Abstract
G9a is a histone methyltransferase that catalyzes the methylation of histone 3 lysine 9 (H3K9), which is involved in the regulation of gene expression. We had previously reported that G9a is expressed in developing tendons in vivo and in vitro and that G9a-deficient tenocytes show impaired proliferation and differentiation in vitro. In this study, we investigated the functions of G9a in tendon development in vivo by using G9a conditional knockout (G9a cKO) mice. We crossed Sox9Cre/+ mice with G9afl/fl mice to generate G9afl/fl; Sox9Cre/+ mice. The G9a cKO mice showed hypoplastic tendon formation at 3 weeks of age. Bromodeoxyuridine labeling on embryonic day 16.5 (E16.5) revealed decreased cell proliferation in the tenocytes of G9a cKO mice. Immunohistochemical analysis revealed decreased expression levels of G9a and its substrate, H3K9me2, in the vertebral tendons of G9a cKO mice. The tendon tissue of the vertebrae and limbs of G9a cKO mice showed reduced expression of a tendon marker, tenomodulin (Tnmd), and col1a1 genes, suggesting that tenocyte differentiation was suppressed. Overexpression of G9a resulted in enhancement of Tnmd and col1a1 expression in tenocytes in vitro. These results suggest that G9a regulates the proliferation and differentiation of tendon progenitor cells during tendon development. Thus, our results suggest that G9a plays an essential role in tendon development.
Keywords: Epigenetics, Histone modification, Tenocytes, Proliferation, Cell differentiation
Subject terms: Developmental biology, Genetics
Introduction
Epigenetic modifications play important roles in development and differentiation by regulating cell lineage-specific genes. Epigenetic modifications include post-translational modifications of histones, which regulate nuclear organization, chromatin structure, and gene expression1. Among histone modifications, methylation of histone 3 lysine 9 (H3K9) is a crucial modification that affects cell differentiation2. The methylated forms of H3K9, namely, H3K9me1, H3K9me2, and H3K9me3, are heterochromatin-associated histone modifications that play a role in genome compartmentalization3. Histone methyltransferases determine the methylated state of H3K94. One of these histone methyltransferases, G9a (also known as Ehmt2), catalyzes the mono- and dimethylation of H3K9 (H3K9me1 and H3K9me2)5. G9a contributes to transcriptional silencing via heterochromatin formation6–8. In vivo study showed that G9a KO embryo died around embryonic day 8 (E8), indicating that it is indispensable for early mouse development9. G9a suppresses germline-specific genes through DNA methylation during mouse embryogenesis10. In mouse oocytes, it regulates chromatin reorganization11. G9a also methylates non-histones proteins and regulates various biological processes. In myogenic differentiation, G9a suppresses functions of the myogenic regulatory factor MyoD by directly methylating it at lysine residue12. G9a regulates gene expression as a co-activator for nuclear hormone receptors such as estrogen receptor α (ERα), glucocorticoid receptors (GR), or critical transcriptional factor13–16. G9a and G9a-like protein (GLP) act as coactivators for a subset of GR target genes and their post-translational methylation are important for coactivator function16. G9a dimethylates ERα and functions as an ERα coactivator to affect hormonal target genes in breast cancer cells17. In erythroid cells, G9a activates β maj-globin gene by stabilizing pre-initiation complex (PIC) formation, and direct G9a -Pol II interaction is involved in this process18. We previously reported that G9a is expressed in the growth plate of the skeleton19, the mesenchyme of tooth germs20, and tendons21. In addition, G9 regulates cranial bone development by activating the function of an osteoblastic transcription factor, Runx215. Thus, G9a plays important biological roles by suppressing or activating gene expression.
Tendons are composed of fibrous connective tissue that connects muscles to bone to transmit the force generated by muscle contraction, thus playing important roles in the locomotor system. During development, tendon progenitor cells are derived from mesenchyme condensation of somite and neural crest. The progenitors express two critical transcription factors, scleraxis (Scx) and SRY-box containing gene 9 (Sox9)22,23. Lineage analysis using Sox9-Cre showed that Sox9-expressing cells differentiate into tendon cells24,25. Scx continues to be expressed in tendon progenitors during development, whereas Sox9 expression shifts from the progenitors to chondrocytes and tendon-bone junctional tissues. Differentiation of tendon cells proceeds through production of extracellular matrix (ECM) components, such as type I, III, V, VI, and XII collagens and proteoglycans, including decorin, fibromodulin, and biglycan26–28. The expression of these ECM proteins is regulated by tendon-specific transcriptional factors such as Scx, Mohawk (Mkx), and Egr129–33. Recent genome-wide study showed the existence of multiple Scx binding sites of Scx-dependent genes including tenomodulin (Tnmd), sine oculis-related homeobox 2 (Six2), and fibromodulin (Fmod)34. The functions of the transcriptional factors may be influenced by epigenetic modifiers, including histone methyltransferases. Enhancer of zeste homolog 2 (EZH2) which catalyzes histone H3 lysine (K) 27 methylation, was shown to be essential for early patterning of all musculoskeletal tissues in EZH2 cKO mice by Prx1-Cre. When EZH2 was deleted by tendon-specific Scx-Cre in the EZH2 cKO mice, formation of tendon collagen matrix was not affected, suggesting that EZH2 is likely to be dispensable for tendon differentiation35. We had previously shown that another epigenetic modifier, G9a, is expressed in the early stage of tendon development, and that the expression of tendon-related genes and proliferation are suppressed in G9a-null tenocytes in vitro21. However, involvement of G9a in tendon formation in vivo have not yet been investigated.
In this study, we wished to determine roles of G9a in tendon formation in vivo. We generated G9a conditional knockout (G9a cKO) mice, in which G9a was deleted from tendon progenitor cells using Sox9-Cre as a Cre-mediating gene deletion24. We first confirmed that Sox9-Cre is functional in the tendon progenitor cells in vivo. We then crossed G9afl/fl mice with Sox9Cre/+ mice to create G9a cKO mice. Our results demonstrated that tendon tissue in G9a cKO mice is hypoplastic, and that this hypoplasia may be associated with inhibition of cell growth and differentiation during tendon development.
Results
Sox9-Cre is active in the tendon progenitor cells
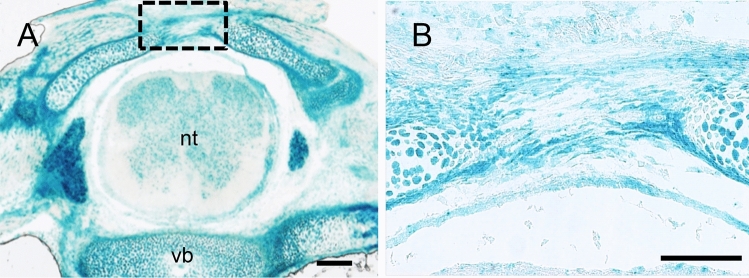
To investigate G9a function in tendon formation in vivo, we generated tendon-specific G9a cKO mice by crossing G9afl/fl mice with Sox9 Cre/+ mice expressing Cre under the control of a specific Sox9 promoter. Tendon progenitor cells are derived from Sox9-expressing cells during development. Therefore, we confirmed the localization of tenocytes derived from Sox9-expressing cells using Sox9Cre/+; R26R mice. The vertebral tendon between the lumbar vertebrae at E16.5 showed β-galactosidase (LacZ) staining (Fig. 1A,B). These results indicated that Cre-recombinase driven by the Sox9 promoter was active in the vertebral tendon tissue.
