Brucella ovis leucine-, isoleucine-, valine-, threonine- and alanine-binding protein structures have a prototypical bacterial periplasmic amino acid-binding protein topology with a conformationally flexible peptide-binding cavity in the absence of peptide.
Keywords: SSGCID, structural genomics, brucellosis, epididymitis, transport protein, drug repurposing
Abstract
Brucella ovis is an etiologic agent of ovine epididymitis and brucellosis that causes global devastation in sheep, rams, goats, small ruminants and deer. There are no cost-effective methods for the worldwide eradication of ovine brucellosis. B. ovis and other protein targets from various Brucella species are currently in the pipeline for high-throughput structural analysis at the Seattle Structural Genomics Center for Infectious Disease (SSGCID), with the aim of identifying new therapeutic targets. Furthermore, the wealth of structures generated are effective tools for teaching scientific communication, structural science and biochemistry. One of these structures, B. ovis leucine-, isoleucine-, valine-, threonine- and alanine-binding protein (BoLBP), is a putative periplasmic amino acid-binding protein. BoLBP shares less than 29% sequence identity with any other structure in the Protein Data Bank. The production, crystallization and high-resolution structures of BoLBP are reported. BoLBP is a prototypical bacterial periplasmic amino acid-binding protein with the characteristic Venus flytrap topology of two globular domains encapsulating a large central cavity containing the peptide-binding region. The central cavity contains small molecules usurped from the crystallization milieu. The reported structures reveal the conformational flexibility of the central cavity in the absence of bound peptides. The structural similarity to other LBPs can be exploited to accelerate drug repurposing.
1. Introduction
Brucellosis is highly contagious and affects both economically important livestock and wild animals (Ducrotoy et al., 2017 ▸; Godfroid, Garin-Bastuji et al., 2013 ▸; Godfroid et al., 2011 ▸; Megersa et al., 2011 ▸; Rossetti et al., 2022 ▸). Even when not resulting in significant zoonotic disease, as in the case of Brucella ovis (sheep and rams), brucellosis is economically devastating globally (Peck & Bruce, 2017 ▸; Franc et al., 2018 ▸). While brucellosis has been eradicated in cattle and small ruminants in some industrialized countries, it remains endemic globally within many animal hosts (Moreno, 2014 ▸). Current control approaches for brucellosis include vaccination, education and basic hygiene. However, these strategies have yet to reduce the disease burden successfully due to the high costs, the ineffectiveness of current antibiotics in the latter stages of brucellosis and other factors (Ariza et al., 2007 ▸; Franc et al., 2018 ▸). Notably, current vaccines are species-specific and are devastating to pregnant livestock, and some animal care practices of rural dwellers and nomadic groups are incompatible with controlling brucellosis in humans and livestock (Ducrotoy et al., 2017 ▸; Godfroid, Al Dahouk et al., 2013 ▸).
B. ovis is nonpathogenic in humans but is devastating globally to sheep, rams, goats, small ruminants and deer by causing ovine epididymitis (Rossetti et al., 2022 ▸). Similarly, B. melitensis, which causes fatal zoonotic disease in humans, also causes ovine epididymitis (Rossetti et al., 2022 ▸). Brucella are classified as category B infectious agents that can be aerosolized, and these small Gram-negative, facultative coccobacilli were the first bacterial agent to successfully be developed for biological warfare by the United States (de Figueiredo et al., 2015 ▸; Riedel, 2004 ▸). There is a continued need for brucellosis treatments in infected people and livestock. New approaches include the rational design or repurposing of small molecules that target proteins that are vital for bacterial survival. Towards these ends, the Seattle Structural Genomics Center for Infectious Disease (SSGCID) has determined the crystal structures of over 120 potential target proteins from different Brucella species. These structures provide a wealth of data for functionally and structurally characterizing Brucella proteins that are potential therapeutic targets and provide insights into fundamental mechanisms that can be used for drug discovery. These structures are used to engage undergraduates in structure analysis and scientific communication (Brooks et al., 2022 ▸; Davidson et al., 2022 ▸; Maddy et al., 2022 ▸; Porter et al., 2022 ▸; Beard, Bristol et al., 2022 ▸; Beard, Subramanian et al., 2022 ▸). Here, we present high-resolution crystal structures of B. ovis leucine-, isoleucine-, valine-, threonine- and alanine-binding protein (BoLBP). BoLBP is a putative periplasmic amino acid-binding protein with less than 29% sequence identity to any previously reported structure. We report high-resolution structures of BoLBP in orthorhombic and monoclinic space groups that reveal a prototypical periplasmic amino acid-binding protein.
