse, affecting their distribution and elimination from the body. In the placenta, ABC transporters significantly reduce fetal exposure to drugs and other foreign substances, potentially influencing therapy for NOWS (Gottesman and Ambudkar, 2001).
Pancreatic Secretion: Altered pancreatic secretion can affect the absorption of certain drugs, potentially impacting their efficacy and bioavailability (Olesen et al., 2013). Pancreatic dysfunction may occur in individuals with substance abuse disorders, affecting drug metabolism and pancreatic health (Jones et al., 2015). Neonatal exposure to drugs affecting pancreatic function can lead to digestive disturbances and contribute to NOWS.
Bile Secretion: Bile secretion can affect the enterohepatic circulation of drugs and their metabolites, impacting drug levels and efficacy. Disruption of bile secretion pathways may contribute to drug-induced liver injury and drug resistance (Li and Apte, 2015). Neonatal exposure to drugs affecting bile secretion can lead to cholestasis and NOWS.
Insulin Resistance: Insulin resistance is associated with metabolic disorders often seen in substance abuse, potentially affecting drug metabolism and response. Substance abuse can contribute to insulin resistance, exacerbating metabolic complications and drug resistance. Neonatal exposure to substances affecting insulin sensitivity may influence fetal growth and contribute to NOWS.
Glutamatergic Synapse. Disruption of the glutamatergic system is linked to addiction and drug tolerance, altering responses to addictive substances (Alasmari et al., 2022). Targeting glutamatergic synapses offers promise in treating drug addiction and reducing drug resistance. Early exposure to drugs impacting glutamatergic neurotransmission in neonates may affect brain development and contribute to NOWS.
Synaptic Vesicle Cycle: Disruption of the synaptic vesicle cycle can alter neurotransmitter release, affecting the rewarding effects of addictive substances (Sulzer, 2011). Drugs of abuse can modulate the synaptic vesicle cycle, leading to long-term changes in synaptic function and drug tolerance (Wang W. et al., 2019). Prenatal exposure to substances affecting the synaptic vesicle cycle may influence neuronal development and contribute to NOWS.
Gastric Acid Secretion: Alterations in gastric acid secretion can affect the absorption and efficacy of certain medications used in addiction treatment. Gastric acid secretion may influence the absorption of drugs of abuse, affecting their onset and duration of action (Bushra et al., 2011). Neonates exposed to maternal substance abuse may experience gastric disturbances contributing to NOWS.
Inflammatory Mediator Regulation of Transient Receptor Potential (TRP) Channels. TRP channels play a role in pain perception and neuroinflammation associated with substance abuse and addiction (Yang et al., 2023). Inflammatory mediators can modulate TRP channel activity, influencing the development of drug tolerance and withdrawal symptoms. Neonatal exposure to drugs affecting TRP channels and inflammatory mediators may impact sensory processing and contribute to NOWS.
Conclusion
In summary, our study enhances our understanding of the epigenetic basis of NOWS, emphasizing the roles of PKC and drug transporters. Methylation changes in these genes may serve as NOWS biomarkers, opening new research and clinical avenues. These findings aim to mitigate maternal OUD and relapse, enhance care for opioid-exposed infants, and support their families. Future steps include investigating the functional impacts of these changes and developing effective therapies for NOWS and OUD management. Exploring epigenetic influences on drug metabolism could improve global patient care by enhancing drug safety and efficacy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Institutional Review Board of Beaumont Health System, Royal Oak, MI, USA (HIC#: 2019-086). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
UR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing−original draft, Writing−review and editing. RuR: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing−original draft, Writing−review and editing. LU: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing−original draft, Writing−review and editing. SM: Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing−original draft, Writing−review and editing. JP: Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing−review and editing. RMR: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing−review and editing, Writing−original draft. RB-S: Conceptualization, Data curation, Investigation, Supervision, Validation, Visualization, Writing−review and editing. SS: Data curation, Formal analysis, Investigation, Resources, Software, Supervision, Validation, Visualization, Writing−original draft, Writing−review and editing.
Funding Statement
The authors declare that financial support was received for the research, authorship, and/or publication of this article. The efforts of UR, SS, and RR are supported by their collaborative NIH grants. The following NIH funding supported the salary and research of SS and RR: R01HD096800 (PI: SS), R43DA058430 (MPI: SS), R01DA059321 (MPI: RR/SS), and U01TR003719 (PI: SS).
