Abstract
Crater lake fishes are common evolutionary model systems, with recent studies suggesting a key role for gene flow in promoting rapid adaptation and speciation. However, the study of these young lakes can be complicated by human-mediated extinctions. Museum genomics approaches integrating genetic data from recently extinct species are, therefore, critical to understanding the complex evolutionary histories of these fragile systems. Here, we examine the evolutionary history of an extinct Southern Hemisphere crater lake endemic, the rainbowfish Melanotaenia eachamensis. We undertook a comprehensive sampling of extant rainbowfish populations of the Atherton Tablelands of Australia alongside historical museum material to understand the evolutionary origins of the extinct crater lake population and the dynamics of gene flow across the ecoregion. The extinct crater lake species is genetically distinct from all other nearby populations due to historic introgression between 2 proximate riverine lineages, similar to other prominent crater lake speciation systems, but this historic gene flow has not been sufficient to induce a species flock. Our results suggest that museum genomics approaches can be successfully combined with extant sampling to unravel complex speciation dynamics involving recently extinct species.
Keywords: DArTseq, diversification, hybridization, Melanotaenia, Melanotaeniidae, threatened species, introgression
With the advent of next-generation sequencing technologies and the development of new protocols, it is becoming increasingly viable to extract DNA from ethanol-preserved and formalin-fixed tissues in a minimally destructive way. These techniques can enable the use of museum specimens for genomic studies (Card et al. 2021; Lalueza-Fox 2022). This historical DNA (hDNA) unlocks new uses for museum and herbarium collections, and its study is emerging as a distinct subdiscipline that bridges the gap between ancient DNA (aDNA) and modern samples. While hDNA is obtained from voucher specimen tissues usually <200 years old, aDNA is normally recovered in trace amounts from naturally preserved organic material and is often significantly older (Raxworthy and Smith 2021). Information from hDNA has the potential to document genetic erosion in endangered or recently extinct species (e.g., Roycroft et al. 2021; Burns et al. 2023); to explore past biodiversity of extant species (e.g., Chattopadhyay et al. 2019); to understand species invasions in the last few centuries (e.g., Lees et al. 2011); to obtain genomes from recently extinct species (e.g., Feigin et al. 2018; Irestedt et al. 2019); to resolve evolutionary relationships (e.g., Baveja et al. 2021; Ciucani et al. 2023); and to resolve taxonomic uncertainties with holotypes (e.g., Wielstra and Arntzen 2014). These applications are of particular use in conservation and in documenting recent biodiversity loss.
Although museum genomics approaches typically emphasize the importance of collecting whole-genome data, few complete palaeogenomes have been sequenced, and still fewer have been sequenced at high coverage (Lalueza-Fox 2022). Whole genomes are difficult to assemble from museum specimens, particularly from older samples, as DNA degrades and fragments with time (Raxworthy and Smith 2021). Furthermore, when coverage is low, it becomes difficult to infer heterozygous loci, which limits the data’s utility in resolving population histories. Mitochondrial DNA may be easier to extract at higher coverage from preserved samples than genomic DNA due to the higher copy number of mtDNA in cells (De Bruyn et al. 2011). However, mitogenome data are unsuitable for reconstructing evolutionary histories and interrelationships that cannot be represented by tree-like structures, that is, histories of population admixture and introgression. The use of short-read nuclear DNA approaches, such as RADseq and ddRADseq, may provide a tractable alternative to whole-genome and mitochondrial DNA approaches. These data provide genome-wide information, and may be more feasible and cost-effective than whole-genome approaches for older, more degraded samples in which DNA is more fragmented. It is also easier to infer heterozygosity with RADseq data than from low coverage whole genome data, so they can also be used to explore population genetic histories including the possibility of hybridization and introgression, a useful component of exploring the past genetic diversity of endangered species with declining ranges.
Hybridization and introgression are important considerations when evaluating relationships between closely related populations, as may be done when considering past and present biodiversity of endangered species. Hybridization and introgression can facilitate diversification and evolutionary innovation (Dowling and Secor 1997; Meier et al. 2017; Marques et al. 2019) or drive homogenization and extinction (Huxel 1999; Seehausen 2006; Seehausen et al. 2008). The prevalence of interspecific hybridization varies across the tree of life, and plants show a higher incidence of hybridization than animals (Mallet 2005). The role of hybridization and subsequent introgression in adaptation and speciation is widely discussed in the literature, and its importance in plant speciation has been accepted for many years (Wong et al. 2022). Increasing evidence shows that it also plays an important role in animal speciation, and that introgression following hybridization events can result in species radiations (Meier et al. 2017; Grant and Grant 2019; Marques et al. 2019) or generate species via the formation of hybrid taxa (DeMarais et al. 1992; Abbott et al. 2013; Hobbs et al. 2022). Although hybridization is typically studied in the context of 2 interacting lineages, more complex forms of hybridization exist in nature. In such cases, multiple species may hybridize directly (e.g., Wu et al. 2018; Natola et al. 2022), or a species may act as a conduit for gene flow between 2 or more other species that do not directly interbreed (e.g., Kronforst et al. 2006; McDonald et al. 2008; Ottenburghs 2019).
Historical DNA may be particularly suited to studying evolution in dynamic but extinction-prone systems like isolated islands and lakes. Lakes play a pivotal role in understanding speciation, both in cases where isolation in lakes leads to impressive adaptive radiations, such as the amphipod and cottoid radiations of Lake Baikal (Kontula et al. 2003; Gurkov et al. 2019), or the large cichlid species flocks of East Africa (Meier et al. 2017, 2023; McGee et al. 2020), but there are also many cases where colonization of lakes does not lead to radiation (Wagner et al. 2012).
While lakes offer a fascinating natural laboratory for evolution, the youth and evolutionary dynamism often found in lake endemics can pose multiple problems. The incomplete lineage sorting and hybridization often seen in lake species flocks can be difficult to quantify using a handful of genes, leading to dramatically different evolutionary interpretations depending on the type of data utilized (Schliewen et al. 1994; Schliewen and Klee 2004; Martin et al. 2015). Additionally, lake systems can be extremely fragile, with many recent and ongoing extinctions due to invasive species and other human-mediated processes (Taylor et al. 2006; Ismail et al. 2014; McGee et al. 2015). The ever-increasing number of extinct lake endemics means that in many cases, the only available material for genetic studies is preserved specimens in museums.
Some of the most exceptional lake systems involve crater lakes. These lakes are often deeper and more isolated than other lake systems of similar sizes, and they frequently host remarkable species flocks (Schliewen and Klee 2004; Pfaender et al. 2010; von Rintelen et al. 2012; Martin et al. 2015; Lemoine et al. 2019; Kautt et al. 2020). Although many crater lake flocks were initially considered as examples of sympatric speciation (Schliewen et al. 1994), evidence suggests that crater lake diversity is more often driven by complex patterns of repeated invasion and gene flow, with both allopatric and sympatric phases (Martin et al. 2015).
Here, we utilize a museum genomics approach to examine the evolutionary history of one of Australasia’s few known crater lake endemics, the now-extinct Lake Eacham rainbowfish, Melanotaenia eachamensis (Allen and Cross 1982). While there are some examples of crater lake endemics in Australasia, such as the cyprinid Rasbora maninjau of Lake Maninjau (Lumbantobing 2014), fish radiations in this region are more commonly associated with ancient lakes, such as the Oryzias ricefish radiation of Lake Poso and the species flocks of sailfin silversides in the genera Telmatherina, Paratherina, and Tominanga found in Lakes Towuti, Mahalona, and Matano (von Rintelen et al. 2012). The Lake Eacham rainbowfish is a member of Melanotaeniidae, a family closely related to Sulawesi silversides and the most speciose clade of freshwater fishes in Australia and New Guinea.
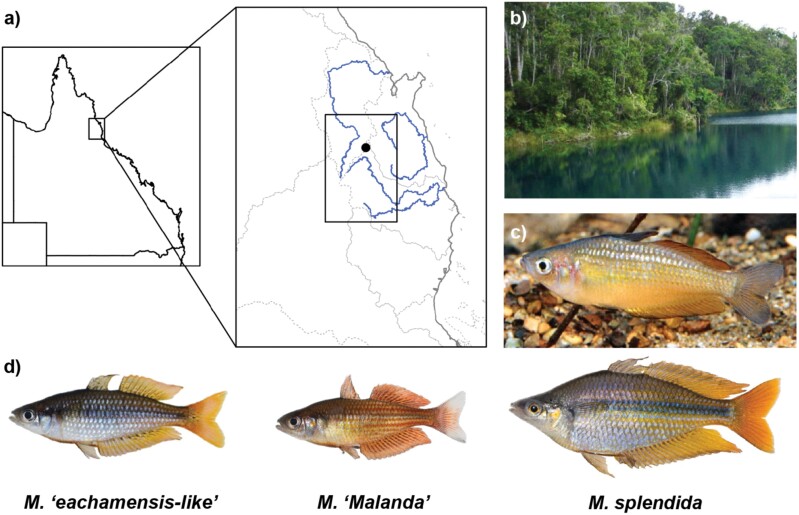
The Lake Eacham rainbowfish was described based on a single locally abundant population in Lake Eacham in the Atherton Tablelands, Queensland (Fig. 1a,b). However, the fish disappeared from the lake in the mid-1980s, concurrent with the appearance of 4 other native species translocated by humans from nearby streams (Barlow et al. 1987). The species is suggested to have disappeared after the introduction of the predatory freshwater cardinalfish Glossamia aprion, with studies suggesting that the Lake Eacham rainbowfish was predator-naïve due to its long isolation from predator species (Brown 1997; Brown and Warburton 1997). After its disappearance from the lake, the species was declared extinct in the wild. However, captive populations (Fig. 1c) had been maintained (Barlow et al. 1987), and the Australia New Guinea Fishes Association (ANGFA) made efforts to breed and conserve these populations as part of Project Eachamensis (Leggett and Merrick 1997). Each captive population was traced back to its original collection to determine the extent of the bottleneck effect on populations (Caughey et al. 1990). Attempts were also made to reintroduce the Lake Eacham rainbowfish to its type locality, but these failed due to the persistence of the introduced species (Lintermans 2013). Any future reintroduction into Lake Eacham is unlikely to succeed whilst introduced predator species are still present (Leggett and Merrick 1997; Pusey et al. 2004). Moreover, a second, larger species of rainbowfish (M. splendida, Fig. 1d) was introduced to the lake between 2000 and 2007 and has now become established (Brown et al. 2012).