2. Materials and methods
2.1. Macromolecule production
Cloning, expression and purification followed standard protocols as described previously (Bryan et al., 2011 ▸; Choi et al., 2011 ▸; Serbzhinskiy et al., 2015 ▸; Brooks et al., 2022 ▸; Davidson et al., 2022 ▸; Maddy et al., 2022 ▸; Porter et al., 2022 ▸). The leucine-, isoleucine-, valine-, threonine- and alanine-binding protein from B. ovis (BoLBP; UniProt A0A0H3ATZ3) encoding amino acids 83–471 was PCR-amplified from cDNA using the primers given in Table 1 ▸ and cloned by ligation-independent cloning (LIC), encoding a noncleavable hexahistidine tag (MAHHHHHH-ORF; Aslanidis & de Jong, 1990 ▸; Choi et al., 2011 ▸). The plasmid DNA was transformed into chemically competent Escherichia coli BL21(DE3)R3 Rosetta cells. The plasmid containing BoLBP underwent expression testing, and 2 l of culture was grown using auto-induction medium (Studier, 2005 ▸) in a LEX Bioreactor (Epiphyte Three Inc.), which allows the controlled expression of proteins, as described previously (Serbzhinskiy et al., 2015 ▸). The expression clone BrovA.17370.a.B2.GE38164 is available at https://www.ssgcid.org/available-materials/expression-clones/.
Table 1. Macromolecule-production information.
| Source organism | Brucella ovis (strain ATCC 25840, 63/290, NCTC 10512) |
| Forward primer | 5′-CTCACCACCACCACCACCATATGGCCGAACCGCTGAAGATCG-3′ |
| Reverse primer | 5′-ATCCTATCTTACTCACTTAGCCCGGACGCTTCATGGAGC-3′ |
| Cloning vector | BG1861 |
| Expression vector | BG1861 |
| Expression host | BL21(DE3)R3 Rosetta |
| Complete amino-acid sequence of the construct produced | MAHHHHHHAEPLKIALVETLSGPQASTGLLYRAAVLYQLGKINEAGGFNGEKIQILEYDNQGGPVGAADRVKAAIADGAQIIVQGSSSAVAGQITEDVRKYNLRNKGKEVLYLNLGAEALELTGSKCHFYHFRFSPNAAIHFKTVAQGMKDKGILGERAYSINQNYSWGVDVENTVVANAKEIGYEVVDKTLHEVNKIQDFSPYVAKIQAANVDTVFTGNWSNDLLLLMKAASGAGLKAKFATSFLDQPGNIGNAGAIAEGHIVSTPFNPEANGEASMAFAEDYKKVTGHYPSYVEPAAVFGLQLFGEALKNVKPGEGKINTTDIALAIENASVKTPMGDYSMRSDDHQAKFPMVVQEVSKKARIKADGTEYGFLPFKTFTGDESIDPVQESCSMKRPG |
BoLBP was purified using the established two-step SSGCID pipeline protocol consisting of an immobilized metal (Ni2+) affinity chromatography (IMAC) step and size-exclusion chromatography (SEC). All chromatography runs were performed on an ÄKTApurifier 10 (GE Healthcare) using automated IMAC and SEC programs (Bryan et al., 2011 ▸). Thawed bacterial pellets (∼25 g) were lysed by sonication in 200 ml buffer consisting of 25 mM HEPES pH 7.0, 500 mM NaCl, 5%(v/v) glycerol, 0.5%(w/v) CHAPS, 30 mM imidazole, 10 mM MgCl2, 1 mM TCEP, 250 mg ml−1 AEBSF, 0.025% sodium azide. After sonication, the crude lysate was clarified with 20 ml Benzonase (25 units ml−1) and incubated while mixing at room temperature for 45 min. The lysate was clarified by centrifugation at 10 000 rev min−1 for 1 h using a Sorvall centrifuge (Thermo Scientific). The clarified supernatant was then passed over a 5 ml Ni–NTA His-Trap FF column (GE Healthcare) which had been pre-equilibrated with loading buffer consisting of 20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 30 mM imidazole, 1 mM TCEP, 0.025%(w/v) sodium azide. The column was washed with 20 column volumes (CV) of loading buffer and was eluted with loading buffer plus 250 mM imidazole in a linear gradient over 7 CV. The peak fractions were pooled and concentrated to 5 ml. A SEC column (Superdex 75, GE Healthcare) was equilibrated with running buffer [20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM TCEP]. The peak fractions were collected and analyzed using SDS–PAGE. BoLBP eluted as a single prominent peak at a molecular mass of ∼49 kDa, and the peak fractions were pooled and concentrated to 49.9 mg ml−1 using an Amicon concentrator (Millipore). Aliquots of 200 µl were flash-frozen in liquid nitrogen and stored at −80°C until use. The purified protein BrovA.17370.a.B2.PS02287 is available at https://www.ssgcid.org/available-materials/ssgcid-proteins/.