Footnotes
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2024.1442915/full#supplementary-material
References
- Abdelatty A., Sun Q., Hu J., Wu F., Wei G., Xu H., et al. (2021). Pan-cancer study on protein kinase C family as a potential biomarker for the tumors immune landscape and the response to immunotherapy. Front. Cell. Dev. Biol. 9:798319. 10.3389/fcell.2021.798319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M. (2011). The roles of ABCA12 in keratinocyte differentiation and lipid barrier formation in the epidermis. Dermatoendocrinol 3 107–112. 10.4161/derm.3.2.15136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Alawi A. M., Majoni S. W., Falhammar H. (2018). Magnesium and human health: Perspectives and research directions. Int. J. Endocrinol. 2018:9041694. 10.1155/2018/9041694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaee E., Moazen P., Pattij T., Semnanian S., Azizi H. (2021). Prenatal exposure to morphine impairs attention and impulsivity in adult rats. Psychopharmacology 238 2729–2741. 10.1007/s00213-021-05888-7 [DOI] [PubMed] [Google Scholar]
- Alasmari F., Sari D. B., Alhaddad H., Al-Rejaie S. S., Sari Y. (2022). Interactive role of acid sensing ion channels and glutamatergic system in opioid dependence. Neurosci. Biobehav. Rev. 135:104581. 10.1016/j.neubiorev.2022.104581 [DOI] [PubMed] [Google Scholar]
- Alves R., Goncalves A. C., Jorge J., Marques G., Ribeiro A. B., Tenreiro R., et al. (2022). Genetic variants of ABC and SLC transporter genes and chronic myeloid leukaemia: Impact on susceptibility and prognosis. Int. J. Mol. Sci. 23:9815. 10.3390/ijms23179815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan S., Mendez M. D. (2023). Neonatal abstinence syndrome. Treasure Island, FL: StatPearls. [PubMed] [Google Scholar]
- Ayka A., Sehirli A. O. (2020). The role of the SLC transporters protein in the neurodegenerative disorders. Clin. Psychopharmacol. Neurosci. 18 174–187. 10.9758/cpn.2020.18.2.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliou S., Adamaki M., Ioannou P., Pappa A., Panayiotidis M. I., Spandidos D. A., et al. (2021). Protective role of taurine against oxidative stress (Review). Mol. Med. Rep. 24:2242. 10.3892/mmr.2021.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar C., Blasco M. A. (2016). Telomeres and telomerase as therapeutic targets to prevent and treat age-related diseases. F1000Res 5:89. 10.12688/f1000research.7020.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril S. A., Gose T., Schuetz J. D. (2023). How Cryo-EM has expanded our understanding of membrane transporters. Drug Metab. Dispos. 51 904–922. 10.1124/dmd.122.001004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht K., Schafer A. M., Meyer, Zu Schwabedissen H. E. (2020). Uptake transporters of the SLC21, SLC22A, and SLC15A families in anticancer therapy-modulators of cellular entry or pharmacokinetics? Cancers 12:2263. 10.3390/cancers12082263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushra R., Aslam N., Khan A. Y. (2011). Food-drug interactions. Oman Med. J. 26 77–83. 10.5001/omj.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbo R., Rodriguez E. (2023). Relevance of sugar transport across the cell membrane. Int. J. Mol. Sci. 24:6085. 10.3390/ijms24076085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput J. P., McHill A. W., Cox R. C., Broussard J. L., Dutil C., da Costa B. G. G., et al. (2023). The role of insufficient sleep and circadian misalignment in obesity. Nat. Rev. Endocrinol. 19 82–97. 10.1038/s41574-022-00747-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Chakraborty S., Mukhopadhyay S., Lee E., Paoliello M. M., Bowman A. B., et al. (2015). Manganese homeostasis in the nervous system. J. Neurochem. 134 601–610. 10.1111/jnc.13170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Le T., Xing G., Johnson L. R., Ursano R. J. (2011). Analysis of kinase gene expression in the frontal cortex of suicide victims: Implications of fear and stress. Front. Behav. Neurosci. 5:46. 10.3389/fnbeh.2011.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri S., Klaassen C. D. (2006). Structure, function, expression, genomic organization, and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) efflux transporters. Int. J. Toxicol. 25 231–259. 10.1080/10915810600746023 [DOI] [PubMed] [Google Scholar]
