Abstract
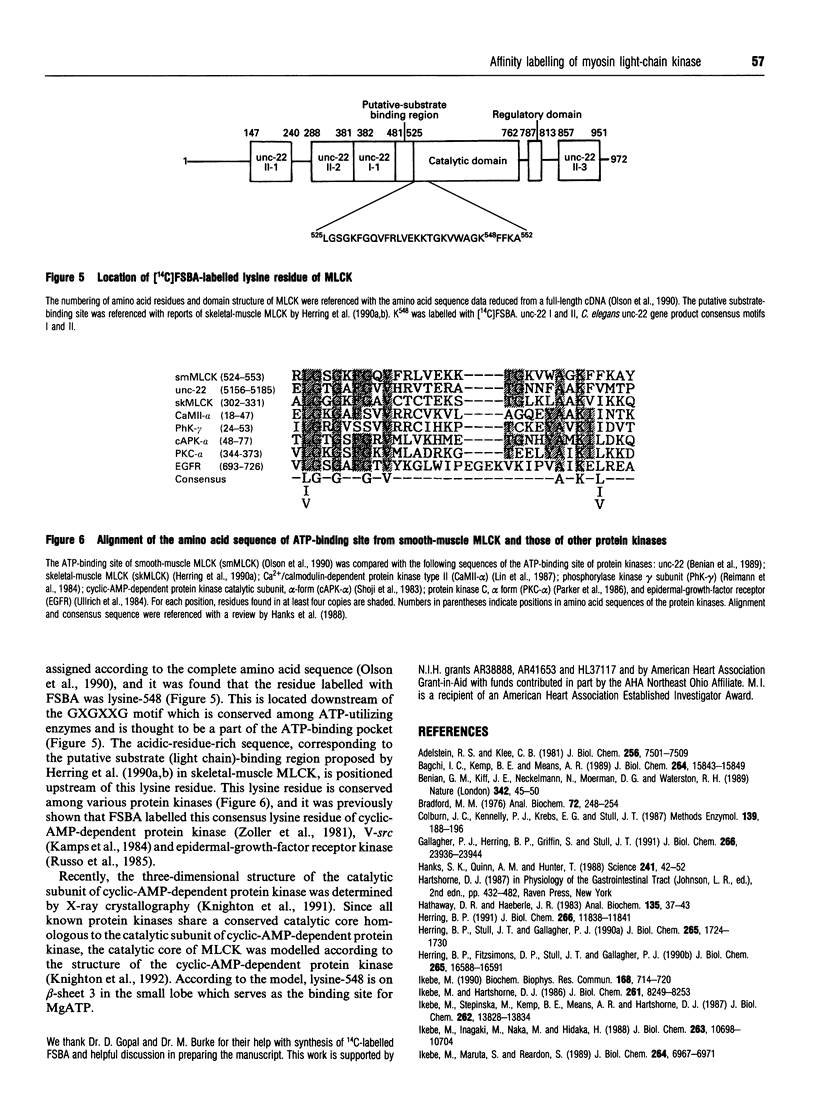
5'-(p-(Fluorosulphonyl)[14C]benzoyl)adenosine (FSBA) was synthesized and used as a probe to study the ATP-binding site of smooth-muscle myosin light-chain kinase (MLCK). FSBA modified both free MLCK and calmodulin/MLCK complex, resulting in inactivation of the kinase activity. Nearly complete protection of the calmodulin/MLCK complex against FSBA modification was obtained by addition of excess ATP whereas MLCK activity alone was lost in a dose-dependent manner even in the presence of excess ATP. These results suggest that FSBA modified ATP-binding sites and ATP-independent sites, and the latter sites are protected by calmodulin binding. The results also suggest that the ATP-binding site is accessible to the nucleotide substrate regardless of calmodulin binding. The FSBA-labelled MLCK was completely proteolysed by alpha-chymotrypsin, and the 14C-labelled peptides were isolated and sequenced. The sequence of the labelled peptide was Ala-Gly-X-Phe, where X is the labelled residue. The sequence was compared with the known MLCK sequence, and the labelled residue was identified as lysine-548, which is located downstream of the GXGXXG motif conserved among ATP-utilizing enzymes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelstein R. S., Klee C. B. Purification and characterization of smooth muscle myosin light chain kinase. J Biol Chem. 1981 Jul 25;256(14):7501–7509. [PubMed] [Google Scholar]
- Bagchi I. C., Kemp B. E., Means A. R. Myosin light chain kinase structure function analysis using bacterial expression. J Biol Chem. 1989 Sep 25;264(27):15843–15849. [PubMed] [Google Scholar]
- Benian G. M., Kiff J. E., Neckelmann N., Moerman D. G., Waterston R. H. Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature. 1989 Nov 2;342(6245):45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Colburn J. C., Kennelly P. J., Krebs E. G., Stull J. T. Affinity labeling of the nucleotide-binding site of myosin light chain kinases. Methods Enzymol. 1987;139:188–196. doi: 10.1016/0076-6879(87)39085-8. [DOI] [PubMed] [Google Scholar]
- Gallagher P. J., Herring B. P., Griffin S. A., Stull J. T. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991 Dec 15;266(35):23936–23944. [PMC free article] [PubMed] [Google Scholar]
- Hanks S. K., Quinn A. M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988 Jul 1;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hathaway D. R., Haeberle J. R. Selective purification of the 20,000-Da light chains of smooth muscle myosin. Anal Biochem. 1983 Nov;135(1):37–43. doi: 10.1016/0003-2697(83)90726-1. [DOI] [PubMed] [Google Scholar]
- Herring B. P. Basic residues are important for Ca2+/calmodulin binding and activation but not autoinhibition of rabbit skeletal muscle myosin light chain kinase. J Biol Chem. 1991 Jun 25;266(18):11838–11841. [PubMed] [Google Scholar]
- Herring B. P., Fitzsimons D. P., Stull J. T., Gallagher P. J. Acidic residues comprise part of the myosin light chain-binding site on skeletal muscle myosin light chain kinase. J Biol Chem. 1990 Sep 25;265(27):16588–16591. [PMC free article] [PubMed] [Google Scholar]
- Herring B. P., Stull J. T., Gallagher P. J. Domain characterization of rabbit skeletal muscle myosin light chain kinase. J Biol Chem. 1990 Jan 25;265(3):1724–1730. [PMC free article] [PubMed] [Google Scholar]