2.2. Crystallization
Purified BoLBP was screened for crystallization in 96-well sitting-drop plates against commercially available screens, including JCSG+ HTS (Rigaku Reagents) and MCSG1 (Microlytic). Vapor-diffusion experiments consisted of equal volumes of protein solution (0.4 µl) and precipitant solution set up at 290 K against an 80 µl reservoir. The crystals were flash-cooled by harvesting and plunging them into liquid nitrogen after passing through Al’s oil or soaking in cryosolution supplemented with 20%(v/v) ethylene glycol (Table 2 ▸). Two crystallization conditions were used for data collection. The orthorhombic crystal form was obtained at basic pH using CHES–NaOH and 30%(w/v) PEG 3000, and was cryoprotected by passing through Al’s oil. Heavy-atom (iodide) phasing was facilitated by the second crystal form, which grew in high salt (1 M LiCl) and 30%(w/v) polyethylene glycol 6000 (PEG 6000). The crystals were subjected to two 30 s soaks in cryo/phasing solution with increasing concentrations of sodium iodide in 20%(v/v) ethylene glycol. A second structure was obtained from soaking crystals grown in polyethylene glycol 3350 (and 200 mM potassium nitrate) overnight with 10 mM threonine in the same buffer. The crystal was briefly dipped into cryosolution comprised of the soaking solution and 20%(v/v) ethylene glycol before vitrification in liquid nitrogen and data collection (Table 2 ▸). Future studies will include co-crystallization and harvesting at different temperatures to identify conditions that may enhance amino-acid binding to BoLBP.
Table 2. Crystallization.
| Crystal 1 | Crystal 2 (phasing) | Crystal 3 | |
|---|---|---|---|
| Method | Vapor diffusion, sitting drop | Vapor diffusion, sitting drop | Vapor diffusion, sitting drop |
| Temperature (K) | 290 | 290 | 287 |
| Protein concentration (mg ml−1) | 25 | 25 | 25 |
| Protein buffer composition | 20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM TCEP | 20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM TCEP | 20 mM HEPES pH 7.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM TCEP |
| Composition of reservoir solution | 100 mM CHES–NaOH pH 9.5, 30%(w/v) PEG 3000 | 1 M LiCl, 100 mM sodium acetate, 30%(w/v) PEG 6000 | 20%(w/v) PEG 3350, 200 mM potassium nitrate |
| Volume and ratio of drop | 0.4 µl:0.4 µl | 0.4 µl:0.4 µl | 0.5 µl:0.5 µl |
| Volume of reservoir (µl) | 50 | 50 | 50 |
| Cryoprotectant | Al’s oil | 30 s soak in cryo/phasing solution 1 [4.5 µl reservoir + 0.5 µl 2.5 M NaI, 20%(v/v) ethylene glycol], 30 s soak in cryo/phasing solution 2 [4 µl reservoir + 1 µl 2.5 M NaI, 20%(v/v) ethylene glycol] | Soak in 10 mM threonine, 20%(w/v) PEG 3350, 200 mM potassium nitrate, 20% ethylene glycol |
2.3. Data collection and processing
For the orthorhombic structure, two data sets were collected: one at 100 K on beamline 21-ID-F at the Advanced Photon Source, Argonne National Laboratory (APS), while the phasing data set was collected on a rotating-anode home source (Table 3 ▸). The monoclinic data set was collected on beamline 21-ID-F at APS. All diffraction data were integrated using XDS and reduced using XSCALE (Kabsch, 2010 ▸). Raw X-ray diffraction images are available from the Integrated Resource for Reproducibility in Macromolecular Crystallography at https://www.proteindiffraction.org.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Crystal 1 (monoclinic) | Crystal 2 (phasing data) | Crystal 3 (orthorhombic) | |
