Abstract
Background
The term gliomatosis cerebri (GC), a radiology-defined highly infiltrating diffuse glioma, has been abandoned since molecular GC-associated features could not be established.
Methods
We conducted a multinational retrospective study of 104 children and adolescents with GC providing comprehensive clinical and (epi-)genetic characterization.
Results
Median overall survival (OS) was 15.5 months (interquartile range, 10.9–27.7) with a 2-year survival rate of 28%. Histopathological grading correlated significantly with median OS: CNS WHO grade II: 47.8 months (25.2–55.7); grade III: 15.9 months (11.4–26.3); grade IV: 10.4 months (8.8–14.4). By DNA methylation profiling (n = 49), most tumors were classified as pediatric-type diffuse high-grade glioma (pedHGG), H3-/IDH-wild-type (n = 31/49, 63.3%) with enriched subclasses pedHGG_RTK2 (n = 19), pedHGG_A/B (n = 6), and pedHGG_MYCN (n = 5), but only one pedHGG_RTK1 case. Within the pedHGG, H3-/IDH-wild-type subgroup, recurrent alterations in EGFR (n = 10) and BCOR (n = 9) were identified. Additionally, we observed structural aberrations in chromosome 6 in 16/49 tumors (32.7%) across tumor types. In the pedHGG, H3-/IDH-wild-type subgroup TP53 alterations had a significant negative effect on OS.
Conclusions
Contrary to previous studies, our representative pediatric GC study provides evidence that GC has a strong predilection to arise on the background of specific molecular features (especially pedHGG_RTK2, pedHGG_A/B, EGFR and BCOR mutations, chromosome 6 rearrangements).
Keywords: chromosome 6, gliomatosis cerebri, H3-wild-type and IDH-wild-type, pedHGG_RTK2, pediatric-type glioma, pediatric-type high-grade glioma
For the podcast associated with this article, please visit ‘https://soc-neuro-onc.libsyn.com/gliomatosis-cerebri-in-children’
Key Points.
The presence of gliomatosis cerebri (GC) phenotype may be considered as an independent dismal prognostic factor in hemispheric pediatric-type diffuse gliomas.
The methylome-based subtypes pediatric-type diffuse high-grade glioma (pedHGG)_RTK2A/B and (provisional) pedHGG_A/B were significantly associated with GC.
EGFR and BCOR alterations and rearrangements of chromosome 6 were the most common genetic features in pediatric GC.
Importance of the Study.
Gliomatosis cerebri (GC) is a rare, highly infiltrative phenotype of a diffuse glioma. Previous studies have failed to identify a molecular signature in GC compared with unselected gliomas in both children and adults. Our case series of more than 100 children and adolescents with GC represents a large GC cohort characterized by central radiological and pathological review. Several clinical (eg, treatment modalities, contrast enhancement), and histomolecular prognostic factors (eg, histopathological grading, presence of TP53 mutations) were detected. For the first time, we were able to identify a molecular profile: the methylation subclasses pediatric-type diffuse high-grade glioma (pedHGG)_RTK2A/B and pedHGG_A/B were significantly associated with pediatric GC. Additionally, EGFR and BCOR alterations as well as rearrangements of chromosome 6 were the most common genetic features across tumor types. Taken together, these results may reopen a discussion on the nature of pediatric GC and provide insight into the disease biology of extensively infiltrating gliomas in children.
It is now widely acknowledged that pediatric diffuse gliomas differ fundamentally in key biological features compared to their adult counterparts.1–5 In recognition of these findings, the fifth edition of the World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS) differentiates a priori between adult- and pediatric-type diffuse gliomas based on established molecular hallmarks.6 In this regard, methylome profiling has emerged as a remarkably robust and reproducible diagnostic tool for the classification of CNS tumors.7,8 However, this molecular-defined approach may neglect the clinical manifestations in various glioma (sub)types. For example, gliomatosis cerebri (GC) was originally defined as a diffuse glioma characterized by an extensively infiltrating growth pattern affecting at least 3 contiguous hemispheric lobes of the brain.9 With the revised fourth edition of the WHO classification, GC was no longer considered a distinct entity, but a phenotype characterized by a specific growth pattern of an underlying diffuse glioma without a distinct molecular signature.10 So far, similar (epi-)genetic characteristics were identified in tumors presenting a GC growth pattern as seen in various adult and pediatric types of non-GC gliomas of corresponding age groups.11,12 Since these data have been derived from adults or small pediatric case series, the biology of GC in children has not yet been conclusively defined. Thus, we conducted a multinational retrospective study of 104 children and adolescents with GC providing a comprehensive radiological, histopathological, clinical, and (epi-)genetic characterization.
Methods
Study Design
Following institutional review board (IRB number: 33–547 ex 20/21) approval by the Medical University of Graz, Austria, a Europe-wide multi-institutional, retrospective initiative was started to collect data of pediatric GC patients fulfilling the following inclusion criteria: (i) age <21 years at diagnosis, (ii) magnetic resonance imaging (MRI) at diagnosis showing a GC phenotype, (iii) neuropathological confirmation of diffuse glioma. Secondary GC following the progression of an initially localized glioma as well as primarily multifocal glioma were excluded. Fourteen European countries contributed to this study. For each included patient, either formalin-fixed paraffin-embedded and/or fresh-frozen tumor tissue was requested for histopathological and molecular analyses. The majority of methylation arrays and whole-exome sequencing (WES) were performed at the Institute of Cancer Research, London, in collaboration with the DKFZ, Heidelberg, for the purpose of this study. A subgroup (methylation array: n = 10; WES: n = 14) was performed at the Gustave Roussy Cancer Research Center, Paris.
Neuroradiological Criteria
Central neuroradiological review was mandatory and performed in accordance with the 2007 WHO classification of CNS tumors,9 the last version in which GC was still described as a distinct entity. Only tumors showing a diffuse infiltrative process involving at least 3 contiguous cerebral lobes of the brain were included. The extent of infiltration was assessed on T2- or fluid-attenuated inversion recovery (FLAIR)-weighted MR-imaging. The lobus insularis was not counted as a separate lobe. All MRI scans were reviewed centrally by one of two experienced neuroradiologists (B.B. and M.W.-M.).
Clinical Data
Apart from basic clinical parameters, clinical data included surgical procedures and nonsurgical oncological treatment. The type of resection was grouped into “biopsy” regardless of whether open or stereotactic, or “partial resection” if the extent exceeded pure diagnostic purposes acknowledging that effective debulking cannot be achieved in GC. Upfront treatment modalities were divided into radiotherapy, chemotherapy, or the combination of both. Because of the retrospective multicenter study design, chemotherapy regimens were highly heterogeneous. Therefore, data were harmonized by dividing them into 3 groups: “TMZ-Mono” if only temozolomide (TMZ) was applied, “TMZ-Multi” if TMZ was used in combination with other cytotoxic drugs or “Other” if chemotherapeutic regimes without TMZ were administered. Additionally, the administration of targeted therapies was recorded. The classification of cytotoxic therapies was performed regardless of dose, number of cycles, treatment duration, etc. Progression status was determined from local clinical and/or radiological reports.
Neuropathological Assessment
Histopathological confirmation of a diffusely infiltrating glioma was mandatory. Data regarding histological tumor type and grade were extracted from local reports based on the WHO classification of tumors of the CNS at the time of diagnosis (WHO CNS classification 20079 or 201610). Therefore, Roman numerals have been retained here. WHO grade II tumors were summarized as “GC with low-grade features” (LGC), WHO grade III and IV tumors as “GC with high-grade features” (HGC). In a second step, all cases with available tumor material were centrally reviewed for the purpose of this study by 1 experienced neuropathologist or in the context of a neuropathological panel (constituted by G.H.G., P.V., T.Pi., F.G., S.Ro., M.A., J.Z., and P.W.) validating glial differentiation and grading. Reclassification according to WHO CNS 20216 was performed retrospectively in synopsis with all molecular findings including DNA methylation profiles and the presence of pathognomonic mutations.
Genome-Wide DNA Methylation Array Profiling
DNA methylation analysis was performed using either Illumina 450K or EPIC BeadArrays. Data were preprocessed using the minfi package [v1.46.0]13 and mnp.v12b6 (DKFZ). The MNP12.5 random forest brain tumor classifier (molecularneuropathology.org) was used to assign a calibrated score to each case, associating it with 1 of 184 tumor types comprising the 2021 WHO Classification of CNS tumors and novel subclasses. DNA copy number was deduced from combined intensities using the conumee package [v1.34.0]14 as processed as combined log2 intensity data based upon an internal median processed using the R packages minfi and conumee. t-Distributed stochastic neighbor embedding (t-SNE) dimensionality reduction was carried out using the MNP12.5 classifier 10 000 training probes data. The t-SNE algorithm was applied using the Rtsne package [v0.16]15 on a distance matrix generated using 1-Pearson correlation, using default Rtsne package parameters except for “theta=0” and “max_iter=1000,” and primed with a numeric seed 12345 prior to execution for reproducibility. A clear allocation in t-SNE and/or a score of at least 0.6 was mandatory for subclass allocation.