Fig. 1.
Analysis of Cre-recombinase expression in tendon tissue using Sox9Cre/+; R26R mice. (A) LacZ staining of vertebral tendons in Sox9Cre/+; R26R mouse embryos at E16.5 (n = 3). Bar = 300 μm. Cre-recombinase driven by the Sox9 promoter was active in the vertebral tendon tissue at E16.5. (B) Enlargement of the boxed area in (A). nt, neural tube; vb, vertebral body. Bar = 50 μm.
G9a and H3K9me2 levels decreased in the tendon tissue of G9a cKO mice
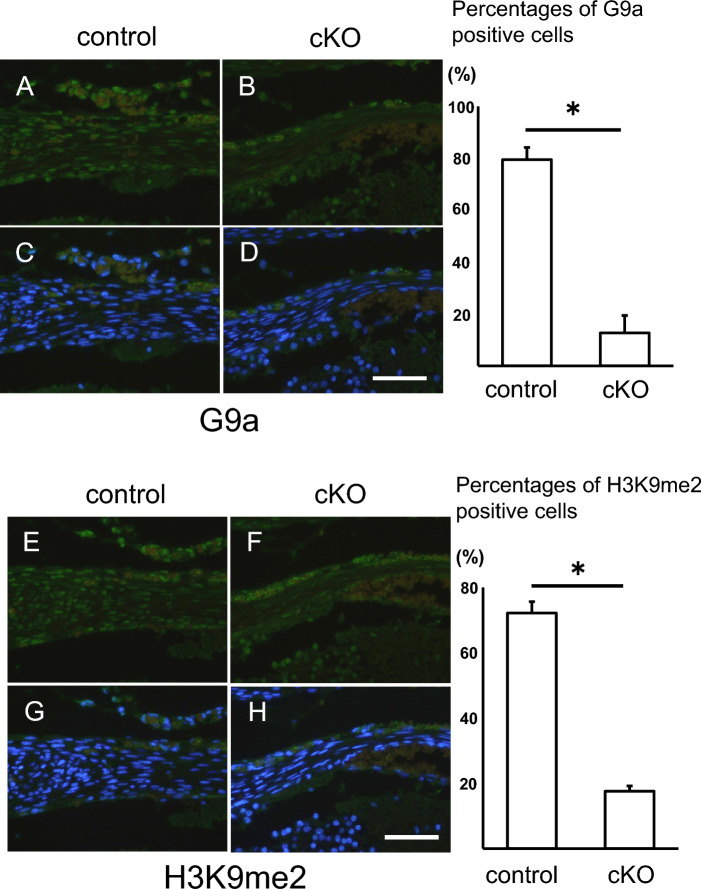
To investigate levels of G9a and H3K9me2 in the tendon tissue of G9a cKO mice, we examined their localization by immunostaining with anti-G9a and H3K9me2 antibodies. G9a expression in the tendon tissues of G9a cKO mice was significantly lower than that in control mice (Fig. 2A–D). The percentage of G9a-positive cells was lower in the tendons of G9a cKO mice (upper right panel in Fig. 2). Similarly, H3K9me2-positive regions were diminished in the tendon tissue of G9a cKO mice (Fig. 2E–H), and the percentage of H3K9me2-positive cells was reduced in the tendon of G9a cKO mice (lower right panel in Fig. 2).
Fig. 2.
Decreased G9a and H3K9me2 levels in tendon tissue of G9a cKO mice. Immunohistochemical analysis of G9a (A–D) and H3K9me2 (E–H) in the vertebral tendon tissue of control (G9afl/+; Sox9Cre/+) and G9a cKO (G9afl/fl; Sox9Cre/+) mouse embryos at E17.5 (control n = 3; cKO n = 3). Immunofluorescent images of staining with anti-G9a (A and B) and anti-H3K9me2 antibodies (E and F) (green), and merged images of fluorescence signals for immunostaining and nuclear staining with DAPI (blue) (C, D, G, and H). Bars = 50 μm. The percentages of G9a-positive cells (upper right panel) and H3K9me2 (lower right panel) were shown in control and G9a cKO mice (*p < 0.05, t-test).
Hypoplastic tendon formation in G9a cKO mice
Next, we examined tendon formation in G9a cKO mice. We previously showed that most of these G9a cKO mice died at the time of birth or within 2 days after birth due to defects of the digestive system36. A few mice survived after birth and could be analyzed at 3 weeks of age. Visual observation of the tissue revealed that the tendons of the limbs and vertebrae in G9a cKO mice were hypoplastic in comparison with those in the control mice at 3 weeks of age (Fig. 3A–F). We then performed histological analysis of the tendon tissue in G9a cKO mice. Sections stained with hematoxylin and eosin revealed that the vertebral tendon was thinner in G9a cKO mice than in the control mice at E16.5 (Fig. 4A–D). Furthermore, Achilles tendon at P0 and masseter tendon at 3 weeks were both hypoplastic in G9a cKO mice (Supplemental Fig. 1). Masseter muscle where Sox9-Cre was not fully active seems to be unchanged at P21. These results suggest that G9a is required for tendon tissue formation during development.
Fig. 3.
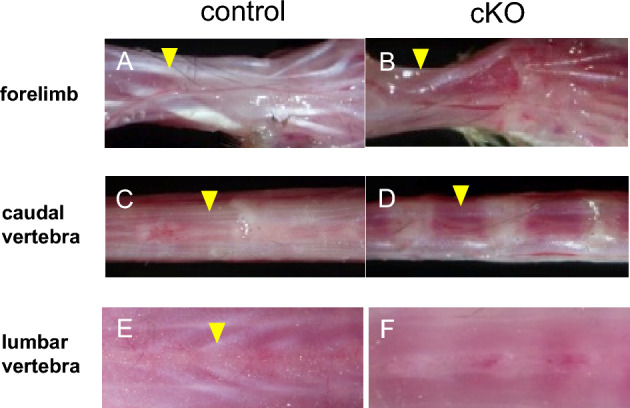
Hypoplastic tendon formation in G9a cKO mice. Tendons of the forelimb (A and B; yellow arrowheads), caudal vertebrae (C and D; yellow arrowheads), and lumbar vertebrae (E and F; yellow arrowheads) in G9a cKO mice (G9afl/fl; Sox9Cre/+) were compared with those in control mice (G9afl/+; Sox9Cre/+) at 3 weeks of age (control n = 3; cKO n = 3). Tendon formation was hypoplastic in all tendons examined.
Fig. 4.