- Coon H., Darlington T. M., DiBlasi E., Callor W. B., Ferris E., Fraser A., et al. (2020). Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide. Mol. Psychiatry 25 3077–3090. 10.1038/s41380-018-0282-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Rzhetsky A., Allikmets R. (2001). The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166. 10.1101/gr.184901 [DOI] [PubMed] [Google Scholar]
- Fan J., To K. K. W., Chen Z. S., Fu L. (2023). ABC transporters affects tumor immune microenvironment to regulate cancer immunotherapy and multidrug resistance. Drug Resist. Updat. 66:100905. 10.1016/j.drup.2022.100905 [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Ambudkar S. V. (2001). Overview: ABC transporters and human disease. J. Bioenerg. Biomembr. 33 453–458. 10.1023/a:1012866803188 [DOI] [PubMed] [Google Scholar]
- Grover T., Gupta R., Arora G., Bal C. S., Ambekar A., Basu Ray S., et al. (2020). Dopamine transporter availability in alcohol and opioid dependent subjects - a (99m)Tc-TRODAT-1SPECT imaging and genetic association study. Psychiatry Res. Neuroimaging 305:111187. 10.1016/j.pscychresns.2020.111187 [DOI] [PubMed] [Google Scholar]
- Gu Z. (2015). ComplexHeatmap: Making complex Heatmaps. R package version 1.6.0. Available online at: https://github.com/jokergoo/ComplexHeatmap [Google Scholar]
- Hasin D. S., O’Brien C. P., Auriacombe M., Borges G., Bucholz K., Budney A., et al. (2013). DSM-5 criteria for substance use disorders: Recommendations and rationale. Am. J. Psychiatry 170 834–851. 10.1176/appi.ajp.2013.12060782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M. A., Clemencon B., Burrier R. E., Bruford E. A. (2013). The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol. Aspects Med. 34 95–107. 10.1016/j.mam.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz A., Kopp J., Bourrat E., Oji V., Sussmuth K., Komlosi K., et al. (2023). Mutational spectrum of the ABCA12 gene and genotype-phenotype correlation in a cohort of 64 patients with autosomal recessive congenital ichthyosis. Genes (Basel) 14:717. 10.3390/genes14030717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Long Y., Song G., Chen S., Xu Z., Li Q., et al. (2023). Dysfunction of Prkcaa links social behavior defects with disturbed circadian rhythm in zebrafish. Int. J. Mol. Sci. 24:3849. 10.3390/ijms24043849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. R., Hall O. M., Kaye A. M., Kaye A. D. (2015). Drug-induced acute pancreatitis: A review. Ochsner J. 15 45–51. [PMC free article] [PubMed] [Google Scholar]
- Joshi A. A., Vaidya S. S., St-Pierre M. V., Mikheev A. M., Desino K. E., Nyandege A. N., et al. (2016). Placental ABC transporters: Biological impact and pharmaceutical significance. Pharm. Res. 33 2847–2878. 10.1007/s11095-016-2028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Inokuchi J., Eto M., Murata M., Kang J. H. (2022). Protein kinase C (PKC) isozymes as diagnostic and prognostic biomarkers and therapeutic targets for cancer. Cancers (Basel) 14:5425. 10.3390/cancers14215425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadka S., Narayanan B., Meda S. A., Gelernter J., Han S., Sawyer B., et al. (2014). Genetic association of impulsivity in young adults: A multivariate study. Transl. Psychiatry 4:e451. 10.1038/tp.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. A., Shimada H., Sano F. K., Itoh Y., Enoki S., Okada Y., et al. (2024). Dimeric transport mechanism of human vitamin C transporter SVCT1. Nat. Commun. 15:5569. 10.1038/s41467-024-49899-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. M., Messing R. O. (2008). Protein kinases and addiction. Ann. N. Y. Acad. Sci. 1141 22–57. 10.1196/annals.1441.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Konigsberg R., Mitra K., Nigam A., Spees K., Hivare P., Liu K., et al. (2023). SLC12A9 is a lysosome-detoxifying ammonium - chloride co-transporter. bioRxiv [Preprint]. 10.1101/2023.05.22.541801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Apte U. (2015). Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv. Pharmacol. 74 263–302. 10.1016/bs.apha.