|---|---|---|---|
| PDB code | 4xfk | 7jfn | |
| Diffraction source | APS beamline 21-ID-F | Rigaku FR-E+ SuperBright | APS beamline 21-ID-F |
| Wavelength (Å) | 0.97872 | 1.54178 | 0.97872 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | RayoniX MX-225 CCD | Rigaku Saturn 944+ CCD | RayoniX MX-300 CCD |
| Crystal-to-detector distance (mm) | 115 | 50 | 250 |
| Rotation range per image (°) | 1.0 | 0.5 | 1.0 |
| Total rotation range (°) | 180 | 360 | 150 |
| Exposure time per image (s) | 1 | 30 | 1 |
| Space group | P21 | P212121 | P212121 |
| a, b, c (Å) | 62.12, 46.39, 62.40 | 60.68, 67.26, 94.49 | 46.77, 69.39, 120.75 |
| α, β, γ (°) | 90, 101.62, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) | 50–1.30 (1.33–1.30) | 50–2.05 (2.10–2.05) | 50–1.70 (1.74–1.70) |
| Total No. of reflections | 314986 (22402) | 337201 (16407) | 243859 (10451) |
| No. of unique reflections | 83917 (6070) | 46709 (3419) | 43048 (2653) |
| Completeness (%) | 97.9 (96.1) | 99.6 (97.7) | 97.7 (82.3) |
| Multiplicity | 3.8 (3.7) | 7.2 (4.8) | 5.7 (3.9) |
| 〈I/σ(I)〉 | 17.96 (2.49) | 17.64 (3.59) | 22.92 (2.57) |
| R r.i.m. | 0.054 (0.605) | 0.095 (0.575) | 0.042 (0.487) |
| Overall B factor from Wilson plot (Å2) | 10.36 | n/a | 33.79 |
2.4. Structure solution and refinement
The structure of the monoclinic conformation was phased de novo by single-wavelength anomalous dispersion (SAD) after iodide ion soaks (Abendroth et al., 2011 ▸). Iterative refinement cycles with Phenix (Adams et al., 2011 ▸) and manual rebuilding using Coot (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸) generated the model coordinates and structure factors deposited in the Protein Data Bank as entry 4xfk. The orthorhombic structure was phased by molecular replacement using the monoclinic structure as the search model and the Phaser software (McCoy et al., 2007 ▸) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▸; Krissinel et al., 2004 ▸; Winn et al., 2011 ▸; Agirre et al., 2023 ▸). After iterative refinement cycles with Phenix (Adams et al., 2011 ▸) and manual rebuilding using Coot, orthorhombic coordinates and structure factors were deposited in the Protein Data Bank as entry 7jfn. Both structures were checked using MolProbity (Williams et al., 2018 ▸). The final refinement data are reported in Table 4 ▸.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| PDB entry 4xfk (monoclinic) | PDB entry 7jfn (orthorhombic) | |
|---|---|---|
| Resolution range (Å) | 30.42–1.30 (1.33–1.30) | 34.82–1.70 (1.74–1.70) |
| Completeness (%) | 97.9 (96.1) | 97.7 (82.3) |
| σ Cutoff | F > 1.35σ(F) | F > 1.35σ(F) |
| No. of reflections, working set | 81905 (5704) | 41102 (2441) |
| No. of reflections, test set | 2009 (139) | 1944 (115) |
| Final Rcryst | 0.137 (0.194) | 0.156 (0.258) |
| Final Rfree | 0.161 (0.248) | 0.194 (0.324) |
| No. of non-H atoms | ||
| Protein | 2921 | 2918 |
| Ion | 10 | 13 |
| Ligand | 0 | 4 |
| Solvent | 540 | 288 |
| Total | 3471 | 3223 |
| R.m.s. deviations from ideal | ||
| Bond lengths (Å) | 0.006 | 0.008 |
| Angles (°) | 1.098 | 0.909 |
| Average B factors (Å2) | ||