Figure 1.
a) Location of the Tablelands and Lake Eacham within north-east Australia. Study region shown in Fig. 2 corresponds to the inset box. Stronger lines (blue in the online version) show major rivers running through the Tablelands, and are labeled in Fig. 2. Fainter lines represent drainage basin boundaries; b) Lake Eacham (photo: Bruceanthro, Attribution-ShareAlike 3.0 Unported (CC BY-SA 3.0); c) M. eachamensis aquarium lineage (photo: Gunther Schmida); d) the 3 rainbowfish lineages of the Atherton Tablelands (photos: Michael Hammer)
Subsequent to the species being declared extinct in the wild, other eachamensis-like populations were discovered in Dirran Creek, a tributary of the North Johnstone River (Allen 1989) and later identified based on morphology in other locations (Pusey et al. 1997). Although some of these populations (Fig. 1d) are still assigned to M. eachamensis today, and the species is currently listed as Endangered by the International Union for the Conservation of Nature (IUCN) (Unmack and Brown 2019), it has long been noted that the extinct Lake Eacham population was morphologically distinct from the extant populations in terms of shape and color (Allen 1989; Crowley and Ivantsoff 1991; Tappin 2011). From this point, we will refer to the extinct type locality population from Lake Eacham as M. eachamensis, and to the morphologically distinct extant populations from the wider Tablelands area as M. sp. “eachamensis-like.” Rainbowfish morphology is known to vary between lake and stream environments (McGuigan et al. 2003), and it is unclear whether the physical differences between extinct M. eachamensis and extant M. sp. “eachamensis-like” are due to adaptive heritable genetic components or due to environment-related phenotypic plasticity. While analyses of mtDNA have been conducted to examine the relationships between extant M. sp. “eachamensis-like” and M. splendida (Zhu et al. 1998), the relationships between the extinct lake M. eachamensis and nearby M. sp. “eachamensis-like” populations remain unclear. Understanding the evolutionary origin of M. eachamensis, and its relationship to nearby M. sp. “eachamensis-like” populations, has implications in species delineation and conservation.
Here, we present a case study utilizing historical DNA from specimens in the Queensland Museum, alongside genomic data from recent surveys in the Atherton Tablelands. We determine the evolutionary relationships between the extinct crater lake population of M. eachamensis and nearby extant populations of M. sp. “eachamensis-like,” addressing the long-standing taxonomic uncertainties associated with these populations. We then place these populations into a broader evolutionary context, exploring the wider population history of rainbowfish species in the Atherton Tablelands and tracking the extent of introgression with the invasive M. splendida. Furthermore, we test for unique variants in the populations using fixed difference analysis to assess the possibility of lost genetic diversity in the extinct crater lake population.
Materials and Methods
Study System
In addition to the extinct M. eachamensis and extant M. “eachamensis-like” populations, the Tablelands region is also home to 2 other rainbowfishes from the “Australis” group: Melanotaenia splendida and Melanotaenia sp. “Malanda” (Fig. 1d). These species are both closely related to M. eachamensis (Brauer et al. 2023), although the 3 are considered distinct due to morphological and genetic differences (Allen and Cross 1982; Zhu et al. 1998; Unmack et al. 2016). Most rainbowfishes are known to readily hybridize in captivity (Unmack et al. 2013), and sympatry between closely related species (those within the same major lineage of Melanotaeniidae; Unmack et al. 2013) is almost non-existent in the wild despite a lack of geographical isolation that would prevent interaction between species (Moy et al. 2019). This suggests that when 2 closely related species come into contact, they either introgress or one is extirpated. In the Tablelands, both M. sp. “eachamensis-like” and M. sp. “Malanda” are known to hybridize with M. splendida in contact zones (Zhu et al. 1998; Unmack et al. 2016; Brauer et al. 2023), and hybridization with M. splendida is considered a threatening process to both species. Melanotaenia splendida is thought to be a relatively recent arrival to the Tablelands (with one estimate placing initial colonization at around 100,000 years ago; Hurwood and Hughes 2001) and may have displaced the other species from parts of their range. This displacement is ongoing due to recent and rapid climate change warming the cooler upland streams, shifting conditions toward those that favor the more generalist lowland species (Brauer et al. 2023). The effects of climate change may be exacerbated by clearing of catchment rainforest and riparian shading due to land use changes following European settlement (Unmack et al. 2016).
A recent assessment showed M. sp. “eachamensis-like” and M. sp. “Malanda” to have very restricted distributions, with areas of occupancy estimated at 72 km2 (Unmack and Brown 2019) and 28 km2 (Brown et al. 2019), respectively, and to be subjected to predicted ongoing declines. Melanotaenia sp. “Malanda” is considered Critically Endangered (Brown et al. 2019) but has yet to be formally described. Its description has been hindered by the complicated taxonomy of fishes in the region and a lack of understanding of the species’ physical characteristics due to widespread hybridization with M. splendida (Unmack et al. 2016). An understanding of the population genetics and patterns of gene flow between lineages is, therefore, critical for robust species description and appropriate conservation management.
Sampling sites and collection
A total of 379 rainbowfishes were sampled from 73 locations in and around the Atherton Tablelands as part of a larger study of Australian rainbowfishes. For the extinct Lake Eacham population, we sampled 5 ethanol-preserved individuals from the collections at the Queensland Museum (registration I.20307, collection date 1 March 1981), plus 5 individuals taken from 2 different aquarium lineages descended from the extinct Lake population. Three individuals were sampled from an Australian aquarium lineage and 2 from a European lineage.
Genotyping using silicoDArT and SNP markers
DNA was extracted by Diversity Arrays Technologies (DArT Pty Ltd, Canberra, Australia) and sequenced for an SNP dataset using DArTseq™, a variation of the double-digest RAD technique that combines next-generation sequencing, complexity reduction using restriction enzymes, and implicit fragment size selection, as described by Kilian et al. (2012). All details of the sequencing methods used follow Georges et al. (2018). This yielded datasets of both SNP and silicoDArT markers. SNPs contain information on allele states within tag sequences at specific positions along the genome, while silicoDArTs are based on the presence or absence of specific sequence tags within the genome. SilicoDArT data provide a complementary source of information to SNPs and increase the amount of data available for analysis. SilicoDArT data are also more appropriate for certain types of analyses because they are explicitly binary (presence/absence), while SNPs may also include heterozygous loci.
Data analysis
Data were filtered using v2.0.4 of the dartR package (Gruber et al. 2018) in R (v4.1.3) (R Core Team 2022) to remove non-informative monomorphic markers and loci for which no data were available for any individuals. We also mapped SNP reads to the genome of Melanotaenia duboulayi, the most closely related rainbowfish species for which an annotated genome is available, using bbmap (Bushnell 2014) to affirm that our panel of SNPs were randomly and evenly distributed across the genome. Pairwise linkage disequilibrium tests of SNPs within 10 kbp of each other were performed using the “ld” function from the genetics package v1.3.8.1.3 (Warnes et al. 2021). These tests confirmed there was relatively little LD in the data even at short distances, and that removal of the few highly correlated SNPs made very little difference to the observed results (Supplementary File S1, https://doi.org/10.5061/dryad.bk3j9kdjw).
ASTRAL
We used the silicoDArT presence/absence dataset to construct individual trees for each marker in R (v4.1.3). ASTRAL requires its input to be binary, and the silicoDArT dataset was therefore more appropriate for this analysis than the SNP dataset, for which we would have had to randomly resolve or exclude all heterozygous loci. The silicoDArT dataset contained 389 individuals and 28,996 markers, and with an additional 6 Melanotaenia trifasciata from the Blyth River in the Northern Territory included as an outgroup to root the tree. Data were not filtered to remove loci or individuals with missing data. ASTRAL is reasonably robust to missing data (Xi et al. 2016; Nute et al. 2018), and filtering based on missing data may reduce mean branch support for true branches (Molloy and Warnow 2018). The silicoDArT trees were then used to construct a species tree in ASTRAL (v5.15.5) (Mirarab et al. 2014). ASTRAL provides a statistically consistent estimation of the true species tree from unrooted gene trees under the multi-species coalescent model. This was performed to test the assumption of shared genetic history and population-like behavior of individuals from each sampling site. When single individuals do not cluster with other individuals from the same sampling site, or when individuals from the same sampling site are not resolved as sister taxa, the assumption of shared genetic history is violated. This may be caused by poor-quality reads (inferred sequences of base pairs) with high error rates in an individual’s data, or by differing levels of hybridization and introgression between individuals from a single sampling site. Subsequent population history analyses with Admixtools relied on the assumption of shared genetic history at each sampling site, to prevent estimates of population history becoming swamped by signals from a genetically distinct individual within a population. Because of this, populations and individuals that did not meet this assumption were pruned from the dataset at this stage to create a subset of populations that would form the core of our subsequent analyses. (However, STRUCTURE results from an analysis of all 389 individuals are presented alongside the ASTRAL results in Supplementary File S3).
STRUCTURE
Based on ASTRAL results, the SNP dataset was pruned to 234 individuals and 23,502 informative markers. The genetic structure of the remaining populations was analyzed using STRUCTURE (v2.3.4) (Pritchard et al. 2000), implemented using the “structureRun” function in the strataG package (v2.5.1) (Archer et al. 2017) in R (v4.1.3). STRUCTURE implements a model-based Bayesian clustering method to infer population structure based on allele frequency data. For this reason, the SNP dataset was used for all STRUCTURE analyses. Individuals were assigned to one of an a priori number of populations (K), or jointly to multiple populations if their genotypes indicated the presence of admixture. Previous genetic and morphological work putatively suggests that 3 rainbowfish lineages are present (see section “Study System”). To test this hypothesis, numbers from 1 to 5 were assumed for K, and the best value of K was determined by plotting an average across all iterations of the estimated natural logarithm of the probability of the data (Ln Pr(X|K)). We also implemented the ΔK method of Evanno et al. (2005) using STRUCTURE HARVESTER (Earl and vonHoldt 2012) for comparison. Five individual Markov Chain Monte Carlo (MCMC) simulations were run for each K-value, with an initial burn-in period of 10,000 followed by 10,000 MCMC iterations per simulation. The admixture and uncorrelated allele frequency models were applied with no population prior estimate. Results for each of the 5 runs were then aggregated using the “clumpp” function in the strataG package.