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu Y., Wei R., Yong V. W., Xue M. (2022). The important role of zinc in neurological diseases. Biomolecules 13:28. 10.3390/biom13010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Aryee M. J., Padyukov L., Fallin M. D., Hesselberg E., Runarsson A., et al. (2013). Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31 142–147. 10.1038/nbt.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiak A., Zajac M. (2021). The distribution and role of the CFTR protein in the intracellular compartments. Membranes (Basel) 11:804. 10.3390/membranes11110804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz P. E., Mechawar N., Turecki G. (2017). Neuropathology of suicide: Recent findings and future directions. Mol. Psychiatry 22 1395–1412. 10.1038/mp.2017.141 [DOI] [PubMed] [Google Scholar]
- Ma Y., Lin W., Ruan Y., Lu H., Fan S., Chen D., et al. (2022). Advances of cobalt nanomaterials as anti-infection agents, drug carriers, and immunomodulators for potential infectious disease treatment. Pharmaceutics 14:2351. 10.3390/pharmaceutics14112351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack J. T., Brown C. B., Tew K. D. (2008). ABCA2 as a therapeutic target in cancer and nervous system disorders. Expert Opin. Ther. Targets 12 491–504. 10.1517/14728222.12.4.491 [DOI] [PubMed] [Google Scholar]
- Mack J. T., Townsend D. M., Beljanski V., Tew K. D. (2007). The ABCA2 transporter: Intracellular roles in trafficking and metabolism of LDL-derived cholesterol and sterol-related compounds. Curr. Drug Metab. 8 47–57. 10.2174/138920007779315044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. (2017). The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 7 339–348. 10.15171/apb.2017.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel I., Sadiq B. A., Blander J. M. (2022). Spotlight on TAP and its vital role in antigen presentation and cross-presentation. Mol. Immunol. 142 105–119. 10.1016/j.molimm.2021.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Fendley G. A., Saxberg A. D., Zoghbi M. E. (2020). Stimulation of the human mitochondrial transporter ABCB10 by zinc-mesoporphrin. PLoS One 15:e0238754. 10.1371/journal.pone.0238754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayati A., Moreau A., Le Vee M., Stieger B., Denizot C., Parmentier Y., et al. (2017). Protein kinases C-mediated regulations of drug transporter activity, localization and expression. Int. J. Mol. Sci. 18:764. 10.3390/ijms18040764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Dong S., Yangyang L., Wang S., Xu X., Liu T., et al. (2022). Adenosine triphosphate-binding cassette subfamily C members in liver hepatocellular carcinoma: Bioinformatics-driven prognostic value. Medicine (Baltimore) 101:e28869. 10.1097/MD.0000000000028869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metpally R. P., Krishnamurthy S., Moran K. M., Weller A. E., Crist R. C., Reiner B. C., et al. (2019). The imperative of clinical and molecular research on neonatal opioid withdrawal syndrome. Mol. Psychiatry 24 1568–1571. 10.1038/s41380-019-0522-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza S., Lima C. N. C., Del Favero-Campbell A., Rubinstein A., Topolski N., Cabrera-Mendoza B., et al. (2024). Blood epigenome-wide association studies of suicide attempt in adults with bipolar disorder. Transl. Psychiatry 14:70. 10.1038/s41398-024-02760-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry C. J., Bawor M., Desai D., Marsh D. C., Samaan Z. (2014). Genetics of opioid dependence: A review of the genetic contribution to opioid dependence. Curr. Psychiatry Rev. 10 156–167. 10.2174/1573400510666140320000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D., Das K., Grimes K. V. (2012). Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 11 937–957. 10.1038/nrd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Levet C. S., Archer S. N., Bucca G., Laing E. E., Slak A., Kabiljo R., et al. (2013). Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl. Acad. Sci. U.S.A. 110 E1132–E1141. 