| Protein | 12.5 | 30.4 |
| Ion | 18.0 | 34.3 |
| Ligand | 0 | 55.2 |
| Water | 26.6 | 41.6 |
| Ramachandran plot | ||
| Most favored (%) | 94.3 | 94.3 |
| Allowed (%) | 5.7 | 5.4 |
| Disallowed (%) | 0 | 0.3 |
3. Results and discussion
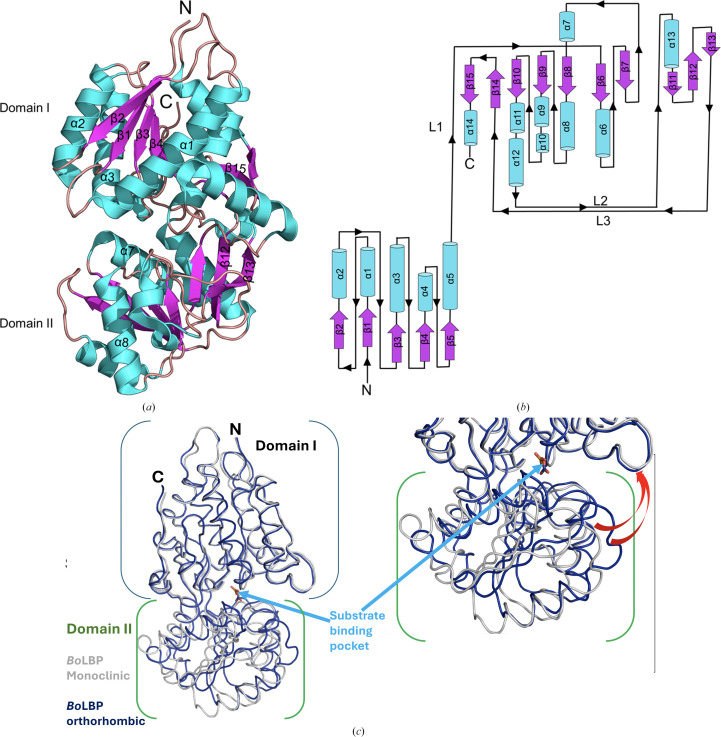
BoLBP resembles a prototypical periplasmic amino acid-binding protein, with a bilobate architecture of two major globular domains forming a Venus flytrap conformation around a large central cleft containing the peptide-binding pocket (Trakhanov et al., 2005 ▸). Both domains (domains I and II) have a characteristic α/β fold consisting of a central antiparallel β-sheet flanked by α-helices (Fig. 1 ▸). β-Strands from each sheet run towards the central cleft, exhibiting the characteristic LBP left-handed propeller twist that connects domains I and II through three interdomain loops: loop 1 (L1; residues 224–229), loop 2 (L2; residues 384–393) and loop 3 (L3; residues 423–426) (Fig. 1 ▸b). Loops 1 and 3 extend from domain I to domain II, while loop 2 traverses in the opposite direction. Loops 1 and 2 are preceded by a β-sheet strand in one domain and succeeded by an α-helix in the other domain, whereas loop 3 spans strands from both domains (Fig. 1 ▸b).
Figure 1.
BoLBP structure. (a) A ribbon diagram of BoLBP shows 15 β-sheets (purple) and 14 α-helices (cyan). (b) The BoLBP topology has two globular domains. α-Helices are labeled α, β-strands are labeled β and the three inter-domain loops are labeled L1, L2 and L3. (c) Superposed BoLBP structures. The orthorhombic monomer (blue) has a more open substrate binding cavity than the monoclinic monomer (gray).
An acetate molecule from the crystallization solution sits in the central cleft in the monoclinic structure determined without soaking with amino acids (PDB entry 4xfk; Supplementary Fig. S1b). Threonine does not bind upon soaking the orthorhombic crystals with threonine; instead, a nitrate from the crystallization solution occupies the central binding cavity (PDB entry 7jfn; Supplementary Fig. S1a). Both ligands have well ordered electron density in 2Fo − Fc maps (Supplementary Figs. S1b and S1c). The two BoLBP structures are similar, with a root-mean-square deviation (r.m.s.d.) value of 1.49 Å on aligning Cα atoms. The main differences in the structures are in domain II, which rotates around the hinge and has a more open central cavity in the orthorhombic structure, with main-chain movements of up to 4.8 Å (Fig. 1 ▸c). The differences in the structures are not as large as the conformational changes that are expected when LBP-like proteins transition from an open to a closed conformation upon binding their peptide ligands (Magnusson et al., 2004 ▸). While neither of the BoLBP structures binds an amino acid, both accommodate different ligands from the crystallization solution.
The two structures were compared using DynDom (https://dyndom.cmp.uea.ac.uk/dyndom/; Lee et al., 2003 ▸, Qi et al., 2005 ▸). DynDom analysis revealed hinge rotation by a 12° angle and conformational plasticity of the ligand-binding cavity in the structures, representing a transition from a ‘closed’ to a ‘semi-closed’ state. Further details of the hinge-bending residues and DynDom analysis results are presented in Section S2.