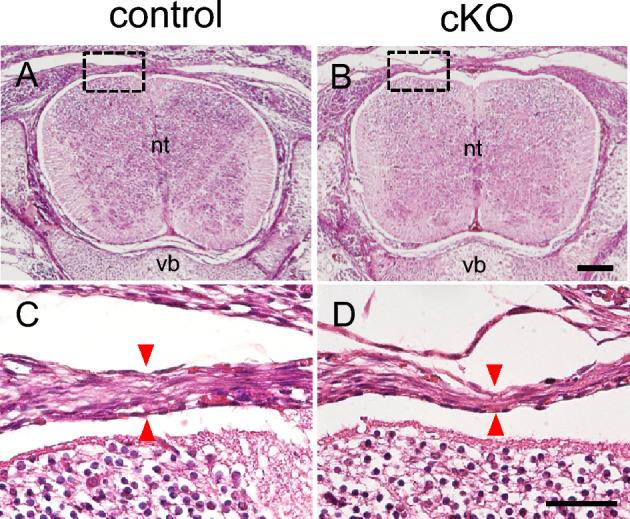
Decreased width of tendon tissue in G9a cKO mice. Histological analysis of vertebral tendons by hematoxylin and eosin staining in control (A and C, G9afl/+; Sox9Cre/+) and G9a cKO (B and D, G9afl/fl; Sox9Cre/+) mouse embryos at E16.5 (control n = 3; cKO n = 3). Red arrowheads indicate the width of the vertebral tendon. (B and D) show enlarged images of the boxed areas in (A and C), respectively. nt, neural tube; vb, vertebral body. Bar = 300 μm (A and B), 50 μm (C and D).
Loss of G9a inhibits the proliferation of tenocytes in the developing tendon
To investigate the effects of G9a on tendon tissue proliferation, we performed BrdU-labeling experiments by injecting BrdU. Immunohistochemical analysis using the BrdU antibody revealed fewer BrdU-positive cells in the tendon tissue of G9a cKO mice (Fig. 5A,B). We confirmed that the percentage of BrdU-positive cells was lower in the vertebral tendon tissue of G9a cKO mice at E16.5 (Fig. 5C). To assess cell proliferation, we further examined expression of proliferating cell nuclear antigen (PCNA) in the tendon tissue sections. Immunohistochemical analysis of PCNA revealed that PCNA-positive cells were decreased in vertebral tendon tissue of G9a cKO mice at E17.5 (Supplemental Fig. 2). We also examined the effects of G9a deletion on tenocyte apoptosis by using the TUNEL assay (Fig. 5D,E). The TUNEL assay revealed that TUNEL-positive cells were not observed in the tendon tissue of both control and G9a KO mice at E16.5. These results suggest that G9a promotes tenocyte proliferation but does not affect apoptosis during tendon formation.
Fig. 5.
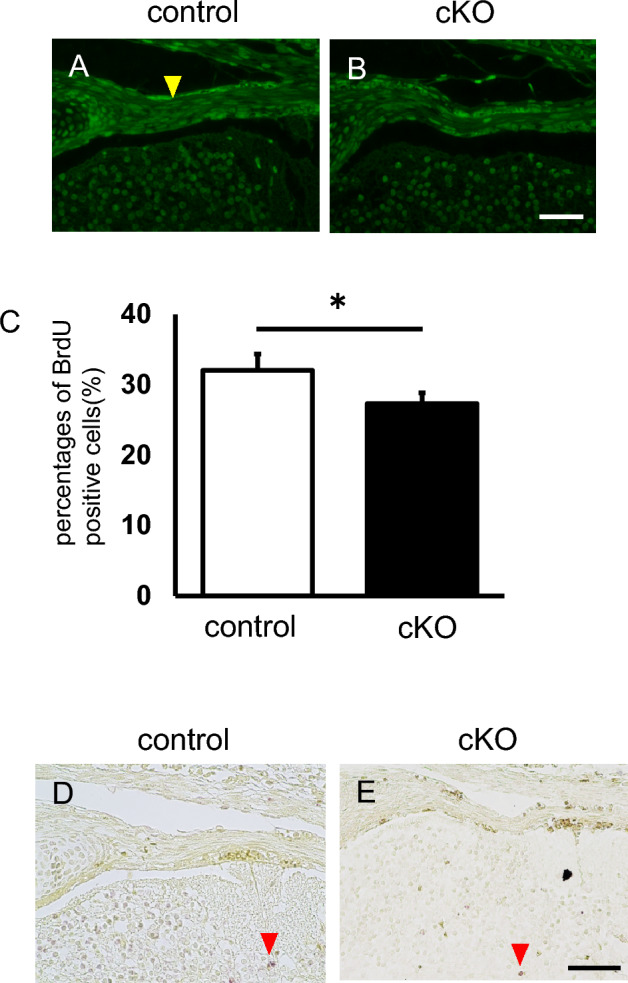
G9a deletion inhibits proliferation but does not affect apoptosis in developing tendons. BrdU was injected at E16.5 and recovered at E17.5. BrdU incorporation was assessed in the vertebral tenocytes of control (A, G9afl/+; Sox9Cre/+) and G9a cKO mice (B, G9afl/fl; Sox9Cre/+) by using an anti-BrdU antibody (control n = 2; cKO n = 2). Immunofluorescence images of BrdU-labeled cells are shown. The yellow arrowheads indicate strongly positive cells. Bar = 50 μm. (C) Comparison of the percentages of BrdU-positive cells (n = 3). We counted 60–64 cells/section and three independent sections were analyzed (*p < 0.05, t-test). (D and E) TUNEL staining in vertebral tenocytes of control (D, G9afl/+; Sox9Cre/+) and G9a cKO (E, G9afl/fl; Sox9Cre/+) mouse embryos at E16.5. Red arrowheads indicate apoptotic nuclei. Bar = 50 μm.
Expression levels of tendon markers decreased in the developing tendons in G9a cKO mice
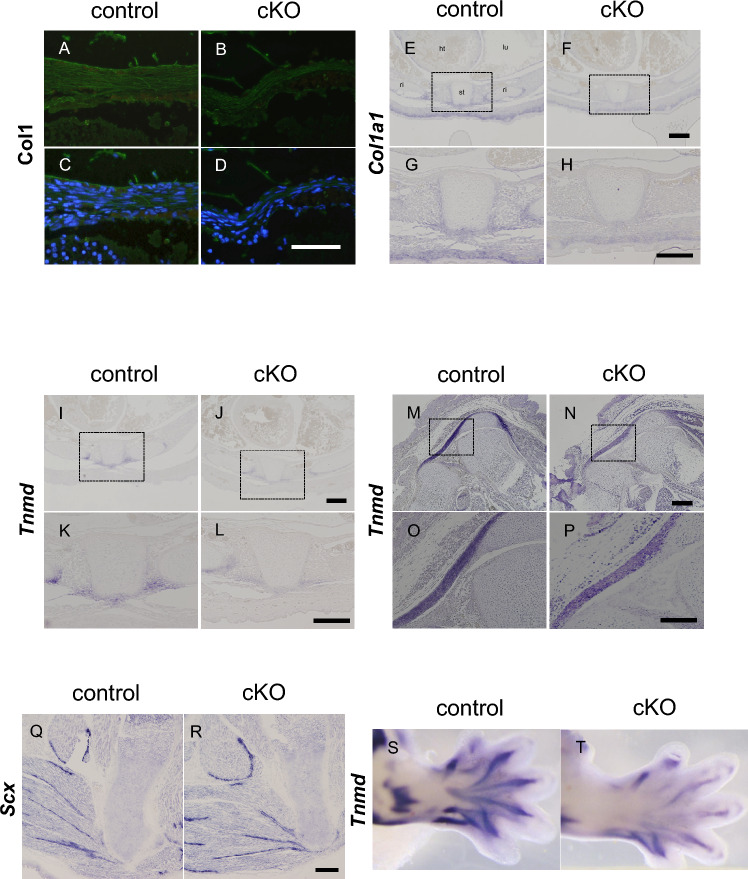
To examine the expression of the molecular markers of tenocyte differentiation, we performed immunostaining for type I collagen, which is the major component of the tendon extracellular matrix. At E17.5, type I collagen was expressed in the tendon tissue of control mice (Fig. 6A,C), whereas its signal was lower in the vertebral tendon tissue of G9a cKO mice (Fig. 6B,D). ISH experiments also showed that Col1a1 mRNA levels were reduced in the tenocytes of G9a mutant embryos by E16.5, in comparison with the levels in control embryos (Fig. 6E–H). We also examined the expression of Tnmd and Scx, markers of tendon differentiation. Tnmd mRNA levels in the tenocytes of G9a-deleted embryos by E16.5 were also lower than those in control embryos (Fig. 6I–P). WISH experiments also showed a reduction in Tnmd mRNA levels in G9a-deleted limbs at E15.5 (Fig. 6S, T). ISH experiments showed that the numbers of cells expressing Scx mRNA seem to be lower in G9a cKO mice, however, its intensity in the tenocytes looks similar between G9a cKO and control embryos (Fig. 6Q,R). By q-PCR experiments, we found that mRNA levels of Col1a1, Tnmd, and Scx were suppressed in hindlimb tendon tissue in G9a cKO mice at E17.5 (Supplemental Fig. 3). We also checked testis-specific genes, which were shown to be activated in G9a KO embryos10, and found that their expressions were not activated in G9a cKO tendons (Supplemental Fig. 4). Thus, the expression of tendon differentiation markers was decreased in G9a cKO mice.