Whole-Exome Sequencing
For whole-exome sequencing, variants were called using GATK/mutect2 using current best practices. In summary, reads were aligned to GRCh37 using bwa [v0.7.17]16 and duplicates were removed with Picard Tools (http://broadinstitute.github.io/picard/) MarkDuplicates [v2.23.8]. After base quality score recalibration with GATK 4.1.9.0 variants were called with Mutect2 in joint calling mode including a panel of normal germline samples. Allelic depth was assigned by GATK VariantsToTable and variants were annotated for functional consequences with the ensembl variant effect predictor [v101.0].17 DNA copy number was calculated from normalized coverage using Picard Tools CollectHsMetrics and segmented with DNAcopy [v1.72.3].18 Integration of mutations and copy-number alterations into oncoprint representations was performed with custom scripts in R Studio 2023.03 and R version 4.2.2.19
Statistical Analysis and Reference Cohorts
Data analysis was carried out with the software SPSS (v27)20 and R (v4.0.3)19 using a survival analysis package.21 Survival analyses were performed via Log-Rank test, Kaplan–Meier plots, and Cox-Regression (multivariate case). Comparisons between two groups regarding qualitative/quantitative variables were conducted using Fisher’s exact test/Mann–Whitney U test, respectively. In all analyses, alpha was set at 0.05 as the threshold for statistical significance. Since interpretations were explorative, adjustment of P-values for multiple testing was not performed. As a reference for comparison of molecular findings in GC, published data of adults and pediatric GC case series by Herrlinger et al.11 and Broniscer et al.12 were used. In both cohorts, methylation-derived data were updated and reclassified according to the MNP12.5 classifier. Published data on a population-based cohort of supratentorial diffuse gliomas in children (n = 80) by Sturm et al.22 served as a reference for comparison of the relative frequency of the different molecular glioma subtypes excluding nondiffuse methylation-defined neuroepithelial tumor subclasses, isolated midline location, and/or calibrated score <0.9. There was an overlap between these two collectives as two cases occurred in both the here-reported GC and this control cohort. As a reference for survival comparison in HGC, a cohort of unselected hemispheric HGGs in children (n = 108) extracted from the prospective German HIT trials (HIT-GBM-A/-B/-C/-D, interimGBM-D, HIT-HGG-2007/-2013) was compiled including tumors which were biopsied or partially resected only. Age limits were adjusted to those of the GC cohort. GC cases in the control group were excluded.
Results
Radiological, Clinical, and Histopathological Analyses Identify Prognostic Factors in Pediatric GC
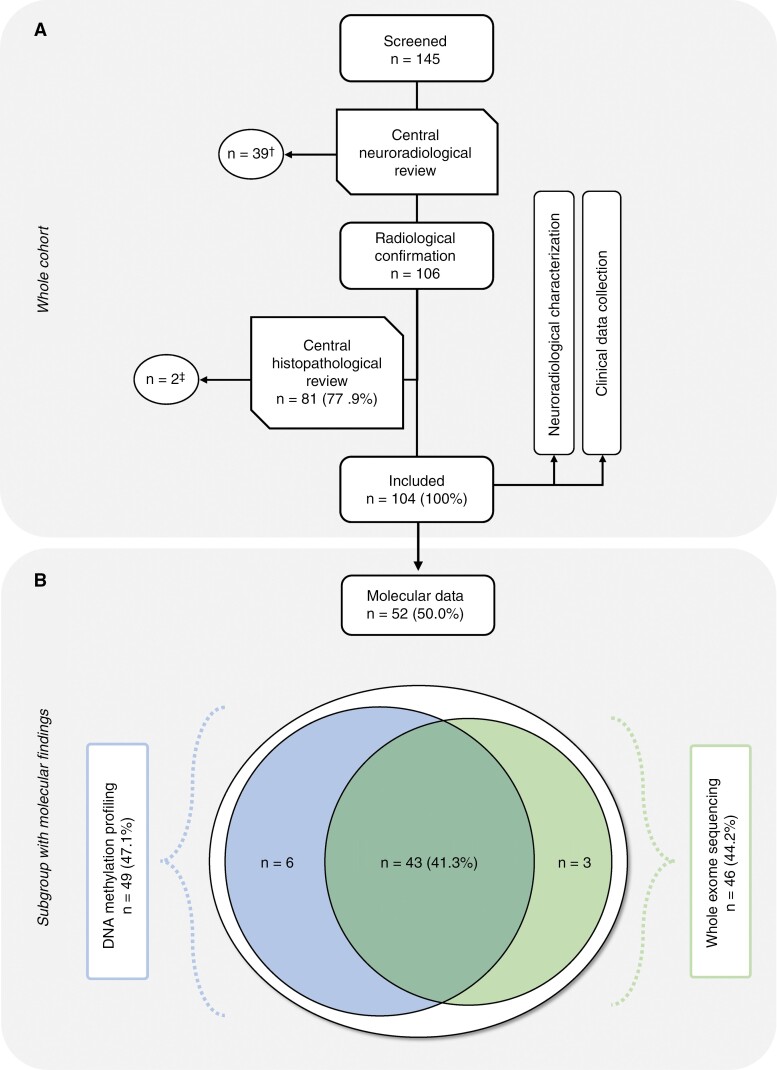
From screening 145 patients, a total of 104 children and adolescents with GC were included (Figure 1A). In most patients (n = 95, 91.5%), 3–5 cerebral lobes were affected (range 3–8). In 56 cases (53.8%), both cerebral hemispheres were involved. An additional uni- or bilateral involvement of the thalamus was observed in 48/30 (46.1%/28.9%) patients, respectively. Contrast enhancement was absent in 50 patients (48.1%) and in 49 children (47.1%) a predominantly focal enhancement was present. A representative MRI is displayed in Figure 2A.
Figure 1:
(A) Flowchart of the study cohort. (A) A total of 145 children and adolescents from 14 European countries were screened for suspected GC. †39 cases did not fulfill the neuroradiological criteria and were excluded. ‡Two tumors were excluded as a glial process could not be confirmed unambiguously through central neuropathological review. (B) Composition of the subgroup with available molecular data comprising DNA methylation profiling and whole exome sequencing.
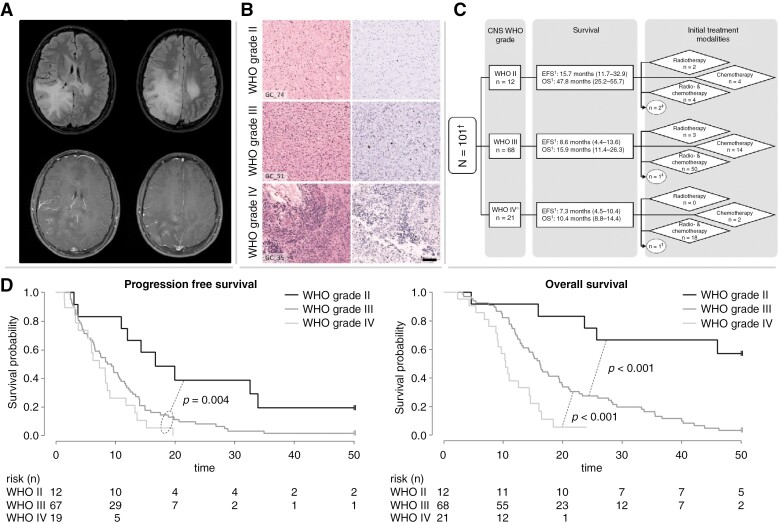
Figure 2:
Collage of clinical data of the whole cohort. (A) Representative MRI of a pediatric patient with GC. Upper row: Fluid-attenuated inversion recovery (FLAIR): high signal as a sign of diffuse tumor infiltration primarily in the right occipital, parietal, and temporal lobe as well as involvement of the contralateral hemisphere with little mass effect. Lower row: contrast-enhanced T1-weighted images: the occipital part of the tumor shows typical mild multifocal enhancement. (B) Histopathological features of GC. Microscopical examination of three representative GC tumors according to histopathological grading: the left side shows HE staining and the right Ki-67 immunohistochemistry of the respective case: GC_74, WHO grade II; GC_51, WHO grade III; GC_35, WHO grade IV. With higher WHO grade, increasing cellularity and proliferative activity can be detected. Scale bar equate to 100 µm in each case. (C) Composition of the treatment groups according to the WHO grade. 1PFS and OS given as median and the interquartile range in parentheses. †Three cases were excluded for this analysis due to absent WHO grading. ‡These patients did not receive an upfront cytotoxic treatment. (D) Kaplan–Meier plot including p-value of the (left) PFS and (right) OS in months according to WHO grading.
The median age at diagnosis was 11.8 years (range 1.3–18.8) with a male predominance of approximately 2:1. Seizures were the most frequent symptom (n = 48, 46.2%). Most tumors (n = 68, 65.4%) were described as histopathological WHO grade III, followed by grade IV (n = 21, 20.2%) and grade II (n = 12, 11.5%) according to the WHO classifications of CNS tumors of 2007 or 2016. Respective photographs of the histopathologic grades in GC are shown in Figure 2B. Overall, central neuropathological review was performed in 81 tumors (77.9%).