- Ikebe M., Hartshorne D. J. Reverse reaction of smooth muscle myosin light chain kinase. Formation of ATP from phosphorylated light chain plus ADP. J Biol Chem. 1986 Jun 25;261(18):8249–8253. [PubMed] [Google Scholar]
- Ikebe M., Inagaki M., Naka M., Hidaka H. Correlation of conformation and phosphorylation and dephosphorylation of smooth muscle myosin. J Biol Chem. 1988 Aug 5;263(22):10698–10704. [PubMed] [Google Scholar]
- Ikebe M., Maruta S., Reardon S. Location of the inhibitory region of smooth muscle myosin light chain kinase. J Biol Chem. 1989 Apr 25;264(12):6967–6971. [PubMed] [Google Scholar]
- Ikebe M. Mode of inhibition of smooth muscle myosin light chain kinase by synthetic peptide analogs of the regulatory site. Biochem Biophys Res Commun. 1990 Apr 30;168(2):714–720. doi: 10.1016/0006-291x(90)92380-i. [DOI] [PubMed] [Google Scholar]
- Ikebe M., Stepinska M., Kemp B. E., Means A. R., Hartshorne D. J. Proteolysis of smooth muscle myosin light chain kinase. Formation of inactive and calmodulin-independent fragments. J Biol Chem. 1987 Oct 5;262(28):13828–13834. [PubMed] [Google Scholar]
- Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992 May 1;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Kamps M. P., Taylor S. S., Sefton B. M. Direct evidence that oncogenic tyrosine kinases and cyclic AMP-dependent protein kinase have homologous ATP-binding sites. Nature. 1984 Aug 16;310(5978):589–592. doi: 10.1038/310589a0. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B., Guerriero V., Jr, Bagchi I. C., Means A. R. The calmodulin binding domain of chicken smooth muscle myosin light chain kinase contains a pseudosubstrate sequence. J Biol Chem. 1987 Feb 25;262(6):2542–2548. [PubMed] [Google Scholar]
- Kennelly P. J., Leng J., Marchand P. The MgATP-binding site on chicken gizzard myosin light chain kinase remains open and functionally competent during the calmodulin-dependent activation-inactivation cycle of the enzyme. Biochemistry. 1992 Jun 16;31(23):5394–5399. doi: 10.1021/bi00138a022. [DOI] [PubMed] [Google Scholar]
- Knighton D. R., Pearson R. B., Sowadski J. M., Means A. R., Ten Eyck L. F., Taylor S. S., Kemp B. E. Structural basis of the intrasteric regulation of myosin light chain kinases. Science. 1992 Oct 2;258(5079):130–135. doi: 10.1126/science.1439761. [DOI] [PubMed] [Google Scholar]
- Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991 Jul 26;253(5018):407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Lin C. R., Kapiloff M. S., Durgerian S., Tatemoto K., Russo A. F., Hanson P., Schulman H., Rosenfeld M. G. Molecular cloning of a brain-specific calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5962–5966. doi: 10.1073/pnas.84.16.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas T. J., Burgess W. H., Prendergast F. G., Lau W., Watterson D. M. Calmodulin binding domains: characterization of a phosphorylation and calmodulin binding site from myosin light chain kinase. Biochemistry. 1986 Mar 25;25(6):1458–1464. doi: 10.1021/bi00354a041. [DOI] [PubMed] [Google Scholar]