Fig. 6.
Decreased expressions of tendon markers in developing tendons in G9a cKO mice. (A–D) Immunohistochemical analysis of type I collagen in the vertebral tendon tissue of control (A and C, G9a+/+; Sox9Cre/+) and G9a cKO mice (B and D, G9afl/fl; Sox9Cre/+) at E17.5 (control n = 3; cKO n = 3). Immunofluorescent images of staining with anti-Col1 antibody (A and B) (green), and merged images of anti-Col1 immunostaining and nuclear staining with DAPI (blue) (C and D). Bar = 50 μm. (E–P) In situ hybridization (ISH) on transverse sections of the trunk (E–L) and sagittal sections of the hindlimb (M–P) at E16.5 (control n = 3; cKO n = 3). Tissue sections from control (E, G, I, K, M, O, and Q, G9a+/+; Sox9Cre/+) and G9a cKO (F, H, J, L, N, P, and R, G9afl/fl; Sox9Cre/+) embryos were analyzed using antisense RNA probes for tendon markers, collagen type I alpha 1 chain (Col1a1), tenomodulin (Tnmd), and screlaxis (Scx) mRNA. Ht, heart; lu, lung; st, sternum; ri, rib. Bar = 100 μm (E–P), 200 µm (Q and R). Whole-mount ISH of Tnmd in forelimbs from control (S, G9a+/+; Sox9Cre/+) and G9aG9a cKO mice (T, G9afl/fl; Sox9Cre/+) was performed at E15.5 (control n = 3; cKO n = 3).
Overexpression of G9a resulted in enhancement of Tnmd and col1a1 expression in tenocytes in vitro
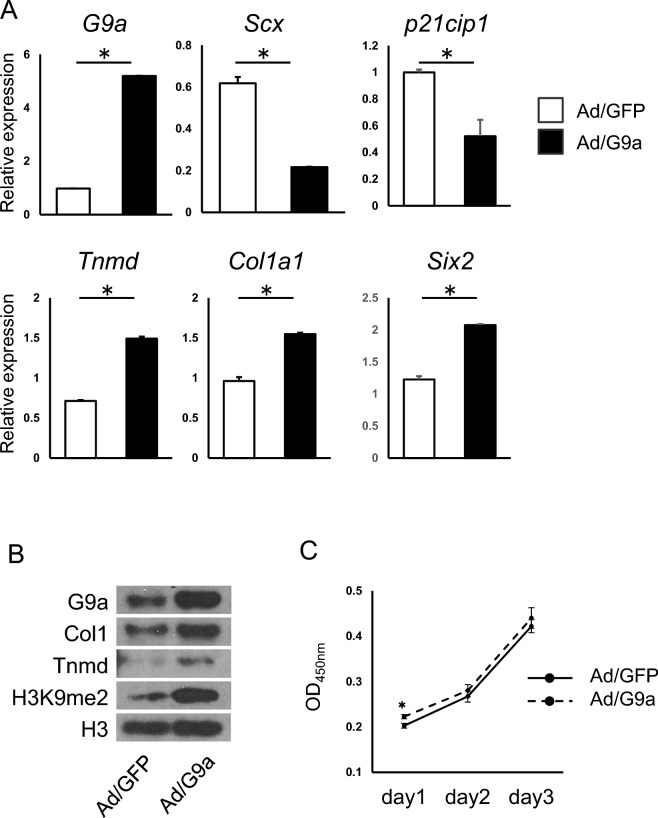
Previously we showed the expression levels of tendon-related genes Tnmd, Col1a1, and Scx were decreased in G9a deleted tenocytes in vitro. From the results of both in vivo and in vitro studies, we have assumed that G9a is involved in the activation of those gene expressions. To test this assumption, we performed overexpression experiment of G9a in tenocytes. Adenovirus expression of G9a in tenocytes resulted in increase of Tnmd, Col1a1, and Six2 gene expressions, whereas Scx expression was suppressed (Fig. 7A). Western blot analysis showed that protein levels of Tnmd and Col1a1 were also increased (Fig. 7B and Supplemental Fig. 5).
Fig. 7.
Overexpression of G9a resulted in enhancement of tendon marker expressions in tenocytes in vitro. Primary tenocytes were infected with adenoviruses expressing G9a (Ad/G9a) or GFP (Ad/GFP). (A) Expression of tendon marker genes at day 4 after the adenovirus infection. Gene expressions of G9a, Scx, p21cip1, Tnmd, Col1a1, and Six2 were shown. Representative results of four independent experiments are shown (*p < 0.05, t-test). (B) Proteins were extracted at day 4 after adenovirus infection. Protein levels of G9a, COL1A1, Tnmd, and H3K9me2 are shown. Histone H3 protein is served as an internal loading control. Representative results of three independent experiments are shown. (C) Measurement of cell proliferation was performed at day1, 2 and 3 after the adenovirus infection. Representative results of two independent experiments are shown (*p < 0.05, t-test).
We confirmed that H3K9me2 level was increased along with G9a overexpression (Fig. 7B). Cell proliferation assay showed that slight increase of proliferation in G9a over-expressed cells on day 1 (Fig. 7C). Expression of a cell cycle negative regulator, p21, was suppressed in those cells (Fig. 7A). These results suggest that G9a activates some of tendon-related genes in tenocytes independent of H3K9me2-mediated gene suppression.
Discussion
In the present study, we showed that conditional deletion of the G9a gene resulted in inhibition of cell growth and a decrease in tendon marker expression, leading to tendon hypoplasia during development.