There is little consensus in the literature regarding optimum data filtering practices, and a range of thresholds and filter types (e.g., site-wise coverage, individual-wise coverage, minimum allele frequency) have been used. To test the effects of filtering by data coverage and minimum allele frequency, we applied 3 different coverage thresholds (80%, 90%, and 95%) to loci and then to individuals, and further filtered these datasets by minimum allele frequency (<1%) to give a total of 6 data subsets. The number of individuals and loci in each data subset is presented in Supplementary File S2. We found no difference in coverage between the 5 ethanol-preserved individuals from the collections at the Queensland Museum and the recently sampled individuals. Missing values leading to lower coverage primarily arise from failure to call a SNP because of a mutation at one or both of the restriction enzyme recognition sites. The similar coverage suggests that the DNA in the preserved samples was not heavily degraded or more prone to sequencing errors.
We also tested the effects of implementing the LOCPRIOR population prior and the correlated allele frequency model on the most stringently filtered dataset (>95% coverage, >1% MAF). The LOCPRIOR population model uses sampling locations as prior information to assist with clustering in cases where population structure may be relatively weak, as in the case of closely related populations (Hubisz et al. 2009). This model does not tend to find spurious population structure and gives similar results to models with no population prior in cases where the population structure is strong (Pritchard et al. 2010). Although the independent allele frequency model works well for many datasets, the correlated allele frequency model (Falush et al. 2003) may improve clustering for closely related populations (Pritchard et al. 2010).
Finally, we re-ran STRUCTURE using only the 8 populations that did not show any significant signs of admixture to test the effects of different K-values on populations thought to correspond to the 3 parental species in the Tablelands. The data were filtered to remove loci and individuals with <80% coverage, and the admixture and uncorrelated allele frequency models were applied with no population prior estimate.
Admixtools
Population histories were inferred using admixture graphs developed with the Admixtools package (v2.0.0) (Maier and Patterson 2022) in R (v4.1.3). Admixture graphs describe relationships among populations allowing for both population divergence and population mixture events. Analyses were performed using the SNP dataset as this is required by Admixtools. We used the “find_graphs” function to produce admixture graphs using loci with >80% coverage and without filtering data by allele frequency (setting options minmaf = 0 and maxmaf = 1 when data were read in with the “extract_f2” function). SNPs were split into blocks of 100 loci by assigning dummy positions to the SNPs in the order in which they appeared in the dataset. The Australian aquarium lineage of M. eachamensis was excluded from Admixtools analyses based on STRUCTURE results, which suggested that breeding with M. splendida had occurred at some point during the population’s history in captivity.
We conducted multiple runs of Admixtools using different population subsets to infer population histories. We used the results from STRUCTURE to identify genetically pure populations of each species, plus populations representing hybrids of 2 species with significant genetic input from both species (>5%). We then ran 3 data subsets: i) non-introgressed M. sp. “eachamensis-like,” non-introgressed M. sp. “Malanda,” and introgressed “eachamensis-like”-“Malanda”; ii) non-introgressed M. sp. “eachamensis-like,” non-introgressed M. splendida, and introgressed “eachamensis-like”-splendida; and iii) non-introgressed M. sp. “Malanda,” non-introgressed M. splendida, and introgressed “Malanda”-splendida. We ran Admixtools multiple times, allowing for different numbers of admixture events.
We compared the fits of different admixture graphs for each population subset. We used the “qpgraph_resample_multi” function to evaluate admixture graphs based on 1000 bootstrap resampled SNP block training and test datasets. We then used the “compare_fits” function to test which graph was a better fit to the data based on the distribution of bootstrap scores. We also implemented ABBA–BABA tests (Durand et al. 2011) using the “qpdstat” function with the option “f4mode = FALSE” to evaluate biological models of admixture in our key results. Further description of ABBA–BABA tests and the populations evaluated with them can be found in Supplementary File S10.
Unique variant testing
We tested each population for unique variants using the silicoDArT dataset, which contains restriction site presence–absence data, and the “gl.fixed.diff” function in the dartR package. We filtered the dataset to include loci with a callrate of >0.95 and a reproducibility of >0.99, leaving a total of 19,142 non-monomorphic loci. We compared individuals from each population, in turn, against all other sampled individuals to determine the number of unique variants, that is, the number of restriction site loci that were present in all members of the focal population but were absent in all others, or that were absent in the focal population but present in every other sampled individual. We set the options “test = TRUE” to calculate the probability that the observed number of unique variants could have arisen due to sampling error alone.
Results
ASTRAL
The species tree estimated by ASTRAL is presented in Supplementary File S3, alongside STRUCTURE results for the full dataset of 389 individuals. Of the 73 sites, 22 had all sampled individuals clustered together and were resolved as distinct from all other sites. Individuals from 23 sampled sites were found to be genetically indistinguishable from at least 1 nearby site; these sites were condensed into 8 distinct populations for subsequent analyses (Table 1). Individuals from 27 sites did not cluster together in the ASTRAL tree, suggesting those sites cannot be treated as populations due to the differing ancestry of individuals at the site; these sites were removed from subsequent analyses. One further site was removed due to a small sample size (n = 2). Five singletons that did not cluster with other members of their otherwise monophyletic site grouping were also removed from the dataset. A total of 234 individuals across 32 populations (30 sampled sites and 2 captive aquarium populations) were retained for subsequent analyses.
Table 1.
Table of populations included in STRUCTURE and Admixtools analyses.
| Site | Population name | River Basin | Population code | Latitude | Longitude | Sample size | Taxon |
|---|---|---|---|---|---|---|---|
| 1 | Tinaroo Creek | Barron | PU16-139 | −17.0797 | 145.5012 | 5 | M. splendida |
| 2 | Emu Creek | Barron | PU16-138 | −17.0995 | 145.5259 | 3 | M. sp. “eachamensis-like” |
| −17.1063 | 145.5205 | 2 | |||||
| 3 | Lake Euramoo | Barron | PU18-20 | −17.1598 | 145.6275 | 5 | M. sp. “eachamensis-like” |
| 4 | Little Mulgrave | Mulgrave | GM14-04, PU21-21a |
−17.1277 | 145.7015 | 5 | M. splendida/M. sp. “eachamensis-like” [<5%] |
| 5 | Barney Springs | Barron | KM051 | −17.1694 | 145.4503 | 5 | M. splendida |
| 6 | Upper Rocky Creek | Barron | KM071 | −17.1931 | 145.4356 | 5 | M. splendida/M. sp. “eachamensis-like” [<5%] |
| 7 | Lake Eacham Australian aquarium lineage | Barron | PU16-AA | −17.2849 | 145.6259 | 3 | M. sp. “eachamensis-like”/M. sp. “Malanda” [20%]/M. splendida [15%] |
| 7 | Lake Eacham European aquarium lineage | Barron | Euro | −17.2849 | 145.6259 | 2 | M. sp. “eachamensis-like”/M. sp. “Malanda” [25%] |
| 7 | Lake Eacham Queensland Museum samples | Barron | EachQM | −17.2849 | 145.6259 | 5 | M. sp. “eachamensis-like”/M. sp. “Malanda” [25%] |
| 8 | Imrie Creek | North Johnstone | KM099 | −17.3009 | 145.6367 | 5 | M. splendida/M. sp. “Malanda”[25%]/M. sp. “eachamensis-like” [<5%] |
| 9 | Gowrie Creek | Barron | PU16-124 | −17.3486 | 145.4855 | 5 | M. sp. “eachamensis-like”/M. splendida [<5%] |
| 10 | Spider Creek | Barron | PU16-123 | −17.3542 | 145.4920 | 5 | M. sp. “eachamensis-like”/M. splendida [5%]/M. sp. “Malanda” [<5%] |
| 11 | Gwynne Creek | Barron | PU15-61 | −17.3408 | 145.5383 | 3 | M. sp. “eachamensis-like”/M. splendida [5%]/M. sp. “Malanda” [<5%] |
| PU16-93 | −17.3408 | 145.5383 | 5 | ||||
| 12 | Nicholas Creek | Barron | PU16-92 | −17.3357 | 145.5578 | 5 | M. splendida/M. sp. “eachamensis-like” [20%] |
| 13 | Cleminson Creek | North Johnstone | PU16-91 | −17.3459 | 145.5774 | 5 | M. splendida |
| 14 | Williams Creek East | North Johnstone | MH18-11 | −17.3659 | 145.5942 | 6 | M. sp. “Malanda”/M. splendida [<5%] |
| PU16-88 | −17.3947 | 145.5967 | 20 | ||||
| 15 | Williams Creek East 2 | North Johnstone | PU19-35 | −17.3757 | 145.6026 | 3 | M. sp. “Malanda”/M. splendida [15%] |
| PU19-37 | −17.3793 | 145.6082 | 4 | ||||
| PU19-36 | −17.3809 | 145.6083 | 4 | ||||
| PU19-50 | −17.3813 | 145.6011 | 4 | ||||
| PU19-33 | −17.3846 | 145.5993 | 3 | ||||
| 16 | Battle Creek | PU16-107 | −17.3491 | 145.6596 | 5 | M. splendida/M. sp. “Malanda [15%] | |
| 17 | Short Creek | North Johnstone | PU15-63 | −17.3819 | 145.6655 | 5 | M. splendida/M. sp. “Malanda” [5%] |
| 18 | Wallace Road | North Johnstone | PU15-64 | −17.3955 | 145.6589 | 2 | M. sp. “Malanda” |
| PU16-100 | −17.3955 | 145.6589 | 8 | ||||
| PU16-101 | −17.3965 | 145.6579 | 7 | ||||
| PU16-111 | −17.4013 | 145.6561 | 3 | ||||
| 19 | Wallace Road lower | North Johnstone | PU16-110 | −17.4101 | 145.6551 | 5 | M. splendida/M. sp. “Malanda” [<5%] |
| 20 | Molo Creek tributary | North Johnstone | MH18-13 | −17.4286 | 145.5710 | 10 | M. sp. “Malanda” |
| PU16-126 | −17.4286 | 145.5710 | 5 | ||||
| 21 | Molo Creek and tributary | North Johnstone | PU19-47 | −17.4160 | 145.5839 | 2 | M. sp. “Malanda”/M. splendida [<5%]/M. sp. “eachamensis-like” [<5%] |
| PU19-46 | −17.4175 | 145.5814 | 3 | ||||
| PU19-49 | −17.4251 | 145.5763 | 3 | ||||
| PU19-48 | −17.4257 | 145.5758 | 4 | ||||
| 22 | Thiaki Creek | North Johnstone | PU15-67 | −17.4136 | 145.5881 | 5 | M. sp. “Malanda”/M. splendida [45%] |
| 23 | Thiaki Creek tributary | North Johnstone | PU16-134 | −17.4197 | 145.5946 | 5 | M. sp. “Malanda” |
| MH18-08 | −17.4239 | 145.5888 | 10 | ||||
| 24 | Gillies Creek | North Johnstone | PU16-96 | −17.4422 | 145.5940 | 5 | M. splendida/M. sp. “Malanda” [<5%] |
| 25 | Dirran Creek | North Johnstone | PU15-66 | −17.4511 | 145.6000 | 5 | M. sp. “eachamensis-like”/M. sp. “Malanda” [25%] |
| 26 | Victoria Creek | North Johnstone | PU16-129 | −17.4546 | 145.7313 | 5 | M. splendida |
| 27 | Beatrice River | North Johnstone | PU15-71 | −17.5522 | 145.6093 | 5 | M. splendida |
| 28 | Middle Brook | North Johnstone | PU16-119 | −17.5699 | 145.6090 | 5 | M. splendida |
| 29 | South Johnstone | South Johnstone | MH18-15 | −17.6536 | 145.7173 | 5 | M. sp. “eachamensis-like” |
| 30 | Charappa Creek | South Johnstone | MH18-14 | −17.7017 | 145.6731 | 5 | M. sp. “eachamensis-like” |
Notes: Site numbers correspond to numbers in Supplementary File S5. Latitude and longitude show sampling localities included in each population. The taxon column shows the lineage to which the population belongs. Where the population has more than a trace introgression signal, approximate contributions are given for the minor parental lineage(s) in square brackets.