Seventy-nine children (75.9%) underwent initial radiotherapy. Ninety-four children (90.4%) received upfront chemotherapy (mono/combined) and, hereof, monotherapy with temozolomide was most frequently used (n = 52, 50.0%). A primary combined modality treatment of chemotherapy and radiotherapy (parallel and/or consecutive) was performed in 73 children (70.2%). Twelve patients (11.5%) received upfront targeted therapies such as anti-EGFR (n = 7) or anti-VEGF (n = 5), mostly in combination with other agents, for example, Vinorelbine or TMZ. Eight children (7.7%) underwent re-irradiation. Comprehensive radiological and clinical characteristics are summarized in Supplementary Tables 1 and 2A.
At the last follow-up, 7 patients (6.8%) were alive with a median follow-up of 83.1 months (range 13.1–138.8), and 4 of them (3.8%) survived >5 years after diagnosis. Ninety-three children (89.4%) died due to disease progression. The median progression-free (PFS) and overall survival (OS) were 8.6 months (interquartile range [Q1–Q3]: 4.3–14.0) and 15.5 months (Q1–Q3: 10.9–27.7) with 1-year and 2-year OS rates of 68.0% and 28.1%, respectively (Supplementary Table 2B).
In univariate analysis, none of the following parameters were significantly associated with PFS or OS: number of affected cerebral lobes (<5 vs. ≥5), bihemispheric or infratentorial involvement, bilateral involvement of basal ganglia, contrast enhancement, sex, age at diagnosis (±4 and ±10 years of age). However, patients with bithalamic involvement showed significantly shorter PFS (median PFS: 6.5 months [Q1–Q3: 3.0–14.0] vs. 10.4 months [Q1–Q3: 5.7–14.9]; P = .025) and a tendency toward decreased overall survival (P = .059). Furthermore, WHO grade was significantly associated with PFS (P = .004) and OS (P < .001) (Figure 2D). Median OS of patients with histological low- (WHO grade II [≙ “LGC”]) and high-grade features (WHO grade III/IV [≙ ‘HGC’]) was 52.4 (Q1–Q3: 23.7–59.0) and 14.6 months (Q1–Q3: 10.4–21.2), respectively. In HGC, contrast enhancement was associated with inferior PFS (median PFS: 6.2 months [Q1–Q3: 3.6–11.7] vs. 10.7 months [Q1–Q3: 6.3–14.9]; P = .044). Patients with high-grade tumors were more commonly treated by combined modality approaches during first-line therapy compared to patients with WHO grade II tumors (P = .017) (Figure 2C). Treatment modalities had a significant impact on PFS in the HGC subgroup: combination of radio- and chemotherapy was associated with longer PFS compared to chemotherapy alone (median PFS: 9.6 months [Q1–Q3: 5.7–14.0] vs. 4.3 months [Q1–Q3: 3.0–7.4]; P < .001), whereas the administration of targeted therapy or total irradiation dose (<50 vs. >50 Gy) did not affect PFS in the whole cohort. By contrast, in terms of treatment modalities, no difference in PFS was observed in the LGC subgroup. However, neither in the HGC nor in the LGC subgroup, OS was significantly influenced by the treatment strategy including re-irradiation or targeted therapies (VEGF or EGFR inhibition) in the respective histopathological subgroup. Multivariate analysis of the whole cohort confirmed the impact of WHO grade on PFS and OS, as well as that of treatment groups and contrast enhancement on PFS (Supplementary Table 3).
To determine whether GC phenotype may represent an independent prognostic parameter, the HGC cohort was compared with a reference collective composed of hemispheric HGGs from the German pedHGG study cohorts. Since GC by nature cannot be completely resected, and the extent of resection in pedHGG is a recognized prognostic parameter,23,24 for better comparability, we only included hemispheric tumors that were partially resected at best in the control group. Compared to this reference cohort (n = 108), HGC showed inferior overall survival (median OS: 14.6 months [Q1–Q3: 10.4–21.2] vs. 16.5 months [Q1–Q3: 11.9–34.8]; P = .007). GC phenotype, extent of resection, and histopathologic grading remained statistically significant in multivariate testing (Supplementary Figure 1 A and 1B).
Accumulation of Certain DNA Methylation-Based Subclasses and Chromosome 6 Alterations
Within our cohort of pediatric GC, tumor material was available from 52 patients (50.0%), of which 49 were subjected to DNA methylation profiling and 46 to whole-exome sequencing (WES); in 43 cases (41.3%), both DNA methylation and WES were conducted (Figure 1B).
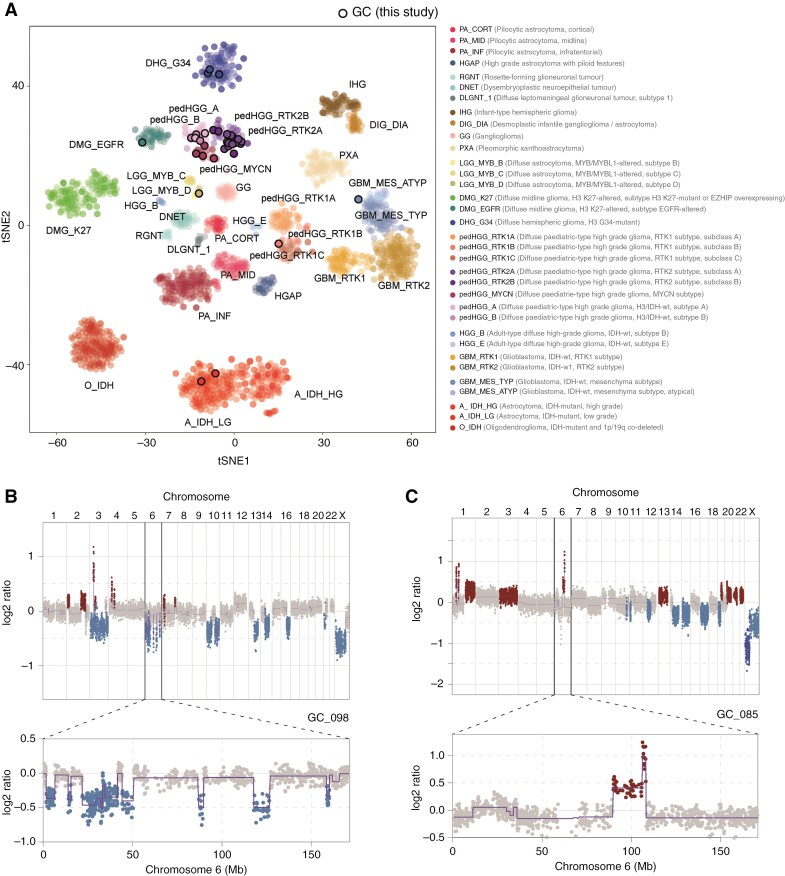
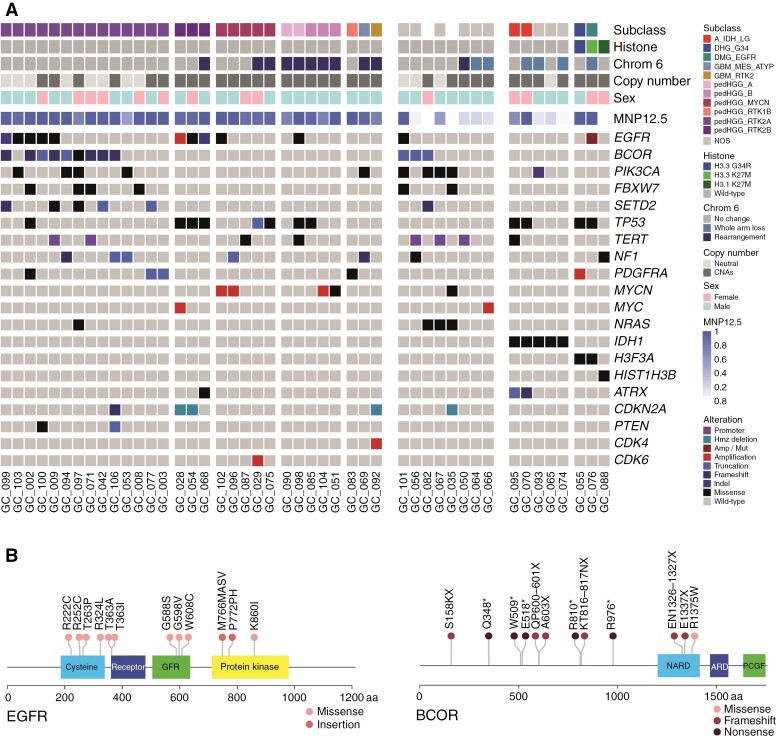
GC tumors analyzed by methylation arrays underwent subclassification by the MNP12.5 classifier and were projected by t-distributed stochastic neighbor embedding (t-SNE) alongside a reference background of n = 2305 gliomas7,25–28 of all ages, grades, classes, and subclasses (Figure 3A). A total of 9 GC cases (9/49, 18.4%) were excluded due to poor classification scores (<0.6), and/or inconsistent clustering by t-SNE, hereafter described as “NEC” (Not Elsewhere Classified). The vast majority of successfully allocated tumors (30/40, 75.0%) were assigned to the closely clustering subclasses of pedHGG_RTK2A/B (n = 16 + 3, 47.5%), pedHGG_MYCN (n = 5, 12.5%), or pedHGG_A/B (n = 2 + 4, 15.0%). The latter are considered novel, not yet published (provisional) molecular subclasses according to MNP12.5.7 There were 3 additional tumors (7.5%) of the pediatric-type DHG_G34, and individual cases (n = 1, 2.5% each) representing the midline DMG_EGFR subclass, or MYB(L1)-altered diffuse glioma, subtype D. Notably, there was only 1 tumor of the pedHGG_RTK1A/B/C subclass of a child, who had received previous CNS irradiation for leukemia. The remaining 4 tumors (10.0%) presented adult-type entities clustering as follows: A_IDH_LG (n = 2), GBM_RTK2 (n = 1), and GBM_MES_ATYP (n = 1).