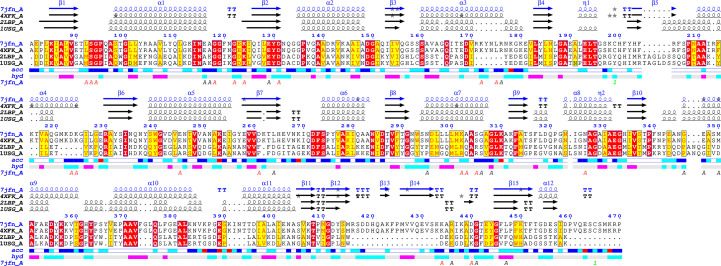
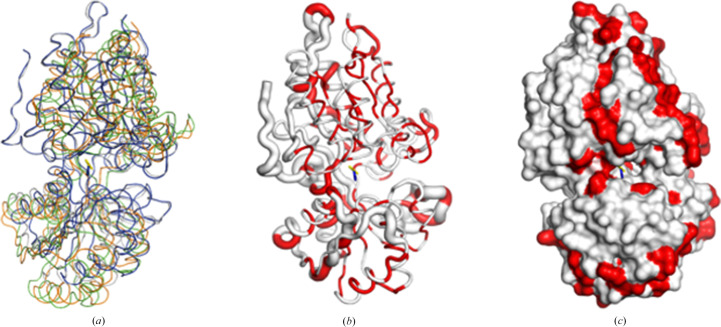
Due to the low sequence similarity of BoLBP to all other reported structures, ENDscript (Gouet et al., 2003 ▸; Robert & Gouet, 2014 ▸) analysis was used to identify its closest structural neighbor (Fig. 2 ▸). The analysis identified the closest structural neighbor of BoLBP to be E. coli LBP (EcLBP; Sack et al., 1989 ▸), and despite sharing less than 29% sequence similarity both have a similar overall topology (Fig. 2 ▸). Additionally, EcLBP and BoLBP share numerous identical residues (Fig. 2 ▸). Interestingly, BoLBP also has key amino-acid insertions resulting in longer helices and additional strands that were not previously observed in EcLBP (Fig. 2 ▸). The superposed structures show the Venus flytrap conformation around a large central cleft containing the peptide-binding pocket. Structural alignment reveals that the peptide-binding site is occupied by components of the crystallization buffer (acetate and nitrate) in our BoLBP structures (Fig. 3 ▸). The presence of these high-concentration molecules may explain the difficulty of soaking threonine into preformed crystals. ENDscript coil analysis shows that the greatest structural difference in the structures lies in the carboxyl-terminus and hinge (Fig. 3 ▸b). Identical residues appear interspersed across both domains (Figs. 2 ▸ and 3 ▸). Nonetheless, the similarities between BoLBP, EcLBP and other bacterial LBPs present unique opportunities for rational drug discovery based on the existing data.
Figure 2.
ENDscript alignment reveals conserved residues between BoLBP and its closest structural neighbors. Identical and conserved residues are highlighted in red and yellow, respectively. The different secondary-structure elements shown are α-helices (α), 310-helices (η), β-strands (β) and β-turns (TT).
Figure 3.
Comparison of BoLBP with its closest structural neighbors. (a) The superposed structures show the Venus flytrap motif with a large central cavity. The superposed structures are monoclinic BoLBP (PDB entry 4xfk, gray), orthorhombic BoLBP (PDB entry 7jfn, blue) and EcLBP (PDB entry 1usg, green; PDB entry 2lbp, orange). The acetate (yellow sticks; from PDB entry 4xfk) and nitrate (blue sticks; from PDB entry 7jfn) are shown. (b) ENDscript coil diagram with thinner ribbons representing more conserved regions and thicker ribbons representing less conserved regions; identical residues in the structures are shown in red. (c) ENDscript surface diagram: identical residues in the structures are shown in red.
4. Conclusion
We report two structures of B. ovis leucine-, isoleucine-, valine-, threonine- and alanine-binding protein (BoLBP). BoLBP is a prototypical bacterial LBP with additional amino acids inserted outside the central cavity and at the carboxyl-terminus. Both structures exhibit conformational flexibility of BoLBP in the absence of bound amino acids. Despite low sequence similarity, the structures have similarities to bacterial LBPs that can be exploited for future drug-discovery efforts.
Supplementary Material
PDB reference: BoLBP, 4xfk
PDB reference: 7jfn
Supplementary Figures. DOI: 10.1107/S2053230X24007027/ir5032sup1.pdf
DynDom movie. DOI: 10.1107/S2053230X24007027/ir5032sup2.mov
Funding Statement
This work was funded by National Institute of Allergy and Infectious Diseases grant 75N93022C00036 to Peter Myler; National Institute of Allergy and Infectious Diseases grant U01GM138433 to Oluwatoyin A. Asojo.