Melanotaenia splendida was found to be the first branching species and was resolved as a monophyletic group, with many of its early branching lineages being hybrid populations. Similarly, all M. sp. “Malanda” populations formed a clade, with hybrid populations appearing as the earliest-branching lineages. Melanotaenia eachamensis and M. sp. “eachamensis-like” together formed a sister clade to M. sp. “Malanda.” The earliest branching lineages of this clade were the extinct Lake Eacham M. eachamensis and the extant M. sp. “eachamensis-like” population from Dirran Creek.
STRUCTURE
The SNP markers were used to estimate the genetic structure of 32 rainbowfish populations in STRUCTURE (Fig. 2; Supplementary File S3). Inspection of mean Ln Pr(X|K) plots across different values of K does suggest an optimal value of K = 3 regardless of data subset, as the addition of extra populations to the model beyond this point did not result in any marked increase of Ln Pr(X|K) (Supplementary File S4). However, STRUCTURE HARVESTER suggests an optimal value of K = 2 for all data subsets based on the ΔK method. This value contradicts what is known about population differences based on prior morphological and phylogenetic work, and we believe it is likely an example of ΔK detecting the uppermost level of a more hierarchical classification (Evanno et al. 2005; Janes et al. 2017). Given the small size of the study region and that populations of the 3 lineages exist in close proximity (Fig. 2), we do not expect there to be a major effect of isolation by distance (Slatkin 1993). The average result across the 5 model runs with K = 3 using the most stringently filtered dataset (95% coverage across loci and individuals, plus MAF > 1%) are presented in Fig. 2. Individual STRUCTURE runs for each filtered data subset were highly consistent, with an average variation of only 0.1% in the estimates of each individual’s population assignment between runs.
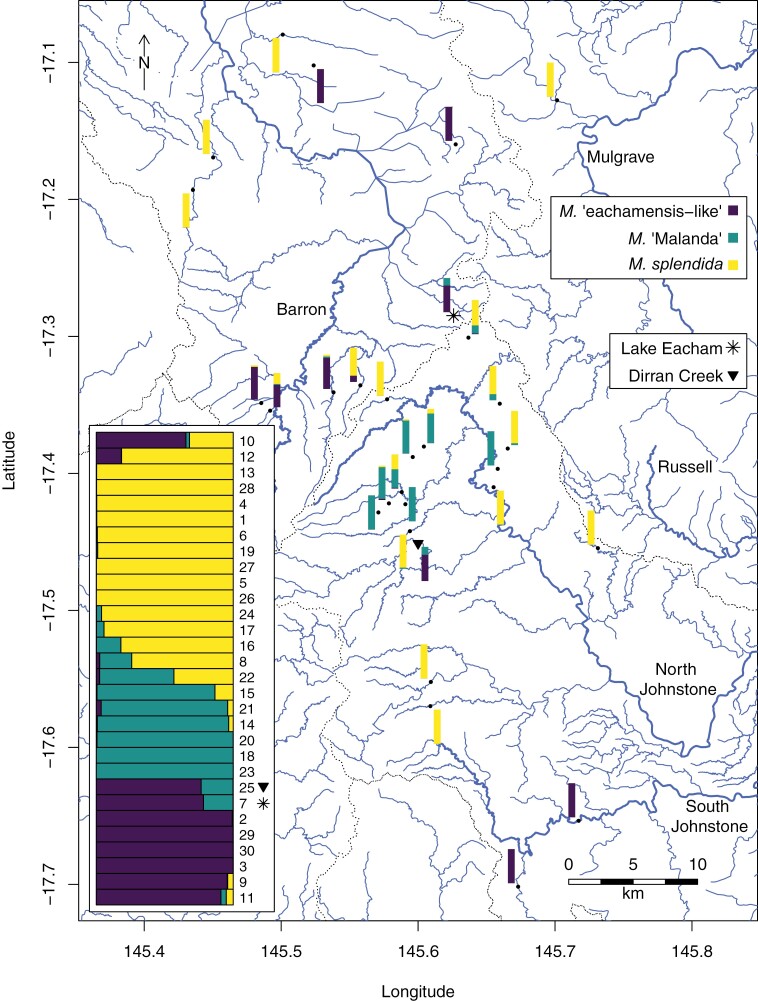
Figure 2.
Map showing sampled locations and admixture proportions of 30 different rainbowfish populations. Major rivers are named and denoted by thicker lines (blue in online version); smaller waterways are denoted by thinner lines. Dotted black lines denote catchment boundaries. The study region corresponds to the inset box in Fig. 1a. A traditional STRUCTURE plot is inset on the bottom left of the figure. Numbers correspond to locality numbers marked in Supplementary File S5.
The STRUCTURE results reveal widespread admixture between all 3 rainbowfish lineages (Fig. 2; Supplementary File S3 and S7). Unexpectedly, we found that crater lake M. eachamensis has a hybrid origin, showing a signal of introgression with extant M. sp. “eachamensis-like” populations and with M. sp. “Malanda” in an approximately 3:1 ratio. This admixture proportion appears to be consistent across all preserved specimens and individuals from the European aquarium lineage and was observed consistently in all individual STRUCTURE runs where K = 3. The Australian aquarium lineage shows evidence of the same “eachamensis-like”-“Malanda” admixture, but additionally shows some admixture by M. splendida that is likely to have accidentally occurred in captivity. The extant Dirran Creek population appears to also be of hybrid origin, although with a slightly different proportion of M. sp. “Malanda” admixture.
In the extant M. sp. “eachamensis-like” samples, admixture with M. splendida is evident in the Gowrie Creek, Gwynne Creek, Nicholas Creek, and Spider Creek populations (Barron system), while the Lake Euramoo (Barron system) and South Johnstone River (section above larger waterfalls) and its tributary Charappa Creek populations appear to remain genetically pure (see Supplementary File S5 for map with locality names). Among the populations of M. sp. “Malanda,” we found that the populations sampled from Wallace Road and from the Thiaki Creek tributary were not introgressed with either M. sp. “eachamensis-like” or M. splendida. We also found that admixture proportions were very low in the Molo Creek tributary, with only 3 individuals from the population showing signs of admixture with M. sp. “eachamensis-like.” Remaining populations show differing levels of introgression, primarily with M. splendida. The Imrie Creek (upper North Johnstone River near Lake Eacham) and Battle Creek populations of M. splendida also showed evidence of genetic admixture with M. sp. “Malanda.”
STRUCTURE plots of the filtered dataset for different K-values are presented in Supplementary File S6. When K = 2, M. splendida and M. sp. “Malanda” are placed into different clusters, and M. sp. “eachamensis-like” clusters largely with M. splendida but show small amounts of M. sp. “Malanda” influence. This occurred whether or not admixed populations were included in the STRUCTURE analysis, and is somewhat unexpected, given that M. sp. “Malanda” and M. sp. “eachamensis-like” are considered closer relatives to each other than to M. splendida. The Lake Eacham and Dirran Creek populations show higher amounts of M. sp. “Malanda” genetic structure than the remainder of the M. sp. “eachamensis-like” populations. Results for K = 2 are highly consistent across individual STRUCTURE runs, but individual run results are more divergent for K = 4 and K = 5. When K = 4, the Lake Eacham and Dirran Creek populations show slightly different admixture proportions in a manner similar to the K = 3 results across 4 of the 5 runs; in the fifth run, the Lake Eacham population is placed in a separate cluster than all other populations, while the Dirran Creek population shows similar admixture proportions as the other 4 runs.