During tendon development, tendon progenitors arise from the sclerotome, lateral plate mesoderm, and neural crest24,37. These progenitor cells migrate and settle in the prospective region, giving rise to the skeletal primordia38. Sox9 is expressed in the mesenchymal condensation of skeletal primordia and gives rise to cartilage, bone, and tendon later in the development23–25. In this study, we confirmed LacZ expression in vertebral tendon tissue, indicating the functionality of Sox9-Cre in tendon progenitor cells (Fig. 1). Previously we showed that G9a is expressed in embryonic tendon progenitor cells21. The results of the present study suggest that G9a deletion in Sox9-expressing tendon progenitor cells inhibited cell proliferation and differentiation, leading to tendon hypoplasia.
Our findings showed that G9a cKO mice exhibit suppressed tenocyte proliferation during development. G9a has been reported to be overexpressed in a number of cancers39–41, and the loss of G9a has been shown to inhibit cell proliferation in various cells42,43. Previous studies have reported that G9a represses the expression of negative regulators of the cell cycle, such as p21Cip1/Waf1 (p21), in a methyltransferase activity-dependent manner in several cell types44–46. We previously showed that the proliferation of G9a-null tenocytes was significantly decreased and the p21 gene was upregulated in vitro21. In this study, we found that overexpression of G9a resulted in down-regulation of p21 gene and slight increase of cell proliferation in tenocytes. Therefore, G9a may promote cell-cycle progression by transcriptionally repressing p21 expression in tenocytes. G9a represses p21 by mediating H3K9me2 marks, whereas it regulates the E2F1 target genes, CyclinD1 and DHFR, leading to cellular proliferation in myoblasts46. Thus, one possible mechanism for the inhibition of tenocyte proliferation in G9a cKO mice is the regulation of cell-cycle modulators by G9a.
Our data also showed that Tnmd expression was suppressed in the developing tendon of G9a cKO mice. A previous study reported that cell growth is suppressed in Tnmd-null mice47. Furthermore, the loss of Tnmd has been shown to result in the suppression of mouse tendon stem/progenitor cells (mTSPCs)48. Tnmd-knockout mTSPCs showed significantly decreased expression of the Cyclin D1 gene and increased p53 mRNA expression. Furthermore, LacZ staining revealed that the number of senescent cells was increased in Tnmd-knockout TSPCs, although apoptosis was not affected. Our data also showed that tenocyte proliferation was inhibited and apoptosis did not change in the G9a cKO mice. Therefore, G9a may indirectly suppress cell growth through Tnmd expression.
G9a mediates methylation of H3K9me2, which is associated with the formation of transcriptionally inactive heterochromatin and gene silencing49. However, the expression of tendon markers such as type I collagen and Tnmd was suppressed in the developing tendons of G9a cKO mice, in accordance with our previous data in vitro21. Furthermore, G9a overexpression in tenocytes increased expressions of Col1a1, Tnmd, and Six2, although H3K9me2 level was increased. Thus, it suggests that G9a activates those tendon genes in tenocytes, independent of H3K9me2 mediated transcriptional repression. How does G9a activate those cell lineage-related genes? Several possibilities were proposed in other type of cells13,14,18,50. In erythroid cells, G9a recruits Mediator to the βmaj promoter of the β globin gene to activate its expression18. In prostate cancer cells and osteoblasts, G9a was recruited to endogenous Runx2 binding sites to activate Runx2 target genes15,50. In tenocytes, it was shown that promoters of Tnmd and Six2 have Scx binding sites and their expressions are Scx-dependent34. Although G9a overexpression suppressed Scx expression, Tnmd and Six2 expressions were increased in tenocytes, raising a possibility that G9a promotes Scx action in tenocytes. Another possibility could be that G9a recruits Mediator to the promoter of Tnmd and Six2 genes in tenocytes, similar to the promoter of β globin gene in erythroid cells.
In contrast to tenocytes, other studies have reported that the suppression of G9a increases Tnmd and Col1a1 expressions in various cells. In the G9a-KO testis and ovaries, Tnmd expression is upregulated51. Col1a1 expression was increased in vascular smooth muscle, and liver cells52,53. Therefore, the effects of G9a deletion on Tnmd and Col1a1 genes are likely context-and/or cell type-dependent. In the G9a KO E8.5 embryos, testis-specific genes are upregulated, in which G9a-guided DNA methylation represses germline genes10. However, germline genes were not activated in tendons in G9a cKO mice (Supplemental Fig. 4), suggesting that suppression of germline genes by G9a-guided DNA methylation is cell type-dependent.
Our results suggest that in tendon development G9a activates tendon-related genes independent of H3K9me2 mediated transcriptional repression, and suppresses p21 expression through H3K9me2. Thus, G9a functions as a repressor or activator depending on its associating partners. Further studies are required to explore this switching mechanism in detail.
Sox9-expressing cells are known to contribute to chondrogenic cells54, and Sox9 and Scx double-positive progenitors reside in the chondrotendinous junction during development and later become tenocytes and chondrocytes55,56. Since tendon progenitors and chondrocytes mutually interact during development, the repression of proliferation of tendon progenitors may be attributable to an indirect mechanism involving G9a deletion in chondrogenic cells. Since Sox9-Cre influences both tendon progenitors and chondrocytes, the findings for our cKO mice could not distinguish the effects on tenocytes from the indirect effects mediated through chondrogenesis, which is a limitation of our study using Sox9-Cre-expressing cKO mice.
In conclusion, our data indicate that G9a plays an essential role in tendon development. These findings suggest that G9a regulates cell growth and gene expression via epigenetic changes during tendon development. These results offer insights into the mechanism underlying tendon development and repair, suggesting potential approaches targeting tendon-related disorders.
Materials and methods
Animals
All animal experiments were approved by the Institutional Animal Care Committee, and the Recombination Experiment and Biosafety Committee of Tsurumi University School of Dental Medicine. Mice were cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and our institutional guidelines. All animal experiments in this study were done in accordance with ARRIVE guidelines.
To generate G9afl/fl (Ehmt2tm2Yshk) mice, G9a deleted in ES cells (TT2 cell line) were injected into ICR mouse embryos51. G9afl/+ mice were sequentially backcrossed with the C57BL/6 background mice, and offsprings later than F15 generation were used for further studies. Sox9Cre/+(Sox9tm3(cre)Crm) mice, which were generously given by Prof. Haruhiko Akiyama24, were intercrossed with G9afl/+ mice to generate a conditional mutant mouse line lacking G9a, G9a-cKO (Sox9-specific G9a gene deletion: G9afl/fl; Sox9Cre/+) mice. G9afl/+ mice were served as control. Polymerase chain reaction genotyping of G9a cKO mice was performed using the following primer pairs: mGE28R (GCTCCAGGGCGATGGCCTCCGCTGAATGC) and mGI27-2F (CGGGACAGGGTTTCTCTGTGTAGTCC) for wild-type G9 allele detection, mGE28R and G3 (GGGCCAGCTCATTCCTCCACTC) for floxed G9a allele detection, and Sox9-Cre-F1 (TCCAATTTACTGACCGTACACCAA) and Sox9-Cre-R1 (CCTGATCCTGGCAATTTCGGCTA) for Sox9Cre allele detection. R26R mutant mice carrying a loxP flanked neo cassette upstream of a β-galactosidase (lacZ) sequence were obtained from the Jackson Laboratory and were intercrossed with Sox9Cre/+ mice to generate Sox9Cre/+; R26R mice to detect cells derived from Sox9-expressing cells.