Figure 3:
DNA methylation profiling in pediatric GC. (A) t-Statistic-based stochastic neighbor embedding (t-SNE) projection of a combined methylation dataset according to the MNP12.5 classifier comprising the pediatric GC cases from this study (circled, n = 40/49) plus a reference set of glioma subtypes (n = 2305). The first 2 projections are plotted on the x and y axes, with samples represented by dots colored by the respective subclass as labeled on the figure. (B, C) DNA copy-number plots for the cases GC_098 and GC_085 derived from methylation array data, with log2 ratios plotted (y axis) against genomic location by chromosome (x axis), and colored red for gain, and blue for loss. Above—whole genome; below—chromosome 6 illustrating representing examples of the structural alteration observed in our cohort.
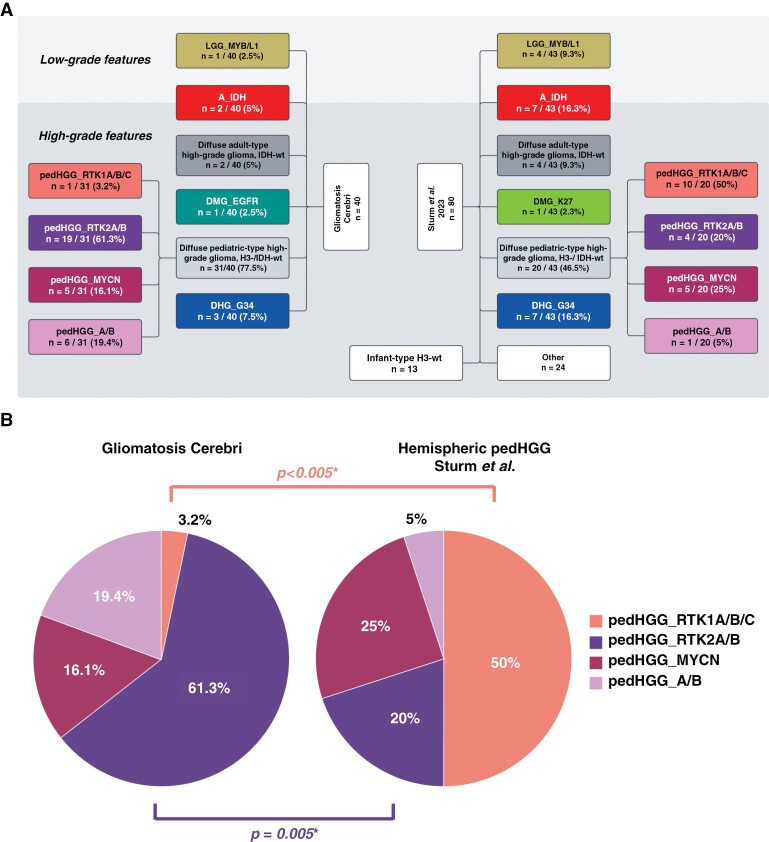
Compared with the population-based cohort of methylome-defined hemispheric diffuse gliomas by Sturm et al.22 (n = 80), there were distinct differences in the subclass distributions between the 2 collectives: apart from the absence of hemispheric infant-type, H3-wild-type (wt) tumors in the GC collective, the reference cohort showed a significantly increased frequency of pedHGG_RTK1 cases (10/20 vs. 1/31, P < .001) while tumors of the pedHGG_RTK2A/B subclass were almost absent (1/20 vs. 19/31, P = .005). Due to the small number of samples, no meaningful comparison regarding the pedHGG_MYCN and pedHGG_A/B subclass could be drawn (Figures 4A and 4B).
Figure 4:
Distribution of methylation-based subclasses. (A) Comparison of DNA methylation data according to MNP12.5 of the GC cohort (n = 40) and the published population-based collective by Sturm et al.22 (n = 80). The subclasses were arranged based on the WHO CNS 2021 classification. In the cohort by Sturm et al., only supratentorial cases with hemispheric location were included. All non-diffuse glioma subclasses, isolated midline location, and with a calibrated score <0.9 were excluded. Infant-type H3-wild-type tumors were not considered in the analysis to approximate the age distribution between the 2 cohorts (median 12 years [1–21] vs. 11.8 years [1.3–18.8]). In the category “others” 22 tumors of “pleomorphic xanthoastrocytoma (-like)” and 2 tumors of the subclass “neuroepithelial tumor, PLAGL1-fused” were present. There was an overlap between these 2 collectives as 2 cases occurred in both the here-reported GC and the control cohort. Therefore, in the subclasses pedHGG_A/B and pedHGG_RTK2A/B, 1 patient is listed once in each of the 2 cohorts. Of note, the only pedHGG_A/B-case in the reference collective was a child from the here-presented GC cohort. (B) Relative frequencies of different subclasses in the diffuse pediatric-type high-grade glioma, H3-wild-type, and IDH-wild-type subgroup of the 2 above-mentioned cohorts. *There was a significant difference in the frequencies of pedHGG_RTK2A/B- and pedHGG_RTK1A/B/C subclasses between the 2 cohorts.
In DNA copy-number profiles generated from the methylation array data, common alterations seen in pedHGG such as gain of chromosome 1q (11/49, 22.4%) were present in several cases of different methylome-defined subclasses. A significant proportion of gliomas harbored structural alterations of chromosome 6 (18/49, 36.7%). These included cases with highly complex rearrangements, partial or whole arm losses, or gains/amplifications (Figure 3B and 3C). These aberrations were found in 5/6 tumors classified as pedHGG_A/B as well as in several other subclasses, but were mutually exclusive with the pedHGG_RTK2A subclass (0/16), which harbored relatively few copy-number alterations in general (Supplementary Figure 2).
Comparison Between Different GC Cohorts Confirms Prevalence of Certain Subclasses
To determine the applicability of these results in other GC datasets, methylation array data from published case series of 18 pediatric12 and 25 adult GC cases11 were reclassified according to MNP12.5: in the pediatric set (median age: 11 years, range: 1–19), a similar pattern of molecular profiles was observed as in the present GC cohort: besides IDH-mutant (n = 3), DHG_G34 (n = 2), and DMG_EGFR (n = 1) subclasses, the remaining classifiable cases clustered as either pedHGG_RTK2A/B (n = 6), pedHGG_A (n = 1), or pedHGG_MYCN (n = 1). Likewise, no cases of pedHGG_RTK1A/B/C group were detected in the pediatric GC-control group (Supplementary Figure 3A). By contrast, in the adult series (median age: 50 years, range 24–77), an enrichment in age-typical subclasses such as A_- or O_IDH (n = 11), GBM_RTK1 (n = 4), and GBM_MES_TYP (n = 3) was present. Interestingly, within the adult cohort, there was 1 case of pedHGG_B in a 39-year-old GC patient (Supplementary Figure 3B). Notably, cases in both series (pediatric 4/18, 22.2%; adult 3/25, 12.0%) were also found to harbor structural chromosome 6 alterations including 1 pedHGG_B tumor, which was considered as chromothripsis, whereas the general pattern of DNA copy-number changes reflected the wider diffuse glioma landscapes in the respective age groups (ie, 1q+ in children, 7+/10− in adults) (Supplementary Figure 4A and 4B).
Enrichment of EGFR and BCOR Alteration in Pediatric GC
Exome sequencing identified subtype-specific alterations enriched in our GC series (Figure 5A). In addition to pathognomonic mutations in specific diffuse glioma subtypes (n = 5 of IDH_R132H, n = 1 each of H3F3A_G34R, H3F3A_K27M, HIST1H3B_K27M), a low frequency of common alterations in hemispheric pedHGG, such as CDKN2A/B deletion (4/46, 8.7%), ATRX mutation/deletion (3/46, 6.5%), or PDGFRA activating mutation/amplification (3/46, 6.5%) was detected. Alterations (mainly missense mutations) in EGFR were found in 12/46 (26.1%) tumors, particularly in the pedHGG_RTK2A/B subgroup (8/17, 47.1%), and were largely found in the extracellular domains, though with no recurrent hotspot mutations (Figure 5B). The equally most common mutated genes were TP53 and BCOR, with predicted inactivating mutations found in 12 cases each (26.1%). Three-quarters of BCOR alterations (n = 9) were present in pedHGG_RTK2A, the other 3 in the NEC subgroup. Additional common mutations in the pedHGG_RTK2A subgroup included PIK3CA (n = 4; plus 4 in NEC), FBXW7 (n = 4 + 2) and SETD2 (n = 5 + 1) (Figure 5A).