Our results were robust to the effects of missing data and data filtering, with all data subsets yielding similar results (Supplementary File S7). The mean difference in population assignment for K = 3 between the strictest and loosest filters was <0.5%. Results were also congruent between runs that did not use any prior population information and runs using the LOCPRIOR models. This is consistent with expectations based on Pritchard et al. (2010), who state that the 2 models “give essentially the same answers when the signal of population structure is very strong.” Finally, the differences in predicted admixture proportions depending on whether the correlated allele frequency model was used were also small (mean difference < 0.1%, 95th percentile difference < 2%).
Admixtools
We modeled a range of possible population histories across different population subsets and numbers of admixture events using Admixtools. The admixture graphs broadly support the results seen in the STRUCTURE plots.
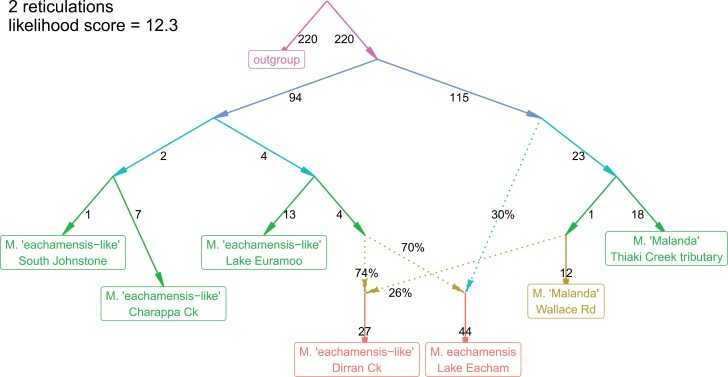
Admixtools identified both the extinct Lake Eacham form and extant Dirran Creek population as having genetic input from both pure M. sp. “eachamensis-like” and M. sp. “Malanda” (Fig. 3; Supplementary File S7). The genetic contributions were estimated to be 26% M. sp. “Malanda” and 74% M. sp. “eachamensis-like” when a single admixture event was modeled (Supplementary File S7). When 2 separate admixture events were modeled, contribution estimates were 74% M. sp. “eachamensis-like,” 26% M. sp. “Malanda” for the Dirran Creek population, and 70% M. sp. “eachamensis-like,” 30% M. sp. “Malanda” for Lake Eacham (e.g., Supplementary File S7). The admixture models with separate hybrid origins for each population fit the data slightly better than models with a single hybrid origin and subsequent divergence of the 2 populations (likelihood scores of 12.3 v 19.8). While bootstrapping results from “compare_fits” showed that the separate-origin model tended to be better (average difference of 3.7), this difference was not significant (P > 0.05). With the inclusion of the Molo Creek tributary population, minus the 3 individuals with significant admixture signal, models with separate hybrid origins for each population fit the data significantly better (P = 0.014) than models with a single hybrid origin and subsequent divergence of the 2 populations (Supplementary File S7). These results fit with other lines of evidence that suggest the Lake Eacham and Dirran Creek populations have separate evolutionary histories (Supplementary File S8). ABBA–BABA testing confirmed that the Lake Eacham and Dirran Creek populations share excess alleles with Malanda populations compared to M. sp. “eachamensis-like” populations (Supplementary File S10). Tests also showed evidence of excess allele sharing between the Molo Creek tributary population of M. sp. “Malanda,” and M. sp. “eachamensis-like” populations, driven by the 3 individuals identified in STRUCTURE plots. When these 3 individuals were removed from the population, ABBA–BABA test results were not significant (Supplementary File S10).
Figure 3.
One of the population histories modeled by Admixtools. This model provides a possible population history of the Lake Eacham and Dirran Creek populations, and a number of M. sp. “eachamensis-like” and M. sp. “Malanda” populations that do not have genetic signatures of admixture. This population model suggests two different hybrid origins for Lake Eacham and Dirran Creek populations, with similar but not identical admixture proportions. Numbers on solid lines are estimated measures of genetic drift shared by descendant populations.
Admixture graphs focusing on interactions between M. splendida and M. sp. “Malanda,” and between M. splendida and M. sp. “eachamensis-like,” also supported the admixture patterns seen in the STRUCTURE plots. These graphs are presented in Supplementary Files S11 and S12, and the wider patterns of gene flow across the Tablelands are discussed in Supplementary File S13.
Unique variant testing
We found a total of 50 unique variants spread among 10 of the 30 populations (Supplementary File S14). The Lake Eacham samples had 8 unique variants, the most of any population with an introgression signal. Only 2 populations had higher numbers of unique variants than this. The Little Mulgrave population of M. splendida had 15 unique variants, the most of any population. This population represents our only sample from the Lower Mulgrave basin, which contains a distinct Lowlands form of M. splendida separate to the invasive form in the Tablelands. The Wallace Road population of M. sp. “Malanda,” and the Lake Euramoo M. sp. “eachamensis-like” population, were found to have 12 and 6 unique variants, respectively. These unadmixed populations contain unique genetic diversity and should be considered as priority populations for conservation.
Discussion
Using historical DNA from museum samples, we found that the extinct type locality population of M. eachamensis was of hybrid origin. The two parent lineages for this hybrid population are the extant M. sp. “eachamensis-like” and M. sp. “Malanda.” The admixture proportions appear to be consistent across all individuals, suggesting a stable natural genetic makeup within the crater lake. The genetic makeup of the extinct crater lake population was unique among our samples, as no extant populations were found to have the same admixture signal as the extinct Lake Eacham fish. This suggests that the Lake Eacham population arose from hybridization within the lake rather than colonization by an introgressed riverine population. Furthermore, our results suggest that this population has a unique origin and is not connected to other hybrid populations in the region (Supplementary Files S8 and S9).
Fixed-difference analysis of silicoDArT data uncovered 8 unique variants in the extinct Lake Eacham fish, more than double that of any other site with signals of introgression (Supplementary File S14). The number of unique variants seen in this population is indicative of unique evolution within the lake environment subsequent to introgression. Lake Euramoo, another nearby crater lake, contains an unadmixed population of M. sp. “eachamensis-like” with 6 unique variants, the most of any M. sp. “eachamensis-like” populations. These results highlight that crater lakes are key environments for unique adaptation and evolution, and suggest that the Lake Euramoo population should represent a priority population for conservation to preserve this unique diversity. The presence of significant numbers of unique variants in the Lake Eacham and Lake Euramoo populations contrasts with sites that have signals of introgression with M. splendida, which have few unique variants, suggesting more recent rather than historic introgression followed by in situ evolution.
There are 2 primary and contrasting interpretations of our findings: i) the type locality of M. eachamensis represents a hybrid population, or ii) crater lake M. eachamensis is a distinct evolutionary species with more ancient hybrid origins. Each scenario has important implications for species delineation and conservation. Type locality (scientific name bearing) populations represented by hybrids are currently not known to be common, but more are likely to be discovered as molecular analyses are applied to different groups. One example is with newts of the genus Triturus, where the type locality for the name T. arntzeni was found to represent a hybrid population of 2 other named species and the name was thus considered invalid and instead assigned as a junior synonym of both its previously described parental species (Wielstra and Arntzen 2014). In the case of M. eachamensis, under interpretation 1—that the extinct Lake Eacham population is a hybrid population, with noted morphological differences reflecting differing parental gene expression and phenotypic plasticity—the scientific name would be invalid, but may be available for the genetically dominant undescribed parental species M. sp. “eachamensis-like.” In this case, redescription of M. eachamensis s.l. would be required to accurately represent this new taxonomic framework (Allen and Cross 1982). Under interpretation 2—that the extinct crater lake M. eachamensis was a distinct evolutionary species where physical differences in shape and colouration reflect heritable traits and adaptation to a unique crater lake environment—the scientific name would remain valid, reaffirming that M. eachamensis was Australia’s first fish species to become extinct in the wild, and M. sp. “eachamensis like” would require description as a new species. Any final decision on the taxonomic status of M. eachamensis is best left until more intensive morphological assessments have been undertaken as part of a full taxonomic revision of all candidate species.
The results from museum hDNA of Lake Eacham specimens are validated by data from modern captive populations. The European aquarium lineage specimens exhibit very similar admixture proportions of M. sp. “eachamensis-like” and M. sp. “Malanda” to the original lake population. Further, all but one of the unique variants present in the museum samples are also seen in the European aquarium lineage specimens. This suggests that the results presented here reflect the true genetic makeup of the extinct lake species, and are not simply the result of DNA degradation in historic samples or an error due to small sample sizes. The Australian aquarium lineage also has similar ratios of M. sp. “eachamensis-like”- to M. sp. “Malanda”-derived alleles, despite evidence of additional admixture with M. splendida in captivity (Supplementary Files S3 and S7).
In contrast to many other crater lakes, we find no evidence of a species flock occurring within Lake Eacham, showing that it is possible to have widespread hybridization that does not necessarily lead to adaptive radiations. While crater lakes host many exceptional fish species flocks, like the Cameroonian crater lakes Barombi Mbo and Lake Bermin (Martin et al. 2015), as well as the Nicaraguan crater lake species flocks of Apoyo and Xiloá in Nicaragua (Kautt et al. 2016), there are many cases of crater lakes where species fail to radiate. In Cameroon, Lake Barombi-ba Kotto houses the singleton lake endemic Coptodon kottae and Nicaragua’s Lake Asosoca Managua contains only a single young Amphilophus species. The Lake Eacham rainbowfish offers another example of a non-radiating lake endemic, even though it possesses a clear signature of hybridization that resembles the patterns seen in exceptional lake species flocks. This suggests that while hybridization may be necessary, it is certainly not sufficient for species flock formation in young lakes.