Antibodies
The following antibodies were used in this study: anti-H3K9me2 (CMA307; mouse monoclonal, prepared by H. Kimura57); anti-G9a (PP-A8620A-00, mouse monoclonal; Perseus Proteomics, Tokyo, Japan); anti-type I collagen (2150-1410, rabbit polyclonal; AbD Serotec, Oxford, UK); anti-tenomodulin (bs-7525R, rabbit polyclonal; Bioss Inc., MA); anti-BrdU (111703760011, mouse monoclonal; Roche, IN); anti-PCNA (2586, mouse monoclonal; Cell Signaling Technology, Danvers, MA); anti-H3 (MABI0301, mouse monoclonal; MAB Institute Inc., Nagano, Japan); anti-mouse IgG-HRP (1858413, goat rabbit polyclonal; Pierce, IL); anti-rabbit IgG-HRP (1858415, goat rabbit polyclonal; Pierce) and anti-mouse IgG (H + L)-Alexa488 (A11001, goat polyclonal; Invitrogen, Carlsbad, CA). The anti-H3K9me2 antibody is specific for histone H3 dimethyl Lys9, which has no cross-reactivity with other histone modifications including H3K27me2 evaluated by ELISA57. By immunofluorescent analysis using this antibody, we showed that levels of H3K9me2 in the G9a-deleted tissues were significantly low, suggesting the specificity of this antibody in immunofluorescence (see Kamiunten et al.36 and results of Fig. 2 in this study).
LacZ activity staining
Staining for LacZ activity was performed as described previously58. Briefly, dissected tissues were fixed with 0.25% glutaraldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature, treated overnight with 20% sucrose in PBS at 4 °C, and embedded in Tissue-Tek® OCT compound (Sakura Finetek Japan, Tokyo, Japan). Frozen sections (12-μm sections) were prepared. Before staining, the sections were treated with fixation solution (0.2% glutaraldehyde, 5 mM EGTA, 2 mM MgCl2) at 4 °C for 5 min. After washing (phosphate buffer containing 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% Nonidet P-40), the sections were incubated with X-gal staining solution (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 1 mg/mL X-gal) for 6 h at room temperature.
Immunohistochemistry
Immunohistochemical analyses were performed using primary antibodies against G9a, H3K9me2, and ColI. At embryonic day 16.5 (E16.5), embryos were fixed with 4% paraformaldehyde (PFA) in PBS for 24 h. PFA-fixed tissue was embedded in paraffin, and 5-μm sections were prepared. After deparaffinization, sections were heated in a microwave in 10 mM citric acid buffer (pH 6) for 20 min. During microwaving, the temperature of the buffer was maintained at 90 °C. Then, the sections were treated with blocking solution in goat serum and incubated overnight at 4 °C with one of the following primary antibodies: anti-H3K9me2 (1:100), anti-G9a (1:400), anti-PCNA (1:400), and anti-Col1 (1:400). After washing, the sections were incubated with secondary antibodies conjugated to Alexa Fluor 488 (1:1000) and DAPI (1:10, CS-2010-06; Cosmo Bio, Tokyo, Japan). Fluorescent images were acquired using a Keyense BZ-9000 microscope. The percentages of G9a-positive and H3K9me2-positive cells were calculated as the number of positive cells/number of cells with DAPI-stained nuclei. These values were compared between the control and G9a cKO mice.
In situ hybridization and whole-mount in situ hybridization
In situ hybridization (ISH) of transverse sections and whole-mount in situ hybridization (WISH) were performed on mouse embryos collected at E15.5, as described previously59. RNA probes for the following regions were used for ISH studies: Col1a1 (AK159285; 4446-4759), Scx (NM_198885; 415-1128), and Tnmd (NM_022322; 654-1299).
Bromodeoxyuridine labeling experiment
Bromodeoxyuridine (BrdU) labeling was performed as described previously60. E16.5 pregnant mice were subcutaneously injected with BrdU at a dose of 50 μg/g body weight in 0.9% NaCl. The mice were euthanized by manual cervical dislocation 24 h after injection and dissected to obtain the tendon tissues. Then, the tissues were fixed in PFA at 4 ℃ overnight, embedded in paraffin, and cut into 5-μm sections. After deparaffinization, the sections were treated with 2 N hydrochloric acid (HCl) for 90 min at 37 ℃ to denature DNA and then neutralized with 0.1 M boric acid for 5 min. The prepared sections were reacted with anti-BrdU monoclonal antibody diluted 1/400 in CanGetSignal® immunostain (TOYOBO, Osaka, Japan) overnight at 4 °C. After washing, the sections were incubated with secondary antibodies conjugated to Alexa Fluor 488 (1:1000). Fluorescent images were acquired using a Keyense BZ-9000 microscope. The percentage of BrdU-positive cells was calculated as the number of BrdU-positive cells divided by the number of cells counted using DAPI staining.
Cell culture
Primary tenocytes were isolated from the tail tendons of 12-week-old mice by collagen gel culture as previously described21. The cells were then plated at 2 × 104 cells/cm2 in alpha minimal essential medium (alpha-MEM) supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 and 95% air. Subsequently, the cells were incubated for the indicated period of time with a change of medium every 2 days.
Preparation of adenovirus vectors and their infection into tenocytes
The production of adenovirus was performed as described previously61. according to the manufacturer's protocol (ViraPower Adenoviral Expression System; Thermo Fisher Scientific). An adenovirus vector expressing enhanced green fluorescent protein (EGFP) (Ad/GFP), which contains CAG-IRES-EGFP, was prepared as described61. Another adenovirus vector expressing G9a (Ad/G9a) was constructed as described previously62. Infection of adenovirus was performed on the day of plating at multiplicity of infection (MOI) 50.
Measurement of cell proliferation
Primary tenocytes infected with Ad/GFP or Ad/G9a at a MOI 50 were seeded at 1.6 × 104 cells/well into 48 well plates. A solution from the Cell Counting Kit-8 (CCK-8) (Dojin Chemistry, Kumamoto, Japan) was added 20 μL per well at 1, 2, and 3 days after plating and incubated for 1 h. Absorbance (450 nm) was measured using a microplate reader (Molecular device, CA).
Western blot analysis
Western blot analysis was performed as described21. Briefly, the proteins were extracted and lysed in 1 × SDS sample buffer (New England Biolabs, Beverly, MA) in which a complete protease inhibitor mixture tablet (Roche Diagnostics, Indianapolis, IN) was dissolved. The lysed proteins were separated on 10% Super Sep™ Ace SDS–PAGE gel (Wako Pure Chemicals, Osaka, Japan) and electro-transferred to PVDF membranes (PALL, Ann Arbor, MI). Primary antibodies [G9a (1:1,000), Col1 (1:1,000), Tnmd (1:1,000), H3K9me2 (1:1000), H3 (1:1,000)] were used in Can Get Signal solution 1 (Toyobo, Osaka, Japan) at room temperature (RT) for 1 h. Secondary antibodies, HRP-conjugated goat anti-rabbit IgG and HRP-conjugated goat anti-mouse IgG, were used at a dilution of 1:5000 in Can Get Signal solution 2 (Toyobo) and hybridized to membranes at RT for 1 h. Immunoreactive bands were detected by chemiluminescence using Western Lightning ECL Pro (Perkin-Elmer, Waltham, MA).
Reverse transcription and real-time PCR
RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA was reverse transcribed using oligo-dT primers and reverse transcriptase (Superscript III; Invitrogen) to synthesize cDNA. After cDNA synthesis, qPCR was performed as previously described61. Three or more independent cultures were subjected to qPCR and representative results are shown. The primer sequences used for each PCR experiment are listed in Table 1.