References
- Abendroth, J., Gardberg, A. S., Robinson, J. I., Christensen, J. S., Staker, B. L., Myler, P. J., Stewart, L. J. & Edwards, T. E. (2011). J. Struct. Funct. Genomics, 12, 83–95. [DOI] [PMC free article] [PubMed]
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Echols, N., Headd, J. J., Hung, L. W., Jain, S., Kapral, G. J., Grosse Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2011). Methods, 55, 94–106. [DOI] [PMC free article] [PubMed]
- Agirre, J., Atanasova, M., Bagdonas, H., Ballard, C. B., Baslé, A., Beilsten-Edmands, J., Borges, R. J., Brown, D. G., Burgos-Mármol, J. J., Berrisford, J. M., Bond, P. S., Caballero, I., Catapano, L., Chojnowski, G., Cook, A. G., Cowtan, K. D., Croll, T. I., Debreczeni, J. É., Devenish, N. E., Dodson, E. J., Drevon, T. R., Emsley, P., Evans, G., Evans, P. R., Fando, M., Foadi, J., Fuentes-Montero, L., Garman, E. F., Gerstel, M., Gildea, R. J., Hatti, K., Hekkelman, M. L., Heuser, P., Hoh, S. W., Hough, M. A., Jenkins, H. T., Jiménez, E., Joosten, R. P., Keegan, R. M., Keep, N., Krissinel, E. B., Kolenko, P., Kovalevskiy, O., Lamzin, V. S., Lawson, D. M., Lebedev, A. A., Leslie, A. G. W., Lohkamp, B., Long, F., Malý, M., McCoy, A. J., McNicholas, S. J., Medina, A., Millán, C., Murray, J. W., Murshudov, G. N., Nicholls, R. A., Noble, M. E. M., Oeffner, R., Pannu, N. S., Parkhurst, J. M., Pearce, N., Pereira, J., Perrakis, A., Powell, H. R., Read, R. J., Rigden, D. J., Rochira, W., Sammito, M., Sánchez Rodríguez, F., Sheldrick, G. M., Shelley, K. L., Simkovic, F., Simpkin, A. J., Skubak, P., Sobolev, E., Steiner, R. A., Stevenson, K., Tews, I., Thomas, J. M. H., Thorn, A., Valls, J. T., Uski, V., Usón, I., Vagin, A., Velankar, S., Vollmar, M., Walden, H., Waterman, D., Wilson, K. S., Winn, M. D., Winter, G., Wojdyr, M. & Yamashita, K. (2023). Acta Cryst. D79, 449–461.
- Ariza, J., Bosilkovski, M., Cascio, A., Colmenero, J. D., Corbel, M. J., Falagas, M. E., Memish, Z. A., Roushan, M. R., Rubinstein, E., Sipsas, N. V., Solera, J., Young, E. J., Pappas, G., International Society of Chemotherapy & Institute of Continuing Medical Education of Ioannina (2007). PLoS Med.4, e317. [DOI] [PMC free article] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res.18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Beard, D. K., Bristol, S., Cosby, K., Davis, A., Manning, C., Perry, L., Snapp, L., Toy, A., Wheeler, K., Young, J., Staker, B., Arakaki, T. L., Abendroth, J., Subramanian, S., Edwards, T. E., Myler, P. J. & Asojo, O. A. (2022). Acta Cryst. F78, 143. [DOI] [PMC free article] [PubMed]
- Beard, D. K., Subramanian, S., Abendroth, J., Dranow, D. M., Edwards, T. E., Myler, P. J. & Asojo, O. A. (2022). Acta Cryst. F78, 45–51. [DOI] [PMC free article] [PubMed]
- Brooks, L., Subramanian, S., Dranow, D. M., Mayclin, S. J., Myler, P. J. & Asojo, O. A. (2022). Acta Cryst. F78, 306–312. [DOI] [PMC free article] [PubMed]
- Bryan, C. M., Bhandari, J., Napuli, A. J., Leibly, D. J., Choi, R., Kelley, A., Van Voorhis, W. C., Edwards, T. E. & Stewart, L. J. (2011). Acta Cryst. F67, 1010–1014. [DOI] [PMC free article] [PubMed]
- Choi, R., Kelley, A., Leibly, D., Nakazawa Hewitt, S., Napuli, A. & Van Voorhis, W. (2011). Acta Cryst. F67, 998–1005. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davidson, J., Nicholas, K., Young, J., Conrady, D. G., Mayclin, S., Subramanian, S., Staker, B. L., Myler, P. J. & Asojo, O. A. (2022). Acta Cryst. F78, 25–30. [DOI] [PMC free article] [PubMed]