The size and age of Lake Eacham also complicate theories of species flock formation. Lake Bermin is of similar size to Lake Eacham (~0.5km2), but supports a species flock (Walker 1999; Martin et al. 2015), suggesting that lake size is not the limiting factor. While Lake Bermin is much older than Lake Eacham, and speciation has, therefore, had more time to occur, other lakes of similar age to Lake Eacham (~10ka, Whitehead et al. 2007) have been found to support species flocks. Lake Ejagham, another Cameroonian lake (although not a crater lake), is of similar size to Lake Eacham and formed approximately 9000 years ago, yet contains 2 distinct radiations of cichlids (Martin et al. 2015; Stager et al. 2018). Lake Xiloá is also of a similar age and supports a young radiation of 4 species (Kautt et al. 2016).
The lack of adaptive radiation in Lake Eacham rainbowfish could involve the presence of other fish species already occupying several niches commonly exploited by young lake species flocks. Rainbowfish feed on a variety of food items (Allen et al. 2002), including algae, terrestrial and aquatic insects, as well as planktonic crustaceans, suggesting that in theory, it would be possible for the lineage to evolve more planktivorous, herbivorous, or benthivorous forms. However, 2 fishes were known to co-inhabit pre-European Lake Eacham: the fly-specked hardyhead (Craterocephalus stercusmusarcum), a species also known to feed on algae and plankton, and possessing a highly protrusible mouth not dissimilar to many limnetic stickleback populations (McGee et al. 2013); and the insectivorous eleotrid species, Mogurnda adspersa (likewise extirpated in the 1980s), which possesses a robust and relatively non-protrusible mouth similar to benthic stickleback populations. The presence of both more “limnetic-like” and “benthic-like” lineages in Lake Eacham may have reduced the likelihood of an ecomorphologically diverse species flock.
Our results also highlight the utility of short-read data in obtaining genetic information from museum specimens. While DNA from ethanol-preserved specimens fragments rapidly within 5–10 years of preservation (Zimmermann et al. 2008), we find that reliable short-read sequence data can still be obtained in the right circumstances using hDNA from samples around 40 years old. This is likely because DArT data rely on short unpaired reads, reducing the impact of breaks in double-stranded DNA that can compromise the sequencing of slightly longer paired reads typically used for whole genome assembly or resequencing.
DArTseq and similar restriction-site based data have previously been demonstrated to be obtainable from museum samples (Ewart et al. 2019), and have been used to explore population structure (e.g., Ryan et al. 2018; Parham et al. 2020; Baveja et al. 2021). However, these short-read data approaches are not often extended to detailed population genetics approaches. Although some studies have demonstrated that museum specimens can be used to infer population histories with admixture graphs (Garg et al. 2022; Salter et al. 2024), these studies did not integrate modern data with historical data. Here, we demonstrate that short-read sequence data obtained from museum specimens can be easily integrated into datasets including modern samples, allowing for population histories to be inferred for species whose ranges may have changed over time or decreased due to local extinction. This integration of modern and historical data allows researchers to examine changes and trends over extended periods, and to assess the impact of human activities, climate change, and other factors on population structure and genetic diversity.
We show that restriction-site data derived from historical samples can be leveraged to explore non-tree-like patterns of evolution. When admixture graphs are developed using both museum and modern samples, they are often done so using data obtained from whole-genome approaches and just a single or few individuals per population (e.g., Stroupe et al. 2022; Ciucani et al. 2023; Hernández‐Alonso et al. 2023; Sun et al. 2023). While these approaches increase the number of markers that can be identified, the samples can often only be sequenced below the level necessary to accurately gauge heterozygosity (Sims et al. 2014). Without accurate heterozygosity information, it can be challenging or impossible to use many population genomic models.
We suggest that when using historical samples, DArTseq and other restriction-site-based methods may represent a better alternative to low-coverage whole-genome resequencing, particularly when inferring population histories under complex scenarios.
Conclusions
Our results show that ethanol-preserved museum specimens have the potential to yield useful quantities of genome-wide SNP and silicoDArT data even 40 years after sample collection. Using short-read data derived from hDNA, we have been able to shed light on the history of the extinct Lake Eacham rainbowfish, revealing its hybrid origin, relationships to extant populations, and the presence of unique variants. The type locality of M. eachamensis has a substantially different genetic make-up and evolutionary history than do the extant M. sp. “eachamensis-like” populations of the Atherton Tablelands, and adds to the exceptional biodiversity known from this endemism hotspot. We suggest that utilizing a museum genomics approach with other recently extinct taxa may provide comparable insights into complex speciation dynamics, particularly in heavily human-impacted systems.
Supplementary Material
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bk3j9kdjw.
Acknowledgements
We acknowledge the traditional owners of the study region, and recognize their deep and ongoing connections to land, water, and fishes. Jeff Johnson kindly facilitated access to historic samples in the Queensland Museum, and members of ANGFA and the International Rainbowfish Group assisted with sourcing samples from captive populations. A range of people contributed to field sampling, especially Keith Martin, Karl Moy, Brendan Ebner, Gary Moores, and Glenn Briggs. Field surveys were conducted under Queensland Fisheries permits (primarily 168221/191126) and a Scientific Purposes Permit for sampling in National Parks (WITK 17362016), in accordance with University of Canberra AEC approvals. We thank associate editor, Dr James Albert, and 2 anonymous reviewers for their helpful comments, which improved our manuscript.
Contributor Information
Amy R Tims, School of Biological Sciences, Monash University, Melbourne, Victoria 3800, Australia; School of Natural Sciences, Macquarie University, Sydney, New South Wales 2109, Australia.
Peter J Unmack, School of Biological Sciences, Monash University, Melbourne, Victoria 3800, Australia; Centre for Applied Water Science, Institute for Applied Ecology, University of Canberra, Australian Capital Territory 2601, Australia.
Michael P Hammer, Museum and Art Gallery of the Northern Territory, Darwin, Northern Territory 0801, Australia.
Culum Brown, School of Natural Sciences, Macquarie University, Sydney, New South Wales 2109, Australia.
Mark Adams, Evolutionary Biology Unit, South Australian Museum, North Terrace, Adelaide, South Australia 5000, Australia; School of Biological Sciences, The University of Adelaide, Adelaide, South Australia 5005, Australia.
Matthew D McGee, School of Biological Sciences, Monash University, Melbourne, Victoria 3800, Australia.
Funding
This project was funded by a Macquarie University COVID Recovery Fellowship to A.R.T.
Data Availability
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bk3j9kdjw.
References
- Abbott R., Albach D., Ansell S., Arntzen J.W., Baird S.J.E., Bierne N., Boughman J., Brelsford A., Buerkle C.A., Buggs R., Butlin R.K., Dieckmann U., Eroukhmanoff F., Grill A., Cahan S.H., Hermansen J.S., Hewitt G., Hudson A.G., Jiggins C., Jones J., Keller B., Marczewski T., Mallet J., Martinez-Rodriguez P., Möst M., Mullen S., Nichols R., Nolte A.W., Parisod C., Pfennig K., Rice A.M., Ritchie M.G., Seifert B., Smadja C.M., Stelkens R., Szymura J.M., Väinölä R., Wolf J.B.W., Zinner D.. 2013. Hybridization and speciation. J. Evol. Biol. 26:229–246. [DOI] [PubMed] [Google Scholar]
- Allen G.R. 1989. Lake Eacham rainbowfish rediscovered? Fish. Sahul 5:217–219. [Google Scholar]
- Allen G.R., Cross N.J.. 1982. Rainbowfishes of Australia and Papua New Guinea. Sydney (Australia): Angus and Robertson. [Google Scholar]
- Allen G.R., Midgley S.H., Allen M.. 2002. Field guide to the freshwater fishes of Australia. Perth (Australia): Western Australian Museum. [Google Scholar]
- Archer F.I., Adams P.E., Schneiders B.B.. 2017. stratag: an r package for manipulating, summarizing and analysing population genetic data. Mol. Ecol. Resour. 17:5–11. [DOI] [PubMed] [Google Scholar]
- Barlow C.G., Hogan A.E., Rodgers L.J.. 1987. Implication of translocated fishes in the apparent extinction in the wild of the lake Eacham rainbowfish, Melanotaenia eachamensis. Mar. Freshw. Res. 38:897–902. [Google Scholar]
- Baveja P., Garg K.M., Chattopadhyay B., Sadanandan K.R., Prawiradilaga D.M., Yuda P., Lee J.G.H., Rheindt F.E.. 2021. Using historical genome-wide DNA to unravel the confused taxonomy in a songbird lineage that is extinct in the wild. Evol. Appl. 14:698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer C.J., Sandoval-Castillo J., Gates K., Hammer M.P., Unmack P.J., Bernatchez L., Beheregaray L.B.. 2023. Natural hybridization reduces vulnerability to climate change. Nat. Clim. Change 13:282–289. [Google Scholar]
- Brown C. 1997. Recovery plan for the Lake Eacham Rainbowfish.
- Brown C., Aksoy Y., Varinli H., Gillings M.. 2012. Identification of the rainbowfish in Lake Eacham using DNA sequencing. Aust. J. Zool. 60:334–339. [Google Scholar]
- Brown C., Hammer M.P., Unmack P.J., Ebner B.C.. 2019. Melanotaenia sp. nov. “Malanda.”. The IUCN Red List of Threatened Species; 2019: e.T123321483A123382516. [Google Scholar]
- Brown C., Warburton K.. 1997. Predator recognition and anti-predator responses in the rainbowfish Melanotaenia eachamensis. Behav. Ecol. Sociobiol. 41:61–68. [Google Scholar]
- Burns P.A., Rowe K.C., Parrott M.L., Roycroft E.. 2023. Population genomics of decline and local extinction in the endangered Australian Pookila. Biol. Conserv. 284:110183. [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner.