Table 1.
Primer lists for qPCR.
| Forward primer (5′-) | Reverse primer (5′-) | |
|---|---|---|
| Gapdh | GCCAAACGGGTCATCATCTC | GTCATGAGCCCTTCCACAAT |
| G9a | CTCTACCGGACTGCCAAGAT | CTCGGCATCAGAGATCAGC |
| Scx | CACCCAGCCCAAACAGATCTGCA | AGTGGCATCACCTCTTGGCTGCT |
| Tnmd | TCCCGCAAGTGAAGGTGGAGAAGA | AGTAAAGGTTCACAGACACGGCGG |
| Col1a1 | GTCTGACTGGTCCCATTGGT | ATCACCAGGTTCACCTTTCG |
| Six2 | ACCACCACGCAAGTCAGCAA | CGACTTGCCACTGCCATTGA |
| p21Cip1 | TTGCACTCTGGTGTCTGAGC | TGCGCTTGGAGTGATAGAAA |
| Brdt | GGTGGACGCCGTGAAACTAAA | GGGCCATAACAACAATGTCATCT |
| Ptpn20 | TCACCCAGGAAGGTTAGAGGA | GAGTTGCGGAGATTCAGGTTAC |
TUNEL assay
The TUNEL assay was conducted using an In Situ Cell Death Detection Kit according to the manufacturer’s instructions (Roche, Penzberg, Germany). Deparaffined sections were treated with proteinase K (1 μg/mL in 10 mM Tris/HCl, pH 7.5) for 40 min at 37 °C. The sections were then rinsed with PBS. Next, the sections were incubated with TUNEL reaction mixture for 1 h at 37 °C and then rinsed with PBS. The sections were further incubated with Converter AP Solution for 30 min at 37 °C and then rinsed with PBS. The sections were stained by incubation with 0.2 mg/mL naphthol AS-MX phosphate (Sigma), 0.4 mg/mL fast red BB salt (Sigma), 0.5% NN-dimethylformamide, 2 mM MgCl2, and 2 mM levamisole in 100 mM Tris-maleic acid (pH 9.2) for 15 min at room temperature.
Statistical analysis
Statistical analysis was performed using an unpaired Student’s t test. P-values < 0.05 were considered to be statistically significant.
Supplementary Information
Acknowledgements
The authors would like to thank Prof. Haruhiko Akiyama and Dr Yoichi Shinkai for providing Sox9Cre/+ mice and G9afl/+ mice, respectively.
Abbreviations
- H3K9
Histone H3 lysine 9
- H3K9MTase
H3K9 methyltransferase
- H3K9me1
monomethylation at Lys 9 of histone H3
- H3K9me2
dimethylation at Lys 9 of histone H3
Author contributions
S.W.: investigation, data analysis, manuscript writing; H.I.: investigation (in vitro experiments, in vivo experiments mouse handling including colony management); K.K. .: investigation; K.N.: provision of mechanistic insight; N.D.: provision of mechanistic insight; H.T.: provision of mechanistic insight; H.K.: methodology, resources; M.T.: methodology, resources; A. N.: conception and design, investigation, provision of mechanistic insight, manuscript writing.
Funding
This study was partially supported by JSPS KAKENHI (Grant Numbers: 16K11797, 21K10199 to S.W. 22390344, 15K15679, 18K09515, 20K09897, 21K09823 to A.N, 23K09158 to H.I).
Data availability
The data that support the findings are available in the methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Satoshi Wada and Hisashi Ideno.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71570-5.
References
- 1.Mozzetta, C., Boyarchuk, E., Pontis, J. & Ait-Si-Ali, S. Sound of silence: The properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol.16, 499–513 (2015). 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- 2.Greer, E. L. & Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet.13, 343–357 (2012). 10.1038/nrg3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padeken, J., Methot, S. P. & Gasser, S. M. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat. Rev. Mol. Cell Biol.23, 623–640 (2022). 10.1038/s41580-022-00483-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkel, P. & Angrand, P. O. The control of histone lysine methylation in epigenetic regulation. Biochimie89, 1–20 (2007). 10.1016/j.biochi.2006.07.009 [DOI] [PubMed] [Google Scholar]
- 5.Shinkai, Y. & Tachibana, M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev.25, 781–788 (2011). 10.1101/gad.2027411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman, N. et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol.8, 188–194 (2006). 10.1038/ncb1353 [DOI] [PubMed] [Google Scholar]
- 7.Roopra, A., Qazi, R., Schoenike, B., Daley, T. J. & Morrison, J. F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell14, 727–738 (2004). 10.1016/j.molcel.2004.05.026 [DOI] [PubMed] [Google Scholar]
- 8.Gyory, I., Wu, J., Fejer, G., Seto, E. & Wright, K. L. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol.5, 299–308 (2004). 10.1038/ni1046 [DOI] [PubMed] [Google Scholar]
- 9.Tachibana, M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev.16, 1779–1791 (2002). 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auclair, G. et al. EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res.26, 192–202 (2016). 10.1101/gr.198291.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.AuYeung, W. K. et al. Histone H3K9 methyltransferase G9a in oocytes is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep.27, 282–293 (2019). 10.1016/j.celrep.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 12.Ling, B. M. et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. U. S. A.109, 841–846 (2012). 10.1073/pnas.1111628109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, D. Y., Northrop, J. P., Kuo, M. H. & Stallcup, M. R. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem.281, 8476–8485 (2006). 10.1074/jbc.M511093200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bittencourt, D. et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U. S. A.109, 19673–19678 (2012). 10.1073/pnas.1211803109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ideno, H. et al. G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development. Bone137, 115332 (2020). 10.1016/j.bone.2020.115332 [DOI] [PubMed] [Google Scholar]
- 16.Poulard, C. et al. A post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP. EMBO Rep.18, 1442–1459 (2017). 10.15252/embr.201744060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, X. et al. G9a-mediated methylation of ERα links the PHF20/MOF histone acetyltransferase complex to hormonal gene expression. Nat. Commun.7, 10810 (2016). 10.1038/ncomms10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaturvedi, C. P. et al. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc. Natl. Acad. Sci. U. S. A.106, 18303–18308 (2009). 10.1073/pnas.0906769106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ideno, H. et al. Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr. Patt.13, 84–90 (2013). 10.1016/j.gep.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 20.Kamiunten, T. et al. Coordinated expression of H3K9 histone methyltransferases during tooth development in mice. Histochem. Cell Biol.143, 259–266 (2015). 10.1007/s00418-014-1284-0 [DOI] [PubMed] [Google Scholar]
- 21.Wada, S. et al. H3K9MTase G9a is essential for the differentiation and growth of tenocytes in vitro. Histochem. Cell Biol.144, 13–20 (2015). 10.1007/s00418-015-1318-2 [DOI] [PubMed] [Google Scholar]
- 22.He, P. et al. Comparison of tendon development versus tendon healing and regeneration. Front. Cell Dev. Biol.10, 821667 (2022). 10.3389/fcell.2022.821667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asou, Y. et al. Coordinated expression of scleraxis and Sox9 genes during embryonic development of tendons and cartilage. J. Orthop. Res.20, 827–833 (2002). 10.1016/S0736-0266(01)00169-3 [DOI] [PubMed] [Google Scholar]
- 24.Akiyama, H. et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A.102, 14665–14670 (2005). 10.1073/pnas.0504750102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soeda, T. et al. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis48, 635–644 (2010). 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spanoudes, K., Gaspar, D., Pandit, A. & Zeugolis, D. I. The biophysical, biochemical, and biological toolbox for tenogenic phenotype maintenance in vitro. Trends Biotechnol.32, 474–482 (2014). 10.1016/j.tibtech.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 27.Riley, G. Tendinopathy-from basic science to treatment. Nat. Clin. Pract. Rheumatol.4, 82–89 (2008). 10.1038/ncprheum0700 [DOI] [PubMed] [Google Scholar]