Figure 5:
Whole-exome sequencing of pediatric GC. (A) Oncoprint representation of an integrated annotation of single-nucleotide variants and DNA copy-number changes for pediatric GC (n = 46). Samples are arranged in columns with genes labeled along rows. Clinicopathological and molecular annotations are provided as bars according to the included key. (B) Lollipop plot of specific variants identified in pediatric GC cases in EGFR (left) and BCOR (right), scaled by number and colored according to the key provided. Abbreviations: cysteine-rich domain (cysteine); receptor L-domain (receptor); growth factor receptor domain 4 (GFR); protein kinase domain (protein kinase); non-ankyrin-repeat domain (NARD); ankyrin repeat domain (ARD); PCGF1-binding domain (PCGF).
Synthesis of Molecular Data According to WHO CNS 2021
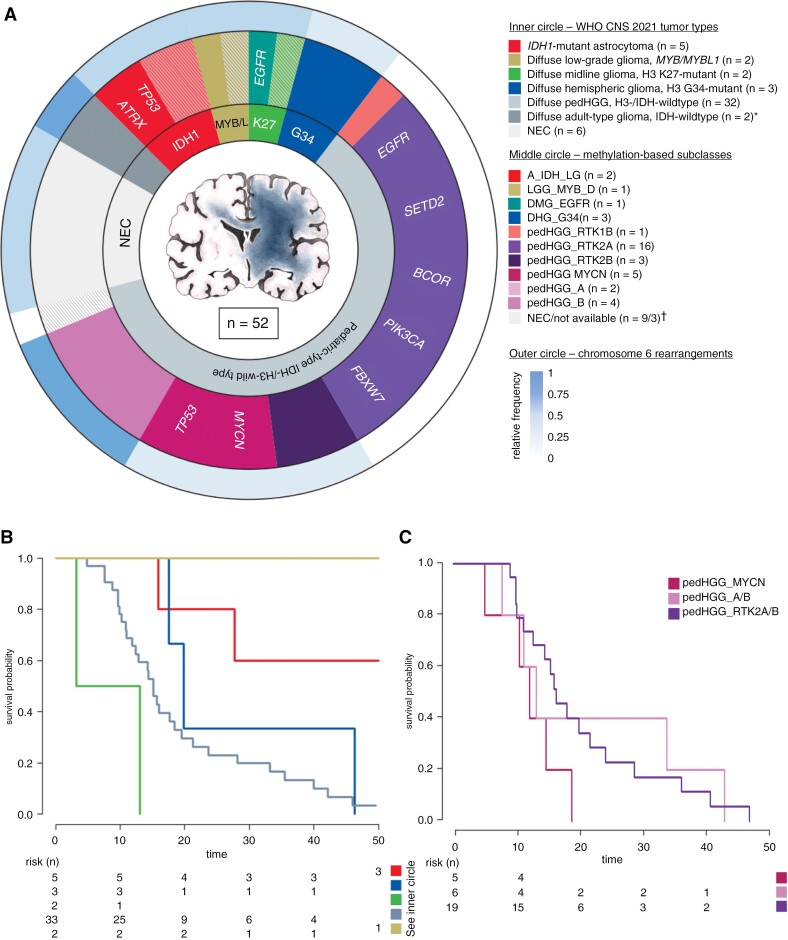
Subsequently, all tumors with molecular data available (n = 52) were reclassified using the integrated diagnostic criteria according to the WHO classification of CNS tumors of 2021,6 based on methylation arrays and the presence of pathognomonic mutations. The majority of tumors (including patients with the provisional pedHGG_A/B subclass) were classified as diffuse pedHGG, H3-wt, and IDH-wt (pedHGG_H3-/IDH-wt) (n = 32/52, 61.5%), followed by astrocytoma, IDH1-mutant (n = 5, 9.6%), and diffuse hemispheric glioma, H3 G34-mutant (n = 3, 5.8%). Two tumors each (3.8%) belonged to the diffuse midline glioma, H3 K27-altered, the diffuse LGG, MYB/MYBL1-altered, and the adult-type glioblastoma, IDH-wt, tumor (sub-)types, respectively. In none of the IDH1-altered tumor samples, oligodendroglial features or 1p/19q-codeletion were present. Six tumors (11.5%) were still considered as NEC. A summary of the molecular findings is shown in Figure 6A. Compared with the pedHGG_H3-/IDH-wt subtype (median OS: 15.2 months [Q1–Q3: 10.9–23.7]), IDH1-mutant gliomas showed prolonged survival (median OS: 54.6 months [Q1–Q3: 27.7–131.2]), whereas the number of cases is too small to draw a statistical conclusion (Figure 6B). Of the 9 LGC tumors available for molecular analysis, 4 cases were IDH1-mutant tumors, 2 cases turned out to be pedHGG_RTK2A, and 1 was MYB/MYBL-altered (2 tumors were still classified as NEC).
Figure 6:
Summary of the molecular findings in a multilayer circle diagram. (A) In total, molecular data was available of 52 tumors. The inner circle represents different tumor types according to WHO CNS 2021 in synopsis with methylation array data and exome sequencing. The middle circle illustrates the DNA methylation-based subtypes according to the MNP12.5 classifier, which clustered to the corresponding WHO CNS types. In addition, the most frequent genetic alterations of the respective subtypes are given. In both circles the (sub-)types are colored as labeled in the given key. Hatched areas represent cases with inconclusive methylation profiling, but subtype allocation was possible through detection of disease-defining genetic alterations in exome sequencing. *adult-type diffuse gliomas, IDH-wild-type (n = 2) including 1 case each of the subtypes GBM_MES_ATYP and GBM_RTK2. †Nine cases were NEC in methylation analyses, and additionally, in 3 cases, no methylation data were available. The outer circle represents the relative frequency of chromosome 6 rearrangements sorted by the respective methylome-based subclass. (B, C) Kaplan–Meier plots of the overall survival in months according to (B) the WHO CNS 2021 and to (C) the methylation-defined subclasses of the pedHGG_H3-/IDH-wt subgroup including pedHGG_A/B. Tumors of the adult-type, IDH-wild-type are not shown in (B).
Characteristics of the pedHGG_H3-/IDH-wt Subgroup With GC Phenotype
In the pedHGG_H3-/IDH-wt subgroup, 29 of the 32 patients (90.6%) succumbed to their disease. Within this cohort, several prognostic factors were observed: with a median age of 12.1 years [Q1–Q3: 8.1–14.2], children under 10 years of age (n = 10) showed decreased OS compared to older patients (median OS 10.3 months [Q1–Q3: 9.7–14.4] vs. 17.7 months [Q1–Q3: 12.4–33.2]; P = .004). With regard to the 3 different subtypes, patients of the pedHGG_MYCN subtype were younger than patients with pedHGG_RTK2A/B- and pedHGG_A/B gliomas (median age 5.3 years [range: 2.9–7.8] vs. 12.8 years [range 6.3–18.0]). Cases of pedHGG_RTK2A showed a tendency for a more extensive growth pattern at diagnosis with a median of 5 affected cerebral lobes (Q1–Q3: 4–5) versus 3 lobes ([Q1–Q3: 3–5] in both other subclasses (P = .073) (Supplementary Figure 5). In univariate analysis, neither the different methylation-based subtypes (pedHGG_RTK2A/B vs. pedHGG_MYCN vs. pedHGG_A/B) (Figure 6C), nor the presence of structural alterations of chromosome 6 were associated with survival. Of all WES-derived molecular alterations tested (including EGFR, BCOR, PIK3CA, FBXW7, SETD2, MYCN), only the presence of TP53 alterations had a significant negative effect on OS (median OS: 9.7 [Q1–Q3: 8.8–12.4] vs. 18.4 months [Q1–Q3: 14.2–28.2]; P < .001). Due to the small number of cases, the statistical analysis of the impact of administrating VEGF or EGFR inhibitors in the respective methylation-based subgroup or in EGFR-altered tumors could not be investigated in a meaningful way. In multivariate analysis of the pedHGG_H3-/IDH-wt subgroup comprising age, methylation subclass, and TP53-status, only the latter remained to affect OS significantly (P = .008) (Supplementary Table 4).
Discussion
This large multi-institutional study on childhood GC aimed at improving the knowledge of this exceptional phenotype of diffuse gliomas. In comparison to hemispheric pedHGG, we showed that GC growth pattern per se is associated with an inferior prognosis. Grading according to the WHO classification of CNS tumors 2007/2016 demonstrated a significant impact on outcome: conventional histological grading was found to be an overall and HGC-related prognostic marker, whereas the absence of histopathological high-grade features was associated with a survival benefit. We also demonstrated that GC with low-grade features does not behave clinically like pediatric low-grade glioma (pedLGG) since the majority of children with LGC succumbed eventually to tumor progression, whereas pedLGGs are associated with low mortality in general.29,30 In molecular analyses, besides MYB/L-altered tumors, no pathognomic pedLGG alteration could be identified. On the contrary, 4 of the 9 LGC tumors available for workup were IDH-altered, which underlines the different molecular origin between pedLGG and LGC. The presence of MYB/L- and IDH-altered tumors, which are associated with more favorable survival in diffuse glioma in children/adolescents,31–34 might contribute to the prolonged survival in the LGC subgroup. In summary, with respect to mortality rates and our scarce molecular data in this subgroup, we support the concept that GC with histological low-grade characteristics—even in the absence of molecular high-grade features—should be considered high-grade entities and approached as such. The adequate treatment strategy for MYB/L or IDH-altered tumors with GC phenotype remains to be determined.