- Card D.C., Shapiro B., Giribet G., Moritz C., Edwards S.V.. 2021. Museum genomics. Annu. Rev. Genet. 55:633–659. [DOI] [PubMed] [Google Scholar]
- Caughey A., Hume S., Wattam A.. 1990. Melanotaenia eachamensis—history and management of the captive stocks. Fish. Sahul 6:241–252. [Google Scholar]
- Chattopadhyay B., Garg K.M., Mendenhall I.H., Rheindt F.E.. 2019. Historic reveals Anthropocene threat to a tropical urban fruit bat. Curr. Biol. 29:R1299–R1300. [DOI] [PubMed] [Google Scholar]
- Ciucani M.M., Ramos-Madrigal J., Hernández-Alonso G., Carmagnini A., Aninta S.G., Sun X., Scharff-Olsen C.H., Lanigan L.T., Fracasso I., Clausen C.G., Aspi J., Kojola I., Baltrūnaitė L., Balčiauskas L., Moore J., Åkesson M., Saarma U., Hindrikson M., Hulva P., Bolfíková B.C., Nowak C., Godinho R., Smith S., Paule L., Nowak S., Mysłajek R.W., Lo Brutto S., Ciucci P., Boitani L., Vernesi C., Stenøien H.K., Smith O., Frantz L., Rossi L., Angelici F.M., Cilli E., Sinding M.-H.S., Gilbert M.T.P., Gopalakrishnan S.. 2023. The extinct Sicilian wolf shows a complex history of isolation and admixture with ancient dogs. iScience 26:107307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley E.L.M., Ivantsoff W.. 1991. Genetic similarity among populations of rainbowfishes (Pisces: Melanotaeniidae) from Atherton Tableland, Northern Queensland. Ichthyol. Explor. Freshwaters 2:129–137. [Google Scholar]
- De Bruyn M., Parenti L.R., Carvalho G.R.. 2011. Successful extraction of DNA from archived alcohol-fixed white-eye fish specimens using an ancient DNA protocol. J. Fish. Biol. 78:2074–2079. [DOI] [PubMed] [Google Scholar]
- DeMarais B.D., Dowling T.E., Douglas M.E., Minckley W.L., Marsh P.C.. 1992. Origin of Gila seminuda (Teleostei: Cyprinidae) through introgressive hybridization: implications for evolution and conservation. Proc. Natl. Acad. Sci. U.S.A. 89:2747–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling T.E., Secor C.L.. 1997. The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28:593–619. [Google Scholar]
- Durand E.Y., Patterson N., Reich D., Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4:359–361. [Google Scholar]
- Evanno G., Regnaut S., Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Ewart K.M., Johnson R.N., Ogden R., Joseph L., Frankham G.J., Lo N.. 2019. Museum specimens provide reliable SNP data for population genomic analysis of a widely distributed but threatened cockatoo species. Mol. Ecol. Resour. 19:1578–1592. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J.K.. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin C.Y., Newton A.H., Doronina L., Schmitz J., Hipsley C.A., Mitchell K.J., Gower G., Llamas B., Soubrier J., Heider T.N., Menzies B.R., Cooper A., O’Neill R.J., Pask A.J.. 2018. Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat. Ecol. Evol. 2:182–192. [DOI] [PubMed] [Google Scholar]
- Garg K.M., Chattopadhyay B., Cros E., Tomassi S., Benedick S., Edwards D.P., Rheindt F.E.. 2022. Island biogeography revisited: museomics reveals affinities of shelf island birds determined by bathymetry and paleo-rivers, not by distance to mainland. Mol. Biol. Evol. 39:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A., Gruber B., Pauly G.B., White D., Adams M., Young M.J., Kilian A., Zhang X., Shaffer H.B., Unmack P.J.. 2018. Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Mol. Ecol. 27:5195–5213. [DOI] [PubMed] [Google Scholar]
- Grant P.R., Grant B.R.. 2019. Hybridization increases population variation during adaptive radiation. Proc. Natl. Acad. Sci. U.S.A. 116:23216–23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber B., Unmack P.J., Berry O.F., Georges A.. 2018. dartr: an r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 18:691–699. [DOI] [PubMed] [Google Scholar]
- Gurkov A., Rivarola-Duarte L., Bedulina D., Fernández Casas I., Michael H., Drozdova P., Nazarova A., Govorukhina E., Timofeyev M., Stadler P.F., Luckenbach T.. 2019. Indication of ongoing amphipod speciation in Lake Baikal by genetic structures within endemic species. BMC Evol. Biol. 19:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Alonso G., Ramos‐Madrigal J., Sun X., Scharff‐Olsen C.H., Sinding M.S., Martins N.F., Ciucani M.M., Mak S.S.T., Lanigan L.T., Clausen C.G., Bhak J., Jeon S., Kim C., Eo K.Y., Cho S., Boldgiv B., Gantulga G., Unudbayasgalan Z., Kosintsev P.A., Stenøien H.K., Gilbert M.T.P., Gopalakrishnan S.. 2023. Conservation implications of elucidating the Korean wolf taxonomic ambiguity through whole-genome sequencing. Ecol. Evol. 13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs J.P.A., Richards Z.T., Popovic I., Lei C., Staeudle T.M., Montanari S.R., DiBattista J.D.. 2022. Hybridisation and the evolution of coral reef biodiversity. Coral Reefs 41:535–549. [Google Scholar]
- Hubisz M.J., Falush D., Stephens M., Pritchard J.K.. 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9:1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwood D.A., Hughes J.M.. 2001. Historical interdrainage dispersal of eastern rainbowfish from the Atherton Tableland, north-eastern Australia. J. Fish Biol. 58:1125–1136. [Google Scholar]
- Huxel G.R. 1999. Rapid displacement of native species by invasive species: effects of hybridization. Biol. Conserv. 89:143–152. [Google Scholar]
- Irestedt M., Ericson P.G.P., Johansson U.S., Oliver P., Joseph L., Blom M.P.K.. 2019. No signs of genetic erosion in a 19th century genome of the extinct Paradise Parrot (Psephotellus pulcherrimus). Diversity 11:58. [Google Scholar]
- Ismail G.B., Sampson D.B., Noakes D.L.G.. 2014. The status of Lake Lanao endemic cyprinids (Puntius species) and their conservation. Environ. Biol. Fishes 97:425–434. [Google Scholar]
- Janes J.K., Miller J.M., Dupuis J.R., Malenfant R.M., Gorrell J.C., Cullingham C.I., Andrew R.L.. 2017. The K = 2 conundrum. Mol. Ecol. 26:3594–3602. [DOI] [PubMed] [Google Scholar]
- Kautt A.F., Kratochwil C.F., Nater A., Machado-Schiaffino G., Olave M., Henning F., Torres-Dowdall J., Härer A., Hulsey C.D., Franchini P., Pippel M., Myers E.W., Meyer A.. 2020. Contrasting signatures of genomic divergence during sympatric speciation. Nature 588:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautt A.F., Machado-Schiaffino G., Meyer A.. 2016. Multispecies outcomes of sympatric speciation after admixture with the source population in two radiations of nicaraguan crater lake cichlids. PLoS Genet. 12:e1006157–e1006133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian A., Wenzl P., Huttner E., Carling J., Xia L., Blois H., Caig V., Heller-Uszynska K., Jaccoud D., Hopper C., Aschenbrenner-Kilian M., Evers M., Peng K., Cayla C., Hok P., Uszynski G.. 2012. Diversity Arrays Technology: a generic genome profiling technology on open platforms. In: Pompanon F., Bonin A., editors. Data production and analysis in population genomics: methods and protocols. Totowa (NJ): Humana Press. [DOI] [PubMed] [Google Scholar]
- Kontula T., Kirilchik S.V., Väinölä R.. 2003. Endemic diversification of the monophyletic cottoid fish species flock in Lake Baikal explored with mtDNA sequencing. Mol. Phylogenet. Evol. 27:143–155. [DOI] [PubMed] [Google Scholar]
- Kronforst M.R., Young L.G., Blume L.M., Gilbert L.E.. 2006. Multilocus analyses of admixture and introgression among hybridizing heliconius butterflies. Evolution 60:1254–1268. [PubMed] [Google Scholar]
- Lalueza-Fox C. 2022. Museomics. Curr. Biol. 32:R1214–R1215. [DOI] [PubMed] [Google Scholar]
- Lees D.C., Lack H.W., Rougerie R., Hernandez-Lopez A., Raus T., Avtzis N.D., Augustin S., Lopez-Vaamonde C.. 2011. Tracking origins of invasive herbivores through herbaria and archival DNA: the case of the horse-chestnut leaf miner. Front. Ecol. Environ. 9:322–328. [Google Scholar]
- Leggett R., Merrick J.R.. 1997. Australia’s Lake Eacham rainbow fish: lessons and outlook. Aquarium Sci. Conserv. 1:37–43. [Google Scholar]
- Lemoine M., Barluenga M., Lucek K., Mwaiko S., Haesler M., Chapman L.J., Chapman C.A., Seehausen O.. 2019. Recent sympatric speciation involving habitat-associated nuptial colour polymorphism in a crater lake cichlid. Hydrobiologia 832:297–315. [Google Scholar]
- Lintermans M. 2013. A review of on-ground recovery actions for threatened freshwater fish in Australia. Mar. Freshw. Res. 64:775–791. [Google Scholar]
- Lumbantobing D.N. 2014. Four new species of Rasbora of the Sumatrana group (Teleostei: Cyprinidae) from northern Sumatra, Indonesia. Zootaxa 3764:1–25. [DOI] [PubMed] [Google Scholar]
- Maier R., Patterson N.. 2022. admixtools: inferring demographic history from genetic data. R package version 2.0.0, 10.3389/fnhum.2015.00181. [DOI]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20:229–237. [DOI] [PubMed] [Google Scholar]
- Marques D.A., Meier J.I., Seehausen O.. 2019. A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34:531–544. [DOI] [PubMed] [Google Scholar]
- Martin C.H., Cutler J.S., Friel J.P., Dening Touokong C., Coop G., Wainwright P.C.. 2015. Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution 69:1406–1422. [DOI] [PubMed] [Google Scholar]