- Ducrotoy, M., Bertu, W. J., Matope, G., Cadmus, S., Conde-Álvarez, R., Gusi, A. M., Welburn, S., Ocholi, R., Blasco, J. M. & Moriyón, I. (2017). Acta Trop.165, 179–193. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Figueiredo, P. de, Ficht, T. A., Rice-Ficht, A., Rossetti, C. A. & Adams, L. G. (2015). Am. J. Pathol.185, 1505–1517. [DOI] [PMC free article] [PubMed]
- Franc, K. A., Krecek, R. C., Häsler, B. N. & Arenas-Gamboa, A. M. (2018). BMC Public Health, 18, 125. [DOI] [PMC free article] [PubMed]
- Godfroid, J., Al Dahouk, S., Pappas, G., Roth, F., Matope, G., Muma, J., Marcotty, T., Pfeiffer, D. & Skjerve, E. (2013). Comput. Immunol. Microbiol. Infect. Dis.36, 241–248. [DOI] [PubMed]
- Godfroid, J., Garin-Bastuji, B., Saegerman, C. & Blasco, J. M. (2013). Rev. Sci. Tech. OIE, 32, 27–42. [DOI] [PubMed]
- Godfroid, J., Scholz, H. C., Barbier, T., Nicolas, C., Wattiau, P., Fretin, D., Whatmore, A. M., Cloeckaert, A., Blasco, J. M., Moriyon, I., Saegerman, C., Muma, J. B., Al Dahouk, S., Neubauer, H. & Letesson, J. J. (2011). Prev. Vet. Med.102, 118–131. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res.31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E. B., Winn, M. D., Ballard, C. C., Ashton, A. W., Patel, P., Potterton, E. A., McNicholas, S. J., Cowtan, K. D. & Emsley, P. (2004). Acta Cryst. D60, 2250–2255. [DOI] [PubMed]
- Lee, R. A., Razaz, M. & Hayward, S. (2003). Bioinformatics, 19, 1290–1291. [DOI] [PubMed]
- Maddy, J., Staker, B. L., Subramanian, S., Abendroth, J., Edwards, T. E., Myler, P. J., Hybiske, K. & Asojo, O. A. (2022). Acta Cryst. F78, 135–142. [DOI] [PMC free article] [PubMed]
- Magnusson, U., Salopek-Sondi, B., Luck, L. A. & Mowbray, S. L. (2004). J. Biol. Chem.279, 8747–8752. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Megersa, B., Biffa, D., Abunna, F., Regassa, A., Godfroid, J. & Skjerve, E. (2011). Trop. Anim. Health Prod.43, 651–656. [DOI] [PubMed]
- Moreno, E. (2014). Front. Microbiol.5, 213. [DOI] [PMC free article] [PubMed]
- Peck, D. & Bruce, M. (2017). Rev. Sci. Tech. OIE, 36, 291–302. [DOI] [PubMed]
- Porter, I., Neal, T., Walker, Z., Hayes, D., Fowler, K., Billups, N., Rhoades, A., Smith, C., Smith, K., Staker, B. L., Dranow, D. M., Mayclin, S. J., Subramanian, S., Edwards, T. E., Myler, P. J. & Asojo, O. A. (2022). Acta Cryst. F78, 31–38. [DOI] [PMC free article] [PubMed]
- Qi, G., Lee, R. & Hayward, S. (2005). Bioinformatics, 21, 2832–2838. [DOI] [PubMed]
- Riedel, S. (2004). Bayl. Univ. Med. Cent. Proc.17, 400–406.
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res.42, W320–W324. [DOI] [PMC free article] [PubMed]
- Rossetti, C. A., Maurizio, E. & Rossi, U. A. (2022). Front. Vet. Sci.9, 887671. [DOI] [PMC free article] [PubMed]
- Sack, J. S., Trakhanov, S. D., Tsigannik, I. H. & Quiocho, F. A. (1989). J. Mol. Biol.206, 193–207. [DOI] [PubMed]
- Serbzhinskiy, D. A., Clifton, M. C., Sankaran, B., Staker, B. L., Edwards, T. E. & Myler, P. J. (2015). Acta Cryst. F71, 594–599. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Trakhanov, S., Vyas, N. K., Luecke, H., Kristensen, D. M., Ma, J. & Quiocho, F. A. (2005). Biochemistry, 44, 6597–6608. [DOI] [PubMed]
- Williams, C. J., Headd, J. J., Moriarty, N. W., Prisant, M. G., Videau, L. L., Deis, L. N., Verma, V., Keedy, D. A., Hintze, B. J., Chen, V. B., Jain, S., Lewis, S. M., Arendall, W. B., Snoeyink, J., Adams, P. D., Lovell, S. C., Richardson, J. S. & Richardson, J. S. (2018). Protein Sci.27, 293–315. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: BoLBP, 4xfk
PDB reference: 7jfn
Supplementary Figures. DOI: 10.1107/S2053230X24007027/ir5032sup1.pdf
DynDom movie. DOI: 10.1107/S2053230X24007027/ir5032sup2.mov