- 28.Huang, A. H., Lu, H. H. & Schweitzer, R. Molecular regulation of tendon cell fate during development. J. Orthop. Res.33, 800–812 (2015). 10.1002/jor.22834 [DOI] [PubMed] [Google Scholar]
- 29.Murchison, N. D. et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development134, 2697–2708 (2007). 10.1242/dev.001933 [DOI] [PubMed] [Google Scholar]
- 30.Ito, Y. et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U. S. A.107, 10538–10542 (2010). 10.1073/pnas.1000525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, W. et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol.30, 4797–4807 (2010). 10.1128/MCB.00207-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lejard, V. et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem.286, 5855–5867 (2011). 10.1074/jbc.M110.153106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerquin, M. J. et al. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest.123, 3564–3576 (2013). 10.1172/JCI67521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, H., Xu, J., Lan, Y., Lim, H. W. & Jiang, R. The scleraxis transcription factor directly regulates multiple distinct molecular and cellular processes during early tendon cell differentiation. Front. Cell Dev. Biol.9, 654397 (2021). 10.3389/fcell.2021.654397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal, D. et al. Ezh2 Is essential for patterning of multiple musculoskeletal tissues but dispensable for tendon differentiation. Stem Cells Dev.30, 601–609 (2021). 10.1089/scd.2020.0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiunten, T. et al. Essential roles of G9a in cell proliferation and differentiation during tooth development. Exp. Cell Res.357, 202–210 (2017). 10.1016/j.yexcr.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 37.Brent, A. E., Schweitzer, R. & Tabin, C. J. A somitic compartment of tendon progenitors. Cell113, 235–248 (2003). 10.1016/S0092-8674(03)00268-X [DOI] [PubMed] [Google Scholar]
- 38.Kardon, G. Muscle and tendon morphogenesis in the avian hind limb. Development125, 4019–4032 (1998). 10.1242/dev.125.20.4019 [DOI] [PubMed] [Google Scholar]
- 39.Casciello, F., Windloch, K., Gannon, F. & Lee, J. S. Functional role of G9a histone methyltransferase in cancer. Front. Immunol.6, 487 (2015). 10.3389/fimmu.2015.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehnertz, B. et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev.28, 317–327 (2014). 10.1101/gad.236794.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong, C. et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest.122, 1469–1486 (2012). 10.1172/JCI57349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda, J. et al. The hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a mechanistic link between angiogenesis and tumor growth. Mol. Cell. Biol.34, 3702–3720 (2014). 10.1128/MCB.00099-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding, J. et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab.18, 896–907 (2013). 10.1016/j.cmet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishio, H. & Walsh, M. J. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. U. S. A.101, 11257–11262 (2004). 10.1073/pnas.0401343101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim, J. K., Esteve, P. O., Jacobsen, S. E. & Pradhan, S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res.37, 493–505 (2009). 10.1093/nar/gkn961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao, V. K. et al. G9a promotes proliferation and inhibits cell cycle exit during myogenic differentiation. Nucleic Acids Res.44, 8129–8143 (2016). 10.1093/nar/gkw483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Docheva, D., Hunziker, E. B., Fassler, R. & Brandau, O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol. Cell. Biol.25, 699–705 (2005). 10.1128/MCB.25.2.699-705.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alberton, P. et al. Loss of tenomodulin results in reduced self-renewal and augmented senescence of tendon stem/progenitor cells. Stem Cells Dev.24, 597–609 (2015). 10.1089/scd.2014.0314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachibana, M., Sugimoto, K., Fukushima, T. & Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem.276, 25309–25317 (2001). 10.1074/jbc.M101914200 [DOI] [PubMed] [Google Scholar]
- 50.Purcell, D. J. et al. Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription. J. Cell. Biochem.113, 2406–2414 (2012). 10.1002/jcb.24114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachibana, M., Nozaki, M., Takeda, N. & Shinkai, Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J.26, 3346–3359 (2007). 10.1038/sj.emboj.7601767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng, X. et al. Sin3B mediates collagen type I gene repression by interferon gamma in vascular smooth muscle cells. Biochem. Biophys. Res. Commun.447, 263–270 (2014). 10.1016/j.bbrc.2014.03.140 [DOI] [PubMed] [Google Scholar]
- 53.Lei, W. et al. Homocysteine induces collagen i expression by downregulating histone methyltransferase G9a. PLoS One10, e0130421 (2015). 10.1371/journal.pone.0130421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori-Akiyama, Y., Akiyama, H., Rowitch, D. H. & de Crombrugghe, B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc. Natl. Acad. Sci. U. S. A.100, 9360–9365 (2003). 10.1073/pnas.1631288100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugimoto, Y. et al. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development140, 2280–2288 (2013). 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- 56.Blitz, E., Sharir, A., Akiyama, H. & Zelzer, E. Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development140, 2680–2690 (2013). 10.1242/dev.093906 [DOI] [PubMed] [Google Scholar]
- 57.Kimura, H., Hayashi-Takanaka, Y., Goto, Y., Takizawa, N. & Nozaki, N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct.33, 61–73 (2008). 10.1247/csf.07035 [DOI] [PubMed] [Google Scholar]
- 58.Shimada, A., Komatsu, K., Nakashima, K., Poschl, E. & Nifuji, A. Improved methods for detection of beta-galactosidase (lacZ) activity in hard tissue. Histochem. Cell Biol.137, 841–847 (2012). 10.1007/s00418-012-0936-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nifuji, A. & Noda, M. Coordinated expression of noggin and bone morphogenetic proteins (BMPs) during early skeletogenesis and induction of noggin expression by BMP-7. J. Bone Miner. Res.14, 2057–2066 (1999). 10.1359/jbmr.1999.14.12.2057 [DOI] [PubMed] [Google Scholar]
- 60.Shimada, A., Shibata, T., Komatsu, K. & Nifuji, A. Improved methods for immunohistochemical detection of BrdU in hard tissue. J. Immunol. Methods339, 11–16 (2008). 10.1016/j.jim.2008.07.013 [DOI] [PubMed] [Google Scholar]
- 61.Ideno, H. et al. Protein related to DAN and cerberus (PRDC) inhibits osteoblastic differentiation and its suppression promotes osteogenesis in vitro. Exp. Cell Res.315, 474–484 (2009). 10.1016/j.yexcr.2008.11.019 [DOI] [PubMed] [Google Scholar]
- 62.Komatsu, K. et al. The G9a histone methyltransferase represses osteoclastogenesis and bone resorption by regulating NFATc1 function. FASEB J.38, e23779 (2024). 10.1096/fj.202400449RR [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings are available in the methods section.