- McDonald D.B., Parchman T.L., Bower M.R., Hubert W.A., Rahel F.J.. 2008. An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proc. Natl. Acad. Sci. U.S.A. 105:10837–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee M.D., Borstein S.R., Meier J.I., Marques D.A., Mwaiko S., Taabu A., Kishe M.A., O’Meara B., Bruggmann R., Excoffier L., Seehausen O.. 2020. The ecological and genomic basis of explosive adaptive radiation. Nature 586:75–79. [DOI] [PubMed] [Google Scholar]
- McGee M.D., Borstein S.R., Neches R.Y., Buescher H.H., Seehausen O., Wainwright P.C.. 2015. A pharyngeal jaw evolutionary innovation facilitated extinction in Lake Victoria cichlids. Science (New York, N.Y.) 350:1077–1079. [DOI] [PubMed] [Google Scholar]
- McGee M.D., Schluter D., Wainwright P.C.. 2013. Functional basis of ecological divergence in sympatric stickleback. BMC Evol. Biol. 13:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan K., Franklin C.E., Moritz C., Blows M.W.. 2003. Adaptation of rainbow fish to lake and stream habitats. Evolution 57:104–118. [DOI] [PubMed] [Google Scholar]
- Meier J.I., Marques D.A., Mwaiko S., Wagner C.E., Excoffier L., Seehausen O.. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.I., McGee M.D., Marques D.A., Mwaiko S., Kishe M., Wandera S., Neumann D., Mrosso H., Chapman L.J., Chapman C.A., Kaufman L., Taabu-Munyaho A., Wagner C.E., Bruggmann R., Excoffier L., Seehausen O.. 2023. Cycles of fusion and fission enabled rapid parallel adaptive radiations in African cichlids. Science 381:eade2833. [DOI] [PubMed] [Google Scholar]
- Mirarab S., Reaz R., Bayzid M.S., Zimmermann T., Swenson M S., Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy E.K., Warnow T.. 2018. To include or not to include: the impact of gene filtering on species tree estimation methods. Syst. Biol. 67:285–303. [DOI] [PubMed] [Google Scholar]
- Moy K.G., Unmack P.J., Lintermans M., Duncan R.P., Brown C.. 2019. Barriers to hybridisation and their conservation implications for a highly threatened Australian fish species. Ethology 125:142–152. [Google Scholar]
- Natola L., Seneviratne S.S., Irwin D.. 2022. Population genomics of an emergent tri-species hybrid zone. Mol. Ecol. 31:5356–5367. [DOI] [PubMed] [Google Scholar]
- Nute M., Chou J., Molloy E.K., Warnow T.. 2018. The performance of coalescent-based species tree estimation methods under models of missing data. BMC Genomics 19:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenburghs J. 2019. Multispecies hybridization in birds. Avian Res. 10:1–11. [Google Scholar]
- Parham J.F., Papenfuss T.J., Sellas A.B., Stuart B.L., Simison W.B.. 2020. Molecular Phylogenetics and Evolution Genetic variation and admixture of red-eared sliders (Trachemys scripta elegans) in the USA. Mol. Phylogenet. Evol. 145:106722. [DOI] [PubMed] [Google Scholar]
- Pfaender J., Schliewen U.K., Herder F.. 2010. Phenotypic traits meet patterns of resource use in the radiation of “sharpfin” sailfin silverside fish in Lake Matano. Evol. Ecol. 24:957–974. [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Wen X., Falush D.. 2010. Documentation for structure software: Version 2.3.:1–37.
- Pusey B.J., Bird J., Kennard M.J., Arthington A.H.. 1997. Distribution of the Lake Eacham rainbowfish in the Wet Tropics region, North Queensland. Aust. J. Zool. 45:75–84. [Google Scholar]
- Pusey B.J., Kennard M.J., Arthington A.H.. 2004. Freshwater Fishes of North-Eastern Australia. Collingwood (Australia): CSIRO Publishing. [Google Scholar]
- R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, url: https://www.R-project.org/. [Google Scholar]
- Raxworthy C.J., Smith B.T.. 2021. Mining museums for historical DNA: advances and challenges in museomics. Trends Ecol. Evol. 36:1049–1060. [DOI] [PubMed] [Google Scholar]
- Roycroft E., MacDonald A.J., Moritz C., Moussalli A., Miguez R.P., Rowe K.C.. 2021. Museum genomics reveals the rapid decline and extinction of Australian rodents since European settlement. Proc. Natl. Acad. Sci. U.S.A. 118:e2021390118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.F., Deines J.M., Scriber J.M., Pfrender M.E., Jones S.E., Emrich S.J., Hellmann J.J.. 2018. Climate-mediated hybrid zone movement revealed with genomics, museum collection, and simulation modeling. Proc. Natl. Acad. Sci. 115:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J.F., Brumfield R.T., Faircloth B.C.. 2024. An island “endemic” born out of hybridization between introduced lineages. Mol. Ecol 33:e16990. [DOI] [PubMed] [Google Scholar]
- Schliewen U.K., Klee B.. 2004. Reticulate sympatric speciation in Cameroonian crater lake cichlids. Front. Zool. 1:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliewen U.K., Tautz D., Pääbo S.. 1994. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 368:629–632. [DOI] [PubMed] [Google Scholar]
- Seehausen O. 2006. Conservation: losing biodiversity by reverse speciation. Curr. Biol. 16:R334–R337. [DOI] [PubMed] [Google Scholar]
- Seehausen O., Takimoto G., Roy D., Jokela J.. 2008. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol. 17:30–44. [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P.. 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15:121–132. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 1993. Isolation by distance in equilibrium and non-equilibrium populations. Evolut. Int. J. Org. Evolut. 47:264–279. [DOI] [PubMed] [Google Scholar]
- Stager J.C., Alton K., Martin C.H., King D.T., Petruny L.W., Wiltse B., Livingstone D.A.. 2018. On the age and origin of Lake Ejagham, Cameroon, and its endemic fishes. Quat. Res. 89:21–32. [Google Scholar]
- Stroupe S., Forgacs D., Harris A., Derr J.N., Davis B.W.. 2022. Genomic evaluation of hybridization in historic and modern North American Bison (Bison bison). Sci. Rep. 12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Liu Y.C., Tiunov M.P., Gimranov D.O., Zhuang Y., Han Y., Driscoll C.A., Pang Y., Li C., Pan Y., Velasco M.S., Gopalakrishnan S., Yang R.Z., Li B.G., Jin K., Xu X., Uphyrkina O., Huang Y., Wu X.H., Gilbert M.T.P., O’Brien S.J., Yamaguchi N., Luo S.J.. 2023. Ancient DNA reveals genetic admixture in China during tiger evolution. Nat. Ecol. Evol. 7:1914–1929. [DOI] [PubMed] [Google Scholar]
- Tappin A. 2011. Rainbowfishes: their care & keeping in captivity, Art Publications, http://rainbowfish.angfaqld.org.au/Book.htm.
- Taylor E.B., Boughman J.W., Groenenboom M., Sniatynski M., Schluter D., Gow J.L.. 2006. Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol. Ecol. 15:343–355. [DOI] [PubMed] [Google Scholar]
- Unmack P.J., Allen G.R., Johnson J.B.. 2013. Phylogeny and biogeography of rainbowfishes (Melanotaeniidae) from Australia and New Guinea. Mol. Phylogenet. Evol. 67:15–27. [DOI] [PubMed] [Google Scholar]
- Unmack P.J., Brown C.. 2019. Melanotaenia eachamensis. The IUCN Red List of Threatened Species; 2019: e.T13054A123378257. [Google Scholar]
- Unmack P.J., Martin K., Hammer M.P., Ebner B.C., Moy K., Brown C.. 2016. Malanda Gold: the tale of a unique rainbowfish from the Atherton Tablelands, now on the verge of extinction. Fish. Sahul 30:1039–1054. [Google Scholar]
- von Rintelen T., von Rintelen K., Glaubrecht M., Schubart C.D., Herder F.. 2012. Aquatic biodiversity hotspots in Wallacea: the species flocks in the ancient lakes of Sulawesi, Indonesia. Biotic evolution and environmental change in southeast Asia. Cambridge: Cambridge University Press, 290–315. [Google Scholar]
- Wagner C.E., Harmon L.J., Seehausen O.. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487:366–369. [DOI] [PubMed] [Google Scholar]
- Walker D. 1999. Some physical and chemical features of two upland crater lakes in tropical north-eastern Australia. Mar. Freshw. Res. 50:159. [Google Scholar]
- Warnes G., Gorjanc G., Leisch F., Man M.. 2021. genetics: population Genetics. R Package version 1.3.8.1.3, https://cran.r-project.org/package=genetics.
- Whitehead P.W., Stephenson P.J., McDougall I., Hopkins M.S., Graham A.W., Collerson K.D., Johnson D.P.. 2007. Temporal development of the Atherton Basalt Province, north Queensland. Aust. J. Earth Sci. 54:691–709. [Google Scholar]
- Wielstra B., Arntzen J.W.. 2014. Kicking Triturus arntzeni when it’s down: large-scale nuclear genetic data confirm that newts from the type locality are genetically admixed. Zootaxa 3802:381–388. [DOI] [PubMed] [Google Scholar]
- Wong E.L.Y., Hiscock S.J., Filatov D.A.. 2022. The role of interspecific hybridisation in adaptation and speciation: insights from studies in senecio. Front. Plant Sci. 13:907363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.D., Ding X.D., Wang S., Wójcik J.M., Zhang Y., Tokarska M., Li Y., Wang M.S., Faruque O., Nielsen R., Zhang Q., Zhang Y.P.. 2018. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat. Ecol. Evol. 2:1139–1145. [DOI] [PubMed] [Google Scholar]
- Xi Z., Liu L., Davis C.C.. 2016. The impact of missing data on species tree estimation. Mol. Biol. Evol. 33:838–860. [DOI] [PubMed] [Google Scholar]
- Zhu D., Degnan S., Moritz C., Zhu D., Degnan S., Moritz C.. 1998. Evolutionary distinctiveness and status of the endangered lake eacham rainbowfish (Melanotaenia eachamensis). Conserv. Biol. 12:80–93. [Google Scholar]
- Zimmermann J., Hajibabaei M., Blackburn D.C., Hanken J., Cantin E., Posfai J., Evans T.C.. 2008. DNA damage in preserved specimens and tissue samples: a molecular assessment. Front. Zool. 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bk3j9kdjw.