We demonstrated that certain subtypes of diffuse gliomas as defined by the WHO CNS 2021 classification are significantly associated with a GC phenotype. Diffuse pediatric-type HGG, H3-wt and IDH-wt, was the most common WHO type, consisting mainly of DNA methylation subclasses pedHGG_RTK2A/B, pedHGG_A/B, and pedHGG_MYCN. Except for single clinical parameters (eg, younger age in pedHGG_MYCN), these subclasses were associated with similar clinical phenotypes and cluster closely on t-SNE. Interestingly, gliomas of the pedHGG_RTK1A/B/C subclass, which may account for approximately one-third in unselected pedHGG_H3-/IDH-wt cases35 and accounted for half in our control group,22 were virtually absent in our series as well as in another independent pediatric GC cohort published by Broniscer et al.12
Besides, our study indicates that the accumulation of some genetic aberrations may promote a GC phenotype as well. For example, gene aberrations in EGFR and BCOR were found at unexpectedly high frequencies in our pediatric GC series. Within published hemispheric cohorts of pedHGG, the frequency of EGFR mutations was 11/326 (3.4%)25 and 2/86 (2.3%).26 Unselected pedHGG_H3-/IDH-wt tumors may show EGFR amplifications,35 but EGFR mutations as described in our series are uncommon outside of the newly recognized DMG-EGFR methylation-based subclass.36,37 These diffusely infiltrating (often bithalamic) pediatric diffuse midline gliomas show phenotypic similarities with GC. However, unlike DMG-EGFR characterized by hotspot EGFR mutations, in our EGFR-mutated GC tumors, the mutations did not cluster in specific regions of the gene. BCOR and FBXW7 alterations have previously been described mainly in H3-altered HGGs,25 and SETD2 alterations in hemispheric H3-wt pedHGG.38 Whether alterations in BCOR, PIK3CA, FBXW7, or SETD2, which occurred mainly in our pedHGG_RTK2A subgroup, are significantly associated with this specific methylome-based subclass or with GC per se, has to be clarified in future studies with a higher number of tumors. Korshunov et al.35 published a small, unselected cohort of pedHGG_RTK2 cases (n = 18), which were primarily supratentorial and frequently carried EGFR amplification, but they did not perform WES to determine the status of the mutations mentioned above. Furthermore, they showed that PDGFRA amplifications are a hallmark of pedHGG_RTK1 tumors, which often arise subsequently to cranial irradiation or in the context of a replication repair deficiency such as Lynch syndrome.39,40 Consistent with the absence of pedHGG_RTK1 gliomas, PDGFRA mutations/amplifications, which are found in up to 16% of hemispheric pedHGG,26,41 were scarce in our cohort. Our pediatric GC collection was characterized by structural aberrations in chromosome 6 across various WHO 2021 CNS tumor (sub-)types as well. Chromosome 6 alterations have not yet been described in pedHGG in general, and their significance needs to be investigated in further studies on the various subtypes. Taken together, the dominance of the methylome-subclasses pedHGG_RTK2A/B and pedHGG_A/B, and the absence of certain other subclasses, which we could reproduce in an independent, reclassified case series by Broniscer et al.,12 as well as the exceptional genetic repertoire of pediatric-type diffuse gliomas with GC phenotype underlines a distinct molecular profile in these tumors. This is in contrast to earlier reports primarily due to smaller case numbers, an earlier version of the methylation classifier, and a lack of comprehensive exome sequencing.
Specific treatment recommendations for GC in children are lacking. Therapy proposals within case reports or small case series were based on approaches in unselected diffuse gliomas, but their impact in GC remained unclear.42,43 In HGC, we observed a marked difference in PFS between upfront therapy modalities favoring a combination of chemo- and radiotherapy versus chemotherapy alone. However, the OS of the whole cohort and of different subgroups was not significantly influenced by the initial treatment modalities, possibly due to the fact that irradiation, which had been omitted primarily, in most cases was subsequently administered after disease progression. Overall, our data showed that children with GC were treated very heterogeneously. Since these tumors behave very aggressively and differ clinically as well as molecularly from unselected hemispheric pedHGG, GC might warrant specific treatment approaches but due to its rarity prospective trials are difficult to conduct. We advocate that GC should be given special consideration in future studies on pedHGG, for example, by introducing an additional radiological label for GC-like diffuse glioma in clinical trials or by focusing on the radiological growth pattern in studies on methylation-based subclasses. By implementing a GC label, more attention would be paid to the clinical impact of this extensively infiltrating growth pattern, and further insights can be gained about this phenotype. In general, therapeutic approaches in GC should consider individual genetic alterations and should evaluate systematically the efficacy of tailored treatments (eg, EGFR inhibitors) as part of the multimodal approach. In our collective, the significance of such a targeted therapy could not be assessed due to the small number of cases. Clinical trial participation based on a GC phenotype, presence of specific genetic alterations, or on methylome would make study results more comparable and provide person-centered treatment options for GC patients, who have often been excluded from participation in clinical trials in the past.44
In conclusion, we provide evidence for molecular signatures enriched in pediatric-type diffuse gliomas with GC phenotype: predominance of pedHGG_RTK2A/B and pedHGG_A/B methylation-defined subclasses as well as of EGFR, BCOR, and chromosome 6 alterations; the absence of the pedHGG_RTK1A/B/C subclasses and of CDKN2A/B and PDGFRA alterations. Furthermore, we assembled comprehensive clinical and radiological characterization and could identify certain prognostic parameters (eg, histopathological grading in the whole cohort, contrast enhancement in HGC, the presence of TP53 mutations in the pedHGG_H3-/IDH-wt subgroup). Taken together, these findings expand the current knowledge of GC and provide insight into disease biology of extensively infiltrating gliomas in children.
Supplementary material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Acknowledgments
We thank Mike Hubank (Royal Marsden Hospital) and Theresa Hammerl (Graz University of Technology) for technical assistance. We remember and thank Prof. Felice Giangaspero for his life-long dedication to childhood brain tumors.
Contributor Information
Gunther Nussbaumer, Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, Medical University of Graz, Graz, Austria.
Martin Benesch, Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, Medical University of Graz, Graz, Austria.
Yura Grabovska, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
Alan Mackay, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
David Castel, U981, Molecular Predictors and New Targets in Oncology, Team Genomics and Oncogenesis of Pediatric Brain Tumors, INSERM, Gustave Roussy, Université Paris-Saclay, Villejuif, France; Department of Pediatric and Adolescent Oncology, Gustave Roussy, Université Paris-Saclay, Villejuif, France.
Jacques Grill, U981, Molecular Predictors and New Targets in Oncology, Team Genomics and Oncogenesis of Pediatric Brain Tumors, INSERM, Gustave Roussy, Université Paris-Saclay, Villejuif, France; Department of Pediatric and Adolescent Oncology, Gustave Roussy, Université Paris-Saclay, Villejuif, France.
Marta M Alonso, Department of Pediatrics, Clínica Universidad de Navarra, Pamplona, Spain; Solid Tumor Program, Center for the Applied Medical Research, Pamplona, Navarra, Spain; Health Research Institute of Navarra (IdiSNA), Pamplona, Navarra, Spain.
Manila Antonelli, Department of Radiological, Oncological and Anatomo-Pathological Sciences, Sapienza University, Rome, Italy.
Simon Bailey, Department of Paediatric Oncology, Sir James Spence Institute of Child Health, Royal Victoria Infirmary Queen Victoria Road, Newcastle upon Tyne, UK.
Joshua N Baugh, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Veronica Biassoni, Pediatric Oncology Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milano, Italy.
Mirjam Blattner-Johnson, Division of Pediatric Glioma Research, German Cancer Research Center (DKFZ) and German Cancer Consortium (DKTK), Heidelberg, Germany; Hopp Children’s Cancer Center (KiTZ), Heidelberg, Germany.
Alberto Broniscer, Division of Hematology-Oncology, Children’s Hospital of Pittsburgh, Pittsburgh, PA, U.S.A.
Andrea Carai, Neurosurgery Unit, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Giovanna Stefania Colafati, Oncological Neuroradiology and Advanced Diagnostics Unit, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Niclas Colditz, Division of Pediatric Hematology and Oncology, University Medical Center Göttingen, Göttingen, Germany.
Selim Corbacioglu, Department of Pediatric Hematology, Oncology and Stem Cell Transplantation, University of Regensburg, Regensburg, Germany.
Shauna Crampsie, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
Natacha Entz-Werle, UMR CNRS 7021—Laboratory of Bioimagery and Pathologies Team tumor signaling and therapeutic targets, University of Strasbourg, Illkirch, France; Pediatric Onco-Hematology Department—Pediatrics III, University Hospital of Strasbourg, Strasbourg, France.
Matthias Eyrich, Pediatric Hematology and Oncology, Department of Pediatrics, University Hospital Würzburg, Würzburg, Germany.
Lea L Friker, Institute of Neuropathology, DGNN Brain Tumor Reference Center, University of Bonn Medical Center, Bonn, Germany.
Michael C Frühwald, Pediatric and Adolescent Medicine, Swabian Children’s Cancer Center, University Medical Center Augsburg, Augsburg, Germany.