10.1073/pnas.1217154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A. M., Watkins S. E., Lok A. S., Firpi-Morell R. J., Trinh H. N., Kupec J. T., et al. (2021). Opioid use is more common in nonalcoholic fatty liver disease patients with cirrhosis, higher BMI, and psychiatric disease. Dig. Dis. 39 247–257. 10.1159/000511074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriithi W., Macharia L. W., Heming C. P., Echevarria J. L., Nyachieo A., Filho P. N., et al. (2020). ABC transporters and the hallmarks of cancer: Roles in cancer aggressiveness beyond multidrug resistance. Cancer Biol. Med. 17 253–269. 10.20892/j.issn.2095-3941.2019.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T. M., Ryan M., Foster T., Kelly C., McClelland R., O’Grady J., et al. (2011). Risk and protective genetic variants in suicidal behaviour: Association with SLC1A2, SLC1A3, 5-HTR1B &NTRK2 polymorphisms. Behav. Brain Funct. 7:22. 10.1186/1744-9081-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. T., Jicha G. A., Wang W. X., Ighodaro E., Artiushin S., Nichols C. G., et al. (2015). ABCC9/SUR2 in the brain: Implications for hippocampal sclerosis of aging and a potential therapeutic target. Ageing Res. Rev. 24 111–125. 10.1016/j.arr.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntimbane T., Comte B., Mailhot G., Berthiaume Y., Poitout V., Prentki M., et al. (2009). Cystic fibrosis-related diabetes: From CFTR dysfunction to oxidative stress. Clin. Biochem. Rev. 30 153–177. [PMC free article] [PubMed] [Google Scholar]
- Oh T. K., Song I. A. (2020). Chronic opioid use and risk of cancer in patients with chronic noncancer pain: A nationwide historical cohort study. Cancer Epidemiol. Biomark. Prev. 29 1962–1967. 10.1158/1055-9965.EPI-20-0206 [DOI] [PubMed] [Google Scholar]
- Olesen A. E., Brokjaer A., Fisher I. W., Larsen I. M. (2013). Pharmacological challenges in chronic pancreatitis. World J. Gastroenterol. 19 7302–7307. 10.3748/wjg.v19.i42.7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Van der Heijden I., Scheffer G. L., Van de Wetering K., Van Deemter E., De Haas M., et al. (2007). Multidrug resistance-associated protein 9 (ABCC12) is present in mouse and boar sperm. Biochem. J. 406 31–40. 10.1042/BJ20070292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram J. F. (2003). HDL apolipoproteins and ABCA1: Partners in the removal of excess cellular cholesterol. Arterioscler. Thromb. Vasc. Biol. 23 720–727. 10.1161/01.ATV.0000054662.44688.9A [DOI] [PubMed] [Google Scholar]
- Pandey G. N., Sharma A., Rizavi H. S., Ren X. (2021). Dysregulation of protein kinase C in adult depression and suicide: Evidence from postmortem brain studies. Int. J. Neuropsychopharmacol. 24 400–408. 10.1093/ijnp/pyab003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Chen L., Ye S., Kang Y., Liu J., Zeng S., et al. (2020). Research and development of drug delivery systems based on drug transporter and nano-formulation. Asian J. Pharm. Sci. 15 220–236. 10.1016/j.ajps.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters F., Ebner L. J. A., Atac D., Maggi J., Berger W., den Hollander A. I., et al. (2022). Regulation of ABCA1 by AMD-associated genetic variants and hypoxia in iPSC-RPE. Int. J. Mol. Sci. 23:3194. 10.3390/ijms23063194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D. H., Kudira R., Xu L., Valencia C. A., Ellis J. L., Shi T., et al. (2021). Deleterious variants in ABCC12 are detected in idiopathic chronic cholestasis and cause intrahepatic bile duct loss in model organisms. Gastroenterology 161:287-300.e216. 10.1053/j.gastro.2021.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. C. (2018). Is ABCA1 a lipid transfer protein? J. Lipid Res. 59 749–763. 10.1194/jlr.R082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson P. S., Howard P., Kleber H. D. (1972). Sudden deaths in infants born to methadone-maintained addicts. JAMA 220 1733–1734. [PubMed] [Google Scholar]