- Olson N. J., Pearson R. B., Needleman D. S., Hurwitz M. Y., Kemp B. E., Means A. R. Regulatory and structural motifs of chicken gizzard myosin light chain kinase. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2284–2288. doi: 10.1073/pnas.87.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker P. J., Coussens L., Totty N., Rhee L., Young S., Chen E., Stabel S., Waterfield M. D., Ullrich A. The complete primary structure of protein kinase C--the major phorbol ester receptor. Science. 1986 Aug 22;233(4766):853–859. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Ito M., Morrice N. A., Smith A. J., Condron R., Wettenhall R. E., Kemp B. E., Hartshorne D. J. Proteolytic cleavage sites in smooth muscle myosin-light-chain kinase and their relation to structural and regulatory domains. Eur J Biochem. 1991 Sep 15;200(3):723–730. doi: 10.1111/j.1432-1033.1991.tb16237.x. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Wettenhall R. E., Means A. R., Hartshorne D. J., Kemp B. E. Autoregulation of enzymes by pseudosubstrate prototopes: myosin light chain kinase. Science. 1988 Aug 19;241(4868):970–973. doi: 10.1126/science.3406746. [DOI] [PubMed] [Google Scholar]
- Reimann E. M., Titani K., Ericsson L. H., Wade R. D., Fischer E. H., Walsh K. A. Homology of the gamma subunit of phosphorylase b kinase with cAMP-dependent protein kinase. Biochemistry. 1984 Aug 28;23(18):4185–4192. doi: 10.1021/bi00313a027. [DOI] [PubMed] [Google Scholar]
- Roush C. L., Kennelly P. J., Glaccum M. B., Helfman D. M., Scott J. D., Krebs E. G. Isolation of the cDNA encoding rat skeletal muscle myosin light chain kinase. Sequence and tissue distribution. J Biol Chem. 1988 Jul 25;263(21):10510–10516. [PubMed] [Google Scholar]
- Russo M. W., Lukas T. J., Cohen S., Staros J. V. Identification of residues in the nucleotide binding site of the epidermal growth factor receptor/kinase. J Biol Chem. 1985 May 10;260(9):5205–5208. [PubMed] [Google Scholar]
- Shoji S., Ericsson L. H., Walsh K. A., Fischer E. H., Titani K. Amino acid sequence of the catalytic subunit of bovine type II adenosine cyclic 3',5'-phosphate dependent protein kinase. Biochemistry. 1983 Jul 19;22(15):3702–3709. doi: 10.1021/bi00284a025. [DOI] [PubMed] [Google Scholar]
- Takio K., Blumenthal D. K., Walsh K. A., Titani K., Krebs E. G. Amino acid sequence of rabbit skeletal muscle myosin light chain kinase. Biochemistry. 1986 Dec 2;25(24):8049–8057. doi: 10.1021/bi00372a038. [DOI] [PubMed] [Google Scholar]
- Togashi C. T., Reisler E. 5'-p-Fluorosulfonylbenzoyladenosine. Inactivation of myosin subfragment 1 and a model reaction with cysteine. J Biol Chem. 1982 Sep 10;257(17):10112–10118. [PubMed] [Google Scholar]
- Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. 1984 May 31-Jun 6Nature. 309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Walsh M. P., Hinkins S., Dabrowska R., Hartshorne D. J. Smooth muscle myosin light chain kinase. Methods Enzymol. 1983;99:279–288. doi: 10.1016/0076-6879(83)99063-8. [DOI] [PubMed] [Google Scholar]
- Watterson D. M., Harrelson W. G., Jr, Keller P. M., Sharief F., Vanaman T. C. Structural similarities between the Ca2+-dependent regulatory proteins of 3':5'-cyclic nucleotide phosphodiesterase and actomyosin ATPase. J Biol Chem. 1976 Aug 10;251(15):4501–4513. [PubMed] [Google Scholar]
- Zoller M. J., Nelson N. C., Taylor S. S. Affinity labeling of cAMP-dependent protein kinase with p-fluorosulfonylbenzoyl adenosine. Covalent modification of lysine 71. J Biol Chem. 1981 Nov 10;256(21):10837–10842. [PubMed] [Google Scholar]