Maria Luisa Garrè, Neuro-Oncology Unit, IRCSS Istituto Giannina Gaslini, Genoa, Italy.
Nicolas U Gerber, Department of Oncology and Children’s Research Centre, University Children’s Hospital, Zurich, Switzerland.
Felice Giangaspero, Department of Radiological, Oncological and Anatomo-Pathological Sciences, Sapienza University, Rome, Italy.
Maria J Gil-da-Costa, Pediatric Oncology Department, University Hospital São João, Porto, Portugal.
Norbert Graf, Department of Pediatric Oncology and Hematology, Saarland University, Homburg, Germany.
Darren Hargrave, Pediatric Oncology, Great Ormond Street Hospital for Children, London, UK.
Peter Hauser, Second Department of Pediatrics, Semmelweis University, Budapest, Hungary.
Ulrich Herrlinger, Division of Clinical Neuro-Oncology, Department of Neurology, University Hospital Bonn, Bonn, Germany.
Marion Hoffmann, Division of Pediatric Hematology and Oncology, University Medical Center Göttingen, Göttingen, Germany.
Esther Hulleman, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Elisa Izquierdo, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
Sandra Jacobs, Department of Pediatric Hematology and Oncology, University Hospitals Leuven, Belgium; Pediatric Oncology, Department of Oncology, KU Leuven, Belgium.
Michael Karremann, Department of Pediatric and Adolescent Medicine, University Medical Center Mannheim, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Antonis Kattamis, Division of Pediatric Hematology-Oncology, First Department of Pediatrics, National and Kapodistrian University of Athens, “Aghia Sophia” Children’s Hospital, Athens, Greece.
Rejin Kebudi, Division of Pediatric Hematology-Oncology, Institute of Oncology, Istanbul University, Istanbul, Turkey.
Rolf-Dieter Kortmann, Department of Radiation Oncology, University of Leipzig, Leipzig, Germany.
Robert Kwiecien, Institute of Biostatistics and Clinical Research, University of Münster, Münster, Germany.
Maura Massimino, Pediatric Oncology Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milano, Italy.
Angela Mastronuzzi, Department of Onco-Hematology, Gene and Cell Therapy, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Evelina Miele, Department of Onco-Hematology, Gene and Cell Therapy, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Giovanni Morana, Department of Neurosciences, University of Turin, Turin, Italy.
Claudia M Noack, Center of Radiology, Department of Diagnostic and Interventional Radiology and Neuroradiology, Hospital of Fulda, Fulda, Germany.
Virve Pentikainen, Division of Hematology-Oncology and Stem Cell Transplantation, Children’s Hospital, Helsinki University Hospital, Helsinki, Finland.
Thomas Perwein, Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, Medical University of Graz, Graz, Austria.
Stefan M Pfister, National Center for Tumor Diseases (NCT) Heidelberg, Heidelberg, Germany; Department of Pediatric Oncology, Hematology & Immunology, Heidelberg University Hospital, Heidelberg, Germany; Division of Pediatric Neurooncology, German Cancer Research Center (DKFZ) and German Cancer Consortium (DKTK), Heidelberg, Germany; Hopp Children’s Cancer Center (KiTZ), Heidelberg, Germany.
Torsten Pietsch, Institute of Neuropathology, DGNN Brain Tumor Reference Center, University of Bonn Medical Center, Bonn, Germany.
Kleoniki Roka, Division of Pediatric Hematology-Oncology, First Department of Pediatrics, National and Kapodistrian University of Athens, “Aghia Sophia” Children’s Hospital, Athens, Greece.
Sabrina Rossi, Pathology Unit, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Stefan Rutkowski, Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Elisabetta Schiavello, Pediatric Oncology Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milano, Italy.
Clemens Seidel, Department of Radiation Oncology, University of Leipzig, Leipzig, Germany.
Jaroslav Štěrba, Department of Pediatric Oncology, University Hospital Brno and Faculty of Medicine, Masaryk University, Brno, Czech Republic.
Dominik Sturm, Department of Pediatric Oncology, Hematology & Immunology, Heidelberg University Hospital, Heidelberg, Germany; Division of Pediatric Glioma Research, German Cancer Research Center (DKFZ) and German Cancer Consortium (DKTK), Heidelberg, Germany; Hopp Children’s Cancer Center (KiTZ), Heidelberg, Germany.
David Sumerauer, Department of Pediatric Hematology and Oncology, 2nd Faculty of Medicine, Charles University in Prague and University Hospital Motol, Prague, Czech Republic.
Anna Tacke, Division of Pediatric Hematology and Oncology, University Medical Center Göttingen, Göttingen, Germany.
Sara Temelso, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
Chiara Valentini, Department of Radiotherapy and Radiation Oncology, Faculty of Medicine and University Hospital Carl Gustav Carus, Technical University Dresden, Dresden, Germany.
Dannis van Vuurden, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Pascale Varlet, GHU-Paris Psychiatry and Neuroscience, Sainte-Anne Hospital, Department of Neuropathology, Paris, France.
Sophie E M Veldhuijzen van Zanten, Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands; Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Maria Vinci, Department of Onco-Hematology, Gene and Cell Therapy, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
André O von Bueren, Department of Pediatrics, Obstetrics and Gynecology, Division of Pediatric Hematology and Oncology, University Hospital Geneva, Geneva, Switzerland.
Monika Warmuth-Metz, Institute of Diagnostic and Interventional Neuroradiology, University of Würzburg, Würzburg, Germany.
Pieter Wesseling, Department of Pathology, Amsterdam UMC, Amsterdam, The Netherlands; Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Maria Wiese, Neurosurgery Unit, Bambino Gesù Children’s Hospital (IRCCS), Rome, Italy.
Johannes E A Wolff, AbbVie Inc, Oncology Development, North Chicago, IL, U.S.A.
Josef Zamecnik, Department of Pathology and Molecular Medicine, 2nd Faculty of Medicine, Charles University in Prague and University Hospital Motol, Prague, Czech Republic.
Andrés Morales La Madrid, Pediatric Neuro-Oncology, Pediatric Cancer Center Barcelona, Hospital Sant Joan de Deu, Barcelona, Spain.
Brigitte Bison, Neuroradiological Reference Center for the Pediatric Brain Tumor (HIT) Studies of the German Society of Pediatric Oncology and Hematology, University Augsburg, Faculty of Medicine, Germany; Diagnostic and Interventional Neuroradiology, Faculty of Medicine, University of Augsburg, Augsburg, Germany.
Gerrit H Gielen, Institute of Neuropathology, DGNN Brain Tumor Reference Center, University of Bonn Medical Center, Bonn, Germany.
David T W Jones, Division of Pediatric Glioma Research, German Cancer Research Center (DKFZ) and German Cancer Consortium (DKTK), Heidelberg, Germany; Hopp Children’s Cancer Center (KiTZ), Heidelberg, Germany.
Chris Jones, Division of Molecular Pathology, Institute of Cancer Research, London, UK.
Christof M Kramm, Division of Pediatric Hematology and Oncology, University Medical Center Göttingen, Göttingen, Germany.
Conflict of interest statement
D.T.W.J. and S.M.P. are shareholders and cofounders of Heidelberg Epignostix GmbH. All remaining authors have declared that they have no competing interests.
Funding
This study including the Neuroradiological Reference Center in Augsburg (B.B.), formerly in Münster (M.W.-M.), the Neuropathological Reference Center in Bonn (T.Pi.), the Reference Center for Radiation Oncology in Dresden (C.V.), and the HIT-HGG/GBM Studies (M.H., C.M.K., and G.N., T.Pe., M.B.) were supported by the German- and Styrian Childhood Cancer Foundation (“Deutsche Kinderkrebsstiftung,” “Steirische Kinderkrebshilfe”), respectively. D.C. and J.G. acknowledge funding from “Etoile de Martin” and “Les Boucles du Cœur de la Fondation Carrefour” for the project RARE, and support from the Necker Imagine Tumor and DNA biobank (BB-033-00065). C.J. acknowledges funding from the “Rudy A Menon Foundation”, “CRIS Cancer Foundation”, the “Ollie Young Foundation”, “Cancer Research UK” (C13468/A23536, DRCRPG-Nov21\100002) and the Joshua Bembo Project, as well as NHS funding to the ICR/RMH Biomedical Research Centre, and is recipient of joint grant support from Izas la princesa guisante, L’union des rayons de soleils and the AYJ Fund. D.H. is supported by funding from the NIHR Great Ormond Street Hospital Biomedical Research Centre. The Pediatric Cancer Center Barcelona (A.M.L.M) acknowledges funding from the “Izas, la Princesa Guisante Foundation.” D.S. acknowledges funding from PRIMUS/19/MED/06, Charles University Grant Agency, Czech Republic. The Bambino Gesù Children’s Hospital (A.C., G.St.C, A.M., E.M., S.Ro., M.V.) acknowledges funding from “Fondazione Heal,” “Il Coraggio dei Bambini,” “Martina e La Sua Luna” and “Il laboratorio di Chiara.” M.M. acknowledges funding from the Italian Ministry of Health and by Regione Lombardia (project ID NET-2019-12371188). M.L.G. acknowledges support by the Italian Ministry of Health and by Regione Liguria (Ricerca Finalizzata di Rete NET-2019-12371188, GLI-HOPE).