- Pramod A. B., Foster J., Carvelli L., Henry L. K. (2013). SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Aspects Med. 34 197–219. 10.1016/j.mam.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzi G., Ursini G., Chen Q., Radulescu E., Tao R., Huuki L. A., et al. (2022). Genetics and Brain Transcriptomics of Completed Suicide. Am. J. Psychiatry 179 226–241. 10.1176/appi.ajp.2021.21030299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puris E., Fricker G., Gynther M. (2023). The role of solute carrier transporters in efficient anticancer drug delivery and therapy. Pharmaceutics 15:364. 10.3390/pharmaceutics15020364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen E. E., Shriver M. D. (2008). SLC24A5: Exchanging genetic and biochemical knowledge. Pigment Cell Melanoma Res. 21 344–345. 10.1111/j.1755-148X.2008.00456.x [DOI] [PubMed] [Google Scholar]
- Radhakrishna U., Nath S. K., Uppala L. V., Veerappa A., Forray A., Muvvala S. B., et al. (2023a). Placental microRNA methylome signatures may serve as biomarkers and therapeutic targets for prenatally opioid-exposed infants with neonatal opioid withdrawal syndrome. Front. Genet. 14:1215472. 10.3389/fgene.2023.1215472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U., Sadhasivam S., Radhakrishnan R., Forray A., Muvvala S. B., Metpally R. P., et al. (2023b). Placental cytochrome P450 methylomes in infants exposed to prenatal opioids: Exploring the effects of neonatal opioid withdrawal syndrome on health horizons. Front. Genet. 14:1292148. 10.3389/fgene.2023.1292148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U., Nath S. K., Vishweswaraiah S., Uppala L. V., Forray A., Muvvala S. B., et al. (2021a). Maternal opioid use disorder: Placental transcriptome analysis for neonatal opioid withdrawal syndrome. Genomics 113 3610–3617. 10.1016/j.ygeno.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Radhakrishna U., Vishweswaraiah S., Uppala L. V., Szymanska M., Macknis J., Kumar S., et al. (2021b). Placental DNA methylation profiles in opioid-exposed pregnancies and associations with the neonatal opioid withdrawal syndrome. Genomics 113 1127–1135. 10.1016/j.ygeno.2021.03.006 [DOI] [PubMed] [Google Scholar]
- Rahimi Mehdi Abad F., Khalili P., Jalali F., Pirsadeghi A., Esmaeili Nadimi A. (2021). Maternal opioid use is reflected on leukocyte telomere length of male newborns. PLoS One 16:e0261013. 10.1371/journal.pone.0261013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnani B., Manivannan P., Jaggernauth S., Malathi K. (2021). ABCE1 regulates RNase L-induced autophagy during viral infections. Viruses 13:315. 10.3390/v13020315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk M. M., Herzog S., Dugad S., Stanley B. (2021). Suicide risk and addiction: The impact of alcohol and opioid use disorders. Curr. Addict. Rep. 8 194–207. 10.1007/s40429-021-00361-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshandel D., Sanders E. J., Shakeshaft A., Panjwani N., Lin F., Collingwood A., et al. (2023). SLCO5A1 and synaptic assembly genes contribute to impulsivity in juvenile myoclonic epilepsy. NPJ Genom. Med. 8:28. 10.1038/s41525-023-00370-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruivo R., Anne C., Sagne C., Gasnier B. (2009). Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim. Biophys. Acta 1793 636–649. 10.1016/j.bbamcr.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Sodani K., Patel A., Kathawala R. J., Chen Z. S. (2012). Multidrug resistance associated proteins in multidrug resistance. Chin. J. Cancer 31 58–72. 10.5732/cjc.011.10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M., Wasserman D. (2020). Genetic origins of suicidality? A synopsis of genes in suicidal behaviours, with regard to evidence diversity, disorder specificity and neurodevelopmental brain transcriptomics. Eur. Neuropsychopharmacol. 37 1–11. 10.1016/j.euroneuro.2020.06.002 [DOI] [PubMed] [Google Scholar]
- Sookoian S., Castano G., Gianotti T. F., Gemma C., Pirola C. J. (2009). Polymorphisms of MRP2 (ABCC2) are associated with susceptibility to nonalcoholic fatty liver disease. J. Nutr. Biochem. 20 765–770. 10.1016/j.jnutbio.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Subbotina E., Yang H. Q., Gando I., Williams N., Sampson B. A., Tang Y., et al. (2019). Functional characterization of ABCC9 variants identified in sudden unexpected natural death. Forensic Sci. Int. 298 80–87. 10.1016/j.forsciint.2019.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D. (2011). How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron 69 628–649. 10.1016/j.neuron.2011.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M., Dejardin J. (2018). Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma 127 3–18. 10.1007/s00412-017-0656-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teafatiller T., Heskett C. W., Agrawal A., Marchant J. S., Baulch J. E., Acharya M. M., et al. (2021). Upregulation of vitamin C transporter functional expression in 5xFAD mouse intestine. Nutrients 13:617. 10.3390/nu13020617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Han X., Tian D. L. (2012). The biological regulation of ABCE1. IUBMB Life 64 795–800. 10.1002/iub.1071 [DOI] [PubMed] [Google Scholar]