Authorship statement
G.N., M.B., A.M.L.M, C.J., and C.M.K. conceived the study. G.N., M.B., C.J., and C.M.K. have written the manuscript. M.M.A., S.B., J.N.B., V.B., A.C., G.St.C., N.C., S.Co., N.E.W., M.E., M.C.F., M.L.G., N.U.G., M.J.G.C., N.G., D.C., D.H., J.G., P.H., M.H., E.H., E.I., S.J., M.K., A.K., R.Ke., R.D.K., M.M., A.M., E.M., G.M., V.P., T.Pe., K.R., S.Ru., C.S., E.S., J.St., D.Su., A.T., C.V., D.v.V., S.E.M.V.v.Z., M.V., A.O.v.B., M.W., J.E.A.W., A.M.LM, and C.M.K. provided samples and the clinical data. C.M.N., M.W.-M., and B.B. collected the MRI scans and performed the radiological reviews. G.H.G., L.L.F., P.V., T.Pi., F.G., S.Ro., M.A., J.Z., and P.W. processed the histopathological samples and carried out the neuropathological review. Y.G., A.Mac., C.J., D.C., D.T.W.J., M.B.J., D.St., and S.M.P. advised methylation data interpretation and provided published data for comparison of unselected hemispheric diffuse glioma. A.B. and U.H. provided published pediatric- and adult GC case series for comparison. Y.G., A.Mac., D.C., S.Cr., S.T., D.T.W.J., and C.J. performed the molecular analyses. G.N., Y.G., A.Mac., and C.J. designed the figures and tables. G.N. and R.Kw. carried out the statistical analyses. All authors approved the final manuscript.
Data Availability
Original data originated in the course of this study, that are not accessible by the publication, will be made available personally upon reasonable request.
Prior presentation
• Preliminary results of this study have in part been presented as a poster at the 20th International Symposium on Pediatric Neuro-Oncology, June 12th –15th, 2022, Hamburg, Germany.
• Results have been presented as a poster in part at the 2023 Society for Neuro-Oncology Neuro-Oncology Pediatric Research Conference, June 22nd –24th, 2023, Washington, DC, USA.
References
- 1. Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 3. Zhang J, Wu G, Miller CP, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones C, Baker SJ.. Unique genetic and epigenetic mechanisms driving signatures of paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10):651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones C, Karajannis MA, Jones DTW, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro-Oncol. 2017;19(2):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO Classification of Tumours Editorial Board. World Health Organization Classification of Tumours of the Central Nervous System. 5th ed. Lyon: International Agency for Research on Cancer; 2021. [Google Scholar]
- 7. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Priesterbach-Ackley LP, Boldt HB, Petersen JK, et al. Brain tumour diagnostics using a DNA methylation-based classifier as a diagnostic support tool. Neuropathol Appl Neurobiol. 2020;46(5):478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.. World Health Organization Histological Classification of Tumours of the Central Nervous System. 4th ed. Lyon: International Agency for Research on Cancer; 2007. [Google Scholar]
- 10. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.. World Health Organization Classification of Tumours of the Central Nervous System. 4th ed., Updated ed. Lyon: International Agency for Research on Cancer; 2016. [Google Scholar]
- 11. Herrlinger U, Jones DTW, Hattingen E, et al. Gliomatosis cerebri: no evidence for a separate brain tumor entity. Acta Neuropathol. 2016;131(2):309–319. [DOI] [PubMed] [Google Scholar]
- 12. Broniscer A, Chamdine O, Hwang S, et al. Gliomatosis cerebri in children shares molecular characteristics with other pediatric gliomas. Acta Neuropathol. 2016;131(2):299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. R package version 1.46.0. https://bioconductor.org/packages/release/bioc/html/minfi.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hovestadt V, Zapatka M.. conumee: Enhanced Copy-Number Variation Analysis Using Illumina DNA Methylation Arrays. 2017. R package version 1.34.0. http://bioconductor.org/packages/conumee/ [Google Scholar]
- 15. Krijthe JH. Rtsne: T-Distributed Stochastic Neighbor Embedding Using Barnes-Hut Implementation. 2015. R package version 0.16. https://cran.r-project.org/web/packages/Rtsne/index.html [Google Scholar]
- 16. Li H, Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seshan VE, Olshen A.. DNAcopy: DNA Copy Number Data Analysis. doi: 10.18129/B9.bioc.DNAcopy, 2023. R package version v1.72.3 https://bioconductor.org/packages/DNAcopy [DOI] [Google Scholar]
- 19. R Core Team. R: A Language and Environment for Statistical Computing. https://www.R-project.org (R Foundation for Statistical Computing, 2021). [Google Scholar]
- 20. IBM Corp. Released 2020. IBM SPSS Statistics for Windows, Version 27.0. Armonk, NY: IBM Corp. [Google Scholar]
- 21. Therneau T. A Package for Survival Analysis in R. 2020. R package version 3.2-7. https://CRAN.R-project.org/package=survival [Google Scholar]
- 22. Sturm D, Capper D, Andreiuolo F, et al. Multiomic neuropathology improves diagnostic accuracy in pediatric neuro-oncology. Nat Med. 2023;29(4):917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finlay JL, Boyett JM, Yates AJ, et al. Randomized phase III trial in childhood high-grade astrocytoma comparing vincristine, lomustine, and prednisone with the eight-drugs-in-1-day regimen. Childrens Cancer Group. J Clin Oncol. 1995;13(1):112–123. [DOI] [PubMed] [Google Scholar]
- 24. Jakacki RI, Cohen KJ, Buxton A, et al. Phase 2 study of concurrent radiotherapy and temozolomide followed by temozolomide and lomustine in the treatment of children with high-grade glioma: a report of the Children’s Oncology Group ACNS0423 study. Neuro-Oncology. 2016;18(10):1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mackay A, Burford A, Molinari V, et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33(5):829–842.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clarke M, Mackay A, Ismer B, et al. Infant High-grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov. 2020;10(7):942–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izquierdo E, Carvalho DM, Mackay A, et al. DIPG harbors alterations targetable by MEK inhibitors, with acquired resistance mechanisms overcome by combinatorial inhibition. Cancer Discov. 2022;12(3):712–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ater JL, Zhou T, Holmes E, et al. Randomized study of two chemotherapy regimens for treatment of low-grade glioma in young children: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(21):2641–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gnekow AK, Falkenstein F, von Hornstein S, et al. Long-term follow-up of the multicenter, multidisciplinary treatment study HIT-LGG-1996 for low-grade glioma in children and adolescents of the German Speaking Society of Pediatric Oncology and Hematology. Neuro-Oncol. 2012;14(10):1265–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 Define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 32. Yeo KK, Alexandrescu S, Cotter JA, et al. Multi-institutional study of the frequency, genomic landscape, and outcome of IDH-mutant glioma in pediatrics. Neuro Oncol. 2023;25(1):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiang J, Harreld JH, Tinkle CL, et al. A single-center study of the clinicopathologic correlates of gliomas with a MYB or MYBL1 alteration. Acta Neuropathol. 2019;138(6):1091–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryall S, Zapotocky M, Fukuoka K, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. 2020;37(4):569–583.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korshunov A, Schrimpf D, Ryzhova M, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134(3):507–516. [DOI] [PubMed] [Google Scholar]
- 36. Mondal G, Lee JC, Ravindranathan A, et al. Pediatric bithalamic gliomas have a distinct epigenetic signature and frequent EGFR exon 20 insertions resulting in potential sensitivity to targeted kinase inhibition. Acta Neuropathol. 2020;139(6):1071–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sievers P, Sill M, Schrimpf D, et al. A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro Oncol. 2021;23(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125(5):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dodgshun AJ, Fukuoka K, Edwards M, et al. Germline-driven replication repair-deficient high-grade gliomas exhibit unique hypomethylation patterns. Acta Neuropathol. 2020;140(5):765–776. [DOI] [PubMed] [Google Scholar]
- 40. Whitehouse JP, Howlett M, Federico A, et al. Defining the molecular features of radiation-induced glioma: a systematic review and meta-analysis. Neurooncol. Adv.. 2021;3(1):vdab109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koschmann C, Zamler D, MacKay A, et al. Characterizing and targeting PDGFRA alterations in pediatric high-grade glioma. Oncotarget. 2016;7(40):65696–65706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Armstrong GT, Phillips PC, Rorke-Adams LB, et al. Gliomatosis cerebri: 20 years of experience at the Children’s Hospital of Philadelphia. Cancer. 2006;107(7):1597–1606. [DOI] [PubMed] [Google Scholar]
- 43. Landi A, Piccirilli M, Mancarella C, Giangaspero F, Salvati M.. Gliomatosis cerebri in young patients’ report of three cases and review of the literature. Childs Nerv Syst. 2011;27(1):19–25. [DOI] [PubMed] [Google Scholar]
- 44. Greenfield Jeffrey P, Alicia CH, Emilie G, et al. Gliomatosis cerebri: a consensus summary report from the First International Gliomatosis cerebri Group Meeting, March 26–27, 2015, Paris, France. Pediatr Blood Cancer. 2016;63(12):2072–2077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data originated in the course of this study, that are not accessible by the publication, will be made available personally upon reasonable request.