- Tolomeo S., Davey F., Steele J. D., Baldacchino A. (2021). Compulsivity and impulsivity in opioid dependence. Drug Alcohol Depend. 229:109018. 10.1016/j.drugalcdep.2021.109018 [DOI] [PubMed] [Google Scholar]
- Toorie A. M., Vassoler F. M., Qu F., Schonhoff C. M., Bradburn S., Murgatroyd C. A., et al. (2021). A history of opioid exposure in females increases the risk of metabolic disorders in their future male offspring. Addict. Biol. 26:e12856. 10.1111/adb.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorie A. M., Vassoler F. M., Qu F., Slonim D., Schonhoff C. M., Byrnes E. M. (2022). Intergenerational effects of preconception opioids on glucose homeostasis and hepatic transcription in adult male rats. Sci. Rep. 12:1599. 10.1038/s41598-022-05528-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V., Vasiliou K., Nebert D. W. (2009). Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 3 281–290. 10.1186/1479-7364-3-3-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishweswaraiah S., Swierkowska J., Ratnamala U., Mishra N. K., Guda C., Chettiar S. S., et al. (2019). Epigenetically dysregulated genes and pathways implicated in the pathogenesis of non-syndromic high myopia. Sci. Rep. 9:4145. 10.1038/s41598-019-40299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Fang T., Chen H. (2023). Zinc and central nervous system disorders. Nutrients 15:2140. 10.3390/nu15092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang H., Chen X. (2019). Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2 141–160. 10.20517/cdr.2019.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zeng F., Hu Y., Li X. (2019). A mini-review of the role of glutamate transporter in drug addiction. Front. Neurol. 10:1123. 10.3389/fneur.2019.01123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm-Benartzi C. S., Koestler D. C., Karagas M. R., Flanagan J. M., Christensen B. C., Kelsey K. T., et al. (2013). Review of processing and analysis methods for DNA methylation array data. Br. J. Cancer 109 1394–1402. 10.1038/bjc.2013.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Arimura H., Fukushige T., Minami K., Nishizawa Y., Tanimoto A., et al. (2014). Abcb10 role in heme biosynthesis in vivo: Abcb10 knockout in mice causes anemia with protoporphyrin IX and iron accumulation. Mol. Cell Biol. 34 1077–1084. 10.1128/MCB.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Sivils A., Cegielski V., Singh S., Chu X. P. (2023). Transient receptor potential (TRP) channels in pain, neuropsychiatric disorders, and epilepsy. Int. J. Mol. Sci. 24:4714. 10.3390/ijms24054714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V., Butelman E. R., Randesi M., van den Brink W., Blanken P., van Ree J. M., et al. (2021). Association of serotonin transporter (SERT) polymorphisms with opioid dependence and dimensional aspects of cocaine use in a caucasian cohort of opioid users. Neuropsychiatr. Dis.Treat. 17 659–670. 10.2147/NDT.S286536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang L., Chen L., Ren W., Mei A., Chen X., et al. (2013). Two novel mutations of the NCSTN gene in Chinese familial acne inverse. J. Eur. Acad. Dermatol. Venereol. 27 1571–1574. 10.1111/j.1468-3083.2012.04627.x [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang N., Ma S., Wei Z., Tao P., Cai H. (2024). SLC25 family with energy metabolism and immunity in malignant tumors. Oncologie 26 65–77. 10.1515/oncologie-2023-0280 [DOI] [Google Scholar]
- Zhang Y. (2018). Overview of transporters in pharmacokinetics and drug discovery. Curr. Protoc. Pharmacol. 82:e46. 10.1002/cpph.46 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.